Abstract
Background&Aims:
Sirtuin (SIRT1) is a NAD+-dependent protein deacetylase that regulates hepatic lipid metabolism by modifying histones and transcription factors. Ethanol exposure disrupts SIRT1 activity and contributes to alcoholic liver disease (ALD) in rodents, but the exact pathogenic mechanism is not clear. We compared mice with liver-specific deletion of Sirt1 (Sirt1LKO) mice with their LOX littermates (controls).
Methods:
We induced alcoholic liver injury in male Sirt1LKO and control mice, placing them on Lieber-DeCarli ethanol-containing diets for 10 days and then administering a single dose of ethanol (5 g/kg body weight) via gavage. Liver and serum samples were collected. We also measured mRNA levels of SIRT1, SFRS10, and lipin-1β and lipin-1α in liver samples from patients with alcoholic hepatitis (AH) and individuals without AH (controls).
Results:
On the ethanol-containing diet, livers of Sirt1LKO mice accumulated larger amounts of hepatic lipid and expressed higher levels of inflammatory cytokines than control mice; serum of Sirt1LKO mice had increased levels of alanine aminotransferase and aspartate aminotransferase. Hepatic deletion of SIRT1 exacerbated ethanol-mediated defects in lipid metabolism, mainly by altering the function of lipin-1, a transcriptional regulator of lipid metabolism. In cultured mouse AML-12 hepatocytes, transgenic expression of SIRT1 prevented fat accumulation in response to ethanol exposure, largely by reversing the aberrations in lipin-1 signaling induced by ethanol. Liver samples from patients with AH had reduced levels of SIRT1 and a higher ratio of Lpin1β:α mRNAs than controls.
Conclusions:
In mice, hepatic deletion of Sirt1 promotes steatosis, inflammation, and fibrosis in response to ethanol challenge. Ethanol-mediated impairment of hepatic SIRT1 signaling via lipin-1 contributes to development of alcoholic steatosis and inflammation. Reagents designed to increase SIRT1 regulation of lipin-1 might be developed to treat patients with alcoholic fatty liver disease.
Keywords: Alcoholic fatty liver, Lipid metabolism, Inflammmation, Signal transduction
Introduction
Alcoholic liver disease (ALD) is a major health problem in the United States and worldwide. Considerable evidence demonstrates that ALD is associated with steatohepatitis as well as more severe forms of liver injury such as fibrosis, cirrhosis, and hepatocellular cancer in humans.1,2 Therefore, efforts to achieve a better understanding of alcoholic steatohepatitis pathogenesis will facilitate development of novel therapeutic strategies.
Mammalian sirtuin (SIRT1) is a nicotinamide adenine dinucleotide (NAD+)-dependent class III histone deacetylase (HDAC).3 The liver is one of the major organs in which SIRT1 plays an essential role in the regulation of lipid metabolism via modification of the acetylation status of a wide range of molecules including histones and transcriptional regulators such as sterol regulatory element-binding protein 1 (SREBP-1), peroxisome proliferator-activated receptor alpha (PPARα), PPAR co-factor (PGC-1α) or forkhead transcription factor O 1 (FoxO1).3 SIRT1 is able to either inhibit or stimulate the activities of these targets, thereby regulating diverse lipid metabolism pathways including lipogenesis, fatty acid β-oxidation, and lipoprotein uptake and secretion in liver. SIRT1 also exerts anti-inflammatory effects by deacetylation of modified lysine residues on transcriptional regulators such as nuclear transcription factor-κB (NF-κB), which is a master transcription factor involved in regulation of pro-inflammatory cytokines.4 Growing evidence from human and rodent studies shows that ALD and non-alcoholic fatty liver disease (NALD) are closely associated with impairment of hepatic SIRT1 signaling.3-7 More importantly, genetic or pharmacological stimulation of SIRT1 protects against both ALD and NALD in animals and humans.3-7 Nevertheless, the causal role of SIRT1 in pathogenesis of ALD has yet to be evaluated.
Lipin-1 is a protein that exhibits dual functions as an Mg2+-dependent phosphatidic acid phosphohydrolase (PAP) in the triglyceride (TG) synthesis pathway and a transcriptional co-activator to promote fat oxidation and suppress de novo lipogenesis.8,9 In the liver, lipin-1 has two major protein isoforms, lipin-1α (891 amino acids) and lipin-1β (924 amino acids), derived from LPIN 1 alternative mRNA splicing.10-12 Lipin-1α and lipin-1β display different subcellular localizations.10-12 In hepatocytes, lipin-1α is predominantly located in the nucleus, whereas lipin-1β is exclusively located in the cytoplasm.13-14 The LPIN 1β isoform is associated with increased expression of lipogenic genes, and excessive hepatic lipid accumulation in animals.11,12 Accumulating evidence demonstrates that ethanol-mediated aberrant lipin-1 signaling contributes to development of alcoholic steatosis in rodents and humans.14-18 Our group found that ethanol significantly accentuates the cytoplasmic pro-lipogenic activity of lipin-1 whereas lipin-1 nuclear entry is attenuated by chronic ethanol exposure in cultured hepatocytes or in mouse livers.14,18
The net consequence of these ethanol-mediated effects on lipin-1 localization is associated with excessive fat accumulation in the liver. To date, there is little knowledge of how SIRT1 regulates lipin-1 signaling in liver. Our recent studies suggest that ethanol-mediated SIRT1 inhibition may alter acetylation/sumoylation status of lipin-1, thereby disrupting lipin-1 signaling in hepatocytes.14,18.
SFRS10 (official gene name, TRA2B), the homolog of Drosophila transformer-2 (Tra2), belongs to the SR-like protein family of splicing factors.12 The alternatively spliced exon 6 of LPIN 1 contains a GGAA sequence motif that binds to SFRS10. Thus, SFRS10 emerges as an important modulator of alternative splicing of LPIN 1.12 The effects of reduced SFRS10 on LPIN1 splicing, favoring the LPIN1β isoform, are sufficient to increase expression of lipogenic genes, activate lipogenesis and cause liver steatosis in mice.12 Of note, SFRS10-mediated LPIN1 splicing is PAP-independent. SFRS10 is found to be down-regulated in the fatty livers of high-fat fed mice as well as obese humans.12
In the present study, using hepatocyte-specific SIRT1 knockout (Sirt1LKO) mice and their LOX littermates (WT), we have demonstrated for the first time that specific ablation of hepatic SIRT1 in mice impairs lipin-1 signaling. Disruption of lipin-1 signaling results in increased expression of genes encoding lipogenic enzymes, reduced expression of genes encoding fatty acid oxidation enzymes, and ultimately leads to aggravated hepatic steatosis and exacerbated inflammation in response to ethanol administration.
Materials and Methods
Mice
Hepatocyte-specific SIRT1 knockout (Sirt1LKO) mice were generated as described previously.5 Briefly, the SIRT1 allele with floxed exon 4 was backcrossed into the C57BL/6 background. It was then bred with mice expressing the Cre recombinase driven by the albumin promoter (Jackson laboratory) to generate Sirt1LKO in over 98% C57BL/6 background. Male Sirt1LKO mice and their age-matched littermate LOX controls (WT) were fed ad libitum a standard laboratory chow diet.
Ethanol feeding Studies
A mouse model of chronic plus single binge ethanol consumption (referred as chronic-binge model) was used as previously described.19,20 Ten to twelve week-old male Sirt1LKO mice and their age-matched LOX littermate (WT) controls were divided into four dietary groups: (1) WT control [low fat (LF) diet alone, 10% of calories as fat (6% cocoa butter and 4% safflower oil)]; (2) WT plus ethanol (E; identical to the control LF diet but with 5% w/v ethanol added); (3) Sirt1LKO control; (4) Sirt1LKO plus ethanol. All mice were first fed a LF liquid control diet for 5 days. Ethanol groups were then fed a liquid diet containing 5% w/v ethanol for 10 days while control mice were pair-fed to their ethanol fed counterparts for 10 days. At day 11, mice in ethanol groups were gavaged a single dose of ethanol (5g/kg body weight, 20% ethanol), while mice in control groups were gavaged an isocaloric dose of dextrin maltose. The gavage was performed in the early morning, and mice had access to the diets after alcohol gavage. The mice were euthanized and blood and tissue samples were collected 9 h post-gavage. Liquid diets were freshly prepared from powder daily. For mice on an ethanol-containing diet, animal cages were placed on heating pads to maintain body temperature to prevent ethanol-induced hypothermia. Food intake was recorded daily. During the entire feeding period, Sirt1LKO mice and their WT counterparts were observed to consume similar volumes of ethanol containing-diets. The studies were approved by the Institutional Animal Care and Use Committee of University of South Florida.
Measurements of Hepatic Lactate, Pyruvate and Lipid Peroxidation
The concentrations of hepatic lactate and pyruvate were determined using deproteinized liver samples with commercial kits from BioVision (Mountain View, CA).21 The lipid peroxidation end-product, malondialdehyde (MDA), in liver was measured based on the formation of thiobarbituric reactive substances and expressed as the extent of MDA production.21
Statistical Analysis
Data are expressed as mean±SEM. Differences between groups were analyzed using a two-tailed unpaired Student’s t-test (for human liver samples) or one-way analysis of variance (ANOVA) test followed by the Newman–Keuls multiple ranges test, with a statistically significant difference defined as a p value < 0.05.
Results
Hepatocyte-Specific Deletion of SIRT1 Alters Lipin-1 mRNA Splicing in Mice
We examined the function of SIRT1 in the liver utilizing hepatocyte-specific SIRT1 knockout (Sirt1LKO) mice on the C57BL/6 background. Consistent with previous findings, Sirt1LKO mice were viable and phenotypically normal under a chow diet.5
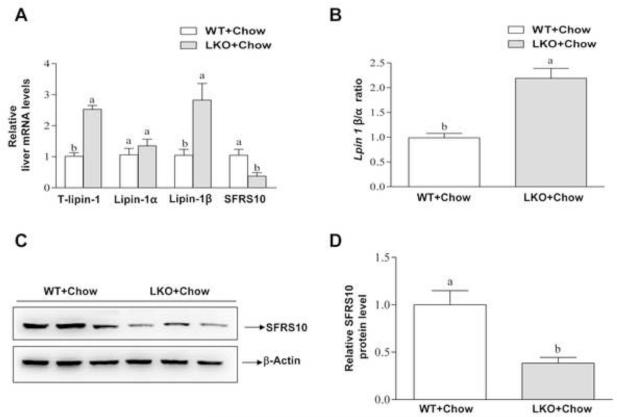
We then examined whether SIRT1 is associated with lipin-1 signaling. Removal of hepatic SIRT1 significantly induced mRNA expression levels of total lipin-1 and lipin-1β by nearly 2.5-fold, respectively, compared with the WT controls (Figure 1A and B). Accordingly, the ratio of Lpin 1 β/α was increased by approximately 2-fold in the livers of Sirt1LKO mice compared to WT controls (Figure 1B). Subsequent analysis revealed that both mRNA and protein expression levels of SFRS10, a splicing factor that regulates lipin-1 mRNA alternative splicing, were severely reduced in Sirt1LKO compared to WT controls (Figure 1C and D). Collectively, these data clearly demonstrate that hepatic SIRT1 deficiency disturbs hepatic lipin-1 mRNA alternative splicing in mice.
Figure 1. Liver-specific deletion of SIRT1 alters lipin-1 signaling in mice.
Sirt1LKO (LKO) and LOX littermates (WT) mice were fed chow diets. (A) Relative liver mRNA levels of total (T) lipin-1, lipin-1α, lipin-1β, and SFRS10. (B) Hepatic Lpin-1β/α ratio. (C) Representative Western analysis of SFRS10 protein expression levels. (D) Relative SFRS10 protein levels. Results are expressed as mean ± SEM (n=4-8 mice). Means without a common letter differ, p< 0.05.
Liver-Specific Deletion of SIRT1 Exacerbates Chronic-Binge Ethanol-Induced Fatty Liver Injury in Mice
We investigated the role of SIRT1 on alcoholic fatty liver by pair-feeding WT and Sirt1LKO mice using a chronic-binge ethanol feeding protocol (Supplementary Figure 1A).19,20 Mice were fed ethanol-containing diets for 10 days followed by a single gavage of ethanol. Control mice were pair-fed control diets without ethanol for 10 days followed by a single gavage of maltose.
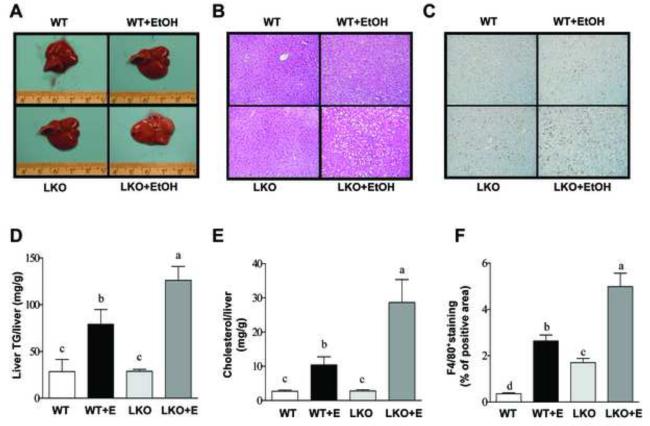
Chronic-binge ethanol significantly increased liver weight to body weight ratio, liver triglyceride, cholesterol and caused mild liver steatosis in WT mice compared with pair-fed WT controls (Figure 2 and Supplementary Table I). Ethanol feeding caused significantly higher immunohistochemical staining of F4/80+, a marker of liver inflammation, in the livers of WT mice compared to WT controls (Figure 2C and F). Interestingly, the serum AST or ALT levels were significantly but mildly elevated in the ethanol-fed WT mice compared to WT controls (Supplementary Figure 1B and C). It has been reported that plasma ALT levels in mice were substantially elevated in response to chronic-binge ethanol challenge.19,20 This discrepancy may be due to the fact that a low fat diet was used in our study.21
Figure 2. Liver-specific deletion of SIRT1 aggravates alcoholic fatty liver in mice.
Sirt1LKO (LKO) mice were fed chronically ethanol containing diets for 10 days followed by single gavage of ethanol. WT Control mice were pair-fed control diets without ethanol for 10 days followed by single gavage of maltose. (A) Pictures of mouse livers. (B) Hematoxylin and eosin (H&E) staining (original magnification×40) of liver sections. (C) Immunohistochemical staining for F4/80+ (original magnification×40). (D) Hepatic triglyceride (TG) levels. (E) Hepatic cholesterol levels. (F) Relative levels of F4/80+ staining. Results are expressed as mean ± SEM (n=4-8 mice). Means without a common letter differ, p< 0.05.
Ethanol administration exacerbated the development of liver injury in Sirt1LKO mice compared to WT mice, as indicated by augmented liver weight to body weight ratio, markedly enhanced hepatic triglyceride and cholesterol contents, significantly elevated serum ALT and AST levels, and substantially enhanced the levels of F4/80+ staining in the livers of ethanol-fed Sirt1LKO mice compared with the livers of ethanol-fed WT mice (Figure 2 and Supplementary Table 1).
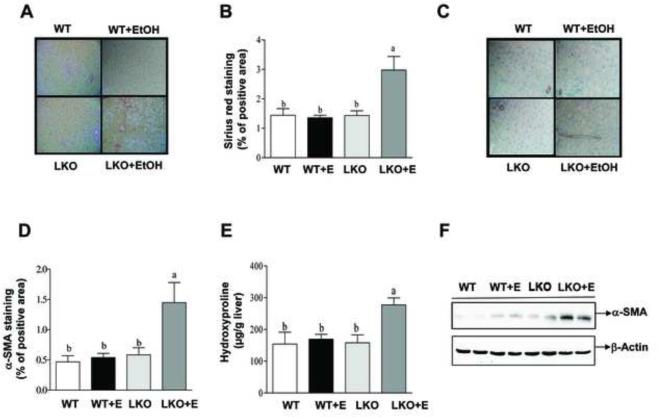
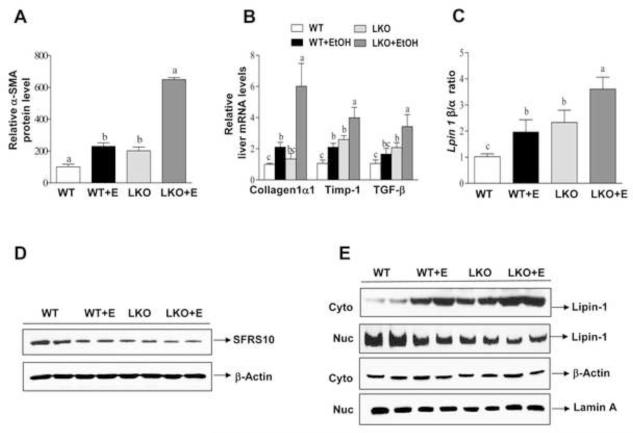
Immunohistochemical staining for Sirius red, α-SMA and collagen revealed higher levels of those indicators of liver fibrosis in the livers of ethanol-fed Sirt1LKO mice compared with the livers of ethanol-fed WT mice (Figure 3 and Figure 4A). Accordingly, ethanol feeding increased the hydroxyproline content and α-SMA levels in Sirt1LKO mice compared to all other groups (Figure 2D). Moreover, the mRNA expression of early markers of hepatic fibrosis such as collagen I, tissue inhibitor of metalloproteinase 1 (Timp-1), or transforming growth factor beta 1 (TGF-β) was augmented in ethanol-fed Sirt1LKO mice compared to ethanol-fed WT mice (Figure 4B). The mRNA levels of fibronectin were significantly increased in ethanol-fed WT and in Sirt1LKO mice with or without receiving ethanol when compared to controls, whereas CD65 mRNA levels were unchanged in all of the groups (Supplementary Figure 2). These results suggest that ethanol-fed Sirt1LKO mice partially progressed from fatty liver to mild fibrotic liver.
Figure 3. Liver-specific deletion of SIRT1 causes liver fibrosis in ethanol-fed mice.
Mice were fed as described in Fig.2. (A) Sirius red staining (original magnification×40). (B) Relative hepatic levels of Sirius red staining. (C) Immunohistochemical staining for α-SMA (original magnification×40). (D) Relative hepatic levels of α-SMA staining. (E) Hepatic hydroxyproline levels. (F) Western analysis of hepatic α-SMA protein expression levels. Results are expressed as mean ± SEM (n=4-8 mice). Means without a common letter differ, p< 0.05.
Figure 4. Liver-specific deletion of SIRT1 promotes ethanol-mediated impairment of lipin-1 signaling in mice.
Mice were fed as described in Fig. 2. (A) Relative levels of α-SMA staining. (B) Relative hepatic mRNA levels of Collagen1α, Timp-1, and TGF-β. (C) Hepatic Lpin-1β/α ratio. (D) Representative Western analysis of hepatic SFRS10 protein expression levels. (E) Representative western blotting analysis of the lipin-1 protein expression levels in nucleus (Nuc) or cytoplasm (Cyto) of mice fed a control or ethanol diet. All data are expressed as mean ± SEM (n =3-8 animals). Means without a common letter differ, p < 0.05.
Taken together, our data clearly demonstrate that liver-specific removal of SIRT1 exacerbates the development of alcoholic fatty liver injury in mice.
Hepatocyte-Specific Deletion of SIRT1 Drastically Exacerbates Oxidative Stress and Promotes the Inflammatory Responses in Mice Fed Ethanol
Analysis of mRNA from livers of Sirt1LKO mice revealed a significant increase in macrophage markers such as macrophage inflammation protein (MIP-1α) and F4/80+, and the macrophage markers were further increased in ethanol-fed Sirt1LKO mice (Supplementary Figure 3A). Accordingly, the mRNA levels of pro-inflammatory cytokines including tumor necrosis factor-alpha (TNF-α) and monocyte chemoattractant protein 1 (MCP-1), two prominent pro-inflammatory cytokines involved in pathogenesis of ALD, were significantly higher in the livers of Sirt1LKO mice fed with or without ethanol compared to WT mice, supporting the role of SIRT1 as a anti-inflammatory regulator (Supplementary Figure 3A).3,4 Consistently, the semi-quantification of histological inflammation showed that removal of hepatic SIRT1 promoted the liver inflammatory responses in ethanol-fed mice (Supplementary Figure 3C).
Interestingly, ethanol feeding dramatically increased the mRNA levels of two pro-inflammatory molecules, lipocaline-2 (Lcn-2) and serum amyloid A-1 (SAA-1), in the livers of WT controls and Sirt1LKO mice compared to their respective controls, suggesting involvement of both SIRT1-dependent and SIRT1-independent inflammatory mechanisms (Supplementary Figure 3B). Note that rremoval of hepatic SIRT1 significantly increased SAA1 mRNA levels in the absence of ethanol.
SIRT1 ablation mildly but significantly generates oxidative stress, as demonstrated by the slight increase in hepatic malondialdehyde (MDA), a lipid peroxidation end-product, compared to WT controls (Supplementary Table I). However, ethanol feeding to Sirt1LKO mice drastically increased MDA levels approximately 5-fold compared with all other groups, demonstrating that oxidative stress is drastically exacerbated in response to ethanol administration in Sirt1LKO mice.
Removal of Hepatic SIRT1 Promotes Ethanol-Mediated Impairment of Lipin-1 Signaling in Mice
We examined the involvement of lipin-1 signaling in exacerbated alcoholic fatty liver injury in Sirt1LKO mice. WT mice fed with ethanol or Sirt1LKO mice fed with a control diet slightly but significantly reduced the mRNA levels of hepatic lipin-1α whereas lipin-1β mRNA expression levels were increased in those mice compared to WT controls (Supplementary Figure 4A). As a result, the ratio of Lpin1 β/α in the livers of ethanol-fed WT mice or Sirt1LKO mice was significantly elevated compared to WT controls (Figure 4C). More importantly, hepatic SIRT1 deficiency significantly augmented the ethanol-mediated increase of Lpin 1 β/α compared to all other groups (Figure 4C). Ethanol administration significantly inhibited mRNA and protein levels of SFRS10 in WT mice compared to WT controls (Figure 4D and Supplementary Figure 4B). Ethanol-mediated SFRS10 inhibition was further slightly but significantly augmented in the livers of ethanol-fed Sirt1LKO mice compared with all other groups (Figure 4D and Supplementary Figure 4B).
Nucleocytoplasmic shutting of lipin-1 regulates its function as a transcriptional co-activator.8-14, 22-24 Ethanol-fed WT mice or Sirt1LKO mice displayed a markedly increased amount of lipin-1 in the cytoplasm whereas the amount of nuclear lipin-1 was significantly reduced in those mice (Figure 4E and Supplementary Figure 4C). Ethanol-mediated lipin-1 nucleocytoplasmic shuttling was greatly augmented in the Sirt1LKO mice compared to all other groups (Figure 4E and Supplementary Figure 4C). These data demonstrate that ethanol feeding exacerbates impairment of hepatic lipin-1 signaling in Sirt1LKO mice.
Hepatic-Specific Removal of SIRT1 Exacerbated Ethanol-Mediated Abnormalities in Hepatic Lipid Metabolism Pathways in Mice
Ethanol-mediated SIRT1 inhibition activates SREBP-1.21,25 As such, we investigated whether SIRT1 deficiency exacerbates ethanol's stimulation of SREBP-1 signaling. Indeed, the amount of active, nuclear form of SREBP-1 (nSREBP-1c) protein was highest in the livers of ethanol-fed Sirt1LKO mice compared to all other groups (Supplementary Figure 5A and B). Immunofluorescent staining for SREBP-1 (green) and DAPI (blue) further confirmed these findings (Supplementary Figure 5C). Additionally, ethanol feeding significantly increased hepatic mRNA expression levels of lipogenic enzymes including SREBP-1c, fatty acid synthase (FAS), stearoyl-coenzyme A desaturase 1 (SCD1) and ACC where these increases were more pronounced in ethanol-fed Sirt1LKO mice (Supplementary Figure 5D).
Ethanol impairs PGC-1α signaling via SIRT1.21 While ethanol significantly reduced mRNA or protein expression of PGC-1α in WT mice, PGC-1α expression levels was further suppressed in the livers of ethanol-fed Sirt1LKO mice (Supplementary Figure 6A and B). Accordingly, SIRT1 ablation further suppressed the mRNA expression levels of several PGC-1α-regulated enzymes involved in fatty acid oxidation, including PPARα, carnitine palmitoyltransferase 1a (CPT1a), acyl-CoA oxidase (AOX), and CD36, to levels significantly lower than all other groups (Supplementary Figure 6C).
It is worthwhile to note that ethanol administration to Sirt1LKO mice substantially increased PPARγ mRNA levels compared to all other groups, suggesting that PPARγ also contributes, at least in part, to aggravated alcohol-induced liver steatosis in Sirt1LKO mice (Supplementary Figure 6C). Taken together, our data suggest that removal of hepatic SIRT1 in mice exacerbates ethanol-mediated abnormalities in hepatic lipid metabolism.
Ethanol Feeding Promotes Nucleocytoplasmic Shuttling of NFATc4 and Aggravates ER Stress in the Livers of SIRT1LKO Mice
Lipin-1 regulates nuclear factor of activated T cells c4 (NFATc4), an important inflammatory factor.26 As shown in Figure 5A and B, in comparison with all other groups, ethanol administration to Sirt1LKO mice robustly induced nuclear accumulation of NFATc4 and significantly decreased the amount of NFATc4 in the cytoplasm, suggesting that exacerbated inflammation in the livers of ethanol-fed Sirt1LKO mice may be mediated, at least in part, by impairment of the hepatic SIRT1-lipin-1-NFATc4 axis.
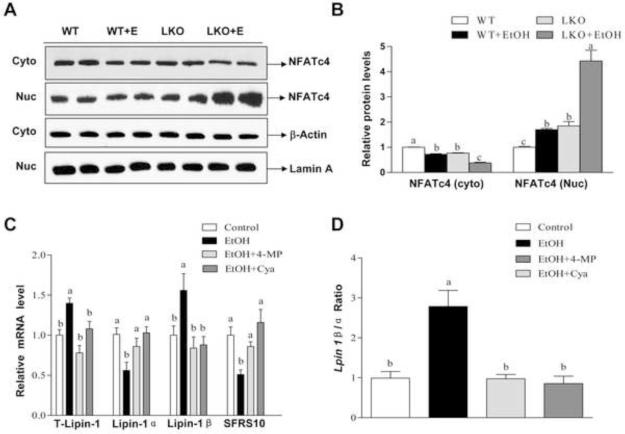
Figure 5. Liver-specific deletion of SIRT1 promotes ethanol-induced nucleocytoplasmic shuttling of NFATc4 in mice.
Mice were fed as described in Fig. 2. (A) Representative western blotting analysis of the NFATc4 protein expression levels in nucleus (Nuc) or cytoplasm (Cyto) of mice fed a control or ethanol diet. (B) Relative hepatic NFATc4 protein levels. (C) AML-12 cells were treated with ethanol (50 mM) in the presence or absence of 4-methylpyrazole (4-MP) (0.1 mM), cyanamide (Cya) (0.1 mM) for 48 h. Relative mRNA levels of total lipin-1, lipin-1α, lipin-1β, and SFRS10. (D) Lpin1 β/α ratio. Results are expressed as mean ± SEM (n=3-6 mice). All in vitro data are from at least 3 experiments. Means without a common letter differ, p< 0.05.
Roles for hepatic excessive fat accumulation, inflammation and oxidative stress are all linked to endoplasmic reticulum (ER) stress in the etiology of alcoholic stestohepatistis.1,2,27 Indeed, XBP-1 mRNA splicing was significantly enhanced in the livers of Sirt1LKO mice fed with or without ethanol compared to WT controls (Supplementary Figure 7). Consistently, significantly increased mRNA of CHOP was detected in ethanol-fed WT or Sirt1LKO mice compared to WT controls (Supplementary Figure 8A). More importantly, the mRNA expression levels of XBP-1 and CHOP were greatly increased in ethanol-fed Sirt1LKO mice compared to all other groups. Accordingly, increased Bip levels, phosphorylation of JNK (p-JNK) and total JNK expression were significantly augmented in the livers of ethanol-fed Sirt1LKO mice compared to other groups (Supplementary Figure 8B-D). Together, these findings demonstrate that aggravated liver injury in Sirt1LKO results in elevated ER stress in the liver in response to ethanol challenge.
Ethanol Metabolism Perturbs Lpin 1 Alternative Splicing Via SFRS10 Inhibition in AML-12 Hepatocytes
We dissected the mechanisms by which ethanol exposure disturbs hepatic lipin-1 signaling using cultured AML-12 hepatocytes. Consistent with in vivo animal studies, the mRNA levels of SFRS10 in AML-12 cells treated with ethanol were significantly lower than controls (Figure 5C). Furthermore, mRNA expression levels of lipin-1β were up-regulated in AML-12 cells exposed to ethanol compared to controls, whereas lipin-1α mRNA expression levels were significantly inhibited by ethanol exposure (Figure 5C). As a result, the Lpin 1 β/α ratio was significantly increased in response to ethanol exposure compared to controls (Figure 5D).
Hepatic ethanol metabolizes via class I (low Km) alcohol dehydrogenase (ADH) and aldehyde dehydrogenase 2 (ALDH2) proteins in AML-12 hepatocytes.14,18 Treatment with either 4-methylpyrazole (an ADH inhibitor) or cyanamide (an ALDH2 inhibitor) blocked the ability of ethanol to interfere with lipin-1 signaling, indicating that ethanol regulates lipin-1 mRNA alternative splicing via its metabolism (Figure 5C and D).
We further investigated whether ethanol-induced aberrant lipin-1 signaling is mediated through inhibition of the SIRT1-SFRS10 axis using a cellular alcoholic steatosis model. Incubation of AML-12 hepatocytes with ethanol increased intracellular lipid accumulation (Supplementary Figure 9 and Figure 10A). This lipid accumulation was largely reversed when AML-12 cells were transfected with SIRT1wt or SFRS10wt. Conversely, overexpression of SIRT1H363Y, SIRT1siRNA or SFRS10siRNA significantly augmented the TG levels in AML-12 hepatocytes (Supplementary Figure 9 and Figure 10A). Accordingly, co-transfection SIRT1wt or SFRS10wt largely blocked the ability of ethanol to disturb the Lpin 1β/α axis (Supplementary Figure 10B and Figure 11). These SIRT1- or SFRS10-medaited protective effects were not seen when SIRT1 or SFRS10 was inhibited by SIRT1H363Y, SIRT1siRNA or SFRS10siRNA (Supplementary Figure 11). Our data demonstrate that increased Lpin 1β/α ratio by ethanol is mediated, at least in part, through SIRT1-SFRS10 inhibition in hepatocytes.
Impairment of SIRT1-SFRS10-Lpin 1β/α Axis in the Livers of Patients with Alcoholic Hepatitis
To investigate the clinical relevance of SIRT1-SFRS10-Lpin1 β/α axis in the pathogenesis of ALD, we examined mRNA expression levels of SIRT1, SFRS10 and Lpin1 β/α in liver samples from patients with alcoholic hepatitis (AH).28-31
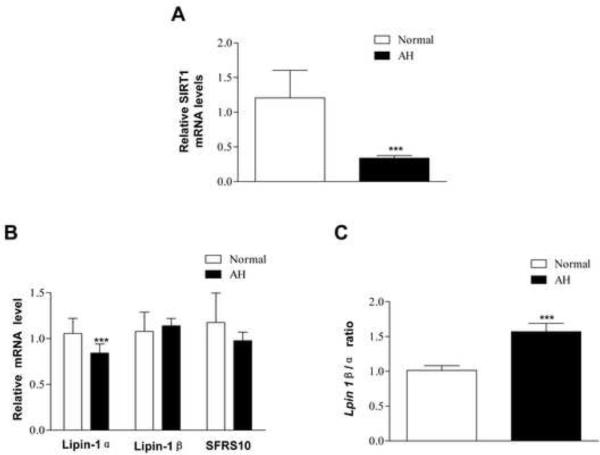
Remarkably, SIRT1 mRNA levels were severely decreased in the livers of patients with AH compared to the normal controls (Figure 6A). The hepatic lipin-1α mRNA levels were significantly lower in patients with AH whereas lipin-1β mRNA expression was not significantly altered in the livers of AH compared to controls. As a result, the ratio of hepatic Lpin1 β/α in the livers of patients with AH was modestly but significantly elevated (Figure 6C). The hepatic mRNA levels of SFRS10 were indeed reduced in patients with AH but did not reach statistical significance compared to controls (Figure 6B). Collectively, our data clearly suggest that impairments of the SIRT1-SFRS10-Lpin 1β/α axis may contribute, at least in part, to the development of human ALD such as AH.
Figure 6. Hepatic mRNA levels of SIRT1, lpin1β/α, SFRS10 from patients with alcoholic hepatitis.
Real-time PCR analyses of liver tissues from normal livers (n=6) and patients with alcoholic hepatitis (AH) (n=34). (A) Relative mRNA levels of SIRT1. (B) Hepatic ratio of Lpin1 β/α. (C) Relative mRNA levels of lipin-1α, lipin-1β and SFRS10. All data are expressed as mean ± SEM (normal liver, n=3; AH, n=34). ***p<0.001.
Discussion
In the present study, utilizing a murine model of chronic-binge ethanol feeding, we clearly demonstrated that genetic ablation of hepatic SIRT1 is associated with the rapid onset and progression of steatosis, inflammation, and fibrosis in response to ethanol challenge compared to WT control mice. We found that ethanol administration to Sirt1LKO mice disrupted lipin-1 signaling, resulting in exacerbated abnormalities in hepatic lipid metabolism which eventually leads to steatosis and inflammation in the liver. Additional mechanistic studies revealed that ethanol metabolism specifically disturbed Lpin1 β/α ratio, largely via SIRT1-SFRS10 inhibition, which results in steatosis in AML-12 hepatocytes. Consistently, the SIRT1-SFRS10-Lpin 1β/α axis was perturbed in the livers from patients with AH. Altogether, our novel findings demonstrate that loss of hepatic SIRT1 aggravates alcoholic steatohepatitis in whole or in part via disrupting lipin-1 signaling in mice (Figure 7).
Figure 7.
Proposed role of SIRT1-lipin-1 signaling in the development of alcoholic steatohepatitis
The present study suggests that hepatic SIRT1-lipin-1 axis represents a major signaling route in ethanol-induced steatosis in hepatocytes and in mice. In recent years, several important transcriptional regulators such as SIRT1, SREBP-1, and PGC-1α have been identified as specific targets of ethanol action in the liver.1,14,18,21,22 Among those molecules, SIRT1 appears to be the most upstream signaling molecule targeted by ethanol in liver. However, the central issue of how ethanol-mediated impairment of SIRT1 leads to subsequent liver steatosis and inflammation is not completely understood. We provide evidence suggesting that lipin-1 is a vital downstream regulator that may be responsible for ethanol-mediated impairment of a hepatic signaling network controlled by SIRT1, and ultimately that dysregulation of this pathway leads to development of steatosis and inflammation in liver.
Lipin-1 mRNA alternative splicing is regulated by SFRS10.12 Reduced SFRS10 alters the generation of LPIN1 splice isoforms, yielding an increase in the β isoform in parallel with a decrease in the α isoform. However, how hepatic SFRS10 is regulated is largely unknown. Our study, for the first time to the best of our knowledge, demonstrates the interplay between SIRT1 and SFRS10. However, the precise mechanism by which SIRT1 regulates SFRS10 gene and protein expression remains to be elucidated. Our data imply that SIRT1 up-regulates SFRS10 at multiple levels, including transcriptional and post-transcriptional levels. It is possible that SIRT1 increases SFRS10 mRNA stability leading to elevated SFRS10 mRNA and subsequently protein levels. Moreover, a recent study demonstrates that SIRT1 up-regulates the H3K9me3 methyltransferase Suv39h1 via increasing its half-life through inhibition of Suv39h1 lysine 87 polyubiquitination by the E3-ubiquitin ligase MDM2.32 Therefore, it is logical to speculate that SIRT1 may physically interact with the SFRS10 protein and modulate the acetylation status of SFRS10 which prevents proteasomal degradation of SFRS10 and subsequently stabilizes and increases SFRS10 protein expression levels in hepatocytes. Surprisingly, we found that the changes in mRNA levels of lipin-1α or SFRS10 in liver samples from patients with AH were modest, suggesting that gene expression of SFRS10 or lpin1 β/α in the livers of patients with AH may correlate with liver disease severity. Further studies may be needed to evaluate if the findings obtained in these patients with severe AH are also found in the livers with mild to moderate fatty liver injury.
We found that ethanol-mediated inhibition of SIRT1 leads to reduced SFRS10 gene and protein expression levels in hepatocytes. On the other hand, ethanol may directly down-regulate SFRS10. Ethanol metabolism via ADH and ALDH2 are required for the inhibitory effect of ethanol on SFRS10 in cultured hepatocytes. Hepatic ethanol metabolism is known to causes a shift in redox state and generates oxidative stress in hepatocytes.1 Therefore, it is possible that SFSR10 is inhibited due to an altered redox state or excessive reactive oxygen species production in hepatocytes exposed to ethanol. The exact mechanism by which ethanol disrupts hepatic SFRS10-Lpin 1 β/α axis merits further investigation.
In addition to the SIRT1-SFRS10-Lpin 1 β/α axis, ethanol likely affects lipin-1 at multiple levels. For instance, we have recently reported that chronic ethanol administration affects acetylation/SUMOylation modifications of lipin-1 and that alteration of these modifications subsequently disturbs lipin-1 signaling in mouse livers.18,33 Therefore, ethanol-mediated SIRT1 inhibition may directly influence acetylation/SUMOylation modifications of lipin-1. Moreover, the impaired SIRT1-lipin-1 signaling can not be solely responsible for alcoholic steatosis. In hepatocytes, SIRT1 interacts with PPARα and is required to activate PGC-1α to induce fatty acid oxidation enzymes.3-5 Ethanol-mediated SIRT1 inhibition may directly perturb PGC-1α/PPAR signaling.1,2 Interestingly, nuclear lipin-1 functions as a transcriptional co-activator by interacting with PGC-1α/PPARα.34 Impaired function of the SIRT1-lipin-1 axis in alcoholic steatosis may lead to inhibited PGC-1α/PPARα, and subsequent incomplete activation of its target genes encoding fatty acid oxidation enzymes.
It is intriguing that removal of SIRT1 in the livers of mice slightly affects AMP-activated kinase (AMPK) and miR-217, two known regulators of SIRT1 (Supplementary Figure 12A and B).14,21 Interestingly, AMPK activity was inhibited in both WT and Sirt1LKO mice receiving ethanol to the same extent. Ethanol feeding also did not further elevate hepatic miR-217 levels compared to WT mice. These findings suggest that the aggravated steatohepatitis in ethanol-fed Sirt1LKO mice may be regulated in an AMPK- or miR-217-independent manner.
The augmented inflammatory response in ethanol-fed Sirt1LKO mice is likely due to increased hepatic fat accumulation and elevated ER stress. Interestingly, we have recently observed that liver specific-deletion of lipin-1 in mice led to the exacerbated inflammatory response via activation of NF-κB and NFATc4, two vital inflammatory regulators (M.You, preliminary observations). It is possible that ethanol-mediated disruption of SIRT-lipin-1 via NFATc4 signaling is partially responsible for the inflammatory response observed in the livers of ethanol-fed Sirt1LKO mice. The role of the SIRT1-lipin-1 axis in ethanol-mediated hepatic inflammation is currently under investigation in our laboratory.
In summary, we have presented novel evidence that ethanol-mediated impairment of SIRT1-lipin-1 signaling is crucial in the development of alcoholic steatohepatitis in mice. Our findings further highlight the SIRT1-SFRS10-Lpin 1 β/α axis as a major signaling route in the pathogenesis of alcoholic liver disease. These findings suggest that nutritional or pharmacological modulation of SIRT1-SFRS10-Lpin1 β/α could be potential therapeutic approaches for treating human alcoholic steaotohepatitis.
Supplementary Material
Acknowledgements
The authors thank Malika Humprhies from the Department of Medicine at University of North Carolina for her technical support.
Grants
This study was supported by grants from National Institute on Alcoholism and Alcohol Abuse (AA013623 and AA015951 to M You and R21AA021247 to SM Stevens Jr.), Center for Gastrointestinal Biology and Disease, University of North Carolina at Chapel Hill and Fundación Española para el Estudio del Hígado (to G Odena).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Huquan Yin, Ming Hu, Xiaomei Liang, Joanne M. Ajmo, Ramon Bataller and Gemma Odena contributed to generating the data presented, making the figures and tables and drafting the procedures. Xiaoling Li contributed to providing Sirt1LKO mice, and editing and revising manuscript. Stanley M. Stevens Jr. contributed to providing conception and design of research, editing and revising manuscript. Min You contributed to designing the experiments, and drafting and the completion of the manuscript through supervision of others, and approving final version of manuscript.
No Conflicts of Interest Exist
REFERENCES
- 1.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–85. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang HJ, Gao B, Zakhari S, et al. Inflammation in alcoholic liver disease. Annu Rev Nutr. 2012;21(32):343–68. doi: 10.1146/annurev-nutr-072610-145138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schug TT, Xu Q, Gao H, et al. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol Cell Biol. 2010;30:4712–21. doi: 10.1128/MCB.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purushotham A, Schug TT, Xu Q, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–38. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Min HK, Kapoor A, Fuchs M, et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15:665–74. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmers S, Konings E, Bilet L, et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14:612–22. doi: 10.1016/j.cmet.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011;22:226–33. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterfy M, Phan J, Xu P, et al. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 10.Han GS, Carman GM. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem. 2010;285:14628–38. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Péterfy M, Phan J, Reue K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J Biol Chem. 2005;280:32883–9. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- 12.Pihlajamäki J, Lerin C, Itkonen P, et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011;14:208–18. doi: 10.1016/j.cmet.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bou Khalil M, Sundaram M, Zhang HY, et al. The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J Lipid Res. 2009;50:47–58. doi: 10.1194/jlr.M800204-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Yin H, Hu M, Zhang R, et al. MicroRNA-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating SIRT1. J Biol Chem. 2012;287:9817–26. doi: 10.1074/jbc.M111.333534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brindley DN, Cooling J, Burditt SL, et al. The involvement of glucocorticoids in regulating the activity of phosphatidate phosphohydrolase and the synthesis of triacylglycerols in the liver. Effects of feeding rats with glucose, sorbitol, fructose, glycerol, and ethanol. Biochem J. 1979;180:195–199. doi: 10.1042/bj1800195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savolainen MJ, Baraona E, Pikkarainen P, et al. Hepatic triacylglycerol synthesizing activity during progression of alcoholic liver injury in the baboon. J Lipid Res. 1984;25:813–820. [PubMed] [Google Scholar]
- 17.Simpson KJ, Venkatesan S, Martin A, et al. Activity and subcellular distribution of phosphatidate phosphohydrolase (EC 3.134) in alcoholic liver disease. Alcohol Alcohol. 1995;30:31–36. [PubMed] [Google Scholar]
- 18.Hu M, Wang F, Li X, et al. Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology. 2012;55:437–46. doi: 10.1002/hep.24708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ki SH, Park O, Zheng M, et al. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291–300. doi: 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertola A, Mathews S, Ki SH, et al. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–37. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajmo JM, Liang X, Rogers CQ, et al. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G833–42. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller JF, Krawczyk SA, Gostilovitch L, et al. Glucose-6-phosphate isomerase deficiency results in mTOR activation, failed translocation of lipin 1α to the nucleus and hypersensitivity to glucose: Implications for the inherited glycolytic disease. Biochim Biophys Acta. 2011;1812:1393–402. doi: 10.1016/j.bbadis.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdearcos M, Esquinas E, Meana C, et al. Subcellular localization and role of lipin-1 in human macrophages. J Immunol. 2011;186:6004–13. doi: 10.4049/jimmunol.1003279. [DOI] [PubMed] [Google Scholar]
- 24.Peterson TR, Sengupta SS, Harris TE, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–20. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You M, Liang X, Ajmo JM, et al. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–8. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 26.Kim HB, Kumar A, Wang L, et al. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol Cell Biol. 2010;30:3126–39. doi: 10.1128/MCB.01671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15(5):623–34. doi: 10.1016/j.cmet.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Colmenero J, Bataller R, Sancho-Bru P, et al. Hepatic expression of candidate genes in patients with alcoholic hepatitis: correlation with disease severity. Gastroenterology. 2007;132:687–97. doi: 10.1053/j.gastro.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez M, Miquel R, Colmenero J, et al. Hepatic expression of CXC chemokines predicts portal hypertension and survival in patients with alcoholic hepatitis. Gastroenterology. 2009;136:1639–50. doi: 10.1053/j.gastro.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 30.Dominguez M, Rincon D, Abraldes JG, et al. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am J Gastroenterol. 2008;103:2747–56. doi: 10.1111/j.1572-0241.2008.02104.x. [DOI] [PubMed] [Google Scholar]
- 31.Affò S, Dominguez M, Lozano JJ, Sancho-Bru P, Rodrigo-Torres D, Morales- Ibanez O, Moreno M, et al. Transcriptome analysis identifies TNF Superfamily Receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–60. doi: 10.1136/gutjnl-2011-301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosch-Presegué L, Raurell-Vila H, Marazuela-Duque A, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42:210–23. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 33.Liu GH, Gerace L. Sumoylation regulates nuclear localization of lipin-1alpha in neuronal cells. PLoS One. 2009;4:e7031. doi: 10.1371/journal.pone.0007031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finck BN, Gropler MC, Chen Z, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.