Abstract
Connective tissue growth factor (CCN2) drives fibrogenesis in hepatic stellate cells (HSC). Here we show that CCN2 up-regulation in fibrotic or steatotic livers, or in culture-activated or ethanol-treated primary mouse HSC is associated with a reciprocal down-regulation of microRNA-214 (miR-214). By using protector or reporter assays to investigate the 3′-untranslated region (UTR) of CCN2 mRNA, we found that induction of CCN2 expression in HSC by fibrosis-inducing stimuli was due to reduced expression of miR-214 which otherwise inhibited CCN2 expression by directly binding to the CCN2 3′-UTR. Additionally, miR-214 was present in HSC exosomes, which were bi-membrane vesicles, 50–150nm in diameter, negatively charged (−26mV), and positive for CD9. MiR-214 levels in exosomes but not in cell lysates were reduced by pre-treatment of the cells with the exosome inhibitor, GW4869. Co-culture of miR-214-transfected donor HSC with CCN2 3′-UTR luciferase reporter-transfected recipient HSC resulted in miR-214- and exosome-dependent regulation of a wild type CCN2 3′-UTR reporter but not of a mutant CCN2 3′-UTR reporter lacking the miR-214 binding site. Exosomes from HSC were a conduit for uptake of miR-214 by primary mouse hepatocytes. Down-regulation of CCN2 expression by miR-214 also occurred in human LX-2 HSC, consistent with a conserved miR-214 binding site in the human CCN2 3′-UTR. MiR-214 in LX-2 cells was shuttled via exosomes to recipient LX-2 cells or human HepG2 hepatocytes, resulting in suppression of CCN2 3′-UTR activity or expression of CCN2 downstream targets, including αSMA or collagen. Experimental fibrosis in mice was associated with reduced circulating miR-214 levels.
Conclusion
Exosomal transfer of miR-214 is a paradigm for the regulation of CCN2-dependent fibrogenesis and identifies fibrotic pathways as targets of epigenetic regulation by exosomal miRs.
Keywords: liver fibrosis, CTGF, CCN2, fibrogenesis, hepatocyte
Fibrogenic pathways are triggered in many tissues as a response to chronic injury and often lead to deposition of insoluble collagen and production of scar. Following liver injury, peri-sinusoidal hepatic stellate cells (HSC) acquire a myofibroblastic phenotype and express increased amounts of alpha smooth muscle actin (αSMA) and fibrillar collagens that facilitate repair by, respectively, promoting wound contraction and providing a provisional matrix for cell repopulation 1. During chronic injury, this process persists causing unrelenting deposition of fibrillar collagens, eventually compromising normal hepatic function 1. Interventions to reduce matrix production and/or increase matrix degradation by HSC are promising anti-fibrotic strategies 2.
Connective tissue growth factor is the second member of the cysteine-rich-61/connective tissue growth factor/nephroblastoma-overexpressed (CCN) family of proteins (CCN2) 3 and a large body of data has accumulated showing that CCN2 directly drives fibrogenesis in HSC 4. CCN2 is over-expressed in fibrotic liver 5–10, and activated HSC exhibit enhanced CCN2 production in livers from patients with hepatitis 5, 10 or fibrosis 11. Quiescent HSC produce only low or non-detectable levels of CCN2, whereas CCN2 mRNA, protein or promoter activity is stimulated in the cells during activation or in response to fibrosis-inducing agents 4. CCN2 is an attractive therapeutic target as shown by the broad anti-fibrotic efficacy of CCN2 antagonists in vitro or in experimental fibrosis models in vivo 12. A humanized anti-CCN2 monoclonal antibody 13 is currently in Phase 2 trials for liver fibrosis and idiopathic pulmonary fibrosis (ClinicalTrials.gov NCT01212187 and NCT01262001).
MicroRNAs (miRs) are non-coding RNAs of ~23 nucleotides that regulate gene expression by interacting with the 3′ untranslated region (3′-UTR) of target gene mRNA to repress translation or enhance mRNA cleavage 14. Thus, increased expression of CCN2 mRNA during HSC activation is likely associated with down-regulation of microRNAs that normally suppress CCN2 expression in quiescent HSC. Here we show that CCN2 expression is regulated directly by miR-214 in experimental fibrosis or during HSC activation. Further, HSC produce nano-sized membranous exosomes which transfer miR-214 to neighboring HSC or hepatocytes in which CCN2 3′-UTR activity is then reduced. These studies highlight exosomal transfer of miR-214 as a paradigm for the regulation of CCN2-dependent pathways between hepatic cells, representing a novel mechanism by which fibrogenic signaling is controlled.
Materials and Methods
Additional experimental procedures are described in the Supporting Materials.
MiR-214 suppression of CCN2 in primary mouse HSC
Passage 6 (P6) primary mouse HSC were transfected with 4μg mouse pLemiR-214 plasmid DNA or scramble plasmid DNA (Open Biosystem, Huntsville, AL) by electroporation (Nucleofector Kit, Lonza, Koln, Germany). Transfected cells were incubated in DMEM/F12/10%FBS medium for 24 hrs. Transfection efficiency was ~40% as determined by co-expression of red fluorescent protein (RFP) contained in the pLemiR-214 vector. Cells were fixed for CCN2, αSMA or collagen α1 immunostaining. For some experiments, HSC transfected for 36 hrs with pLemiR-214 were sorted using a Beckman Coulter Epics Altra cell sorter (Beckman, HighWycombe, UK) for RNA analysis. At least 20,000 events were acquired for RNA isolation.
Target protector for miR-214 binding site in CCN2 3′-UTR
A CCN2 3′-UTR protector sequence was designed to bind to the mRNA sequence that was complementary to the miR-214 seed region (5–8 nt miRNA target site) and flanking sequences in the CCN2 3′-UTR (Supporting Fig. S1A). Sequence specificity was verified using the Basic Local Alignment Search Tool (BLAST, National Center for Biotechnology, Bethesda MD). CCN2 target protector or a randomly generated negative control target protector (100nM; Qiagen, Valencia, CA, USA) were co-transfected with pLemiR-214 plasmids (3 μg) as described above into P6 primary mouse HSC for 24 hrs prior to detection of RFP by auto-fluorescence, CCN2 by immunofluorescence, or CCN2 or collagen α1(I) mRNA by real-time polymerase chain reaction (RT-PCR).
Transfection of mouse primary HSC with miR-Selection Fire-Ctx lentivector-CCN2 3′-UTR
Potential binding sites for miR-214 in human or mouse CCN2 3-UTRs were identified using BLAST (Supporting Fig. S1A). The full-length 997bp 3′-UTR of mouse CCN2 (Genbank SEQ ID: BC006783.1) was amplified by PCR from primary mouse HSC genomic DNA using forward primer 5′-GAGGGATCCGTCACACTCTCAACAAATAAACTGCCC-3′ and reverse primer 5′-GAGGAATTCAGCCAGGAAGTAAGGGAACCGAACTCA -3′. The PCR fragment was digested with BamH I and EcoR I, subcloned into Fire-Ctx sensor lentivector (SBI, Mountain View, CA, USA), downstream of the Firefly luciferase reporter and cytotoxin (CTX) drug sensor genes, and verified by DNA sequencing (Supporting Fig. S1B). A mutant CCN2 3′-UTR containing a 5-base point mutation (GTCCG → ACAAT; see Supporting Fig. S1A) in the predicted miR-214 binding site was amplified from the wild-type mouse CCN2 3′-UTR using forward primer 5′-CTGGCTCAGGGTAAGACAATATTCCTACCAGGAAG-3′ and reverse primer 5′-CTTCCTGGTAGGAATATTGTCTTACCCTGAGCCAG -3′, and verified by DNA sequencing.
Primary mouse HSC up to P6 were co-transfected by electroporation (Nulceofector, Lonza) with 100nM of the hairpin precursor of miR-214 (pre-mir-214; Life Technologies, Carlsbad, CA USA) and 3 μg Fire-Ctx sensor lentivectors containing CCN2 wild-type or mutant 3′-UTR, or vector alone. To control for transfection efficiency, cells were also transfected with 0.8 μg pRL-CMV vector (Promega, Madison WI, USA) containing Renilla luciferase reporter gene. After 24 hrs, luciferase activity was measured in triplicate using an E1910 Dual Luciferase Reporter Assay System (Promega). Renilla luciferase activity was used for normalization, and Firefly luciferase activity in pre-mir-214 transfected cells was compared to that in mock-transfected cells. Alternatively, 24 hours after transfection, the cells were treated with CTX (1:1000; Clontech, Mountain View CA) for 3–4 days after which cell viability was assessed using a CytoSelect™ assay (Cell Biolabs Inc., San Diego, CA, USA).
Results
Direct targeting of the CCN2 3′-UTR by miR-214
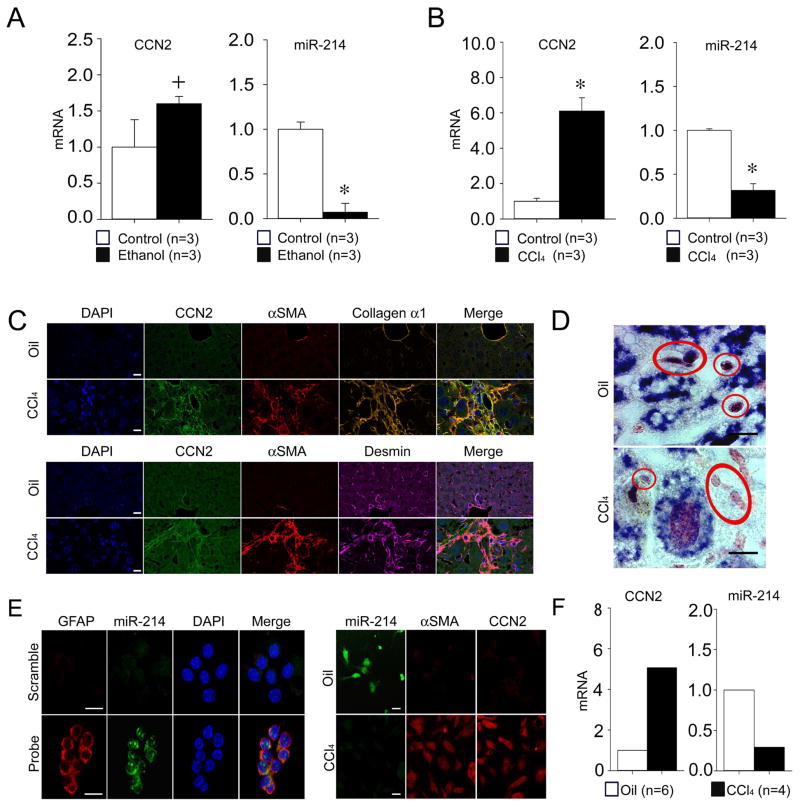
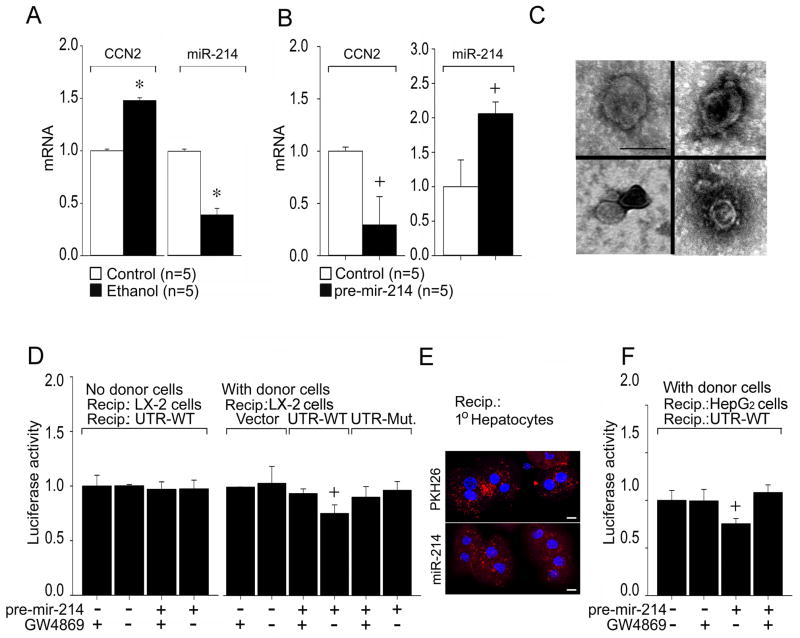
We have previously shown that ethanol stimulates CCN2 expression in mouse or human HSC 11. MiR-214 emerged as a candidate regulator of CCN2 because miR-214 expression was shown to be down-regulated in livers of rats with alcoholic steatosis 15 and we determined that it has a potential but hitherto unrecognized binding site in the CCN2 3′-UTR which is evolutionarily conserved between human and mouse (see below). Upon directly exploring the relationship between CCN2 and miR-214, we found that elevated levels of hepatic CCN2 mRNA but diminished levels of hepatic miR-214 were present in mice after chronic ethanol feeding or exposure to carbon tetarchloride (CCl4) or thioacetamide (TAA) (Figure 1A,B; Supporting Fig. S2A,B). Presumptive quiescent HSC (positive for desmin) in normal mouse livers expressed high levels of miR-214 as assessed by ISH whereas presumptive activated HSC (positive for desmin, CCN2, αSMA and collagen) in the fibrous tracts of mice with experimental hepatic fibrosis expressed highly diminished miR-214 levels (Figure 1C,D; Supporting Fig. S2C,D). These in vivo findings were supported by the observation that quiescent HSC isolated from normal livers contained high levels of miR-214 and low levels of CCN2 mRNA as assessed by ISH or RT-PCR, whereas transitionally activated HSC from CCl4-injured livers contained low levels of miR-214 and high levels of CCN2 mRNA (Fig. 1E,F). The identity and activation status of the isolated HSC was confirmed by their common staining with oil red but exclusive expression of activation markers (CCN2, αSMA) only in HSC isolated from the CCl4-exposed livers (Supporting Fig. S3A–C). Moreover, primary HSC cultures from normal mice expressed increased amounts of CCN2 mRNA and concomitantly decreased amounts of miR-214 in response to ethanol (Fig. 2A) or during their autonomous activation over 3 weeks in vitro (Fig. 2B). Although miR-199a-5p and miR-322 were reported to be suppressed in livers of rats with alcoholic steatosis 15, we were unable to verify this finding by RT-PCR in our mouse model of chronic ethanol feeding. In view of the central role of CCN2 in HSC-mediated fibrogenic pathways and the reciprocity in expression of CCN2 and miR-214 in HSC in the CCl4 or TAA fibrosis models in vivo or during HSC activation in vitro, our subsequent studies focused on the CCN2-miR-214 axis in HSC.
Fig. 1. Reciprocal expression of miR-214 and CCN2 in liver tissues or HSC.
Hepatic expression of CCN2 mRNA or miR-214 assessed by RT-PCR and normalized to GAPDH mRNA after (A) 4-week administration of ethanol or (B) 5-week administration of CCl4 versus carrier controls (n=3 independent experiments performed in triplicate, *P <0.001, +P <0.05 vs. no treatment). (C) Immunohistochemical detection of CCN2, αSMA, desmin or collagen α1 in oil-treated control or CCl4-treated mice. Specimens were also stained with DAPI nuclear stain (blue) (D) ISH for miR-214 in the livers of oil-treated control mice or in the vicinity of fibrous deposits in CCl4-treated mice; presumptive HSC are circled and show diminished miR-214 (purple) staining in the fibrotic mice. Scale bar: 10μm. (E) ISH of HSC cultured for one day after isolation from normal mice (left) or 36 hrs after the last of two oil or CCl4 injections spaced 48 hrs apart (right). Labeled probes were used to detect miR-214 (green), GFAP (red), αSMA (red) or CCN2 (red) (lower left, upper right, or lower right) or a scrambled sequence (upper left). Specimens were also stained with DAPI nuclear stain (blue; left). ISH images are representative of 3 independent experiments. Scale bar: 20μm. (F) RT-PCR, performed in triplicate, of mRNA extracted from HSC on Day 1 of culture after isolation from the oil- or CCl4-treated mice in (E) (n=6 mice per group).
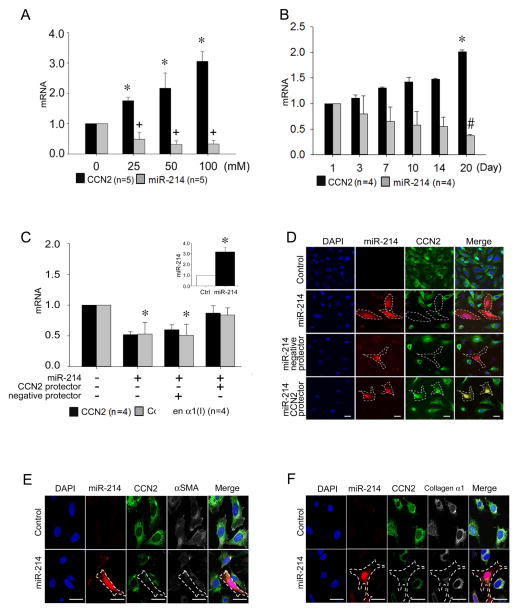
Fig. 2. MiR-214-dependency of CCN2 production in mouse HSC.
(A) RT-PCR of CCN2 mRNA or miR-214, normalized to GAPDH mRNA, after Day 3 primary mouse HSC were incubated in 1% serum for 24 hrs prior to 48-hr treatment with 0–100mM ethanol (n=5 independent experiments performed in triplicate, *P <0.001 vs. non treatment, +P <0.01 vs. non treatment). (B) CCN2 mRNA or miR-214 expression, assessed by RT-PCR and normalized to GAPDH mRNA, over 20 days of primary culture of HSC isolated from normal mouse liver (n=4 independent experiments performed in triplicate, *P <0.001 vs. Day 1 CCN2 expression, #P <0.001 vs. Day 1 miR-214 expression). (C) RT-PCR of CCN2 or collagen α1(I) mRNA or miR-214 (inset) relative to GAPDH mRNA in RFP-positive HSC collected by cell sorting or (D) detection of CCN2 protein by indirect immunofluorescence (green) or RFP by direct fluorescence (red) in P6 HSC transfected for 24 hrs with pLemiR-214 either alone or with a co-transfected CCN2 3′-UTR protector. A negative protector was used as a control (n=4 independent experiments performed in triplicate; *P<0.001 vs control). RFP fluorescence (red) and indirect immunofluorescence for CCN2 (green) and (E) αSMA (white) or (F) collagen α1 (white) in P6 control HSC or after transfection for 24 hrs with pLemiR-214. In (D–F), blue is DAPI nuclear stain. Scale bar: 20μm.
A functional link between CCN2 and miR-214 was demonstrated by transfection of P6 activated mouse HSC with pLemiR-214 which resulted in enhanced miR-214 levels (Fig. 2C, inset), decreased CCN2 mRNA expression and protein production (Fig. 2C–F), and decreased production of CCN2 downstream markers such as αSMA (Figure 2E) or collagen α1 (Fig. 2C,F). Inspection of the CCN2 3′-UTR revealed an unreported potential miR-214 binding site that was conserved between mouse and human sequences (Supporting Fig. S1A). To determine if this region was indeed a binding site for miR-214, it was first targeted with a protector sequence that was complementary to the presumptive miR-214 binding domain in the CCN2 3′-UTR. The ability of miR-214 to inhibit CCN2 mRNA expression in HSC was blocked by the protector but not by a negative protector lacking complementarity to the target sequence (Fig. 2C). Similarly, the production of CCN2 protein persisted in activated HSC when they were co-transfected with pLemiR-214 and the CCN2 3′-UTR protector whereas CCN2 production was inhibited in control cells transfected with pLemiR-214 alone or in combination with the negative protector (Fig. 2D). Although the collagen α1(I) 3′-UTR does not contain a miR-214 binding site, the protector also inhibited production of collagen α1(I) mRNA in activated HSC (Fig. 2C), consistent with previous data showing that expression of collagen α1(I) in HSC is CCN2-dependent 11. Since a single miR often regulates multiple targets in a common biological pathway, we performed differential expression analysis of mRNA from pre-mir-214-transfected HSC versus control HSC using a Fibrosis RT2 Profiler PCR Array. MiR-214 affected the expression of multiple fibrosis-related genes (Supporting Table S1), including integrin β8 mRNA which was down-regulated and has a miR-214 binding site in its 3′-UTR. The expression of other genes including members of the integrin, interleukin, matrixmetalloprotease and platelet-derived growth factor families were also inhibited in miR-214-transfected cells which possibly reflected indirect downstream effects of miR-214 because the individual genes were not predicted direct targets of miR-214.
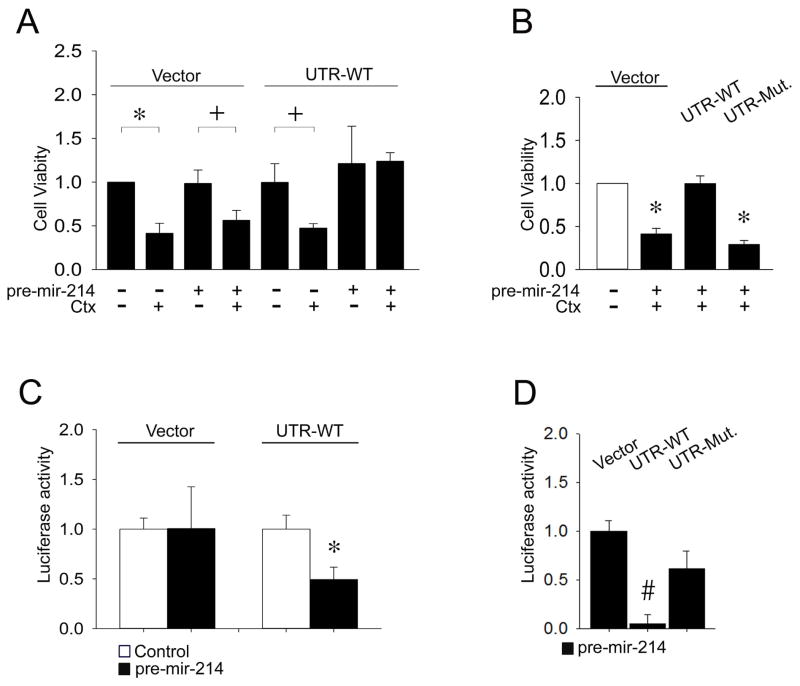
Next, we analyzed the cytotoxic or luciferase reporter activities of a Fire-Ctx lentivector parental vector versus the same vector harboring either a wild-type CCN2 3′-UTR or a mutant form of the 3′-UTR containing a substitution mutation in the presumptive miR-214 binding site (Supporting Fig. S1). In HSC transfected with the parental lentivector, CTX-induced cell death occurred whether or not miR-214 was present since the vector lacked a miR-214 binding site for regulation of CTX translation (Fig. 3A and Supporting Fig. S5A). In HSC transfected with the Fire-Ctx vector containing the wild-type CCN2 3′-UTR, CTX treatment resulted significant cell death but this was reversed by the presence of miR-214 which restored cell viability to control levels, consistent with its suppression of 3′-UTR activity leading to reduced activity of the CTX gene product and an inability to drive production of cytotoxic drug metabolites (Fig. 3A and Supporting Fig. S5A). MiR-214 inhibition of CTX-induced cell death was not evident in HSC containing the mutated CCN2 3′-UTR (Fig. 3B and Supporting Fig. S5B), showing that the wild-type CCN2 3′-UTR was a direct target of miR-214. Finally, as compared to cells transfected with parental Fire-Ctx vector, the firefly luciferase activity of the vector containing the full length wild type CCN2 3′-UTR was diminished by 50–90% in the presence of miR-214 (Fig. 3C,D), whereas miR-214 did not significantly suppress luciferase reporter activity when the CCN2 3′-UTR contained the substitution mutation (Fig. 3D). Thus, CCN2 transcriptional activity is suppressed by the direct binding of miR-214 to a unique site in the CCN2 3′-UTR.
Fig. 3. Identification of CCN2 3′-UTR as a direct target of miR-214.
(A) P6 mouse HSC were transfected with parental Fire-Ctx (“Vector”) or Fire-Ctx containing wild-type mouse CCN2 3′-UTR (“UTR-WT”). Some cells were co-transfected with pre-mir-214 and/or treated with CTX. Cell viability, assessed by fluorescence of cell lysates at 480nm/520nm, was determined on Day 4 with CytoSelect™ (n=4 independent experiments performed in triplicate, *P <0.001, +P <0.05 vs. no CTX treatment). (B) P6 HSC were treated and assayed as in (A) except that some cultures were transfected with Fire-Ctx containing a CCN2 3′-UTR with a substitution mutation in the miR-214 binding site (“UTR-Mut.”). (n=4 independent experiments performed in triplicate, *P <0.001 vs. no CTX treatment). (C,D) P6 HSC were treated as in (B) and the activities of the dual luciferase reporters in the Fire-Ctx plasmids were measured 36 hrs after transfection. Firefly luciferase activity in cell lysates was normalized to that of Renilla luciferase (n=4 independent experiments performed in triplicate, *P <0.001 vs. no pre-mir-214 transfection, #P <0.001 vs. vector).
Exosome characterization
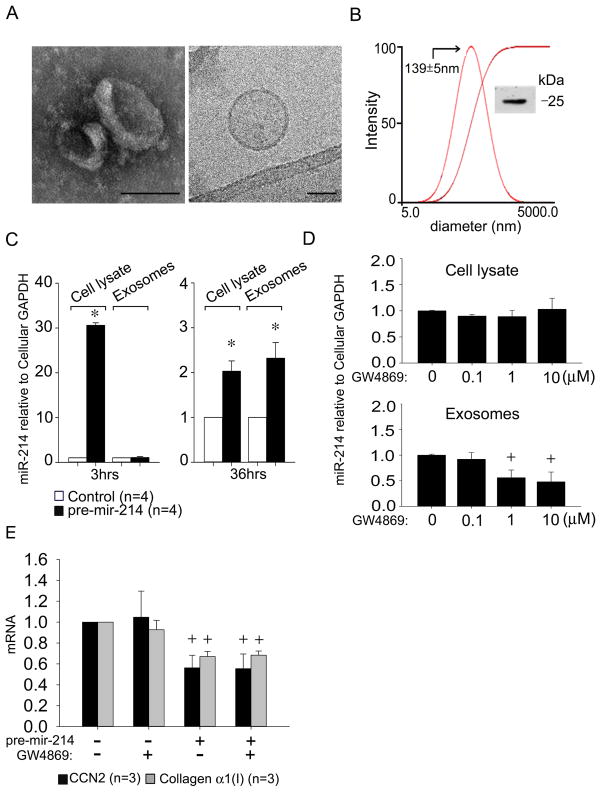
The presence of exosomes in conditioned medium from cultures of activated mouse HSC was confirmed by several criteria including (i) appearance as 50–150 nm bi-membrane vesicles assessed by TEM or cryogenic TEM; (ii) high negative charge (−26mV) and diameter of ~140 nm as assessed by zeta potential analysis and dynamic light scattering respectively; and (iii) Western blot detection of CD9, a well characterized marker of exosomes of murine origin (Fig. 4A,B). Despite the relatively low abundance of miR-214 produced by highly activated HSC (Fig. 2B), RT-PCR demonstrated that miR-214 was nonetheless detectable in the exosomes from these cells (Fig. 4C). The exosomal concentration of miR-214 was significantly increased (~2-fold) 36 hours after transfection of the cells with pre-mir-214 and reached levels that were comparable to those in cell lysates at the same time point (Fig. 4C). Whereas cellular miR-214 levels were ~30-fold increased within 3 hrs after transfection, there was no discernable increase in exosomal miR-214 levels (Fig. 4C) showing that miR-214-enriched exosomes were not yet present at this early time point. MiR-214 levels in exosomes but not in cell lysates were reduced by pre-treatment of the cells with GW4869, a well characterized exosome inhibitor 16, 17 that blocks neutral sphingomyelinase2 (nSMase2) which is required for the biosynthesis of ceramide on which exosome production is dependent (Fig. 4D). Collectively, these data confirmed that miR-214 was a bone fide constituent of exosomes from primary mouse HSC. Treatment of pre-miR-214-transfected cells with GW4869 did not significantly alter the inhibited levels of CCN2 or collagen α1 in these cells (Fig. 4E), showing that, as expected, miR-214 exerted its effects in transfected cell cultures (such as those documented in Figs 2–3) principally by its direct action in the transfected cells themselves as opposed to a fully exosome-dependent export mechanism that was subsequently characterized in the experiments below.
Fig. 4. Identification and characterization of mouse HSC-derived exosomes.
Exosomes were isolated by sequential centrifugation of conditioned medium from P6 mouse HSC. (A) TEM (left) and cryogenic TEM (right). Scale bar: 100nm (left), 50nm (right). (B) Dynamic light scattering and zeta potential analysis, showing the presence of ~140nm particles (−26mV). Inset: CD9 Western blot. (C) RT-PCR of cellular or exosomal microRNA isolated from control or pre-mir-214-transfected cells after 3hrs or 36hrs, normalized to cellular GAPDH. (n=4 independent experiments performed in triplicate, *P <0.001 vs. control group). Ct value = 38 for endogenous exosomal miR-214 in control cells at 3hrs. (D) Cellular or exosomal miR-214 levels assessed by RT-PCR in pre-mir-214-transfected HSC after 24 hrs treatment with GW4869, with data normalized to cellular GAPDH. (n=4 independent experiments performed in triplicate, +P <0.05 vs. Control group). (E) CCN2 or collagen α1(I) expression in control or pre-mir-214-transfected HSC in the presence or absence of 10 μM GW4869.
MiR-214 is transferred between HSC by exosomes
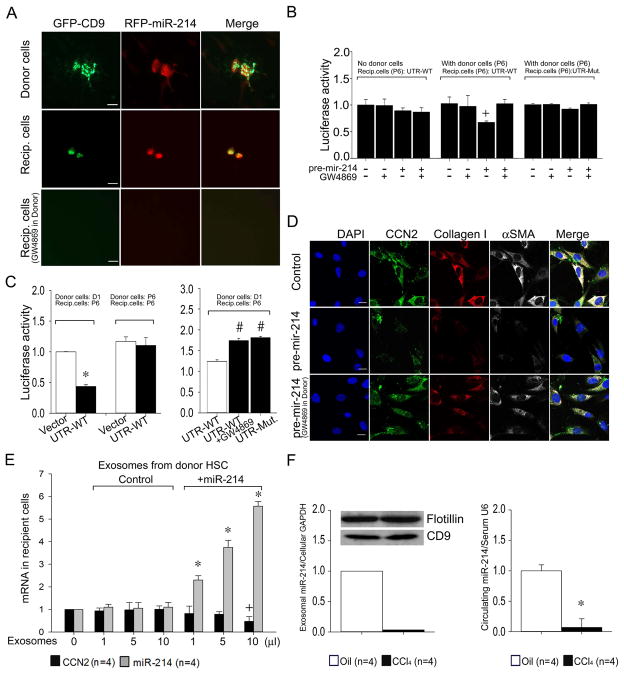
Co-culture experiments were performed to establish whether exosomal miR-214 was transferred intercellularly between neighboring HSC and whether it was functionally active after uptake into recipient cells. A silicon culture device was used in which donor and recipient cells were initially seeded in adjoining micro-wells but subsequently allowed to directly communicate by removal of the wall separating them. Mouse HSC donor cells transfected with GFP-CD9 or pLemiR-214 were co-incubated with non-transfected HSC for 24 hrs. GFP or RFP were detected by their respective fluorescence in the transfected cells as expected but they were also detected in the non-transfected cells HSC as well (Fig. 5A) showing that GFP and RFP had transferred from the transfected (donor) cells to the non-transfected (recipient) cells. The involvement of exosomes in this process was supported by the detection of the GFP-CD9 in recipient cells and the finding that treatment of donor cells with GW4869 resulted in a complete absence of GFP or RFP in recipient HSC (Fig. 5A).
Fig. 5. HSC-HSC intercellular signaling via exosomal transport of miR-214.
(A) HSC donor and recipient co-culture experiments were performed as shown in Supporting Fig. S4. Donor P6 mouse HSC were co-transfected with plasmids for GFP-CD9 (green) and RFP-miR-214 (pLemiR-214; red) and cultured with or without GW4869. Recipient cells were non-transfected P6 mouse HSC. Representative immunofluorescence images of the donor and recipient cells are shown (n=3 independent experiments.) Scale bar: 20μm. (B) Recipient P6 mouse HSC transfected with Fire-Ctx plasmids containing either wild-type or mutant CCN2 3′-UTR (Fig. 3A,B, Supporting Figs. S1A, S4B) were co-incubated with control or pre-mir-214-transfected donor P6 HSC in the presence or absence of GW4869. In control experiments, some recipient HSC were exposed to the donor side of the micro-wells which lacked donor HSC but contained pre-mir-214 in the culture medium at the same concentration (100nM) as used for donor HSC transfection. The activity of firefly luciferase in lysates of recipient cells, normalized to that of Renilla luciferase, is shown 24 hrs after the central divider had been removed from the micro-wells. (n=4 independent experiments performed in triplicate, +P <0.05 vs. control group). (C) HSC co-culture experiments were set up and evaluated as in (B) except donor HSC had been freshly isolated from normal mouse livers and placed in culture for 1 day prior to treatment with GW4869 (n=4 independent experiments performed in triplicate, *P <0.001 vs. vector, # P <0.001 vs. UTR-WT). (D) Co-cultures were established for 12 hrs between control or pre-mir-214-transfected P6 donor HSC (± GW4869) and P6 recipient cells (Supporting Fig. S4B), the latter of which were then evaluated for immunocytochemical detection of CCN2 (green), collagen I (red) or αSMA (white). Blue depicts DAPI nuclear stain. Scale bar: 20μm. (E) Expression of CCN2 or miR-214 in P6 HSC after addition for 24hrs of exosomes isolated from 48-hr conditioned medium of control P6 HSC or P6 HSC transfected with pre-mir-214. (F) MiR 214 in exosomes from 24-hr conditioned medium of HSC isolated from Swiss Webster mice receiving two injections of oil or CCl4 (Western blots show detection of flotillin or CD9 in exomosal extracts) (left) or serum from Swiss Webster mice that were administered oil or CCl4 for 5 weeks (right) (*P < 0.001 vs oil group).
To demonstrate definitive exosomal transfer of miR-214 (versus the RFP tag in pLemiR-214) and its subsequent ability to bind to its target CCN2 3′-UTR, recipient activated HSC were transfected with wild-type or mutant CCN2 3′-UTR luciferase reporter (see above) and co-cultured with donor activated HSC transfected with pre-mir-214. Luciferase activity in recipient cells was significantly diminished only when they were co-cultured in the presence of donor cells transfected with pre-mir-214, an effect that was reversed by exposure of the donor cells to GW4869 (Fig. 5B). This effect was not seen when recipient cells were exposed to reagent controls in the absence of donor cells or when the recipient cells harbored the mutant CCN2 3′-UTR (Fig. 5B). Collectively, these data showed that miR-214 was transferred between neighboring HSC in an exosomal-dependent fashion and that it bound to its target CCN2 3′-UTR in recipient cells. GW4869 treatment of non-transfected activated cells did not increase CCN2 3′-UTR activity in recipient cells (Fig. 5B), likely reflecting the insensitivity of the assay to potential exosomal transfer of the very low levels of miR-214 produced by highly activated donor cells (Figs. 2B, 4C). To address this issue, we instead used donor HSCs within 1 day of isolation from normal livers, at which time their intrinsic miR-214 levels are much higher (Fig. 2B). Luciferase activity in highly activated recipient HSC was significantly suppressed for the wild-type CCN2 3′-UTR compared to the vector control when they were co-cultured with Day 1 donor HSC but not with highly activated donor HSC (Fig. 5C). However, this suppressed activity was not seen when donor Day 1 HSC were treated with GW4869, nor when activated recipient HSC were transfected with the mutated CCN2 3′-UTR (Fig. 5C). Collectively these data support the intercellular transfer of exosomes and miR-214 from quiescent HSC to activated HSC in which CCN2 3′-UTR activity is subsequently down-regulated. In addition, when pre-mir-214-transfected donor HSC were co-cultured with recipient activated HSC, the latter exhibited a GW4869-dependent decrease in immunocytochemical staining for CCN2 as well as its downstream targets, collagen α1 or αSMA (Fig. 5D). Thus exosomes serve as a conduit for the regulation of CCN2-dependent fibrogenic signaling in activated HSC, a phenomenon that was attributable, at least in part, to the intercellular transfer and action of exosomal miR-214, including its targeting of the CCN2 3′-UTR in activated HSC.
MiR-214-enriched exosomes isolated from the medium from miR-214-transfected HSC elicited a dose-dependent increase in miR-214 expression and concomitant down-regulation of CCN2 expression in recipient HSC, an effect that was not evident with exosomes isolated from control HSC (Figure 5E). In addition, exosomes collected from the medium of 24-hour primary cultures of HSC demonstrated substantially lower miR-214 levels when the HSC were isolated from the livers of CCl4-treated mice as compared to those of control mice (Fig. 5F). Finally, the circulating concentration of miR-214 was >10-fold higher in normal mice versus mice with CCl4-induced hepatic fibrosis (Fig. 5F).
Functional properties of miR-214 produced by human HSC
We determined that the miR-214 binding site is present in the human CCN2 3′-UTR (Supporting Fig. S1A), prompting us to examine if the mechanisms described above were evolutionarily conserved. Compared to control cells, ethanol-treatment of the human HSC LX-2 line resulted in an up-regulation of CCN2 mRNA expression as previously reported 11 and this was associated with a concomitant decrease in miR-214 expression (Fig. 6A). A functional link between miR-214 and CCN2 was shown by transfection of the cells with pre-mir-214, which resulted in significant decrease in CCN2 mRNA expression (Fig. 6B). The presence in LX-2 cell conditioned medium of bi-membrane vesicles, 50–150nm in diameter, was confirmed by TEM (Fig. 6C). In LX-2 cells transfected with wild type CCN2 3′-UTR, luciferase reporter activity was significantly diminished when the cells were co-cultured with donor LX-2 cells transfected with pre-mir-214, but this effect was not apparent when the donor cells were also exposed to GW4869, when recipient cells were exposed to reagent controls without donor cells present, or in recipient cells transfected with the mutant CCN2 3′-UTR (Fig. 6D). Collectively, these data support the presence of exosomal transfer of miR-214 between neighboring LX-2 cells and the ability of miR-214 to target the CCN2 3′-UTR in recipient HSC.
Fig. 6. Functional characterization of miR-214 produced by human HSC.
(A) Reciprocal expression of CCN2 and miR-214 after serum-starved LX-2 HSC were treated for 48 hrs with 25mM ethanol. RNA was subjected to RT-PCR and expression of CCN2 or miR-214 was normalized to that of GAPDH. (n=5 independent experiments performed in triplicate, * P<0.001 vs non treatment). (B) Down-regulation of CCN2 expression in LX-2 cells after transfection with pre-mir-214, determined as in (A). (n=5 independent experiments performed in triplicate, +P<0.05 vs Control). (C) TEM of exosomes purified from LX-2 cell conditioned medium. Scale bar: 100nm. (D) Donor LX-2 cells were transfected with pre-mir-214, in the presence or absence of GW4869. Some donor wells contained pre-mir-214 in the medium but no LX-2 cells. Recipients were LX-2 cells transfected with Fire-Ctx plasmids that were parental or contained wild-type or mutant CCN2 3′-UTRs (Supporting Fig. S4B). Firefly luciferase activity was measured in cell lysates 24 hrs after co-culture and normalized to that of Renilla luciferase. (n=4 independent experiments performed in triplicate, +P <0.05 vs. control group). (E) Confocal microscopy showing uptake of exosomes by primary mouse hepatocytes after 4-hour incubation with P6 HSC-secreted exosomes pre-labeled with PKH26 (upper) or that were isolated from the medium of P6 HSC transfected with pLemiR-214 (lower); Scale bar: 10μm. (F) Co-cultures were established for 24 hrs between pre-mir-214-transfected donor LX-2 cells (± GW4869) and recipient HepG2 cells transfected with Fire-Ctx plasmid containing wild-type CCN2 3′-UTR (Supporting Fig. S4B). Luciferase was measured in cell lysates as in (D). (n=4 independent experiments performed in triplicate, +P <0.05 vs. control group).
Finally, since CCN2 is also produced in response to injury by hepatocytes which lie in intimate association with HSC in vivo, we next assessed if exosomal miR-214 transfer occurred between HSC and hepatocytes. Primary mouse hepatocytes were shown to uptake mouse HSC-derived exosomes that had either been labeled with PKH26 fluorescent dye after collection or which contained RFP-miR-214 by transfection of the donor HSC with pLemiR-214 prior to collection (Figure 6E). Since primary mouse hepatocytes were not amenable to transfection with the CCN2 3′-UTR reporter, we used HepG2 cells as an alternative recipient cell as they have proven utility for transfection studies. We thus established co-cultures of control or pre-mir-214-tranfected LX-2 donor cells and HepG2 hepatocyte recipients transfected with the wild-type CCN2 3′-UTR luciferase reporter. Luciferase activity in the HepG2 cells was diminished in the presence of pre-miR-214-transfected LX-2 cells but this effect was not apparent when these donor LX-2 cells were treated with GW4869 (Fig. 6F). Overall, these data showed that HSC-derived miR-214 was delivered via exosomes to hepatocytes in which it then regulates its target CCN2 3′-UTR.
Discussion
Hepatic fibrosis is a highly debilitating pathology which affects millions of people globally and is caused by excessive deposition of fibrous scar tissue during chronic liver injury. Recent reports have indicated that miRs are important regulators of gene expression during liver fibrosis or HSC activation 18 but the underlying mechanisms have not been elucidated. In this study, we have directly addressed this question by identification of the CCN2 3′-UTR as an evolutionarily conserved target of miR-214, providing a mechanistic basis for the reciprocal pattern of expression between CCN2 mRNA and miR-214 observed in models of experimental liver disease or HSC activation. MiR-214 is located within the intron of the dynamin 3 gene and is encoded with miR-199a, producing a 7.9 kb non-coding DNM3 opposite strand transcript 19. Consistent with our data, hepatic miR-199a/214 is down-regulated in rodent models of alcoholic steatohepatitis 15 while expression of miR-214-5p, comprising the complementary sequence from the 5′ arm of the miR-214 hairpin, is enhanced in fibrotic liver and activated HSC 20. Our data show that miR-214 impacts fibrogenic pathways in HSC by direct effects on CCN2 expression and indirect effects on collagen expression downstream of CCN2. Moreover, we found that miR-214 regulates expression of a variety of other fibrosis-related genes in HSC (Supporting Table S1) but the involvement, if any, of the CCN2-miR-214 axis will require further study.
Exosomes are nano-sized membranous vesicles that arise by inward budding from the limiting membranes of multivesicular bodies and are released extracellularly providing for a novel mode of intercellular communication 21. Signaling occurs by surface-expressed ligands that directly stimulate target cells or by the transfer of receptors, functional proteins, infectious agents, or genetic information into target cells. Exosomes are an evolutionary adaptation by which signaling molecules are protected as they traverse the potentially hostile extracellular environment. Here we showed that miR-214 is intercellularly transported via exosomes from HSC donor cells to neighboring recipient HSC or hepatocytes in which the CCN2 3′-UTR is subsequently regulated. Exosomal delivery of miR-214 to activated HSC resulted in decreased expression of endogenous CCN2 mRNA, as well as of mRNAs for αSMA and collagen, both of which are CCN2-dependent 11. Our findings reveal novel mechanisms by which fibrogenic pathways are regulated by exosomes which act as conduits for miR transfer between hepatic cells involved in the fibrotic response. Previous studies of hepatic exosomes have focused on their production by parenchymal (epithelial) cells rather than mesenchymal cells: (i) unidentified components in biliary exosomes caused changes in ERK1/2 phosphorylation, proliferation, or miR-15A expression when added to rat cholangiocytes 22; (ii) exosomes from Hep3B hepatocarcinoma cells regulated expression or down-stream signaling of TGF-β-activated kinase 1 when added to the same cells, an effect consistent with the predicted Hep3B exosomal miR content 23; and (iii) the ability of Huh7 human hepatocytes to inhibit viral replication and gene expression in their HCV replicon counterparts was attributed to exosome-mediated transfer of unidentified small inhibitory RNAs 24.
Production and exchange of exosomal miR-214 among different hepatic cells may contribute to the suppressed levels of hepatic CCN2 expression which is hallmark of many cells in normal liver. Alternatively, exosomal transfer of miR-214 from quiescent or weakly activated HSC may dampen the fibrogenic pathways in more fully activated HSC thus limiting the magnitude of the fibrotic response, a possibility supported by the reduced expression of fibrogenic markers in activated HSC when co-cultured in the presence of pre-miR-214-transfected HSC. Enhanced CCN2 production by HSC, such as that seen during chronic injury, likely reflects diminishing endogenous levels of miR-214 in the cells themselves coupled with diminished concentrations of miR-214 in exosomes produced by activated HSC. However, determination of the in vivo role of exosomes in these processes must await much-anticipated development of instrumentation and animal models that permit exosome imaging, tracking, and functional characterization in the liver in a cell-specific manner.
Fibrosis-related miRs such as miR-214 may have potential utility as non-invasive biomarkers for fibrosis progression especially as circulating exosomes and other microvesicles can be readily harvested and analyzed for their miRs content 25. Emerging data suggests that analysis of circulating miRs may permit discrimination of healthy from injured livers or detection and severity of drug-induced liver injury, hepatic necroinflammation, biliary atresia, hepatic fibrosis, or hepatocellular carcinoma 26–31, but the involvement of exosomes as carriers of these miRs has not yet been addressed. While we detected miR-214 in the serum of normal mice, its cellular origin(s) and presence in circulating exosomes remain(s) to be determined, as does the potential prognostic or diagnostic value of the reduced serum miR-214 levels in mice with hepatic fibrosis. Future studies will address these questions and whether other constituents of the molecular cargo (miR, mRNA and protein) in exosomes produced by HSC are dynamically regulated as a function of the activation status of the cells.
In conclusion, elevated expression of CCN2 during liver fibrosis or HSC activation is due, in part, to reduced expression of miR-214 which otherwise suppresses CCN2 production via its direct binding to the CCN2 3′-UTR. MiR-214 exerts an evolutionarily conserved role in inhibiting CCN2 in HSC and may offer new opportunities as a fibrosis marker or anti-fibrotic agent. MiR-214 is exported extracellularly from HSC by exosomes whereupon it is delivered to neighboring cells, including HSC or hepatocytes, in which it then down-regulates its CCN2 3′-UTR target and suppresses expression of CCN2 and its downstream targets. This is the first description of exosome-dependent delivery of miRs from HSC to adjacent cells and is a paradigm for the epigenetic control of fibrogenic signaling by intercellular miR transfer between various hepatic cell types.
Supplementary Material
Acknowledgments
This study was supported by NIH grants R01 AA021276 (DRB), R01 AA016003 (DRB) and P50AA011099 (H.T.), the latter of which supported the tissue sharing program that provided liver samples from the mouse intragastric feeding model. The authors thank Sherri Kemper and Ben Meacham for technical support, Melissa Piper and Leni Moldovan (Ohio State University, Columbus, OH) for helpful suggestions, Scott Friedman (Mount Sinai Hospital, New York, NY) for providing LX-2 cells, Cindy McAllister (Biomorphology Core, Nationwide Children’s Hospital, Columbus OH) for help with TEM, and Dr Min Gao (Liquid Crystal Institute, Kent State University, Kent OH) for help with cryogenic TEM.
Abbreviations
- CCl4
carbon tetrachloride
- CCN
cysteine-rich-61/connective tissue growth factor/nephroblastoma-overexpressed
- CCN2
connective tissue growth factor
- CTX
cytotoxin
- GFP
green fluorescent protein
- HSC
hepatic stellate cell
- ISH
in situ hybridization
- miR
microRNA
- RFP
red fluorescent protein
- RT-PCR
real time polymerase chain reaction
- TAA
thioacetic acid
- UTR
untranslated region
- αSMA
alpha smooth muscle actin
- TEM
transmission electron microscopy
Footnotes
Conflicts of interests: None of the authors have any conflicts to declare.
Author Contributions: LC: acquire & analyze data, manuscript draft, AC, YZ, RC, BY & KA: acquire & analyze data, HT: material support, funding; LJL & MEP: material support; DRB: study design, funding, manuscript prep.
References
- 1.Friedman SL. Hepatic fibrosis -- overview. Toxicology. 2008;254:120–129. doi: 10.1016/j.tox.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Ghiassi-Nejad Z, Friedman SL. Advances in antifibrotic therapy. Expert Rev Gastroenterol Hepatol. 2008;2:803–816. doi: 10.1586/17474124.2.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brigstock DR, Goldschmeding R, Katsube KI, Lam SC, Lau LF, Lyons K, et al. Proposal for a unified CCN nomenclature. Mol Pathol. 2003;56:127–128. doi: 10.1136/mp.56.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang G, Brigstock DR. Regulation of hepatic stellate cells by connective tissue growth factor. Front Biosci. 2012;17:2495–2507. doi: 10.2741/4067. [DOI] [PubMed] [Google Scholar]
- 5.Abou-Shady M, Friess H, Zimmermann A, di Mola FF, Guo XZ, Baer HU, et al. Connective tissue growth factor in human liver cirrhosis. Liver. 2000;20:296–304. doi: 10.1034/j.1600-0676.2000.020004296.x. [DOI] [PubMed] [Google Scholar]
- 6.Hora C, Negro F, Leandro G, Oneta CM, Rubbia-Brandt L, Muellhaupt B, et al. Connective tissue growth factor, steatosis and fibrosis in patients with chronic hepatitis C. Liver Int. 2008;28:370–376. doi: 10.1111/j.1478-3231.2007.01608.x. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi H, Hayashi N, Hayashi K, Yamataka A, Lane GJ, Miyano T. Connective tissue growth factor and progressive fibrosis in biliary atresia. Pediatr Surg Int. 2005;21:12–16. doi: 10.1007/s00383-004-1254-z. [DOI] [PubMed] [Google Scholar]
- 8.Rachfal AW, Brigstock DR. Connective tissue growth factor (CTGF/CCN2) in hepatic fibrosis. Hepatol Res. 2003;26:1–9. doi: 10.1016/s1386-6346(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 9.Williams EJ, Gaca MD, Brigstock DR, Arthur MJ, Benyon RC. Increased expression of connective tissue growth factor in fibrotic human liver and in activated hepatic stellate cells. J Hepatol. 2000;32:754–761. doi: 10.1016/s0168-8278(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 10.Paradis V, Dargere D, Vidaud M, De Gouville AC, Huet S, Martinez V, et al. Expression of connective tissue growth factor in experimental rat and human liver fibrosis. Hepatology. 1999;30:968–976. doi: 10.1002/hep.510300425. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Charrier AL, Leask A, French SW, Brigstock DR. Ethanol-stimulated differentiated functions of human or mouse hepatic stellate cells are mediated by connective tissue growth factor. J Hepatol. 2011;55:399–406. doi: 10.1016/j.jhep.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brigstock DR. Strategies for blocking the fibrogenic actions of connective tissue growth factor (CCN2): From pharmacological inhibition in vitro to targeted siRNA therapy in vivo. J Cell Commun Signal. 2009;3:5–18. doi: 10.1007/s12079-009-0043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipson KE, Wong C, Teng Y, Spong S. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenesis Tissue Repair. 2012;5 (Suppl 1):S24. doi: 10.1186/1755-1536-5-S1-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, et al. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M. microRNA are Central Players in Anti- and Profibrotic Gene Regulation during Liver Fibrosis. Front Physiol. 2012;3:49. doi: 10.3389/fphys.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loebel DA, Tsoi B, Wong N, Tam PP. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–789. doi: 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Iizuka M, Ogawa T, Enomoto M, Motoyama H, Yoshizato K, Ikeda K, et al. Induction of microRNA-214-5p in human and rodent liver fibrosis. Fibrogenesis Tissue Repair. 2012;5:12. doi: 10.1186/1755-1536-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masyuk AI, Huang BQ, Ward CJ, Gradilone SA, Banales JM, Masyuk TV, et al. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol. 2010;299:G990–999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Q, Ramakrishnaiah V, Henry S, Fouraschen S, de Ruiter PE, Kwekkeboom J, et al. Hepatic cell-to-cell transmission of small silencing RNA can extend the therapeutic reach of RNA interference (RNAi) Gut. 2012;61:1330–1339. doi: 10.1136/gutjnl-2011-300449. [DOI] [PubMed] [Google Scholar]
- 25.Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 26.Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. doi: 10.1371/journal.pone.0026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starkey Lewis PJ, Merz M, Couttet P, Grenet O, Dear J, Antoine DJ, et al. Serum microRNA biomarkers for drug-induced liver injury. Clin Pharmacol Ther. 2012;92:291–293. doi: 10.1038/clpt.2012.101. [DOI] [PubMed] [Google Scholar]
- 28.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 30.Zahm AM, Hand NJ, Boateng LA, Friedman JR. Circulating microRNA is a biomarker of biliary atresia. J Pediatr Gastroenterol Nutr. 2012;55:366–369. doi: 10.1097/MPG.0b013e318264e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.