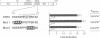Abstract
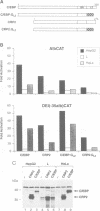
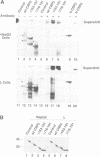
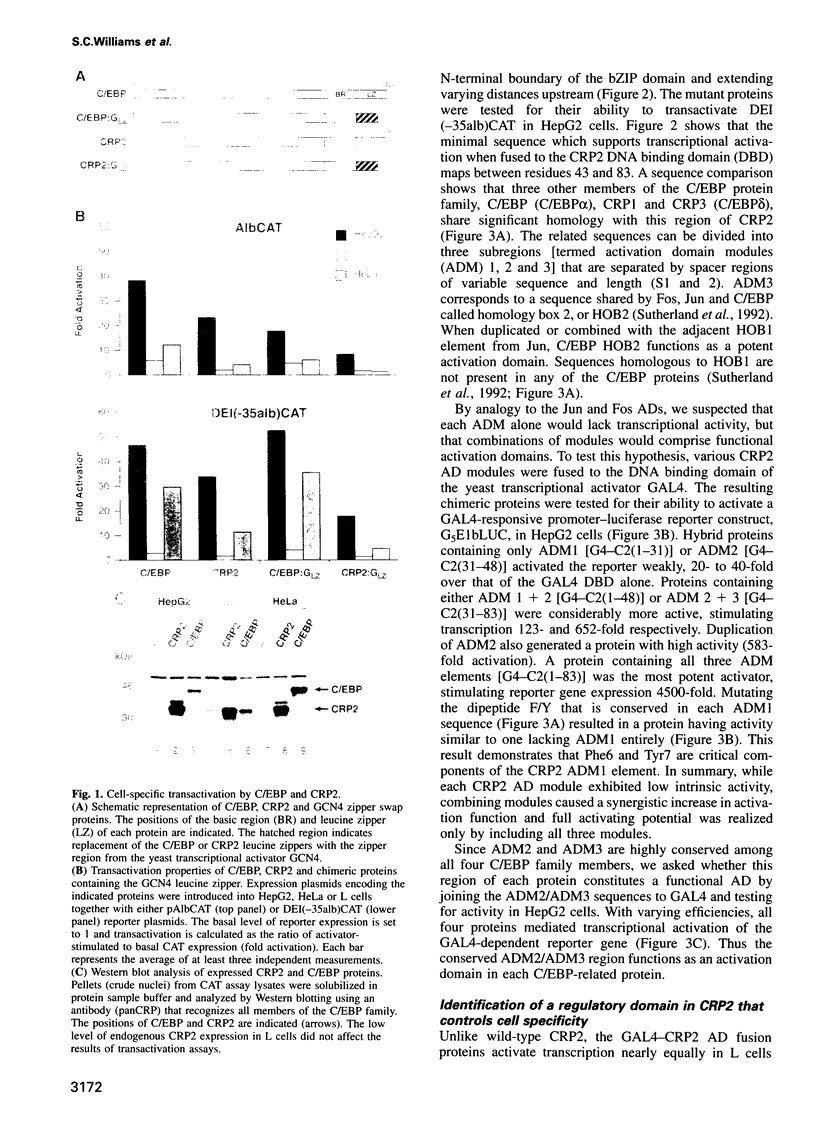
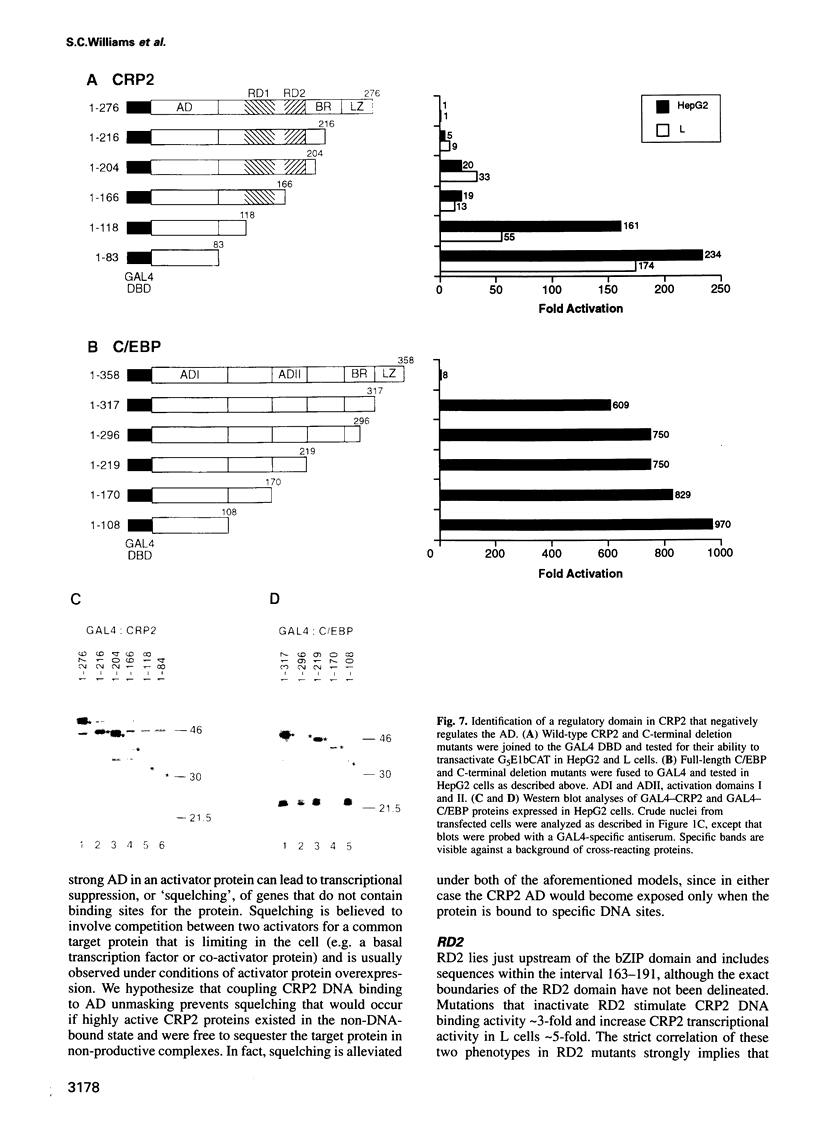
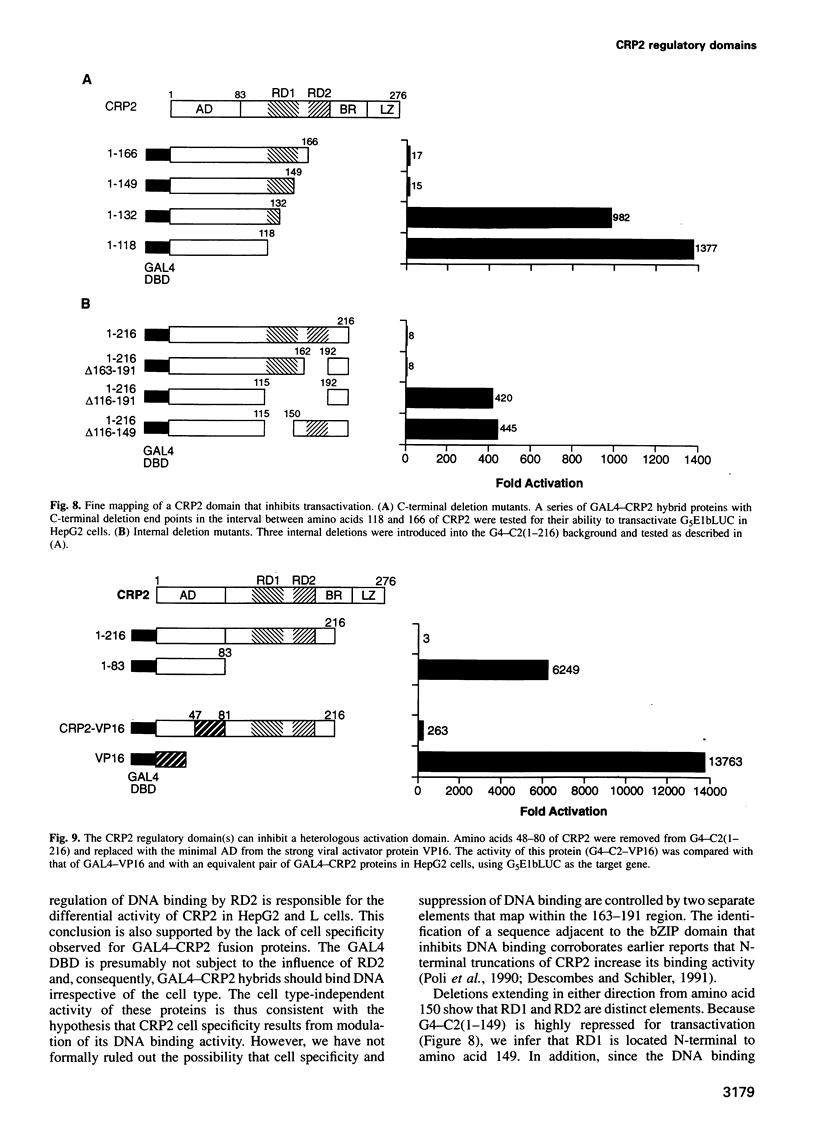
Two members of the C/EBP family of basic region-leucine zipper proteins enriched in the liver, C/EBP (C/EBP alpha) and CRP2 (C/EBP beta), were previously shown to transactivate the albumin promoter in a cell type-dependent manner. These proteins function efficiently in HepG2 hepatoma cells, but inefficiently in HeLa (epithelial) and L (fibroblastic) cells. Here we have investigated the mechanism for cell-specific control of CRP2 activity. We show that CRP2 contains a negative regulatory region composed of two elements, RD1 and RD2. Deletions of RD2 relieve the inhibition of CRP2 activity in L cells, but do not affect CRP2 function in HepG2 cells. These deletions also increase the DNA binding activity of CRP2 approximately 3-fold, suggesting that RD2-mediated repression of DNA binding activity is responsible for CRP2 inhibition in L cells. The adjacent RD1 element functions independently of RD2 and modulates the CRP2 activation domain, which we show to be composed of three subdomains that are conserved within the C/EBP protein family. RD1 does not affect cell type specificity, but inhibits the transactivation potential of GAL4-CRP2 hybrid proteins by 50-fold. These findings suggest that CRP2 assumes a tightly folded conformation in which the DNA binding and activation domains are masked by interactions with the regulatory domain and that to function efficiently in HepG2 cells the protein must undergo an activation step. We propose that relief of inhibition conferred by the regulatory domains also accounts for CRP2 activation in response to extracellular signals.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990 Jun;9(6):1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann H., Gauldie J. The acute phase response. Immunol Today. 1994 Feb;15(2):74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- Bretz J. D., Williams S. C., Baer M., Johnson P. F., Schwartz R. C. C/EBP-related protein 2 confers lipopolysaccharide-inducible expression of interleukin 6 and monocyte chemoattractant protein 1 to a lymphoblastic cell line. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7306–7310. doi: 10.1073/pnas.91.15.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Umek R. M., McKnight S. L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991 Sep;5(9):1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990 May 24;345(6273):361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Raymondjean M., Carranca A. G., Herbomel P., Yaniv M. Factors involved in control of tissue-specific expression of albumin gene. Cell. 1987 Aug 14;50(4):627–638. doi: 10.1016/0092-8674(87)90036-5. [DOI] [PubMed] [Google Scholar]
- Chang C. J., Chen T. T., Lei H. Y., Chen D. S., Lee S. C. Molecular cloning of a transcription factor, AGP/EBP, that belongs to members of the C/EBP family. Mol Cell Biol. 1990 Dec;10(12):6642–6653. doi: 10.1128/mcb.10.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. M., Takiff H. E., Barber A. M., Dubois G. C., Bardwell J. C., Court D. L. Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J Biol Chem. 1990 Feb 15;265(5):2888–2895. [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Descombes P., Chojkier M., Lichtsteiner S., Falvey E., Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990 Sep;4(9):1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- Descombes P., Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991 Nov 1;67(3):569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Friedman A. D., Landschulz W. H., McKnight S. L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989 Sep;3(9):1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- Friedman A. D., McKnight S. L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990 Aug;4(8):1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Janknecht R., Hipskind R. A., Houthaeve T., Nordheim A., Stunnenberg H. G. Identification of multiple SRF N-terminal phosphorylation sites affecting DNA binding properties. EMBO J. 1992 Mar;11(3):1045–1054. doi: 10.1002/j.1460-2075.1992.tb05143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S., Kowenz-Leutz E., Müller C., Meese K., Ness S. A., Leutz A. The NF-M transcription factor is related to C/EBP beta and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 1993 Apr;12(4):1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Court D., Papas T. S. High-level expression in Escherichia coli of the carboxy-terminal sequences of the avian myelocytomatosis virus (MC29) v-myc protein. Gene. 1983 Jul;23(1):75–84. doi: 10.1016/0378-1119(83)90218-4. [DOI] [PubMed] [Google Scholar]
- LeClair K. P., Blanar M. A., Sharp P. A. The p50 subunit of NF-kappa B associates with the NF-IL6 transcription factor. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Yano M., Liu S. Y., Matsunaga E., Johnson P. F., Gonzalez F. J. A novel cis-acting element controlling the rat CYP2D5 gene and requiring cooperativity between C/EBP beta and an Sp1 factor. Mol Cell Biol. 1994 Feb;14(2):1383–1394. doi: 10.1128/mcb.14.2.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. M., Tsai W. H., Lai M. Y., Chen D. S., Lee S. C. Induction of liver alpha-1 acid glycoprotein gene expression involves both positive and negative transcription factors. Mol Cell Biol. 1993 Jan;13(1):432–442. doi: 10.1128/mcb.13.1.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtsteiner S., Wuarin J., Schibler U. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell. 1987 Dec 24;51(6):963–973. doi: 10.1016/0092-8674(87)90583-6. [DOI] [PubMed] [Google Scholar]
- Maire P., Wuarin J., Schibler U. The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science. 1989 Apr 21;244(4902):343–346. doi: 10.1126/science.2711183. [DOI] [PubMed] [Google Scholar]
- Manak J. R., de Bisschop N., Kris R. M., Prywes R. Casein kinase II enhances the DNA binding activity of serum response factor. Genes Dev. 1990 Jun;4(6):955–967. doi: 10.1101/gad.4.6.955. [DOI] [PubMed] [Google Scholar]
- Matsusaka T., Fujikawa K., Nishio Y., Mukaida N., Matsushima K., Kishimoto T., Akira S. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R., Ziff E. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to trans-locate to the nucleus and induce c-fos transcription. Genes Dev. 1991 Oct;5(10):1754–1766. doi: 10.1101/gad.5.10.1754. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Kinoshita S., Sasagawa T., Sasaki K., Naruto M., Kishimoto T., Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsuka S., Akira S., Nishio Y., Hashimoto S., Sugita T., Isshiki H., Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992 Jan 15;79(2):460–466. [PubMed] [Google Scholar]
- Nerlov C., Ziff E. B. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes Dev. 1994 Feb 1;8(3):350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- Ness S. A., Kowenz-Leutz E., Casini T., Graf T., Leutz A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993 May;7(5):749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- Nishio Y., Isshiki H., Kishimoto T., Akira S. A nuclear factor for interleukin-6 expression (NF-IL6) and the glucocorticoid receptor synergistically activate transcription of the rat alpha 1-acid glycoprotein gene via direct protein-protein interaction. Mol Cell Biol. 1993 Mar;13(3):1854–1862. doi: 10.1128/mcb.13.3.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordeen S. K. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques. 1988 May;6(5):454–458. [PubMed] [Google Scholar]
- O'Neil K. T., Shuman J. D., Ampe C., DeGrado W. F. DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry. 1991 Sep 17;30(37):9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- Patel L., Abate C., Curran T. Altered protein conformation on DNA binding by Fos and Jun. Nature. 1990 Oct 11;347(6293):572–575. doi: 10.1038/347572a0. [DOI] [PubMed] [Google Scholar]
- Patterson T. A., Costantino N., Dasgupta S., Court D. L. Improved bacterial hosts for regulated expression of genes from lambda pL plasmid vectors. Gene. 1993 Sep 30;132(1):83–87. doi: 10.1016/0378-1119(93)90517-7. [DOI] [PubMed] [Google Scholar]
- Pei D. Q., Shih C. H. An "attenuator domain" is sandwiched by two distinct transactivation domains in the transcription factor C/EBP. Mol Cell Biol. 1991 Mar;11(3):1480–1487. doi: 10.1128/mcb.11.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli V., Mancini F. P., Cortese R. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell. 1990 Nov 2;63(3):643–653. doi: 10.1016/0092-8674(90)90459-r. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri P., Bagchi S., Nevins J. R. DNA-binding activity of the adenovirus-induced E4F transcription factor is regulated by phosphorylation. Genes Dev. 1989 May;3(5):620–627. doi: 10.1101/gad.3.5.620. [DOI] [PubMed] [Google Scholar]
- Rivera V. M., Miranti C. K., Misra R. P., Ginty D. D., Chen R. H., Blenis J., Greenberg M. E. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol Cell Biol. 1993 Oct;13(10):6260–6273. doi: 10.1128/mcb.13.10.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989 Sep 25;17(18):7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman J. D., Vinson C. R., McKnight S. L. Evidence of changes in protease sensitivity and subunit exchange rate on DNA binding by C/EBP. Science. 1990 Aug 17;249(4970):771–774. doi: 10.1126/science.2202050. [DOI] [PubMed] [Google Scholar]
- Sutherland J. A., Cook A., Bannister A. J., Kouzarides T. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 1992 Sep;6(9):1810–1819. doi: 10.1101/gad.6.9.1810. [DOI] [PubMed] [Google Scholar]
- Trautwein C., Caelles C., van der Geer P., Hunter T., Karin M., Chojkier M. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature. 1993 Aug 5;364(6437):544–547. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Kerckaert J. P., Wasylyk B. A novel modulator domain of Ets transcription factors. Genes Dev. 1992 Jun;6(6):965–974. doi: 10.1101/gad.6.6.965. [DOI] [PubMed] [Google Scholar]
- Wegner M., Cao Z., Rosenfeld M. G. Calcium-regulated phosphorylation within the leucine zipper of C/EBP beta. Science. 1992 Apr 17;256(5055):370–373. doi: 10.1126/science.256.5055.370. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Ellenberger T., Wobbe C. R., Lee J. P., Harrison S. C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990 Oct 11;347(6293):575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- Williams S. C., Cantwell C. A., Johnson P. F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991 Sep;5(9):1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]