Abstract
In eukaryotes, the 26S proteasome degrades ubiquitinylated proteins in an ATP-dependent manner. Archaea mediate a form of post-translational modification of proteins termed sampylation that resembles ubiquitinylation. Sampylation was identified in Haloferax volcanii, a moderate halophilic archaeon that synthesizes homologs of 26S proteasome subunits including 20S core particles and regulatory particle triple-A ATPases (Rpt)-like proteasome-associated nucleotidases (PAN-A/1 and PAN-B/1). To determine whether sampylated proteins associate with the Rpt subunit homologs, PAN-A/1 was purified to homogeneity from Hfx. volcanii and analyzed for its subunit stoichiometry, nucleotide hydrolyzing activity and binding to sampylated protein targets. PAN-A/1 was found associated as a dodecamer (630-kDa) with a configuration in TEM suggesting a complex of two stacked hexameric rings. PAN-A/1 had high affinity for ATP (Km of ∼0.44 mM) and hydrolyzed this nucleotide with a specific activity of 0.33 ± 0.1 μmol Pi/h per mg protein and maximum at 42°C. PAN-A1 was stabilized by 2M salt with a decrease in activity at lower concentrations of salt that correlated with dissociation of the dodecamer into trimers to monomers. Binding of PAN-A/1 to a sampylated protein was demonstrated by modification of a far Western blotting technique (derived from the standard Western blot method to detect protein-protein interaction in vitro) for halophilic proteins. Overall, our results support a model in which sampylated proteins associate with the PAN-A/1 AAA+ ATPase in proteasome-mediated proteolysis and/or protein remodeling and provide a method for assay of halophilic protein-protein interactions.
Keywords: archaea, protein modification, post-translational modification, AAA ATPases, proteasomes, ubiquitylation, sampylation
Introduction
Energy-dependent proteolysis is a vital process in all domains of life. Proteasomes are well-conserved nanocompartmentalized proteases distributed throughout Archaea, Eukarya, and bacterial actinomycetes that are important in energy-dependent proteolysis (Maupin-Furlow, 2012). In eukaryotes, proteins are targeted for degradation by 26S proteasomes through covalent linkage of ubiquitin chains (Hershko and Ciechanover, 1992). 26S proteasomes can be separated into two subparticles: a 20S proteasome peptidase core particle (CP) and a 19S ATP-dependent regulatory particle (RP) (Voges et al, 1999). The subunits of 20S proteasomes cluster to related α and β superfamilies (Coux et al, 1994) and respectively form two outer and two inner heptameric rings that create a barrel-shaped complex (Löwe et al, 1995). Access to the inner chamber of 20S proteasomes, which house the catalytically active Thr residues, is through two small pore openings further restricted by axial gates (Stadtmueller and Hill, 2011). Two conditions must be met in order for substrate translocation to the proteolytic active sites to occur: the protein must be unfolded and the 20S CP gate must be opened. Both of these events appear controlled by AAA+ ATPases (Bar-Nun and Glickman, 2012). The six regulatory particle triple-A (AAA) type I ATPase (Rpt1-6) subunits of the 19S RP associate with the outermost a rings of 20S proteasomes (Beck et al, 2012) and are required for degradation of ubiquitin-conjugates (Glickman et al, 1998).
Archaea possess simplified versions of the proteasome made of a 20S core particle of one to two types of α and β subunits and an AAA+ ATPase of Cdc48, Ama, or proteasome activating nucleotidase (PAN) families with the latter a close homolog of the 19S Rpt subunits (Forouzan et al, 2012; Humbard and Maupin-Furlow, 2013). Considering its simplicity, the archaeal 20S proteasome and PAN have served as a paradigm to study the basic mechanism of 26S proteasome function. The Methanocaldococcus jannaschii (Mj) PAN, purified from recombinant Escherichia coli, was the first archaeal proteasome-associated ATPase to be characterized and is required for the energy-dependent degradation of proteins by 20S proteasomes (Benaroudj and Goldberg, 2000; Wilson et al, 2000; Zwickl et al, 1999). During this process, MjPAN functions in substrate unfolding, opening the 20S CP gate and substrate translocation (Smith et al, 2007; Smith et al, 2005; Yu et al, 2010; Zhang et al, 2009b). The C-terminal Hb[Y/F]X motif of MjPAN is required for docking and opening the 20S CP gate for substrate entry (Smith et al, 2007; Yu et al, 2010), while the N-terminal coiled coil domains of PANs recognize substrate protein (Djuranovic et al, 2009; Zhang et al, 2009a and b).
The biological significance of the PAN-proteasome system in archaeal cell physiology and environmental and extremophilic adaptation remains poorly documented. Characterization of PANs from native organisms is limited to halophilic archaea with Hfx. volcanii and Halobacterium salinarium known to synthesize two PANs (PAN-A1/-B2, also denoted as PAN-A/-B) that are distinct in structure, post-translational modification, regulation and biological function (Chamieh et al, 2008; Chamieh et al, 2012; Humbard et al, 2010a; Humbard et al, 2010b; Kirkland et al, 2008; Kirkland and Maupin-Furlow, 2009; Reuter et al, 2004; Zhou et al, 2008).
In eukaryotic cells, the conjugation of ubiquitin and ubiquitin-like proteins to protein targets plays an integral role in a wide variety of processes including proteasome-mediated proteolysis. Although universal in eukaryotes, the presence of protein conjugation systems in prokaryotes is less clear. Three small archaeal modifier proteins (SAMPs) are differentially conjugated to protein targets in the archaeon Haloferax volcanii (Humbard et al, 2010a; Miranda et al, in press). Sampylation is thought to target proteins for degradation by proteasomes, based on the increased level of SAMP1-modified proteins in strains with deletion of PAN-A/1 and 20S core particle α1 subunit encoding genes (Humbard et al. 2010a) as well as the increased levels of SAMP2-modified proteins in cells treated with proteasome inhibitor VELCADE (bortezomib) (Miranda et al., in press). SAMP3 was recently showed to be involved in regulation of MoCo biosynthesis (Miranda et al, in press). It is unclear how the PAN system is integrated with the SAMP-based protein tagging system and the proteasomes.
Here, for the first time, we have expressed, purified and characterized a PAN to homogeneity from its native archaeal host. PAN-A/1 was purified from an Hfx. volcanii strain devoid of PAN-B/2 and was found associated as a dodecamer and able to catalyzed the hydrolysis of ATP with high affinity for ATP. The presence of PAN-B/2 was not required for PAN-A/1 association or ATPase activity. However, the biochemical properties of PAN-A/1 were highly dependent on molar concentrations of salt with significant loss of ATPase activity and complex dissociation detected at 0.75 M NaCl. Here we also adapted the far Western blotting procedure for halophilic proteins and used this method to screen for PAN-A/1 partners. With this approach, we found that PAN-A/1 specifically bound SAMP1-MoaE conjugates (but not SAMP1, MoaE or BSA alone), thus, providing an insight into how archaeal proteasomes may associate with sampylated substrates.
Experimental Procedures
DNA isolation, analysis, and strain construction
Plasmids used in this study are summarized in Table S1. PCR was performed according to standard methods with Hfx. volcanii DS70 genomic DNA or appropriate plasmid DNA as a template and primer pairs as indicated in Table S2. Phusion DNA polymerase (New England Biolabs, Ipswich, MA) was used for high-fidelity PCR-based cloning, and Taq DNA polymerase (Bioline) was used for colony screening. PCR generated-DNA fragments of appropriate size were isolated from 0.8% (w/v) SeaKem GTG agarose (FMC Bioproducts, Rockland, ME) gels in TAE [40 mM Tris, 20 mM acetic acid, and 1 mM ethylenediaminetetraacetic acid (EDTA)] buffer at pH 8.0 using the QIAquick gel extraction kit (Qiagen, Valencia, CA) as needed. The fidelity of all DNA plasmid constructs was verified by Sanger DNA Sequencing (UF ICBR DNA sequencing core, Gainesville, FL).
Strains
Strains used in this study are summarized in Table S1. E. coli TOP10 was used for routine recombinant DNA experiments. E. coli GM2163 was used for replication of plasmid DNA prior to its transformation into Hfx. volcanii strains according to standard methods (Dyall-Smith, 2009). E. coli strains were grown in Luria–Bertani (LB) medium at 37°C with rotary shaking (200 rpm). LB medium was supplemented with ampicillin (Amp, 100 μg·ml−1) or kanamycin (30 μg·ml−1) for E. coli strains carrying pJAM plasmids. Hfx. volcanii strains were grown in ATCC974 complex medium (ATCC) at 42°C with rotary shaking (200 rpm). ATCC974 medium was supplemented with novobiocin (Nov, 0.2 μg·ml−1) to maintain pJAM plasmids in Hfx. volcanii.
Protein purification
For protein purification, Hfx. volcanii strains were grown to stationary phase in 1 L ATCC Nov medium in 2.8 L Fernbach flasks at 42°C (200 rpm), and E. coli Rosetta (DE3) strains freshly transformed with pJAM1131 plasmids were grown in 1L LB Amp in 2.8 L Fernbach flasks at 37°C (200 rpm). For E. coli strains, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM at log phase (OD600 of 0.4-0.6 units), and cultures were shifted to 25°C for 4.5 h (200 rpm) prior to harvest. All cells were harvested by centrifugation (4000 × g, 4°C, 15 min), washed once with ice-chilled wash buffer, and stored at −80°C. Wash buffer was 20 mM Tris-HCl (pH 8) supplemented with supplemented with 2M NaCl (buffer A) for Hfx. volcanii strains and 150 mM NaCl (buffer B) for E. coli strains. Purified proteins were detected by Western blotting and/or Coomassie Blue R-250 staining after separation by 10-12% SDS-PAGE (reducing gels).
PAN-A/1-StrepII
Hfx. volcanii GZ108-pJAM2001 (a ΔpanB/2 strain) was used for purification of PAN-A/1-StrepII in the absence of PAN-B/2. Cells (16-20 g wet weight) were resuspended in 35-40 mL of buffer A and broken by passage three times by French Press at a pressure of 2000 psi. After 15 min of centrifugation at 3000 × g (4 °C) to remove unbroken cells, the supernatant was filtered by a 0.8 μM cellulose acetate membrane (Fisher Scientific, USA). The supernatant was applied to a Strep-Tactin column (1 mL, Qiagen) equilibrated with buffer A at room temperature (RT). PAN-A/1-StrepII protein was eluted from the column with buffer A supplemented with 2.5 mM D-desthiobiotin. The protein was concentrated 10-fold to a volume of 500 μL (at 4 °C) using a Vivaspin centrifugal concentrator (30,000 molecular weight cut off membrane, Sartorius Stedim Biotech, Bohemia, NY) and applied to a Superose 6 HR 10/30 gel filtration column (FPLC, GE Healthcare, Piscataway, NJ) equilibrated at RT with buffer A at a flow of 0.3 mL·min-1. Fractions were collected every 0.5 mL. PAN-A/1-StrepII fractions were pooled, dialyzed against buffer A, and stored at 4°C for up to 1 week.
MoaE-StrepII
Hfx. volcanii HM1052–pJAM1119 was used for purification of MoaE-StrepII by Strep-Tactin chromatography similarly as described for PAN-A/1-StrepII with the following modifications. Cell pellet was resuspended in lysis buffer (2 M NaCl, 1 mM PMSF, 20 mM Tris-HCl, pH 8.0) and lysed thrice using French press at 2000 psi. Whole-cell lysate was clarified by centrifugation (twice at 17,000 × g at 4°C for 20 min) and filtration (0.45 μm). Clarified cell-free extract was loaded into the Strep-Tactin Superflow Plus affinity column (Qiagen), pre-equilibrated with Strep Wash buffer (2 M NaCl, 50 mM Tris-HCl, pH 8.0). MoaE-StrepII was recovered from the column upon addition of Strep elution buffer (2 M NaCl, 5 mM desthiobiotin, 50 mM Tris-HCl, pH 8.0). The eluate was dialyzed twice against high-salt buffer (2 M NaCl, 20 mM Tris-HCl, pH 7.5) and concentrated using Ultracel centrifugal filter (10 kD, Millipore) prior to final purification by Superdex 75 HR 10/30 chromatography (pre-equilibrated in high-salt buffer).
Flag-Samp1-MoaE-StrepII linear fusion
Hfx. volcanii HM1052-pJAM1796 was used for tandem affinity purification of the Flag-SAMP1-MoaE–StrepII linear fusion by Strep-Tactin and α-Flag affinity chromatography steps as described previously (Hepowit et al, 2012). Purified protein was dialyzed against buffer A and stored at 4°C for up to one week.
Flag-His6-SAMP1
E. coli Rosetta (DE3)-pJAM1131 was used for purification of Flag-His-SAMP1. Cell pellets were resuspended in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 40 mM imidazole, 1 mM PMSF) (4 ml per 1 g wet weight) and lysed thrice by French Press (2000 psi). An equal volume of dilution buffer (20 mM Tris-HCl, pH 7.5, 4 M NaCl) was added. Cell lysate was clarified by centrifugation (twice for 20 min at 17,000 × g and 4°C) and filtration (0.45 μm, cellulose acetate membrane, Fisher). The proteins were applied to a HisTrap HP column (1 mL, 71-5027-68AH, GE Healthcare) pre-equilibrated in buffer A with 40 mM imidazole and washed with the same buffer. The Flag-His6-SAMP1 was eluted using 500 mM imidazole in 20 mM Tris-HCl, pH 7.5, 2 M NaCl. The eluate was concentrated to a volume final of 500 μl using a Vivaspin centrifugal concentrator (similarly to described for PAN-A/1-StrepII) and applied to a Superdex 75 HR 10/30 gel filtration column (FPLC, GE Healthcare) equilibrated with buffer A at a flow of 0.3 mL·min-1. Fractions were collected every 0.5 mL. Flag-His6-SAMP1 fractions were pooled and dialyzed twice against buffer A (12 h per dialysis) to storage at 4°C.
Analytical Procedures
Protein concentration
Protein concentrations were determined by the bicinchoninic acid method (Smith et al, 1985) (Thermo Scientific Pierce BCA Protein Assay Kit, Rockford, IL) with bovine serum albumin (BioRad) as the protein standard.
Electrophoresis
Protein samples were mixed in equal volume ratio with 2× loading buffer [containing 125 mM Tris-HCL pH 6.8, 20 mM β-mercaptoethanol, 4% (w/v) SDS, 20% (v/v) glycerol and 0.01% (w/v) bromophenol blue] and boiled for 20 min. Samples were separated by 10-12% SDS-PAGE (reducing gels), using a mini-Protean III cell electrophoresis apparatus (Bio-Rad) at 20 mA constant at RT in a running buffer of 25 mM Tris and 190 mM glycine at pH 8.3 with 0.1% (w/v) SDS. After migration, proteins were stained in-gel with Coomassie Blue R-250 or were detected by Western blotting.
Western blotting
Proteins separated by SDS-PAGE were electroblotted onto PVDF membranes (Amersham, GE Healthcare). PAN-A/1 protein was detected by immunoblotting using anti-PAN-A/1 polyclonal antibody at a dilution 1:5000 followed by goat anti-rabbit IgG (H + L)-alkaline phosphatase-linked antibody (SouthernBiotech) as previously described (Reuter et al, 2004). C-terminal StrepII tagged proteins were detected by immunoblotting using anti-StrepII alkaline phosphatase-conjugated antibody (Qiagen) at a dilution of 1:5000. Alkaline phosphatase activity was detected by chemiluminescence using CDP-Star (Applied Biosystems) with X-ray film (Hyperfilm; Amersham Biosciences).
Far Western blotting
Proteins transferred to PVDF membranes (as described in the sections above) were analyzed by far Western blotting (Wu et al, 2007) with the following adaptations for halophilic proteins (see Table S3 for details). In general, the proteins were denatured and renatured on the membrane by gradually reducing the guanidine-HCl concentration from 6 to 0 M and increasing the NaCl concentration from 0.1 to 2 M. The membrane was blocked with 5% (w/v) nonfat instant dry milk (Publix, Gainesville, FL) in 20 mM HEPES buffer (pH 8) supplemented with 2 M NaCl (buffer C) for 1 h at RT. The membrane was incubated without or with PAN-A/1 (bait protein; 8 μg·ml-1) in buffer C in the presence or absence of MgATP (1 mM ATP and 9 mM MgCl2) or MgADP (1 mM ADP and 9 mM MgCl2) for 2 h at 42°C. Unbound bait protein was removed by washing (2× 10 min) with buffer C in the presence or absence of MgATP or MgADP. PAN-A/1 complexes were crosslinked with 3 mM N-ethyl-N′-[3-(dimethylamino)propyl]-carbodiimide hydrochloride (EDC) in buffer C with or without MgATP or MgADP. The crosslinking reaction was stopped by incubation for 20 min at RT with Tris-HCl at a final concentration of 100 mM. The membrane was washed 2 times 10 min in Tris-buffered saline (TBS; 50 mM Tris-HCl buffer (pH 7.5), 150 mM NaCl). The presence of PAN-A/1 was detected by Western blotting as described in the section above.
Nucleotide-hydrolyzing Activity
Nucleotide-hydrolyzing activity assays were performed with PAN-A/1 (3 to 13 μg, estimated at 0.048 to 0.21 μM for dodecamer) and nucleotide (1 mM ATP, GTP or TTP) in a final volume of 100 μl in 20 mM Tris-HCl buffer (pH 8) supplemented with 2M NaCl and 9.6 mM MgCl2. Reactions were carried out for 5, 10, 20 and 40 min. When used to inhibit the reaction, the EDTA was added at final concentration of 1 mM. To avoid phosphate contamination, all plastic ware was new and non-autoclaved, and deionized double distilled reagent grade water (Ricca Chemical Co., Arlington, TX) was used to prepare all buffers. Nucleotide hydrolysis was monitored as the production of inorganic phosphate (Pi) by a method adapted from (Kodama et al, 1986). In brief, a portion of the reaction products (50 μl) was mixed with an equal volume of a malachite green/molybdate reagent (2 g of Na2MoO4, 0.3 g of malachite green and 0.5 g of Triton X-100 in 1 liter of 0.7 M HCl) and incubated for 20 min at RT. Absorbance was measured at 630 nm using a BioTek Synergy HT microtiter plate reader (Bio-Tek Instruments, Inc., Winooski, VT). All activities were measured at least three times experimentally to ensure reproducibility. A calibration curve was established using KH2PO4 at 0.54 to 40 μM diluted in buffer A. Effects of different concentrations of salt on PAN-A/1 activity were monitored by ATPase activity assay after 1 h preincubation of PAN-A/1 (6 or 13 μg) on ice in 100 μl of 20 mM Tris-HCl buffer (pH 8) supplemented with either NaCl or KCl at final concentrations of 0.25 to 2 M. ATP was added after incubation on ice to start the reaction.
Molecular mass determination
Dependence of the molecular mass of PAN-A/1 complexes on salt concentration was determined by Superose 6 HR 10/30 (Pharmacia) chromatography with a sample loading volume of 200 μl and flow rate of 0.3 ml.min-1. PAN-A/1 was incubated at 1.64 mg·ml-1 in 20 mM Tris-HCl buffer (pH 8) supplemented with either 2 M or 0.75 M NaCl (for 1 h on ice) and applied to the gel filtration column equilibrated in similar buffer and salt concentrations. The Superose 6 HR 10/30 column was calibrated with apoferritine (480 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa) and albumin (66 kDa) (Sigma-Aldrich, USA) using 20 mM Tris-HCl buffer (pH 8) supplemented with 150 mM NaCl.
Transmission Electron Microscopy
PAN-A/1 complexes were dialyzed overnight against 20 mM HEPES (pH 8) supplemented with 2M NaCl. Proteins were deposited onto carbon coated Formvar 400-mesh copper grids, fixed with 2% HEPES-buffered glutaraldehyde, floated on water to remove salts and negative stained with 1% aqueous uranyl acetate. Stain was removed with filter paper. Sample was air dried and examined with a Hitachi H-7000 TEM operated at 100kV (Hitachi High Technologies America, Inc. Schaumburg, IL). Digital images were acquired with a Veleta 2k × 2k camera and iTEM software (Olympus Soft-Imaging Solutions Corp, Lakewood, CO).
Results
Purification of PAN-A/1
PAN-A/1 was isolated from Hfx. volcanii (GZ108-pJAM2001, a ΔpanB/2 strain encoding PAN-A/1-StrepII) by sequential Strep-Tactin and Superose 6 HR 10/30 column chromatography (Fig 1A). No proteins were detected when the same two-step purification method was used for the vector control (GZ108-pJAM809). Based on migration by SDS-PAGE, PAN-A/1 was purified to apparent homogeneity as a 47-kDa protein (Fig. 1B, lane 1). The identity of PAN-A/1 was confirmed by Western blotting using antibodies raised against the StrepII tag (WSHPQFEK) (Fig. 1B, lane 2) and against PAN-A/1 (Fig. 1B, lane 3). PAN-A/1 was found to associate as a homooligomeric dodecamer of 630 kDa based on elution by gel filtration chromatography (Fig. 1A). Transmission electron micrographs of the PAN-A/1 revealed particles of 13 to 15 nm in diameter that were ring-like but not distinct hexamers or symmetrical complexes (Fig. 1C). The PAN-A/1 complex isolated from Hfx. volcanii was stable in the absence of ATP. This PAN-A/1 association did not require the presence of PAN-B/2. The results demonstrate that PAN-A/1 alone is able to form a complex of ∼12 subunits.
Figure 1.
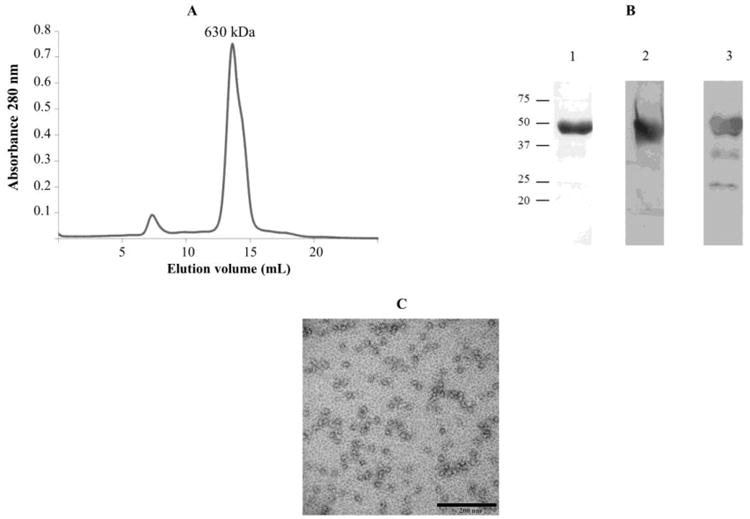
PAN-A/1 purified as a 630 kDa complex of 47 kDa subunits from Hfx. volcanii suggesting a dodecameric configuration. A: Superose 6 HR 10/30 chromatography of PAN-A/1 complex. B: PAN-A/1 (4 μg) separated by reducing 10% SDS-PAGE was detected by staining with Coomassie Blue R-250 (lane 1) and by Western blotting with antibodies raised against StrepII-tag and (lane 2) and PAN-A/1 (lane 3). C: Transmission electron micrograph of PAN-A/1 complex. Proteins were fixed with 2% HEPES-buffered glutaraldehyde and negative stained with 1% aqueous uranyl acetate.
PAN-A/1 nucleotide hydrolyzing activity
The PAN-A/1 complex purified from Hfx. volcanii differentially catalyzed the hydrolysis of nucleotides (ATP, GTP and TTP) (Table 1). The nucleotide hydrolyzing activities of PAN-A/1 were determined by measuring the formation of Pi from different nucleoside triphosphates at 42°C (the physiological growth temperature for Hfx. volcanii). PAN-A/1 was found to have specific activity for the hydrolysis of ATP at 0.33 ± 0.1 μmol Pi/h per mg protein, and this activity was significantly inhibited (3-fold) by EDTA to a level of 0.11 ± 0.05 μmol Pi/h per mg protein. Maximum ATPase activity of PAN-A/1 was detected at 42°C (Fig. 2A). The activity is 1.5 fold decreased at 25°C and 37°C, 4.7 fold at 20°C. The protein is not active at 50°C. The activity was not stimulated by addition of putative protein substrates (5 μM casein or 5 μM Flag-His-SAMP-MoaE-StrepII linear fusion) (data not shown). The kinetics constants of PAN-A/1 were determined. The Km for ATP was 438 μM ± 30, and the Vm was 6.12 ± 1 nmole Pi/h for 13 μg of PAN-A/1 (Fig. 2B). PAN-A/1 was found to cleave other nucleoside triphosphates including GTP and TTP but the rate of hydrolysis was 4- to 5-fold slower than for ATP (Table 1).
Table 1.
| Nucleotides | Relative activity (%) |
|---|---|
| ATP | 100%a± 10 |
| GTP | 24% ± 6.4 |
| TTP | 18.2% ± 7 |
| CTP | N.D. |
100% activity defined as 0.33 μmoles Pi/h per mg protein. PAN-A/1 was assayed at 42 °C using 13 μg protein per 100 μl reaction volume according to Materials and Methods. N.D. not determined due to non-enzymatic hydrolysis of CTP during assay.
Figure 2.
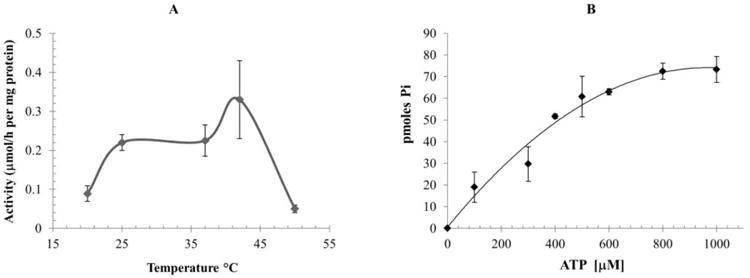
PAN-A/1 ATPase activity as a function of temperature (A) were determined at 20°C, 25°C, 37°C, 42°C and 50°C. Activity as function of ATP concentration at 42°C (B). ATPase activities were determined by measurement of Pi formed in presence of 13 μg of PAN-A/1.
The PAN-A/1 protein was found to require high salt for optimal ATPase activity and complex stability (Fig. 3A). At 1.4 M NaCl, the ATPase activity of PAN-A/1 was reduced to only 43% of activity in the presence of 2 M NaCl. Further reduction of NaCl concentrations to levels of 0.75 to 1 M NaCl resulted in PAN-A/1 protein with only 5-8% of the ATPase activity detected at 2 M NaCl. These results are in line with the findings that Hfx. volcanii is a moderate halophile that requires a salt concentration between 1.7-2.5M NaCl for optimal growth (Mullakhanbhai and Larsen, 1975) and uses a ‘salt-in’ strategy to maintain osmotic homeostasis (Pérez-Fillol and Rodríguez-Valera, 1986). The loss of activity was not due to an aggregation of PAN-A/1 protein. Based on gel filtration chromatography (Fig. 3B), PAN-A/1 protein did not aggregate and instead eluted as a complex of 112 kDa (dimeric to trimeric) instead of 630 kDa (dodecameric) (Fig. 1A). Thus, oligomeric stability of PAN-A/1 was affected by the salt concentration. Addition of glycerol (10 to 20% v/v final) or sorbitol (30% w/v final) to the low salt buffers did not stabilize the PAN-A/1 complex based on ATPase activity assay (data not shown). Compared to NaCl, the ATP hydrolyzing activity of PAN-A/1 was increased several-fold in buffers with KCl with specific activity of 1.0 ± 0.1 μmol Pi/h per mg protein at 2 M KCl. The PAN-A/1 complex was found to be more stable in low-salt buffer with KCl compared to NaCl based on ATPase activity assay (Fig. 3A).
Figure 3.
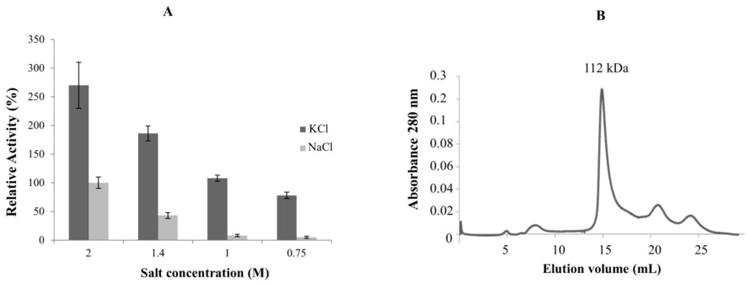
Effect of salt on PAN-A/1 activity and structure. A: Relative ATPase activity of PAN-A/1 after incubation of enzyme for 1 h at 4°C in different concentrations of NaCl (light grey) and KCl (grey). 100% corresponds to 0.33 μmoles Pi/h per mg protein. ATPase activities were determined at 42°C in presence of 6 and 13 μg PAN-A/1. B: Superose 6 HR 10/30 gel filtration chromatography of PAN-A/1 in 20 mM Tris-HCl buffer (pH 8) supplemented with 0.75 M NaCl after incubation of PAN-A/1 for 60 min at 4°C in buffer of the same composition.
In vitro interaction of PAN-A/1 with SAMP1-MoaE
To study the interaction of PAN-A/1 with potential protein substrates including those covalently linked to SAMP1, a far Western blotting technique was used. SAMP1 (Hepowit et al, 2012) and more recently SAMP3 (Miranda et al, in press) were found conjugated to MoaE. We adapted the protocol of (Wu et al, 2007) for analysis of halophilc proteins, since PAN-A/1 activity and stability was dependent on high concentrations of salt. The fold of SAMP1 is also high-salt dependent (Ye et al, 2013). To preserve as much as possible the native conformation of the halophilic proteins under study, 2 M NaCl was included in the renaturing and binding buffers used in far Western blotting (Table S3). To stabilize weak interactions and prevent dissociation of PAN-A/1 complexes during incubation with antibodies in low salt buffer, complexes were cross-linked with EDC after renaturation as described by (Sato et al, 2011). BSA was included as a negative control (Fig. 4, lane 1). Integrity of target (prey) protein was confirmed by Coomassie brilliant blue R250 staining (Fig. 4A) and Western blotting (Fig. 4B-C). Lack of cross-reactivity with anti-PAN-A/1 antibody was also confirmed for target (prey) protein (Fig. 4D). Based on far Western blotting, PAN-A/1 was found to bind specifically to the SAMP1-MoaE linear fusion protein (Fig. 4E-F, lane 4). The presence of ATP or ADP was not necessary for this interaction. PAN-A/1 did not bind to SAMP1 or MoaE alone or to the BSA used as a negative control (Fig. 4E-F, lanes 1-3). The binding of PAN-A/1 to MoaE required the covalently linked SAMP1. No interaction between SAMP1 and PAN-A/1 was detected in our studies. Furthermore, PAN-A/1 did not interact with BSA under the far Western blotting conditions even when it was present at 4-fold higher level than other target (prey) proteins. While the mesohalic BSA is not anticipated to be in a native conformation after renaturation in high salt buffer, we cannot exclude the possibility that PAN-A/1 interacted with SAMP1-MoaE due to improper folding of this linear protein fusion on the membrane. However, the far Western blotting technique used in this study was highly reproducible and could be used in the future to screen potential partners of PAN-A/1.
Figure 4.
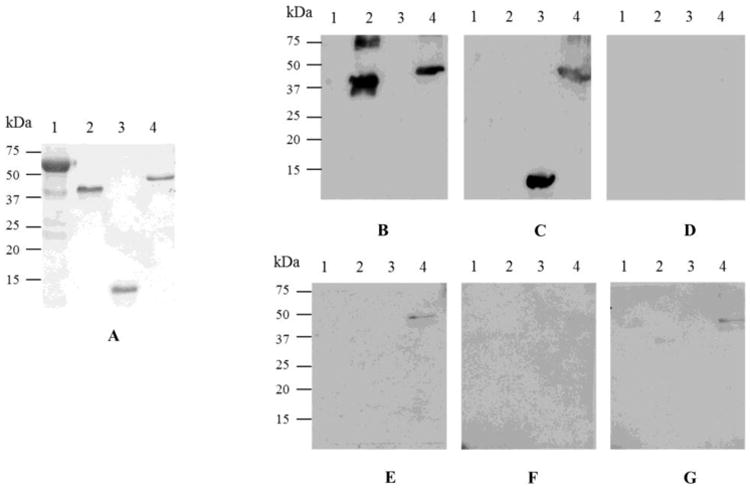
PAN-A/1 associates with SAMP1-MoaE but not the free forms of SAMP1 or MoaE. Prey proteins. Lane 1, BSA (3 μg); 2, MoaE (0.75 μg); 3, SAMP1 (0.75 μg); and 4, SAMP1-MoaE (0.75 μg) were separated by reducing SDS-PAGE (12% polyacrylamide). Proteins were detected by Coomassie Blue R-250 staining (A) and Western blotting with antibodies raised against the StrepII tag (B), Flag tag (C), and PAN-A/1 (D). BSA prey and Western blotting with anti-PAN-A/1 antibodies served as negative controls. Far western blotting was performed by denaturing, renaturing and incubating prey proteins with PAN-A/1 (bait protein) in 20 mM HEPES buffer (pH 8), 2 M NaCl (buffer C) with no nucleotide (E), 1 mM ADP and 9 mM MgCl2 (F), and 1 mM ATP and 9 mM MgCl2 (G). After washing, bound PAN-A/1 was detected with anti-PAN-A/1 antibody. See Methods section for details.
Discussion
Despite the importance of AAA+ proteins for the regulation of intracellular proteolysis, little information is available regarding their physical states and mechanisms of regulating protein degradation in the cell. In general, AAA+ proteins need to be assembled to perform their mechanical actions on substrate proteins and to associate with the peptidase complexes (Sauer and Baker, 2011). However, the different physical states of proteasome-associated AAA+ proteins from native organisms are not well studied. The archaeal PAN systems represent a paradigm to address these questions.
Here, we report biochemical and structural properties of a PAN-A/1 complex with ATPase and SAMP1-protein conjugate binding activity purified from Hfx. volcanii, PAN-A/1 purified as a dodecamer in a proposed two stacked hexameric ring configuration based on TEM and previous work finding that most AAA+ proteins are crystallized as stable ring-shaped oligomers (Snider and Houry, 2008). This dodecameric configuration is similar to what has been observed for PAN of Methanocaldococcus jannaschii (formerly Methanococcus jannaschii) (Wilson et al, 2000) and other related AAA+ ATPases including mammalian NSF (Hanson et al, 1997), Thermoplasma VAT (Pamnani et al, 1997) and Rhodococcus ARC (Wolf et al, 1998). The PAN-A/1 dodecamer was stable in the absence of ATP similar to what has been observed for M. jannaschii PAN (Wilson et al, 2000). However, unlike its mesohalic counterparts, which do not require molar concentrations of salt for activity, the PAN-A/1 dodecamer was found to dissociate into complexes of 3 subunits or less when the concentration of NaCl was reduced from 2 M to 0.75 M. These low molecular mass complexes of PAN-A/1 were significantly less active in hydrolyzing ATP even when assayed in buffer supplemented with 2 M KCl or NaCl. Once inactivated/dissociated by low salt, PAN-A/1 could not be reactivated by dialysis against high salt buffers (data not shown). This result contrasts with the 20S proteasomes of Hfx. volcanii, which similarly dissociate into monomers at low salt but can be reactivated and re-associated into peptide hydrolyzing 20S core particles by dialysis against high salt buffers (Wilson et al, 1999). Hfx. volcanii SAMP1 is recently shown to undergo conformational conversion from disorder to an ordered β-grasp fold with increasing ion concentration (Ye et al, 2013). Likewise, the Hfx. volcanii HvJAMM1 desampylase is inactivated by exposure to low concentrations of salt (Hepowit et al, 2012). Protein stability arises from a combination of many factors, which each contribute to various extents in different proteins. Salt concentration appears to be a dominant factor in stabilizing the proteomes of halophilic archaea (Mevarech et al, 2000).
With exception of the requirement of extremely high concentrations of salt, the observed biochemical properties of PAN-A/1 were comparable to other AAA+ATPases. Similarly to M. jannaschii PAN (Wilson et al, 2000; Zwickl et al, 1999), the Hfx. volcanii PAN-A/1 was found to release Pi from various nucleoside triphosphates (ATP, GTP, TTP) with the highest rate of hydrolysis for ATP, was found to be inhibited by EDTA, and was found to have relatively high affinity for ATP with a Km of ∼440 μM. Km values of 500 to 550 μM for ATP have been observed for M. jannaschii PAN (Wilson et al, 2000) and the related E.coli ClpX (Wawrzynow et al, 1995) and yeast Cdc48 (Fröhlich et al, 1995). Unlike other AAA+ATPases that have been characterized, the ATPase activity of PAN-A/1was increased several-fold in buffers with molar concentrations of salt with preference for KCl over NaCl. Similarly, the type II chaperonins (CCTs) purified from Hfx. volcanii are more active in molar concentrations of KCl compared to NaCl (Large et al, 2002). Salt preference by these enzymes correlates well with the intracellular environment of many halophilic archaea where K+ is the most prominent ion (between 1.9 and 5.5 M) and far exceeds that of Na+ (Pérez-Fillol and Rodríguez-Valera, 1986).
Currently, it is unclear how the PAN system is integrated with the SAMP-based protein tagging system, whose conjugates are increased (SAMP1) or decreased (SAMP2) in a genetic background deficient in synthesis of PAN-A/1 and α1 (Humbard et al, 2010a). Here we studied the interaction of PAN-A/1 with potential protein substrates including those covalently linked to SAMP1. Based on atomic structure and protein binding assays, the N-terminal coiled-coil domain of the PANs appears crucial for substrate recognition in energy-dependent proteolysis by proteasomes (Djuranovic et al, 2009; Zhang et al, 2009a; Zhang et al, 2009b). PAN-A/1 and PAN-B/2 are proposed to bind distinct sets of protein substrates based on significant structural differences in their N-terminal coiled-coil domains (Reuter et al, 2004) and phenotypic distinctions between Δpan-A/1 and Δpan-B/2 mutant strains (with no apparent cross-complementation) (Zhou et al, 2008).
Addition of a StrepII tag with a GT linker (-GTWSHPQFEK) to the C-terminus of PAN-A/1 enabled us to study the interaction of PAN with potential protein substrates, since this type of modification should not disrupt the N-terminal substrate binding domain. Our far Western experiments have demonstrated that the StrepII tagged PAN-A/1 binds MoaE, but only when this substrate is covalently linked to a SAMP1 moiety, suggesting that sampylation triggers association of PAN-A/1 with protein targets. In contrast, Ye et al. (2013) show by NMR spectroscopy that SAMP1 can bind an N-terminal peptide of PAN-B/2 (residues 1-74) with weak affinity over a wide range of salt concentrations, even those that disrupt SAMP1 structure. Thus, the PANs may form weak associations with the SAMPs alone. However, our results suggest that at least PAN-A/1 has a higher affinity for SAMP1 when covalently bound to a target protein compared to SAMP1 in its free form. The far Western blotting technique could be used in the future to screen for different PAN substrates.
The association state between various AAA+ ATPases and their corresponding peptidase particles remains poorly documented in the three domains of life. The C-terminal Hb[Y/F]-X motif of the M. jannaschii PAN is important for its association with 20S core particles (Smith et al, 2007; Yu et al, 2010). The PAN-A/1 protein was tagged in C-terminal tail and, thus, did not allow us to study its interaction with the 20S proteasomes of Hfx. volcanii. Similar modification of the C-terminus of Rpt1 (i.e., addition of a FlagHis6 epitope tag to the AAA ATPase subunit of the 19S regulatory particle) inhibits its association with the 20S proteasome core particle in yeast (Verma et al, 2000). Further studies are needed to understand the regulation of the degradation of proteins by proteasomes in archaea and the link between the SAMP system and this degradation.
Supplementary Material
Acknowledgments
Thanks to S. Shanker and the staff at UF ICBR for Sanger DNA sequencing. The authors acknowledge funding awarded to JM-F through the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (Grant DE-FG02-05ER15650) for the analysis of the biological function of SAMP-MoaE and development of halophilic protein technologies and the National Institutes of Health (Grant R01 GM57498) for the purification and biochemical characterization of PANs.
Nonstandard abbreviations
- UbaA
ubiquitin activating E1 enzyme homolog of archaea
- SAMP
small ubiquitin-like archaeal modifier protein
- Rpt
regulatory particle triple-A ATPase
- PAN
proteasome activating nucleotidase (Rpt homolog)
- CP
20S proteasome core particle
- RP
19S ATP-dependent regulatory particle
Contributor Information
Laurence Prunetti, Email: lprunetti@ufl.edu.
Christopher J. Reuter, Email: creuter@ospreybiotechnics.com.
Nathaniel L. Hepowit, Email: nathaniellhepowi@ufl.edu.
Yifei Wu, Email: lushafei333@163.com.
Luisa Barrueto, Email: lbarrueto@ufl.edu.
Hugo V. Miranda, Email: manson7@ufl.edu.
Karen Kelly, Email: vau@ufl.edu.
Julie A. Maupin-Furlow, Email: jmaupin@ufl.edu.
References
- Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: structure and function. Biochim Biophys Acta. 2012;1823:67–82. doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, Förster F. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj N, Goldberg AL. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat Cell Biol. 2000;2:833–839. doi: 10.1038/35041081. [DOI] [PubMed] [Google Scholar]
- Chamieh H, Guetta D, Franzetti B. The two PAN ATPases from Halobacterium display N-terminal heterogeneity and form labile complexes with the 20S proteasome. Biochem J. 2008;411:387–397. doi: 10.1042/BJ20071502. [DOI] [PubMed] [Google Scholar]
- Chamieh H, Marty V, Guetta D, Perollier A, Franzetti B. Stress regulation of the PAN-proteasome system in the extreme halophilic archaeon Halobacterium. Extremophiles. 2012;16:215–225. doi: 10.1007/s00792-011-0421-0. [DOI] [PubMed] [Google Scholar]
- Coux O, Nothwang HG, Silva Pereira I, Recillas Targa F, Bey F, Scherrer K. Phylogenic relationships of the amino acid sequences of prosome (proteasome, MCP) subunits. Mol Gen Genet. 1994;245:769–780. doi: 10.1007/BF00297284. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Dyall-Smith M. The Halohandbook: Protocols for Halobacterial Genetics. 2009;Version 7.2 http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2009_v7.2mds.pdf. [Google Scholar]
- Forouzan D, Ammelburg M, Hobel CF, Ströh LJ, Sessler N, Martin J, Lupas AN. The archaeal proteasome is regulated by a network of AAA ATPases. J Biol Chem. 2012;287:39254–39262. doi: 10.1074/jbc.M112.386458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KU, Fries HW, Peters JM, Mecke D. The ATPase activity of purified CDC48p from Saccharomyces cerevisiae shows complex dependence on ATP-, ADP-, and NADH-concentrations and is completely inhibited by NEM. Biochim Biophys Acta. 1995;1253:25–32. doi: 10.1016/0167-4838(95)00136-i. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hepowit NL, Uthandi S, Miranda HV, Toniutti M, Prunetti L, Olivarez O, De Vera IM, Fanucci GE, Chen S, Maupin-Furlow JA. Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol Microbiol. 2012;86:971–987. doi: 10.1111/mmi.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Humbard M, Miranda H, Lim J, Krause D, Pritz J, Zhou G, Chen S, Wells L, Maupin-Furlow J. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010a;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Maupin-Furlow JA. Prokaryotic proteasomes: nanocompartments of degradation. J Mol Microbiol Biotechnol. 2013;23:321–334. doi: 10.1159/000351348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Reuter CJ, Zuobi-Hasona K, Zhou G, Maupin-Furlow JA. Phosphorylation and methylation of proteasomal proteins of the haloarchaeon Haloferax volcanii. Archaea. 2010b;2010:481725. doi: 10.1155/2010/481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland P, Gil M, Karadzic I, Maupin-Furlow J. Genetic and proteomic analyses of a proteasome-activating nucleotidase a mutant of the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland P, Maupin-Furlow J. Stabilization of an archaeal DNA-sliding clamp protein, PCNA, by proteasome-activating nucleotidase gene knockout in Haloferax volcanii. FEMS Microbiol Lett. 2009;294:32–36. doi: 10.1111/j.1574-6968.2009.01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T, Fukui K, Kometani K. The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J Biochem. 1986;99:1465–1472. doi: 10.1093/oxfordjournals.jbchem.a135616. [DOI] [PubMed] [Google Scholar]
- Large AT, Kovacs E, Lund PA. Properties of the chaperonin complex from the halophilic archaeon Haloferax volcanii. FEBS Lett. 2002;532:309–312. doi: 10.1016/s0014-5793(02)03685-2. [DOI] [PubMed] [Google Scholar]
- Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat Rev Microbiol. 2012;10:100–111. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevarech M, Frolow F, Gloss LM. Halophilic enzymes: proteins with a grain of salt. Biophys Chem. 2000;86:155–164. doi: 10.1016/s0301-4622(00)00126-5. [DOI] [PubMed] [Google Scholar]
- Miranda HV, Antelmann H, Hepowit N, Chavarria NE, Krause DJ, Pritz JR, Basell K, Becher D, Humbard MA, Broccheri L, Maupin-Furlow JA. Archaeal ubiquitin-like SAMP3 is isopeptide-linked to proteins by a UbaA-dependent mechanism. Mol Cell Proteomics. doi: 10.1074/mcp.M113.029652. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullakhanbhai MF, Larsen H. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch Microbiol. 1975;104:207–14. doi: 10.1007/BF00447326. [DOI] [PubMed] [Google Scholar]
- Pamnani V, Tamura T, Lupas A, Peters J, Cejka Z, Ashraf W, Baumeister W. Cloning, sequencing and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum. FEBS Lett. 1997;404:263–268. doi: 10.1016/s0014-5793(97)00138-5. [DOI] [PubMed] [Google Scholar]
- Pérez-Fillol M, Rodríguez-Valera F. Potassium ion accumulation in cells of different halobacteria. Microbiologia. 1986;2:73–80. [PubMed] [Google Scholar]
- Reuter C, Kaczowka S, Maupin-Furlow J. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J Bacteriol. 2004;186:7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Kameya M, Arai H, Ishii M, Igarashi Y. Detecting weak protein-protein interactions by modified far-western blotting. J Biosci Bioeng. 2011;112:304–307. doi: 10.1016/j.jbiosc.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Snider J, Houry WA. AAA+ proteins: diversity in function, similarity in structure. Biochem Soc Trans. 2008;36:72–77. doi: 10.1042/BST0360072. [DOI] [PubMed] [Google Scholar]
- Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H, Aldrich H, Maupin-Furlow J. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: Purification, characterization, and gene sequence analysis. J Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H, Ou M, Aldrich H, Maupin-Furlow J. Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J Bacteriol. 2000;182:1680–1692. doi: 10.1128/jb.182.6.1680-1692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Nagy I, Lupas A, Pfeifer G, Cejka Z, Müller SA, Engel A, De Mot R, Baumeister W. Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J Mol Biol. 1998;277:13–25. doi: 10.1006/jmbi.1997.1589. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by Far western blotting. Nat Protoc. 2007;2:3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
- Ye K, Liao S, Zhang W, Fan K, Zhang X, Zhang J, Xu C, Tu X. Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii. Sci Rep. 2013;3:2136. doi: 10.1038/srep02136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y. Interactions of PAN's C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 2010;29:692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009a;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009b;34:485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Kowalczyk D, Humbard M, Rohatgi S, Maupin-Furlow J. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


