Abstract
Hepatocellular carcinoma (HCC) is among the leading causes of cancer-related death. Despite the advances in diagnosis and management of HCC, the biology of this tumor remains poorly understood. Recent evidence highlighted long non-coding RNAs (lncRNAs) as crucial determinants of HCC development. In this study, we report the lncRNA HOTTIP as significantly up-regulated in HCC specimens. HOTTIP gene is located in physical contiguity with HOXA13 and directly controls the HOXA locus gene expression via interaction with the WDR5 / MLL complex. HOX genes encode transcription factors regulating embryonic development and cell fate. We previously described HOX genes deregulation to be involved in hepatocarcinogenesis. Indeed, we observed the marked up-regulation of HOXA13 in HCC. Here, by correlating clinico-pathological and expression data, we demonstrate that the levels of HOTTIP and HOXA13 are associated with HCC patients’ clinical progression and predict disease outcome. In contrast to the majority of similar studies, our data are obtained from snap-frozen needle HCC biopsies (n=52) matched with their non-neoplastic counterparts collected from patients that had not yet received any HCC-tailored therapeutic treatments at the time of biopsy. In addition, taking advantage of gain and loss of function experiments in liver cancer-derived cell lines (HuH-6 and HuH-7), we uncover a novel bidirectional regulatory loop between HOTTIP / HOXA13. Conclusion: our study highlights the key role of HOTTIP and HOXA13 in HCC development by associating their expression to metastasis and survival in HCC patients, provides novel insights on the function of lncRNA-driven hepatocarcinogenesis and paves the way for further investigation about the possible role of HOTTIP as predictive biomarker of HCC.
Keywords: Hepatocellular carcinoma, lncRNAs, HOX genes, HOTTIP, HOXA13
INTRODUCTION
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide and the principal cause of mortality among cirrhotic patients (1, 2). Unlike most malignancies, mortality from liver cancer has increased significantly over the past 20 years and epidemiologic evidence indicates that the medical and economic burden of liver cancer will still increase significantly in Western populations during the next decades (1, 2). HCC mostly arises in a cirrhotic liver (3) and around 70% of new cases worldwide (with a higher prevalence in Eastern countries) are related to chronic infection with either Hepatitis B (HBV) or C (HCV) virus (4, 5). Moreover, a constantly rising proportion of HCCs in Western countries is ascribed to alcohol abuse and metabolic disorders (6, 7). Hepatocarcinogenesis has often been described as a multi-step process involving a number of genetic alterations eventually leading to the malignant transformation of the hepatocytes (8). Despite significant advances in diagnosis and management (9), the biology of HCC remains poorly understood mainly because of the complex genomic landscape of this tumor type (9). Besides well-known molecular alterations involving regulatory pathways such as Wnt/β-catenin, MAPK, p14ARF/p53, p16INK4A/Rb, transforming growth factor-β (TGF-β) and PTEN/Akt (8, 10–13), recent evidence has identified additional key players in hepatocarcinogenesis including: i) small non-coding RNAs (typically microRNAs) that regulate the translation of liver-specific mRNAs (14); ii) long non-coding RNAs (lncRNAs), mRNA-like polyadenylated transcripts ranging in length from 200 nucleotides up to ≈ 100 kb (kilobase) acting as molecular scaffold to organize and direct ribonucleoprotein (RNP) complexes (15); iii) epigenetic alterations caused by the deregulation of the gene system made of Polycomb (H3K27me3/H2AK119ub1), Trithorax (H3K4me3) and the Hox gene network (10, 16–19). Class I homeobox genes (Hox in mouse, HOX in human) encode 39 transcription factors initially described as master regulators of embryonic development (20). HOX genes represent the most repeat-poor-regions within the human genome (21) and display a unique gene network organization (4 chromosomal loci containing 9 to 11 genes aligned into 13 paralogous groups) (22, 23). During development, Hox gene expression controls the identity of body regions according to the rules of spatio-temporal colinearity (24). In adult tissues, Hox genes regulate tissue homeostasis and preserve the spatio-temporal coordinates established during embryonic development (25). At the cellular level, Hox genes regulate cell differentiation and are involved in neoplastic transformation (26). We recently screened the entire HOX gene network expression levels in a series of liver biopsies demonstrating that the HOXA locus represents the prevalent locus involved in hepatocarcinogenesis (27). Within HOXA genes, we have identified HOXA13 as the most up-regulated in HCC (27). HOXA13 is a marker of gut primordia posteriorization during development, plays a crucial role in extra-embryonic vascularization (28) and is involved in leukemogenesis (29).
Up to date, 231 lncRNAs have been annotated within the 4 HOX loci (30). Among these newly described ncRNAs, a lncRNA named HOTTIP (HOXA transcript at the distal tip) located in physical contiguity (chr 7p15.2) with the HOXA13 gene has been recently functionally characterized (31). Consistent with its genomic location 5’ to HOXA13, HOTTIP is expressed from development to adulthood in lumbo-sacral anatomic regions (31). Via interacting with the WDR5/MLL complex, HOTTIP is able to directly coordinate and control the activation of several 5’ HOXA genes (31). The depletion of HOTTIP in mice induces defects resembling HoxA11 (32, 33) and HoxA13 inactivation (34), suggesting the in vivo control of HOXA genes expressed in lumbo-sacral regions by HOTTIP RNA.
In the present study, we elucidate the involvement of the lncRNA HOTTIP in HCC tumorigenesis and further investigate the role of HOXA13 in this process. The correlation of clinico-pathological and expression data obtained from HCC liver biopsies matched with their non-neoplastic counterparts provides evidence that both HOTTIP and HOXA13 play a major role in HCC being associated with disease progression and predicting patients’ clinical outcome. In addition, our results uncover a novel bidirectional regulatory loop between HOTTIP and HOXA13 in liver cancer cells.
MATERIALS AND METHODS
Patient samples
All patients were recruited between 2002 and 2012 in the Hepatology Outpatient Clinic, University Hospital of Basel, Switzerland. All patients gave written informed consent to the study, which was approved by the Ethics Committee of the University Hospital of Basel (EKKB). Patients’ specimens and related clinico-pathological data including complete follow-up were obtained from the Institute of Pathology and from the Hepatology Outpatient Clinic, University Hospital of Basel and are summarized in Table 1. No selection bias was introduced in HCC samples collection for this study. Sex-distribution in our cohort reflects the frequencies of our geographical region. All patients in this study met the following criteria: HCC diagnosis was verified by pathological examination, no anti-cancer treatments were given before biopsy collection and exhaustive clinical-pathologic and follow-up data were available. Additional details are provided in the Supporting Information.
Table 1.
Clinico-pathological data of the HCC studied cohort.
| Factors | Frequency | % | |
|---|---|---|---|
| Age | |||
| <65 | 20 | 33.5 | |
| >65 | 32 | 61.5 | |
| Median (range) | 70.5 (26–89) | ||
| Gender | |||
| Female | 5 | 9.6 | |
| Male | 47 | 90.4 | |
| Foci | |||
| Multifocal | 30 | 57.7 | |
| Singular | 20 | 38.5 | |
| No Data | 2 | 3.8 | |
| AFP(>250 ng/ml) | |||
| Negative | 36 | 69.2 | |
| Positive | 13 | 25 | |
| No Data | 3 | 5.8 | |
| Fibrosis grade | |||
| F0 | 1 | 1.9 | |
| F1 | 3 | 5.8 | |
| F2 | 2 | 3.8 | |
| F3 | 2 | 3.8 | |
| F4 | 4.3 | 82.7 | |
| No Data | 1 | 1.9 | |
| Edmondson grade | |||
| 2 | 37 | 712 | |
| 3 | 13 | 25 | |
| 4 | 2 | 3.8 | |
| Etiology | |||
| No Virus | 30 | 57.7 | |
| Virus | 22 | 42.3 | |
| Metastasis | |||
| No | 40 | 76.9 | |
| Yes | 12 | 23.1 |
Relative expression of HOXA13 and HOTTIP by qRT-PCR
Quantitative reverse-transcription PCR (qRT-PCR) analysis to assess HOXA13 and HOTTIP expression levels in collected specimens was performed as previously described (27, 35). Additional details are provided in the Supporting Information.
Cell lines
Liver cancer-derived cell lines SNU182, SNU449, SNU423, SNU387, SNU475, SNU398, Hep-G2, HEP3B, HEP3B-RT, PLC, WRL68, SK-HEP1, Hep40, HLF, FOCUS, 7703 and the immortalized human normal liver cells THLE-2 and THLE-3 were derived from the cell depository of the laboratory of experimental carcinogenesis (LEC) (Bethesda, MD, USA). HuH-6, and HuH-7 were obtained from RIKEN Cell Bank (Ibaraki, Japan), and HLE, HuH-1 and HLF from the Health Science Research Resources Bank (Osaka, JP). All cell lines were maintained under the condition as recommended by the provider. Total RNAs were extracted from cells at 75% confluence using the RNeasy Mini kit (Qiagen, Hilden, DE).
siRNA transfection and HOXA13 overexpression
HuH-6 and HuH-7 cells have been transfected as described by Wang et al., 2011 (31). Additional information about the performed experiments is provided in the Supporting Information.
Proliferation, Migration and Apoptosis assay
Cells proliferation was assayed using the xCELLigence system (Roche, Basel, CH) (36) and Ki67 staining (37). Apoptotic levels have been evaluated using fluorescence activated cell sorter (FACS). Migration has been assessed using in vitro scratch assay as described by Liang et al, 2007 (38). Comprehensive information is given in the Supporting Information.
Statistical analysis
For statistical analysis, the Chi-square test (χ2 test) and Fisher's exact test for nonparametric variables and Student’s t-test for parametric variables were used (two-tailed). Differences in patient survival were assessed using the Kaplan–Meier method and analysed using the log-rank test in univariate analysis. To assess the relative risk for each factor, univariate and multivariate Cox regression analysis were performed. All tests were two-sided and p-values <0.05 considered to be statistically significant. Cut-off scores to discriminate between High and Low HOTTIP / HOXA13 expressing samples were selected by evaluating the receiver-operating characteristic (ROC) curves. The point on the curve with the shortest distance to the coordinate (0,1) was selected as the threshold value to classify cases as ‘positive/high-expressing’ or ‘negative/low-expressing’ (39). Analysis was performed using the SPSS software (IBM, New York, USA).
RESULTS
HOTTIP and HOXA13 are up-regulated in HCC
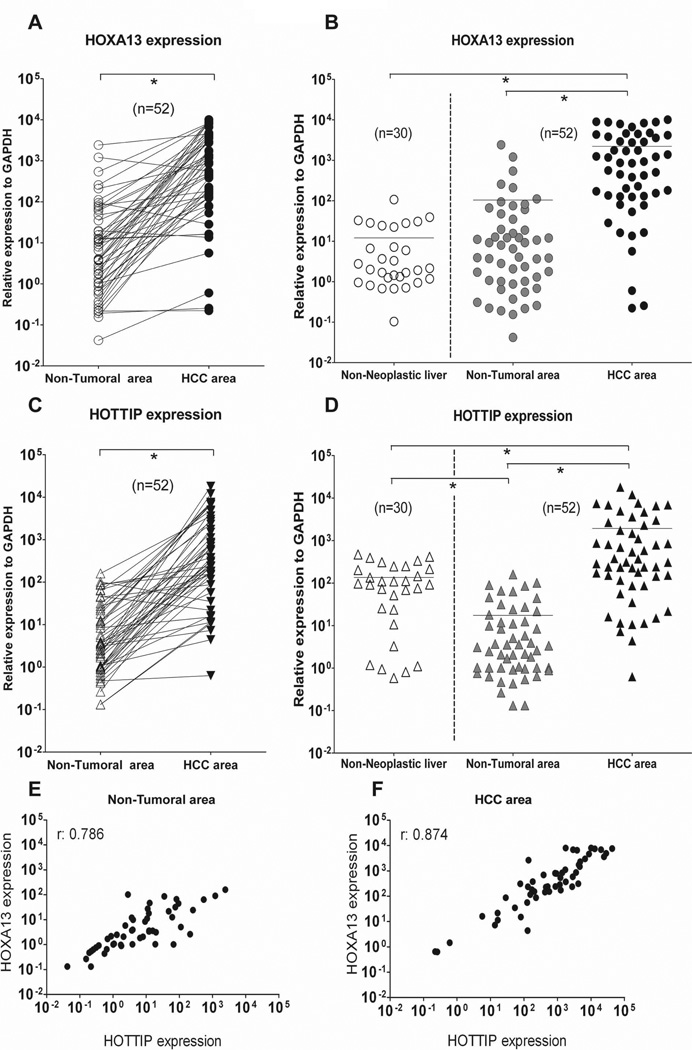
To investigate HOTTIP and HOXA13 expression levels in HCC, we performed qRT-PCR analysis on total RNA extracted from 52 HCC liver needle biopsies and their matched non-neoplastic counterpart. Clinico-pathological data of our HCC patients’ cohort are summarized in Table 1. In addition, we evaluated HOTTIP and HOXA13 expression in 30 liver biopsies obtained from patients with non-neoplastic liver disease including: steatosis (n=6), liver inflammation (total of n=6, of which n=2 drug induced liver hepatitis plus n=4 non-alcoholic steatohepatitis), alcohol and virus related cirrhosis (n=5), HCV infection without signs of cirrhosis (n=4), HCV infection with signs of cirrhosis (n=5) and normal liver donors (n=4). Using a larger cohort of HCC liver biopsies (n=52) with respect to our previous study (n=12) (32), we confirm here that HOXA13 is significantly overexpressed in HCC samples compared to their non-tumoral counterpart (Fig. 1A) and to non-neoplastic liver disease specimens (Fig. 1B). In addition, we find that HOTTIP expression levels are also increased in HCC samples compared to their non-tumoral counterpart (Fig. 1C) as well as to non-neoplastic liver disease specimens (Fig. 1D). We observe increased HOTTIP expression also in most of the evaluated non-neoplastic liver disease (20 out of 30 samples, 66.7%) except steatosis (Suppl. Fig. 1A). Conversely, none of the investigated non-neoplastic liver disease samples is characterized by HOXA13 overexpression (Suppl. Fig. 1B). Etiological samples’ stratification reveals that alcohol-related and viral infection-related (both HCV or HBV) HCCs present similar levels of HOTTIP (Suppl. Fig. 2). The same is observed for HOXA13 (Suppl. Fig. 2). Spearman analyses performed using qRT-PCR expression data indicate that HOTTIP and HOXA13 have a high correlation score both in non-tumoral and HCC area (r: 0.786 and r: 0.874, respectively) (Fig. 1E-F). These results suggest a potential interrelated role of HOTTIP and HOXA13 in HCC. No correlation between patient’s clinical features and HOTTIP or HOXA13 expression is found (Table 2) beside metastasis and survival, as described in the next paragraph.
Fig. 1.
Comparison of HOXA13 and HOTTIP expression in non-neoplastic liver samples and HCC specimens. Total RNA was extracted from liver biopsies to assess gene expression levels by qRT-PCR. (A-B) As we previously reported, expression data confirm HOXA13 up-regulation in HCC samples compared to their matched non-tumoral counterpart (A) and to non-neoplastic liver specimens (B). (C-D) HOTTIP is also up-regulated in HCC samples compared to their matched non-tumoral counterpart (C) as well as to non-neoplastic liver specimens (D). (E-F) Correlation scatter plot (Spearman test) of HOTTIP and HOXA13 expression in non-tumoral area (E) and HCC area (F) of tested biopsies. GAPDH has been used as reference gene. *p: ≤ 0.05
Table 2.
Correlation of HOTTIP and HOXA13 expression levels (Low and High) to clinico-pathological data.
| Factors | HQTTIP | HOXA13 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | Chi-square | p-value | Low | High | Chi-square | p-value | ||
| Age | |||||||||
| ≤65 | 7 | 13 | 0.165 | 0.685 | 7 | 13 | 1.123 | 0.289 | |
| >65 | 13 | 19 | 16 | 16 | |||||
| Gender | |||||||||
| Female | 3 | 2 | 1.084 | 0.298 | 4 | 1 | 2.869 | 0.09 | |
| Male | 17 | 30 | 19 | 28 | |||||
| Foci | |||||||||
| Multifocal | 10 | 20 | 0.807 | 0.668 | 13 | 17 | 0.042 | 0.979 | |
| Singular | 9 | 11 | 9 | 11 | |||||
| AFP (>250 ng/ml] | |||||||||
| Negative | 16 | 20 | 1.878 | 0.391 | 19 | 17 | 3.569 | 0.168 | |
| Positive | 3 | 10 | 3 | 10 | |||||
| Fibrosis grade | |||||||||
| F0/F1/F2 | 1 | 5 | 2.849 | 0.241 | 1 | 5 | 3.217 | 0.2 | |
| F3/F4 | 18 | 27 | 21 | 24 | |||||
| Edmondson grade | |||||||||
| I/II | 16 | 21 | 1.239 | 0.266 | 19 | 18 | 2.636 | 0.104 | |
| III/IV | 4 | 11 | 4 | 11 | |||||
| Etiology | |||||||||
| No-Virus | 12 | 18 | 0.071 | 0.79 | 16 | 14 | 2.382 | 0.123 | |
| Virus | 8 | 14 | 7 | 15 | |||||
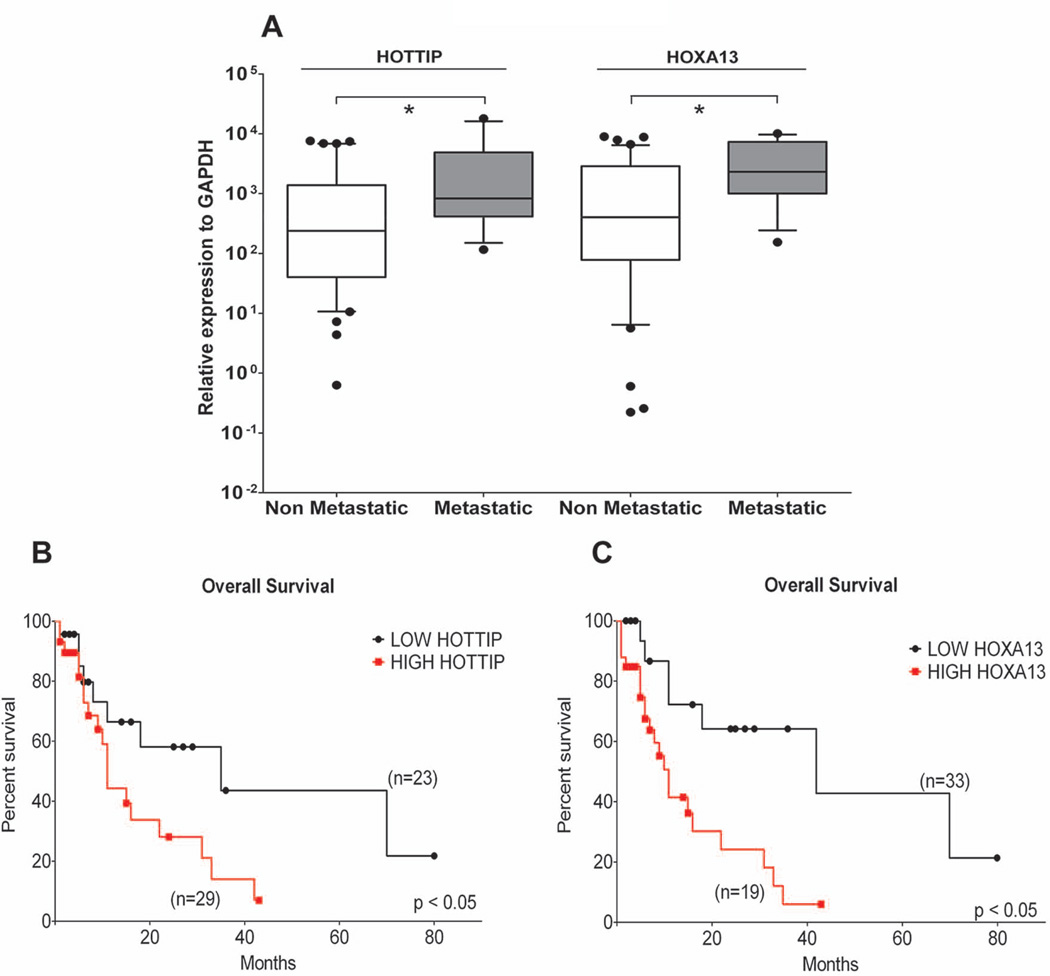
High HOTTIP and HOXA13 expression is associated with metastasis formation and poor patient survival in HCC
To explore whether HOTTIP and HOXA13 expression levels determine the clinical progression and outcome of HCC patients, we examined the incidence of metastasis and the patient overall survival rates by using Kaplan-Maier analysis. The metastatic status was defined as either regional lymph node invasion and/or distant organ involvement. Patients with high HOTTIP / HOXA13 expression show increased metastasis formation (Fig. 2A) and decreased overall survival (median of 11 versus 35 months for HOTTIP and 11 versus 42 months for HOXA13) (Fig. 2B-C), independently of whether they did or did not receive any HCC-tailored therapeutic treatment. The same holds true for patients exposed to HCC-tailored treatment (Suppl. Fig. 3). In addition, the univariate analysis of the overall survival reveals multiple intrahepatic HCC foci and HOXA13 expression as prognostic indicators (relative risk: 2.740, p=0.014 and relative risk: 2.181, p=0.050, respectively) (Suppl. Table 1). Multivariate analysis further identified HOXA13 expression as an independent prognostic indicator (relative risk: 2.345, p=0.042) (Suppl. Table 1). These data outline HOTTIP/HOXA13 as possible markers of clinical outcome in HCC patients.
Fig. 2.
High HOTTIP and HOXA13 expression is associated with metastasis formation and predicts poor HCC patients’ survival. (A) Patients developing metastasis present the highest expression levels of HOTTIP and HOXA13. (B-C) Survival plots of 52 HCCs including both untreated and HCC-treated patients analysed using the Kaplan-Maier method. ROC analysis was used to discriminate between HIGH and LOW expressing samples. HIGH HOTTIP and HOXA13 expression results in shorter patient survival. GAPDH has been used as reference gene. *p: ≤ 0.05
HOTTIP / HOXA13 expression in human liver cancer-derived cell lines
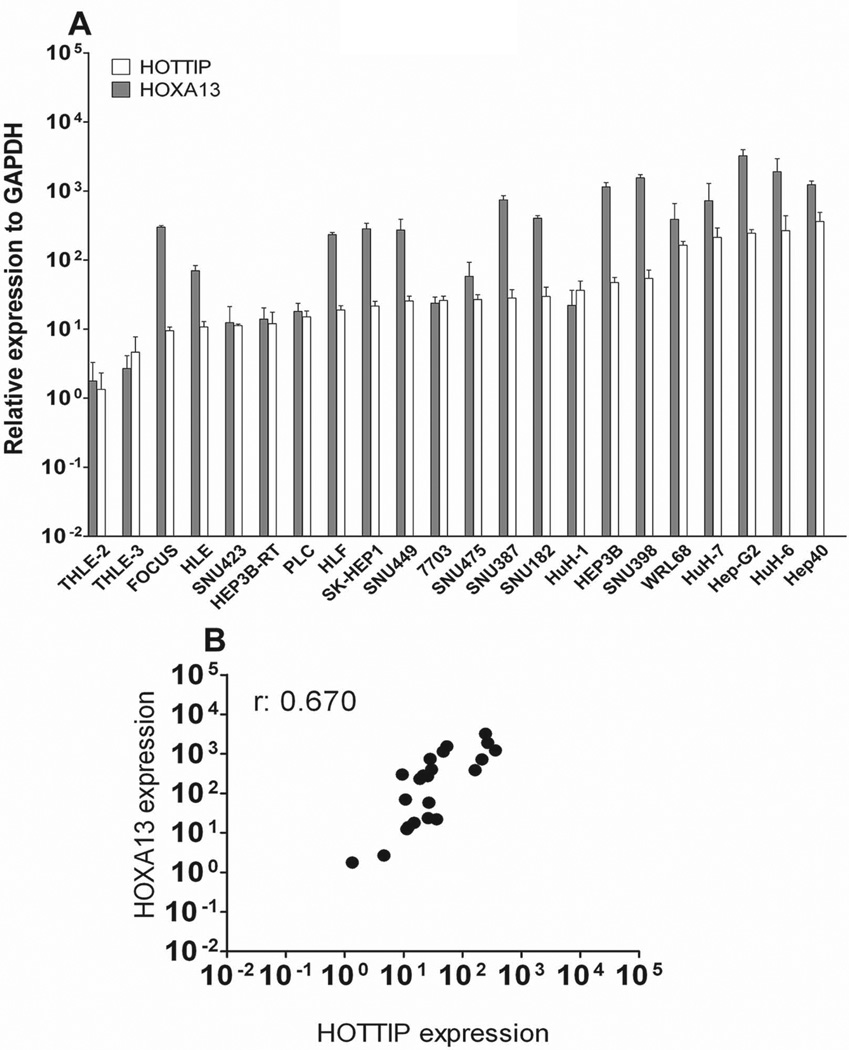
HOTTIP and HOXA13 expression was further determined in twenty liver cancer-derived cell lines by qRT-PCR (Fig. 3A). All liver cancer-derived cell lines showed higher HOTTIP and HOXA13 levels than the non-tumoral liver cell lines THLE-2 and THLE-3. The expression levels of HOTTIP and HOXA13 displayed a broad spectrum of variability with 10- to 100-fold expression change among the different tested cell lines. In accordance with the results obtained from human samples, HOTTIP and HOXA13 expression data exhibited a high correlation score (r: 0.670 - Spearman analysis) also in liver cancer-derived cell lines (Fig. 3B).
Fig. 3.
HOTTIP and HOXA13 expression levels in liver-cancer derived cell lines. (A) qRT-PCR data reveal a broad spectrum of expression among the 20 tested cell lines. Non-tumoral liver cell lines THLE-2 and THLE-3 display the lowest expression of both HOTTIP and HOXA13. Data are presented as mean ± SEM. (B) Correlation scatter plot (Spearman test) of HOTTIP and HOXA13 expression in liver-cancer derived cell lines. GAPDH has been used as reference gene.
HOTTIP regulates HOXA13 expression in HCC-derived cell lines
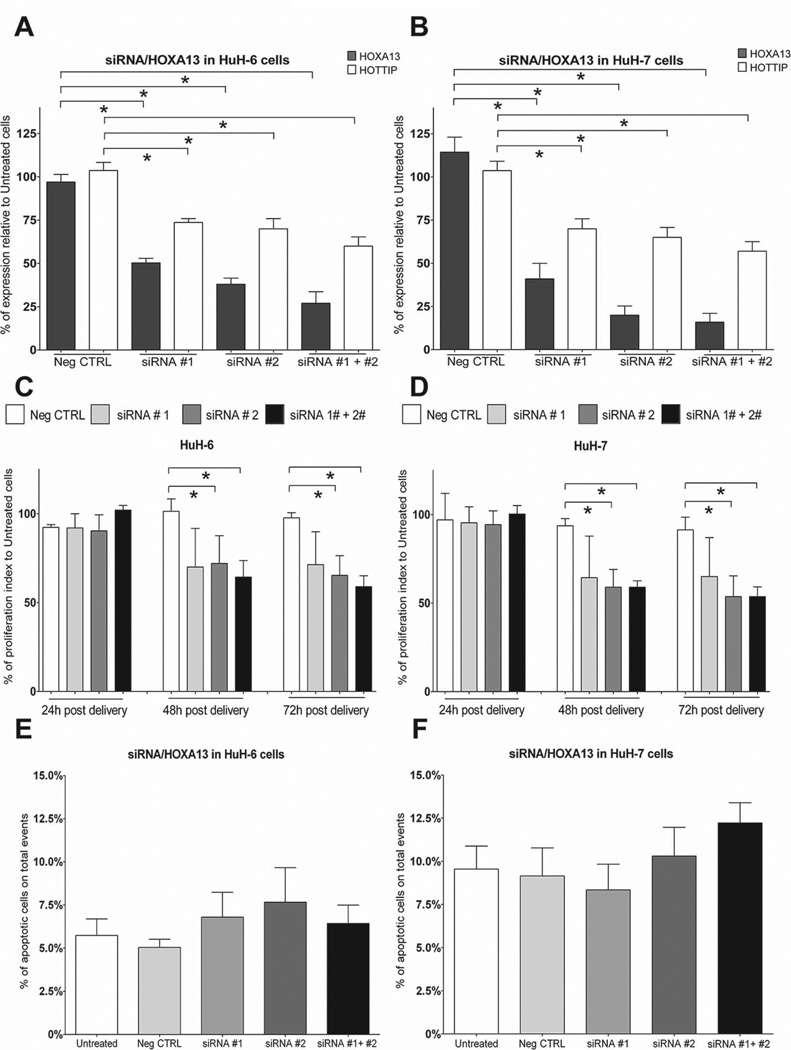
The siRNA-mediated knockdown of HOTTIP resulted in a clear reduction of HOXA13 expression in primary human fibroblast (31). To explore whether the same holds true in liver cancer-derived cell lines, we silenced HOTTIP expression through siRNAs in HuH-6 and HuH-7 cells. In HuH-6 cells, 48h post siRNA delivery, HOTTIP expression is reduced to 53.3% (Fig. 4A, siRNA#1 + siRNA#2). This also results in 42.3% reduction of HOXA13 levels (Fig. 4A, siRNA#1 + siRNA#2). In HuH-7 cells, 48h post siRNA delivery, HOTTIP levels are reduced by 74.6% (Fig. 4B, siRNA#1 + siRNA#2). HOXA13 expression is as well reduced by 45% (Fig. 4B). Next, we sought to evaluate the consequences of HOTTIP knockdown on tumour cell physiology by evaluating cell proliferation, apoptosis and migratory behaviour. Proliferation index measurement at 48h and 72h after siRNA delivery in HuH-6 and HuH-7 cells shows a significant reduction in both cell lines as compared to controls (Fig. 4C-D). These results are confirmed as well by using Ki67 staining as marker of cell proliferation (Suppl. Fig. 4A-B). No difference in apoptotic levels (Fig. 4E-F) and in cell migratory behaviour (Suppl. Fig. 5A-B) is observed in siRNA-treated cells versus controls. We conclude that HOTTIP can regulate HOXA13 expression in liver cancer-derived cells and that HOTTIP levels influence HCC cell proliferation rates.
Fig. 4.
Knockdown of HOTTIP results in decreased HOXA13 levels and impaired liver-cancer derived cell lines proliferation. Cells were treated with two siRNAs targeting HOTTIP RNA and adequate controls. (A-B) Efficiency of HOTTIP knockdown in HuH-6 (A) and HuH-7 (B) cells 48h after siRNA treatment. HOTTIP knockdown results in reduced HOXA13 expression. GAPDH has been used as reference gene. (C-D) Proliferation rate analysis of HuH-6 (C) and HuH-7 (D) cells upon siRNA treatment reveals that HOTTIP knockdown reduces cell proliferation. (E-F) 72h post siRNA treatment apoptotic index was analysed by Fluorescence Activated Cell Sorting (FACS) revealing no increase in cell apoptosis upon HOTTIP knockdown in neither HuH-6 (E) nor HuH-7 (F) cells. Data are presented as mean ± SEM. *p: ≤ 0.05
HOXA13 knockdown reduces HOTTIP expression
To gain further insights into the regulation of the HOTTIP / HOXA13 gene axis in HCC, we knocked down HOXA13 using siRNAs. In HuH-6 cells 48h post siRNA delivery, we observe a 73% reduction of HOXA13 levels (Fig. 5A siRNA#1 + siRNA#2). Interestingly, HOTTIP expression is also reduced by 40% (Fig. 5A). The same observation is made for single siRNA-mediated knockdown (Fig. 5A). As observed in HuH-6 cells, HOXA13 knockdown in HuH-7 cells results in up to 84% reduction of HOXA13 expression levels and concomitant reduction by 43% of HOTTIP (Fig. 5B, siRNA#1 + siRNA#2). Effective HOXA13 knockdown is confirmed at the protein level by western blotting (Suppl. Fig. 6A). As for HOTTIP knockdown, HOXA13 knockdown causes a significant reduction of proliferation both in HuH-6 and HuH-7 cells at 48h and 72h after siRNA delivery (Fig. 5C-D). These results are confirmed using Ki67 staining (Suppl. Fig. 4C-D). No difference in apoptotic levels (Fig. 5E-F) and in cell migratory behaviour (Suppl. Fig. 5C-D) is observed in siRNA-treated cells versus controls.
Fig. 5.
Knockdown of HOXA13 reduces HOTTIP levels and diminishes liver-cancer derived cell lines proliferation. Cells were treated with two siRNAs targeting HOXA13 mRNA and adequate controls. (A-B) Efficiency of HOXA13 knockdown in HuH-6(A) and HuH-7 (B) cells 48h upon siRNA treatment. HOXA13 knock downed cells show reduced HOTTIP expression. GAPDH has been used as reference gene. (C-D) Proliferation rate measurement of HuH-6 (C) and HuH-7 (D) cells upon siRNA treatment reveals that HOXA13 knockdown also reduces cell proliferation. (E-F) Apoptotic index analysis by FACS at 72h post siRNA treatment reveals, as for HOTTIP knockdown, no increase in cell apoptosis upon HOXA13 knockdown in neither HuH-6 (E) nor HuH-7 (F) cells. Data are presented as mean ± SEM. *p: ≤ 0.05
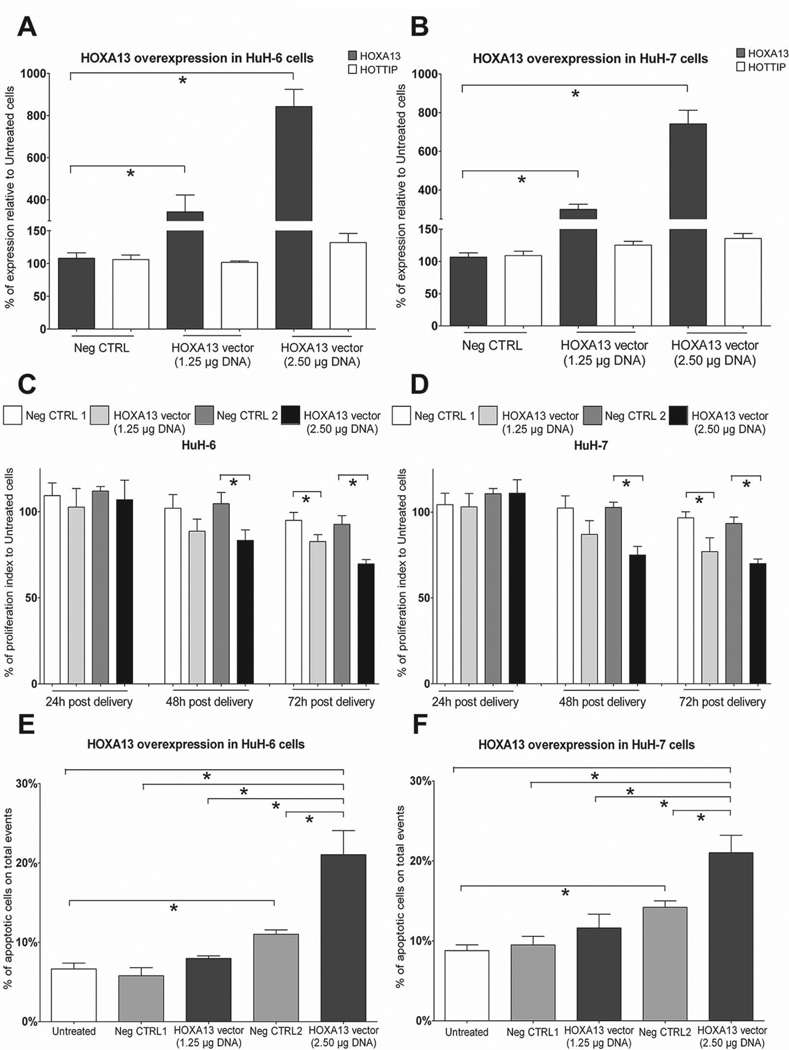
In a parallel approach, we addressed the impact of HOXA13 overexpression on HOTTIP levels in liver cancer-derived cell lines. Upon robust HOXA13 overexpression (Figure 6 A-B), confirmed at the protein level by western blotting (Suppl. Fig. 6B), we could not detect any significant (p: 0.16 in HuH-6 and p: 0.07 in HuH-7) increase of HOTTIP levels. Nonetheless, cells overexpressing HOXA13 show no difference in cell migratory behaviour (Suppl. Fig. 5E-F) but reduced proliferative index (Figure 6C-D and Suppl. Fig. 4E-F) and higher apoptotic levels (Fig. 6E-F), thus suggesting that further increased HOXA13 levels in liver cancer-derived cell lines might result in cell toxicity. Altogether, these results further substantiate the presumptive role of HOTTIP and HOXA13 in HCC progression and add new information supporting their interdependently regulated expression.
Fig. 6.
HOXA13 overexpression in liver cancer-derived cells has marginal effect on HOTTIP levels. Cells were transfected with HOXA13-expression vector at two different concentrations. Untreated and empty vector transfected cells (Neg CTRL) were used as controls. (A-B) Efficiency of HOXA13 overexpression in HuH-6 (A) and HuH-7 (B) cells 48h upon plasmid transfection. Higher amount of transfected plasmid DNA/cells results in more robust HOXA13 overexpression. HOXA13 increased levels only marginally influence HOTTIP expression. GAPDH has been used as reference gene. (C-D) Proliferation rate measurement of transfected HuH-6 (C) and HuH-7 (D) cells reveals that HOXA13 overexpression (2.50 µg DNA/10.000 cells) reduces cell proliferation. (E-F) Apoptotic index analysis by FACS performed at 72h post transfection reveals increased cell apoptosis in HOXA13 overexpressing cells (2.50 µg DNA/10.000 cells). Lipofectamine transfection using empty vector also has a marginal influence on cell apoptosis. Data are presented as mean ± SEM. *p: ≤ 0.05
DISCUSSION
In this study we report that the lncRNA HOTTIP is significantly up-regulated in HCC specimens. In addition, as previously described (27), we confirm the marked up-regulation of HOXA13 in HCC. Our data indicate that the expression levels of these two genes independently predict HCC tumor progression and disease outcome. Of note, our data were obtained from snap-frozen needle biopsies samples collected from patients that did not received any HCC-tailored therapeutic treatments prior to the biopsy. This increases the value of our work in contrast to the majority of similar studies employing surgical resected specimens often exposed to ischemic or therapy-induced damage.
The discovery of numerous non-coding RNA transcripts in humans has dramatically altered our understanding of the biology of complex diseases such as cancer (15). Recent studies have found that dysregulated expression of lncRNAs in solid cancer pinpoints the spectrum of disease progression (40) and may independently predict patient outcome (41). Among the first reported lncRNAs to be specifically up-regulated in HCC there was HULC (42). A number of lncRNAs, such as MALAT1(43) and TUC338 (44), have been since described to be involved in HCC disease development and progression (15, 43).
We previously reported HOXA13 to be significantly up-regulated in HCC(27). Since then, a lncRNA, named HOTTIP, has been described to be located in physical contiguity to HOXA13(31). Thus, we attempted to investigate the role of HOTTIP in hepatocarcinogenesis. HOTTIP plays an important role in regulating gene expression through chromatin modification similar to other lncRNAs like Xist (X-inactive specific transcript) and HOTAIR (HOX transcript Antisense Intergenic RNA) (45). Xist and HOTAIR interact with chromatin remodelling complexes to induce heterochromatin formation in specific genomic loci leading to reduce target gene expression (41, 45). HOTAIR hyper-expression was shown to act as an independent prognostic factor to predict tumor recurrence in HCC patients after liver transplant (46). Once activated, HOTAIR is capable of repressing in trans the expression of 5’ HOXD genes on chromosome 2q31-33 as well as the nearby located gene SATB2, whose target gene miR-31 is up-regulated in HCCs (30, 47).
In contrast to Xist and HOTAIR, HOTTIP exhibits H3K4me3 and H3K27me3 bivalent histone marks and is usually not transcribed (31). However, when transcribed, HOTTIP RNA directly interacts with the Trithorax protein WDR5 inducing an open DNA-chromatin configuration to target WDR5/MLL complexes driving histone H3 lysine 4 trimethylation and thus regulating the transcription of 5’ end HOXA locus genes including HOXA13 (31). We here provide further evidence supporting this proposed mechanism: we find that HOTTIP and HOXA13 expression strongly positively correlate in HCC samples. In the analysed specimens, high expression of HOTTIP is always coupled with increased HOXA13 levels and conversely low HOTTIP levels correlate with low HOXA13 expression. Interestingly, even if to a lower extent, we observe HOTTIP up-regulation also in non-neoplastic liver diseases such as in cirrhotic and HCV-infected cirrhotic liver. Higher HOTTIP expression in non-neoplastic liver compared to non-tumoral area tissue is difficult to explain merely on the basis of our data. However, considering that many samples among the non-neoplastic liver tissues were characterized by cirrhosis with moderate to severe inflammatory activity whereas non-tumoral samples displayed absence or a relative lesser degree of inflammation, it is tempting to speculate that this could in part explain such observation. On the other hand, a possible role of HOTTIP as an early predictive marker of HCC development deserves further investigation. Conversely, no increased HOXA13 levels are observed in the analysed non-neoplastic liver diseases, suggesting that HOXA13 up-regulation is specific for HCC. Altogether, these data suggest that HOTTIP deregulation might be an early step in hepatocarcinogenesis followed by HOXA13 during disease onset. HOTTIP is similarly expressed in viral infection-related (both HCV and HBV) and non-viral infection-related HCC patient samples. The same is observed for HOXA13, thus indicating that their deregulation is not restricted to an etiological subgroup of patients, but rather represents a common feature of HCCs.
In this study, the combination of clinico-pathological and expression data indicates that high levels of HOTTIP / HOXA13 expression are associated with metastasis formation and predict HCC patients’ clinical outcome. Our data are consistent with other studies demonstrating that deregulated expression of lncRNAs correlates with poor outcome in HCC patients (42, 48). For example, the overexpression of H19, encoding for a 2.3 kb lncRNA playing an important role in genomic imprinting during development and among the first lncRNAs to be described as involved in HCC (48), is linked to advanced tumor stages and unfavourable disease outcome (49). However, differently to H19 data, we find that the overexpression of HOTTIP is not significantly correlated to any of the clinical features analysed, such as tumor or fibrosis grade, except for metastasis and survival.
As above-mentioned, pioneering work in the field demonstrated HOTTIP-targeted regulation of HOXA13 expression (31). Primary human fibroblast manipulation via siRNAs directed against HOTTIP RNA resulted in a concomitant reduction of HOXA13 levels (31). In accordance, we observe HOXA13 downregulation upon siRNA-mediated knockdown of HOTTIP in two different liver cancer-derived cell lines. Thus, the regulatory impact of HOTTIP on the HOXA locus is preserved also during hepatocarcinogenesis. In addition, our work uncovers a previously unknown regulatory loop between HOTTIP and its target HOXA13. We observe that the knockdown of HOXA13 induces also a decrease in HOTTIP expression. Conversely, forced HOXA13 overexpression has a marginal impact on HOTTIP levels. We speculate that, as reported for ectopic HOTTIP expression driven by retroviral insertion sites scattered randomly in the genome (31), merely ectopic HOXA13 expression is not sufficient to promote HOTTIP / HOXA13 interaction. As well, it is conceivable that the effect of HOXA13 overexpression on HOTTIP levels is masked as a consequence of cell toxicity caused by high levels of HOXA13. Indeed not surprisingly, we observe that further forced HOXA13 overexpression results not only in reduced cell proliferation but also in markedly increased apoptosis in liver cancer-derived cell lines.
Altogether these results highlight a tightly controlled bidirectional gene expression loop regulating HOTTIP and HOXA13 levels. Of note, knocking down either HOTTIP or HOXA13 results in reduced cell proliferation in the tested liver cancer-derived cell lines. Our data further underline the importance of HOTTIP and HOXA13 in HCC progression, providing direct evidence of their role in tumor cell growth. Our observation is in agreement with previously published data demonstrating that forced reduction of HOXA13 expression in esophageal squamous carcinoma cell lines also leads to reduced proliferation (50). Overall the data we obtained from the gain or the loss of function experiments in vitro set the rationale to perform additional work aiming to monitor in vivo tumour cell behaviour to gather further insights into the role of HOTTIP and HOXA13 in hepatocarcinogenesis.
In conclusion, using a combination of human-derived data and in vitro approaches, our study highlights the molecular axis comprising HOTTIP and HOXA13 as a key player in HCC progression. Future work will validate HOTTIP as a predictive biomarker for HCC development. A deeper characterization of the function and downstream signalling pathways influenced by HOTTIP and HOXA13 deregulation may provide novel insights into the mechanisms of hepatocarcinogenesis possibly leading to the development of new therapeutic agents. Finally, this work further supports the importance of lncRNA-driven hepatocarcinogenesis emphasizing the need to expand our knowledge about lncRNAs function in liver diseases.
Supplementary Material
ACKNOWLEDGEMENTS
The study was supported by grants from the University of Basel and ONCOSUISSE (KLS-2867-08-2011 to L.M.T.). M.S.M. is supported by Schweizerische Stifung für Medizinisch-Biologische Stipendien (PASMP-3_140071). HCC research in the lab of S.D. is supported by the German Research Foundation (DFG TRR77 TP B03). We are gratefully to Prof. HY Chang and Dr. K Wang for the siRNA sequences information. We acknowledge Tanja Ditche and Alex Rufle for their excellent technical support.
Abbreviation
- HCC
Hepatocellular carcinoma
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- lncRNA
long non-coding RNA
- HOTTIP
HOXA transcript at the distal tip
- RNP
ribonucleoprotein
- AFP
α-fetoprotein
- FACS
Fluorescence Activated Cell Sorting
Footnotes
AUTHOR CONTRIBUTIONS
L.Q. conceived, performed and supervised all the experiments and wrote the manuscript; M.S.M. conceived and performed cell lines experiments, performed clinico-pathological assessment of HCC samples and wrote the manuscript; S.P. performed RNA extractions and statistical analysis; L.A. performed siRNA experiments; M.K., Z.M, F.M., J.B.A., C.R. and M.H. provided vital reagents and materials; L.T. performed clinico-pathological assessment of HCC samples; T.B and M.H. provided the HCC samples and the clinical information; S.D. conceived experiments and wrote the manuscript; C.C. conceived and supervised experiments, coordinated the project and wrote the manuscript, L.M.T. conceived experiments, performed clinico-pathological assessment of HCC samples, provided financial support, coordinated the project and wrote the manuscript. All authors agreed to the final version of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Han ZG. Functional genomic studies: insights into the pathogenesis of liver cancer. Annu Rev Genomics Hum Genet. 2012;13:171–205. doi: 10.1146/annurev-genom-090711-163752. [DOI] [PubMed] [Google Scholar]
- 3.Mair RD, Valenzuela A, Ha NB, Ayoub WS, Daugherty T, Lutchman GA, Garcia G, et al. Incidence of Hepatocellular Carcinoma Among US Patients With Cirrhosis of Viral or Nonviral Etiologies. Clin Gastroenterol Hepatol. 2012 doi: 10.1016/j.cgh.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14:4300–4308. doi: 10.3748/wjg.14.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, Kleeff J, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. doi: 10.1002/hep.22033. [DOI] [PubMed] [Google Scholar]
- 6.Grewal P, Viswanathen VA. Liver cancer and alcohol. Clin Liver Dis. 2012;16:839–850. doi: 10.1016/j.cld.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 7.White DL, Kanwal F, El-Serag HB. Association Between Nonalcoholic Fatty Liver Disease and Risk for Hepatocellular Cancer, Based on Systematic Review. Clin Gastroenterol Hepatol. 2012 doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 9.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 10.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis. 2011;31:173–187. doi: 10.1055/s-0031-1276646. [DOI] [PubMed] [Google Scholar]
- 12.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longerich T, Mueller MM, Breuhahn K, Schirmacher P, Benner A, Heiss C. Oncogenetic tree modeling of human hepatocarcinogenesis. Int J Cancer. 2012;130:575–583. doi: 10.1002/ijc.26063. [DOI] [PubMed] [Google Scholar]
- 14.Toffanin S, Hoshida Y, Lachenmayer A, Villanueva A, Cabellos L, Minguez B, Savic R, et al. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140:1618–1628. doi: 10.1053/j.gastro.2011.02.009. e1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 17.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 18.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 19.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 21.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 22.Scott MP. Vertebrate homeobox gene nomenclature. Cell. 1992;71:551–553. doi: 10.1016/0092-8674(92)90588-4. [DOI] [PubMed] [Google Scholar]
- 23.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 24.Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 25.Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 26.Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- 27.Cillo C, Schiavo G, Cantile M, Bihl MP, Sorrentino P, Carafa V, M DA, et al. The HOX gene network in hepatocellular carcinoma. Int J Cancer. 2011;129:2577–2587. doi: 10.1002/ijc.25941. [DOI] [PubMed] [Google Scholar]
- 28.Shaut CA, Keene DR, Sorensen LK, Li DY, Stadler HS. HOXA13 Is essential for placental vascular patterning and labyrinth endothelial specification. PLoS Genet. 2008;4:e1000073. doi: 10.1371/journal.pgen.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 30.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small KM, Potter SS. Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Dev. 1993;7:2318–2328. doi: 10.1101/gad.7.12a.2318. [DOI] [PubMed] [Google Scholar]
- 33.Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 34.Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- 35.de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–159. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- 36.Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, Zhang JD, et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. 2012;32:633–651. doi: 10.1128/MCB.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jabari S, Meissnitzer M, Quint K, Gahr S, Wissniowski T, Hahn EG, Neureiter D, et al. Cellular plasticity of trans- and dedifferentiation markers in human hepatoma cells in vitro and in vivo. Int J Oncol. 2009;35:69–80. doi: 10.3892/ijo_00000314. [DOI] [PubMed] [Google Scholar]
- 38.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 39.Piscuoglio S, Lehmann FS, Zlobec I, Tornillo L, Dietmaier W, Hartmann A, Wunsch PH, et al. Effect of EpCAM, CD44, CD133 and CD166 expression on patient survival in tumours of the ampulla of Vater. J Clin Pathol. 2012;65:140–145. doi: 10.1136/jclinpath-2011-200043. [DOI] [PubMed] [Google Scholar]
- 40.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 43.Gutschner T, Hammerle M, Diederichs S. MALAT1 - a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013 doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 44.Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, Terracciano L, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108:786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. Bioessays. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 47.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 49.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 50.Gu ZD, Shen LY, Wang H, Chen XM, Li Y, Ning T, Chen KN. HOXA13 promotes cancer cell growth and predicts poor survival of patients with esophageal squamous cell carcinoma. Cancer Res. 2009;69:4969–4973. doi: 10.1158/0008-5472.CAN-08-4546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.