Abstract
Fas and its ligand (FasL) play an important role in apoptosis and carcinogenesis. Therefore, the potential association of polymorphisms in the Fas (-670A>G, rs1800682; -1377G>A, rs2234767) and FasL (-844C>T, rs763110) with cancer risk has been widely investigated. However, all the currently available results are not always consistent. In this work, we performed a meta-analysis to further determine whether carriers of the polymorphisms in Fas and FasL of interest could confer an altered susceptibility to cancer. All relevant data were retrieved by PubMed and Web of Science, and 52 eligible studies were chosen for this meta-analysis. There was no association of the Fas -670A>G polymorphism with cancer risk in the pooled data. For the Fas -1377G>A and FasL -844C>T polymorphisms, results revealed that the homozygotes of -1377A and -844C were associated with elevated risk of cancer as a whole. Further stratified analysis indicated markedly increased risk for developing breast cancer, gastric cancer, and esophageal cancer, in particular in Asian population. We conclude that carriers of the Fas-1377A and the FasL -844C are more susceptible to the majority of cancers than non-carriers.
Introduction
With new cases and mortality increased dramatically, cancer has become the major public health burden worldwide. For this reason, novel diagnostic markers are needed urgently for early detection and prevention of cancer. However, carcinogenesis is a complicated biological process that is not fully understood. It is generally believed that interactions of low-penetrance susceptibility genes with environmental factors might contribute to carcinogenesis [1]. As one of the important low-penetrance genes, Fas is considered to be a potential cancer susceptibility gene. This is because Fas (TNFSF6, CD95, or APO-1) is a cell surface receptor involved in apoptotic signal transmission in many cell types and interacts with its natural ligand Fas ligand (also known as FasL) to initiate the death signal cascade that leads to apoptotic cell death [2], [3]. Furthermore, in these two genes, there are several functionally significant polymorphisms, such as the −670A>G and −1377G>A in the Fas promoter region, and the −844C>T in the FasL promoter region, because they might be associated with cancer risk, including cervical cancer [4]–[9], gastric cancer [10]–[15], breast cancer [16]–[21], lung cancer [22]–[25] and so on. However, all available results are not always consistent with one another, partially because of the small sample size of some published studies, different ethnic backgrounds, publication bias, and little effect of the polymorphisms on cancer risk. Therefore, it's necessary to retrieve and pool all eligible data to further determine whether these genetic polymorphisms could be at increased risk for developing cancer and to what extent heterogeneity existed across all the studies.
Materials and Methods
Identification and eligibility of relevant studies
Two online medical databases, PubMed, and Web of Science, were searched (updated February 2013), using the search terms “Fas/CD95/TNFSF6/APO-1”, “FasL/CD95L”, “polymorphism/genetic variation” and “cancer/carcinoma/tumor”). The literature search was limited to English articles. In addition, more studies were also identified by manual search based on the references provided in the retrieved studies. The inclusion criteria were prespecified as below: (1) be a case-control study, (2) evaluate association between the Fas and/or FasL polymorphisms and cancer risk, (3) present sufficient data to calculate an odds ratio (OR) with 95% confidence interval (CI), and (4) list genotype frequency. Moreover, the studies without raw data, or those that were case-only studies, case reports, editorials, and review articles (including meta-analyses) were eliminated.
Data extraction
Information was extracted carefully from all eligible articles independently by two authors (Yeqiong Xu and Bangshun He) according to the above inclusion and exclusion criteria. Discrepancies were resolved by extensive discussion in our research team. The characteristics of enrolled studies were extracted as below: the first author's last name, year of publication, country of subjects, ethnicity, type of cancer, the source of controls, genotyping method (whether PCR was performed using a dual-labelled TaqMan probe with a specific 3'base to detect the SNPs or whether an RFLP method was used), the number of matched cases and controls, polymorphism sites, and P value for Hardy–Weinberg equilibrium (HWE) as summarized in Table 1.
Table 1. Characteristics of studies included in the meta-analysis.
| Cancer type | Year | First author | Country | Ethnicity | Source of control | Genotyping method | Polymorphism sites | Cases | Controls | HWE |
| Cervical cancer | ||||||||||
| 2009 | Zucchini [52] | Maton Grosso do Sul, Brazil | African | PB | PCR-RFLP | Fas -670A>G | 91 | 176 | 0.545 | |
| 2008 | Tamandani [35] | Northern India | Asian | HB | PCR-RFLP | Fas -670A>G | 200 | 200 | 0.001 | |
| 2008 | Kang [4] | Korea | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 154 | 160 | 0.264, 0.233, 0.327 | |
| 2007 | Ivansson [53] | Sweden | Caucasian | PB | TaqMan | FasL -844C>T | 1284 | 280 | 0.738 | |
| 2006 | Ueda [34] | Japan | Asian | PB | PCR-RFLP | Fas -670A>G | 83 | 95 | 0.172 | |
| 2005 | Zoodsma [5] | Netherlands | Caucasian | PB | TaqMan | Fas -670A>G | 670 | 607 | 0.274 | |
| 2005 | Sun [6] | China | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 314 | 615 | 0.641, 0.304, 0.002 | |
| 2005 | Lai [7] | China | Asian | HB | TaqMan | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 318 | 318 | 0.736, 0.293, 0.920 | |
| 2004 | Dybikowska [8] | Poland | Caucasian | PB | PCR-RFLP | Fas -670A>G | 51 | 65 | 0.638 | |
| 2003 | Lai [9] | China | Asian | HB | PCR-RFLP | Fas -670A>G | 176 | 176 | 0.444 | |
| Gastric cancer | ||||||||||
| 2012 | Zhang [10] | China | Asian | HB | PCR-RFLP | Fas -1377G>A, FasL -844C>T | 375 | 496 | 0.064, 0.112 | |
| 2011 | Liu [12] | China | Asian | PB | PCR-RFLP | Fas -1377G>A, FasL -844C>T | 344 | 324 | 0.424, 0.083 | |
| 2011 | Kupcinskas [11] | Mixed | Caucasian | PB | TaqMan | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 114 | 238 | 0.199, 0.492, 0.715 | |
| 2010 | Zhou [13] | China | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 262 | 524 | 0.133, 0.062, 0.899 | |
| 2009 | Wang [14] | China | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 332 | 324 | 0.806, 0.870, 0.554 | |
| 2008 | Hsu [15] | China | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 86 | 101 | 0.736, 0.914, 0.612 | |
| 2006 | Ikehara [54] | Japan | Asian | PB | PCR-CTPP | Fas -670A>G | 271 | 271 | 0.504 | |
| Breast cancer | ||||||||||
| 2013 | Hashemi [16] | Iranian | Caucasian | PB | T-ARMS-PCR | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 134 | 152 | 0.045, 0.000, 0.183 | |
| 2012 | Wang [17] | China | Asian | HB | PCR-RFLP | Fas -1377G>A, FasL -844C>T | 375 | 496 | 0.064, 0.112 | |
| 2012 | Mahfoudh [18] | Tunisia | African | PB | PCR-RFLP | FasL -844C>T | 438 | 332 | 0.334 | |
| 2007 | Crew [19] | America | Caucasian | PB | TaqMan | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 1051 | 1101 | 0.754, 0.069, 0.602 | |
| 2007 | Zhang [20] | China | Asian | HB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 836 | 834 | 0.797, 0.700, 0.110 | |
| 2004 | Krippl [21] | Austria | Caucasian | PB | TaqMan | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 499 | 495 | 0.924, 0.610, 0.418 | |
| Lung cancer | ||||||||||
| 2008 | Ter-Minassian [22] | America | Caucasian | HB | TaqMan | Fas -1377G>A, FasL -844C>T | 2174 | 1497 | 0.751, 0.254 | |
| 2007 | Gormus [23] | Turkey | Caucasian | PB | PCR-RFLP | Fas -1377G>A | 94 | 50 | 0.000 | |
| 2006 | Park [24] | Korea | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 582 | 582 | 0.132, 0.024, 0.570 | |
| 2005 | Zhang [25] | China | Asian | PB | PCR-RFLP | Fas -1377G>A, FasL -844C>T | 1000 | 1270 | 0.046, 0.180 | |
| 2003 | Wang [55] | America | Mixed | PB | PCR-RFLP | Fas -670A>G | 68 | 74 | 0.481 | |
| Esophageal cancer | ||||||||||
| 2011 | Bye [32] | Eastern or Western Cape | African | PB | TaqMan | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 343 | 466 | 0.027, 0.670, 0.097 | |
| 2011 | Bye [32] | Western Cape | Mixed | PB | TaqMan | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 195 | 420 | 0.170, 0.469, 0.741 | |
| 2007 | Jain [56] | Northern India | Asian | PB | PCR-RFLP | Fas -670A>G | 151 | 201 | 0.140 | |
| 2003 | Sun [57] | China | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 588 | 648 | 0.130, 0.218, 0.061 | |
| Skin cancer | ||||||||||
| 2010 | Qureshi [58] | Britain | Caucasian | PB | NA | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 779 | 842 | 0.210, 0.916, 0.427 | |
| 2007 | Zhang [59] | Sweden | Caucasian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 229 | 351 | 0.380, 0.009, 0.609 | |
| 2006 | Li [60] | America | Caucasian | HB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 602 | 603 | 0.453, 0.951, 0.071 | |
| 2001 | Nelson [61] | America | Caucasian | PB | PCR-RFLP | Fas -670A>G | 776 | 435 | 0.117 | |
| Ovarian cancer | ||||||||||
| 2012 | Li [62] | China | Asian | PB | Allele-specific multiple ligase detection | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 342 | 344 | 0.357, 0.972, 0.547 | |
| 2007 | Gormus [63] | Turkey | Caucasian | PB | PCR-RFLP | Fas -1377G>A, FasL -844C>T | 47 | 41 | 0.272, 0.678 | |
| 2006 | Ueda [34] | Japan | Asian | PB | PCR-RFLP | Fas -670A>G | 68 | 95 | 0.172 | |
| Prostate cancer | ||||||||||
| 2012 | Mandal [51] | Northern India | Asian | HB | PCR-RFLP | Fas -670A>G, Fas -1377G>A | 192 | 224 | 0.296, 0.035 | |
| 2011 | Shao [38] | China | Asian | HB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 602 | 703 | 0.579, 0.099, 0.801 | |
| 2008 | Lima [64] | Portugal | Caucasian | PB | PCR-RFLP | Fas -670A>G | 657 | 247 | 0.365 | |
| Nasopharyngeal cancer | ||||||||||
| 2010 | Cao [65] | China | Asian | PB | PCR-RFLP | Fas -1377G>A, FasL -844C>T | 576 | 608 | 0.984, 0.015 | |
| 2010 | Zhu [66] | China | Asian | PB | PCR-RFLP | Fas -670A>G | 237 | 264 | 0.478 | |
| 2006 | Jrad [37] | Tunisia | African | PB | PCR-RFLP | Fas -670A>G | 170 | 224 | 0.585 | |
| Bladder cancer | ||||||||||
| 2010 | Gangwar [67] | Northern India | Asian | PB | PCR-RFLP | Fas -670A>G | 212 | 250 | 0.384 | |
| 2006 | Li [68] | China | Asian | HB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 216 | 252 | 0.409, 0.970, 0.234 | |
| Other cancers | ||||||||||
| 2010 | Zhu [69] | China | Asian | HB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 353 | 365 | 0.831, 0.777, 0.278 | |
| 2010 | Wang [70] | China | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 294 | 333 | 0.034, 0.628, 0.271 | |
| 2008 | Yang [39] | China | Asian | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 397 | 907 | 0.653, 0.062, 0.986 | |
| 2007 | Koshkina [71] | America | Mixed | PB | PCR-RFLP | Fas -670A>G, Fas -1377G>A | 123 | 510 | 0.786, 0.210 | |
| 2007 | Erdogan [72] | Turkey | Caucasian | HB | PCR-RFLP | Fas -670A>G, FasL -844C>T | 45 | 100 | 0.812, 0.727 | |
| 2007 | Hoa [33] | America | Mixed | HB | PCR-RFLP | Fas -1377G>A | 279 | 510 | 0.210 | |
| 2007 | Hob [33] | America | Mixed | HB | PCR-RFLP | Fas -1377G>A | 154 | 510 | 0.210 | |
| 2006 | Zhang [36] | America | Caucasian | HB | PCR-RFLP | Fas -670A>G, Fas -1377G>A, FasL -844C>T | 721 | 1234 | 0.481, 0.268, 0.411 | |
| 2006 | Ueda [34] | Japan | Asian | PB | PCR-RFLP | Fas -670A>G | 108 | 95 | 0.172 |
The Ho(a) investigated thyroid cancer, and the Ho(b) investigated salivary gland cancer.
PB: population based; HB: hospital based; T-ARMS-PCR:tetra-primeramplification refractory mutation system PCR; PCR-RFLP: restriction fragment length polymorphism; HWE: Hardy-Weinberg equilibrium.
Genotype-gene expression correlation analysis
The International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) was used to obtain data of the Fas and FasL genotypes determined in 270 enrolled subjects. Meanwhile, the mRNA expression data of these enrolled subjects were available online from SNPexp (http://app3.titan.uio.no/biotools/help.php?app=snpexp) as described in the previous studies [26], [27]. In brief, these data were obtained from the HapMap phase II release 23 data set consisting of 3.96 million SNP genotypes from 270 subjects of three populations, including 90 European (CEU), 90 Asian (45 Chinese, 45 Japanese), and 90 Yoruba (YRI) subjects [28]. Additionally, the mRNA expression data were derived from the lymphoblastic cell lines from the same 270 subjects [29].
Statistical analysis
Crude ORs with 95% CIs were used to assess the strength of association between the polymorphisms in Fas-670A>G, Fas -1377G>A, and FasL -844T/C and cancer risk. The pooled ORs were estimated for dominant model (variant homozygotes + heterozygous vs homozygous reference), recessive model (variant homozygotes vs heterozygous + homozygous reference), homozygote comparison (variant homozygotes vs homozygous reference), heterozygote comparison (heterozygous vs homozygous reference) and allelic comparison in the polymorphisms, respectively. Stratified analyses were performed by the type of cancer (that with only one study was grouped together as ‘other cancers’), ethnicity, source of controls and genotyping method. Heterogeneity across the studies was evaluated by using the Chi-square test based Q-statistic test, and it was considered statistically significant when Pheterogeneity (Ph)<0.05. The data were combined using random-effects model (the DerSimonian and Laird method) [30] in the presence of heterogeneity (P<0.05 or I2>50%), or fixed-effects model (the Mantel-Haenszel method) models [31] was chosen to use in the absence of heterogeneity (P>0.05 or I2<50%). Moreover, sensitivity analyses were performed to assess the stability of the results. Publication bias was evaluated graphically by using funnel plots and statistically by the Egger's linear regression test. HWE of the three polymorphisms was assessed using a web-based program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). All statistical tests were performed with STATA 11.0 and SPSS 20.0. All the P values were two-sided.
Results
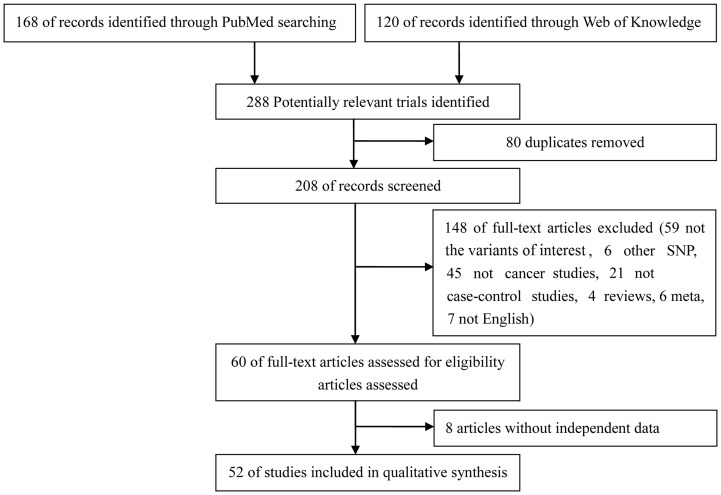
A total of 52 studies were enrolled in this meta-analysis (Figure 1). The major characteristics of the 52 selected studies are summarized in Table 1. The study carried out by Bye et al [32] analyzed individuals of African or Mixed ethnicity, and thus was divided into two studies. Similarly, the studies reported by Ho et al [33] and Ueda et al [34] investigated two and three types of cancer, and therefore, these two studies were cited as two studies and three studies, respectively (Table 1).
Figure 1. Flow chart of studies identified according to inclusion and exclusion criteria.
For the Fas -670A>G polymorphism, there was no association in the pooled analysis. In the subgroup analysis, statistically significantly decreased risk was observed in prostate cancer and melanoma for GG+AG vs AA comparison model, whereas there was significantly increased risk among those of African ancestry for GG+AG vs AA models (all data shown in Table 2).
Table 2. Stratified analyses of the Fas -670A>G (rs1800682) polymorphism and cancer.
| Variables | na | GG+AG vs AA | GG vs AG+AA | G vs A | ||||||
| OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | ||
| Total | 44 | 1.01(0.94, 1.09)c | <0.0001 | 47.1 | 1.04(0.96, 1.12)c | 0.003 | 40.9 | 1.02(0.97,1.06)c | 0.005 | 39.4 |
| Cancer type | ||||||||||
| Cervical cancer | 9 | 1.05(0.79,1.40)c | <0.0001 | 74.5 | 0.92(0.69, 1.22)c | 0.006 | 62.8 | 0.99(0.86,1.14)c | 0.013 | 58.5 |
| Gastric cancer | 5 | 1.08(0.91,1.28) | 0.340 | 11.6 | 0.97(0.79,1.21) | 0.978 | 0.0 | 1.03(0.91,1.15) | 0.735 | 0.0 |
| Esophageal cancer | 4 | 1.02(0.85,1.21) | 0.459 | 0.0 | 1.21(0.86,1.69)c | 0.017 | 70.4 | 1.10(0.99,1.23) | 0.215 | 32.9 |
| Breast cancer | 4 | 1.01(0.90,1.14) | 0.325 | 13.4 | 1.03(0.90,1.18) | 0.062 | 59.1 | 1.02(0.94,1.10) | 0.259 | 25.5 |
| Prostate cancer | 3 | 0.83(0.70,0.98) | 0.155 | 46.4 | 0.82(0.66,1.01) | 0.346 | 5.8 | 0.87(0.77,0.97) | 0.163 | 44.8 |
| Ovarian cancer | 2 | 0.87(0.66,1.15) | 0.952 | 0.0 | 0.85(0.57,1.28) | 0.622 | 0.0 | 0.90(0.74,1.09) | 0.745 | 0.0 |
| Bladder cancer | 2 | 1.01(0.77,1.33) | 0.588 | 0.0 | 1.00(0.47,2.16)c | 0.043 | 75.6 | 1.03(0.85,1.24) | 0.491 | 0.0 |
| Skin cancer | 2 | 1.08(0.91,1.27) | 0.414 | 0.0 | 1.02(0.86,1.23) | 0.483 | 0.0 | 1.04(0.93,1.16) | 0.902 | 0.0 |
| Nasopharyneal cancer | 2 | 1.55(0.75,3.24)c | 0.017 | 82.4 | 1.39(0.69,2.79)c | 0.042 | 75.8 | 1.33(0.80,2.19)c | 0.008 | 85.6 |
| Melanoma | 2 | 0.79(0.64,0.97) | 0.765 | 0.0 | 0.96(0.77,1.21) | 0.790 | 0.0 | 0.90(0.78,1.02) | 0.725 | 0.0 |
| Lung cancer | 2 | 0.82(0.65,1.04) | 0.852 | 0.0 | 1.07(0.82,1.40) | 0.906 | 0.0 | 0.94(0.81,1.10) | 0.984 | 0.0 |
| Other cancers | 7 | 1.08(0.96,1.22) | 0.373 | 7.3 | 1.15(0.99,1.32) | 0.747 | 0.0 | 1.08(1.00,1.17) | 0.528 | 0.0 |
| Ethnicity | ||||||||||
| Asian | 25 | 0.97(0.88,1.06)c | 0.004 | 48.3 | 1.01(0.89, 1.15)c | 0.003 | 49.3 | 0.99(0.93,1.05)c | 0.030 | 37.8 |
| Caucasian | 13 | 1.03(0.95, 1.12) | 0.120 | 32.8 | 1.00(0.92, 1.09) | 0.277 | 16.5 | 1.01(0.96,1.06) | 0.277 | 16.6 |
| African | 3 | 1.72(1.24,2.38) | 0.288 | 19.6 | 1.23(0.78,1.95)c | 0.039 | 69.1 | 1.25(0.90,1.74)c | 0.022 | 73.9 |
| Mixed | 3 | 1.10(0.82, 1.48) | 0.607 | 0.0 | 1.28(0.99, 1.65) | 0.803 | 0.0 | 1.15(0.97,1.37) | 0.610 | 0.0 |
Number of comparisons.
P value of Q-test for heterogeneity test.
Random-effect model was applied when P value for heterogeneity < 0.05; otherwise, fixed-effect model was applied.
Statistically significant results were in bold.
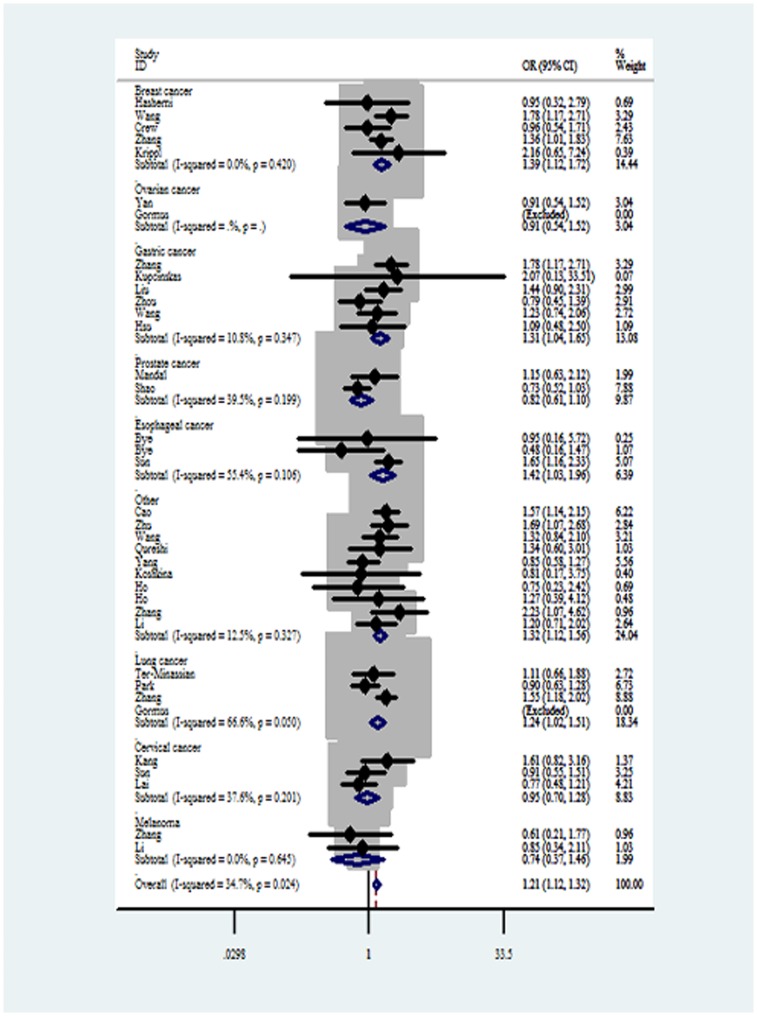
For Fas -1377G>A polymorphism, significantly increased cancer risks were observed in AA vs GG (Figure 2) and AA vs GA+GG comparison models in the overall analysis. In the subgroup analysis by cancer type, a significantly increased risk was observed in breast cancer for all comparison models. Meanwhile, increased risks were found for the comparison of AA vs GG and AA vs GA+GG in gastric cancer and esophageal cancer. In addition, a borderline decreased cancer risk was found in melanoma for GA vs GG and AA+GA vs GG comparison models (all data shown in Table 3).
Figure 2. Forest plots of effect estimates for Fas -1377G>A polymorphism (AA vs GG).
For each of the studies, the estimation of OR and its 95% CI is plotted with a box and a horizontal line. Filled diamond pooled OR and its 95% CI.
Table 3. Stratified analyses of the Fas -1377G>A (rs2234767) polymorphism and cancer.
| Variables | na | AA vs GG | GA vs GG | AA+GA vs GG | AA vs GA+GG | A vs G | ||||||||||
| OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | OR(95%CI) | Pb | I2 | OR(95%CI) | Pb | I2 | ||
| Total | 37 | 1.19(1.06, 1.34) c | 0.024 | 34.7 | 1.00(0.94,1.06)c | 0.033 | 32.2 | 1.03(0.97,1.10)c | 0.012 | 37.8 | 1.21(1.09, 1.34) c | 0.048 | 30.3 | 1.06(1.00,1.11) c | 0.002 | 44.8 |
| Cancer type | ||||||||||||||||
| Gastric cancer | 6 | 1.31(1.05, 1.65) | 0.934 | 0.0 | 0.99(0.85,1.14) | 0.810 | 0.0 | 1.04(0.91,1.20) | 0.659 | 0.0 | 1.32(1.07,1.64) | 0.328 | 13.6 | 1.09(0.99,1.21) | 0.372 | 7.0 |
| Breast cancer | 5 | 1.39(1.12,1.72) | 0.420 | 0.0 | 1.15(1.02,1.30) | 0.246 | 26.3 | 1.18(1.06,1.32) | 0.253 | 25.3 | 1.28(1.05,1.56) | 0.236 | 27.8 | 1.15(1.06,1.26) | 0.186 | 35.3 |
| Lung cancer | 4 | 1.18(0.82,1.70)c | 0.050 | 66.6 | 0.97(0.87,1.08) | 0.743 | 0.0 | 1.01(0.91,1.12) | 0.687 | 0.0 | 1.23(0.86,1.74)c | 0.044 | 68.0 | 1.06(0.97,1.14) | 0.279 | 21.8 |
| Esophageal cancer | 3 | 1.42(1.03,1.96) | 0.106 | 55.4 | 0.96(0.66,1.37)c | 0.031 | 71.2 | 1.00(0.72,1.39)c | 0.043 | 68.1 | 1.58(1.16,2.13) | 0.089 | 58.7 | 1.05(0.76,1.45)c | 0.022 | 73.8 |
| Cervical cancer | 3 | 0.95(0.70, 1.28) | 0.201 | 37.6 | 0.85(0.70,1.04) | 0.149 | 47.4 | 0.88(0.73,1.06) | 0.165 | 44.6 | 1.08(0.81,1.42) | 0.215 | 35.0 | 0.95(0.83,1.09) | 0.195 | 38.8 |
| Prostate cancer | 2 | 0.82(0.61,1.10) | 0.199 | 39.5 | 0.91(0.53,1.54)c | 0.042 | 75.9 | 0.90(0.54,1.50)c | 0.042 | 75.8 | 0.91(0.70,1.19) | 0.698 | 0.0 | 0.88(0.77,1.01) | 0.099 | 63.2 |
| Ovarian cancer | 2 | 0.91(0.54,1.52) | NA | NA | 1.04(0.77,1.40) | 0.286 | 12.1 | 1.02(0.77,1.36) | 0.270 | 17.9 | 0.91(0.56,1.50) | NA | NA | 1.00(0.80,1.24) | 0.308 | 3.8 |
| Melanoma | 2 | 0.74(0.37,1.46) | 0.645 | 0.0 | 0.79(0.62,1.00) | 0.614 | 0.0 | 0.78(0.62,0.98) | 0.748 | 0.0 | 0.77(0.39,1.52) | 0.617 | 0.0 | 0.80(0.65,0.98) | 0.916 | 0.0 |
| Other cancers | 10 | 1.32(1.12,1.56) | 0.327 | 12.5 | 1.07(0.97,1.17) | 0.437 | 0.0 | 1.10(1.01,1.21) | 0.364 | 8.5 | 1.28(1.10,1.49) | 0.351 | 10.0 | 1.12(1.04,1.20) | 0.244 | 21.7 |
Number of comparisons.
P value of Q-test for heterogeneity test.
Random-effect model was applied when P value for heterogeneity <0.05; otherwise, fixed-effect model was applied.
Statistically significant results were in bold.
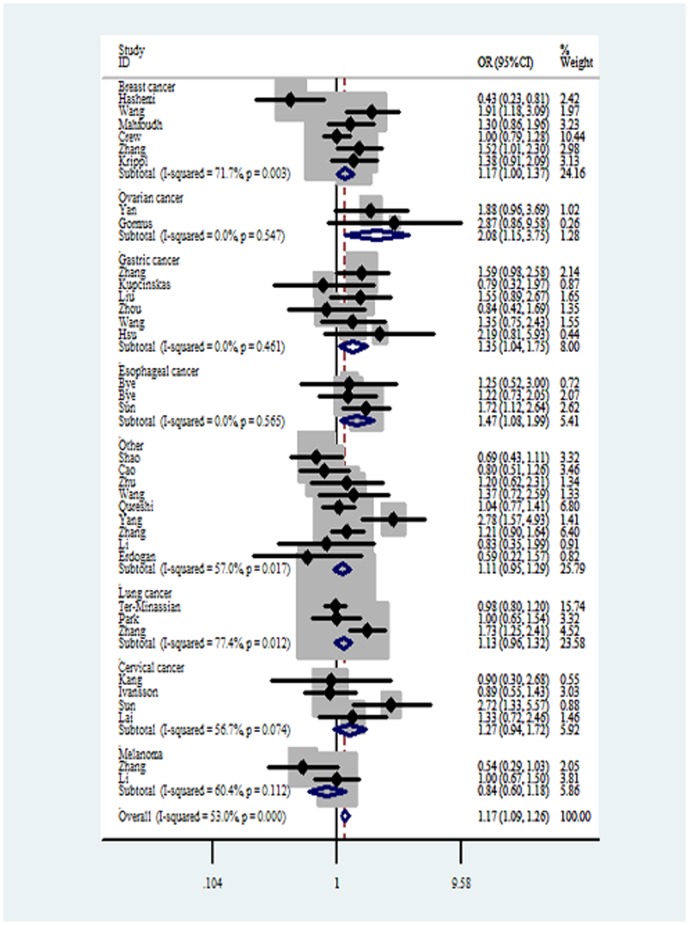
For FasL -844C>T polymorphism, significantly increased cancer risks were observed in CC vs TT (Figure 3), CC+CT vs TT and CC vs CT+TT in the overall analysis. When the analysis was stratified by genotyping method, an increased cancer risk was observed in studies carried out by PCR-RFLP (shown in Table 4).
Figure 3. Forest plots of effect estimates for FasL-844C>T polymorphism (CC vs TT).
For each of the studies, the estimate of OR and its 95% CI is plotted with a box and a horizontal line. Filled diamond pooled OR and its 95% CI.
Table 4. Stratified analyses of the FasL-844C>T (rs763110) polymorphism and cancer.
| Variables | na | CC vs TT | CT vs TT | CC+CT vs TT | CC vs CT+TT | C vs T | ||||||||||
| OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | OR(95%CI) | P b | I2 | ||
| Total | 35 | 1.19(1.06, 1.35) c | <0.0001 | 53.0 | 1.02(0.95,1.09) | 0.135 | 21.2 | 1.09(1.00,1.20) c | 0.046 | 30.6 | 1.20(1.08, 1.34) c | <0.0001 | 81.3 | 1.13(1.05,1.22) c | <0.0001 | 78.2 |
| Genotype | ||||||||||||||||
| PCR-RFLP | 24 | 1.28(1.09,1.51) c | 0.001 | 53.4 | 0.97(0.88,1.08) | 0.113 | 26.8 | 1.14(1.03,1.25) | 0.063 | 32.7 | 1.30(1.13,1.49) c | <0.0001 | 83.3 | 1.19(1.08,1.31) c | <0.0001 | 79.6 |
| TaqMan | 8 | 1.04(0.92,1.18) | 0.758 | 0.0 | 1.10(0.98,1.23) | 0.842 | 0.0 | 1.07(0.97,1.19) | 0.793 | 0.0 | 0.97(0.89,1.06) | 0.839 | 0.0 | 1.01(0.95,1.07) | 0.761 | 0.0 |
PCR-RFLP: restriction fragment length polymorphism.
Number of comparisons.
P value of Q-test for heterogeneity test.
Random-effect model was applied when P value for heterogeneity <0.05; otherwise, fixed-effect model was applied.
Statistically significant results were in bold.
Overall effects for alleles
Allele comparisons were also conducted in the meta-analysis. However, no significant associations were found in Fas -670A>G polymorphism and cancer risks (shown in Table 2).
There was borderline association between Fas -1377G>A polymorphism and cancer risks for A allele vs G allele in the overall analysis. In the subgroup analysis by cancer type, opposite results were shown between breast cancer and melanoma (shown in Table 3).
For FasL -844C>T polymorphism, in the subgroup analysis of genotyping method, an increased cancer risk was found in the studies carried out by PCR-RFLP (shown in Table 4).
The Fas and FasL mRNA expression by genotypes and population
The Fas and FasL mRNA expression levels were stratified by genotype (shown in Table 5) and population (shown in Table 6) groups. In the genotype subgroup analysis, significant associations between mRNA expression levels and Fas -670A>G were observed in all populations (GA: P = 0.043), especially in Asian population (GG: P = 0.0003; dominant: P = 0.003; recessive: P = 0.001). Meanwhile, significant differences between mRNA expression levels and FasL -844C>T were observed in Asian population (recessive: P = 0.001). In the population-subgroup analysis, decreased expression of Fas was found in YRI (Yoruba in Ibadan) population than in the CEU population (P = 0.002).
Table 5. Fas and FasL mRNA expression by the genotypes of SNPs, using data from the HapMap1.
| Fas -670A>G | FasL -844C>T | ||||||||
| Population | Genotypes | No. | Mean ± SD | P 2 | Ethnicity | Genotypes | No. | Mean ± SD | P 2 |
| CEU3 | AA | 23 | 8.79±0.36 | CEU3 | CC | 76 | 5.94±0.07 | ||
| GA | 46 | 8.87±0.28 | 0.321 | CT | 5 | 5.89±0.07 | 0.137 | ||
| GG | 12 | 8.74±0.36 | 0.687 | TT | 0 | — | — | ||
| Dominant | 58 | 8.84±0.30 | 0.511 | Dominant | 5 | 5.89±0.07 | 0.137 | ||
| Recessive | 69 | 8.84±0.31 | 0.292 | Recessive | 81 | — | — | ||
| YRI3 | AA | 6 | 8.58±0.33 | YRI3 | CC | 0 | — | — | |
| GA | 25 | 8.70±0.31 | 0.402 | CT | 28 | 5.94±0.06 | — | ||
| GG | 53 | 8.67±0.30 | 0.450 | TT | 53 | 5.95±0.06 | — | ||
| Dominant | 78 | 8.58±0.33 | 0.410 | Dominant | 81 | 5.95±0.06 | — | ||
| Recessive | 31 | 8.67±0.31 | 0.987 | Recessive | 28 | 5.94±0.06 | 0.493 | ||
| Asian3 | AA | 28 | 8.65±0.29 | Asian3 | CC | 0 | — | — | |
| GA | 36 | 8.78±0.26 | 0.059 | CT | 50 | 5.96±0.06 | — | ||
| GG | 21 | 8.98±0.30 | 0.0003 | TT | 33 | 5.91±0.06 | — | ||
| Dominant | 57 | 8.85±0.29 | 0.003 | Dominant | 83 | 5.94±0.06 | — | ||
| Recessive | 64 | 8.72±0.28 | 0.001 | Recessive | 50 | 5.96±0.06 | 0.001 | ||
| All3 | AA | 57 | 8.70±0.33 | All3 | CC | 76 | 5.94±0.07 | ||
| GA | 107 | 8.80±0.28 | 0.043 | CT | 83 | 5.95±0.06 | 0.163 | ||
| GG | 86 | 8.76±0.33 | 0.297 | TT | 86 | 5.94±0.06 | 0.913 | ||
| Dominant | 193 | 8.78±0.30 | 0.081 | Dominant | 169 | 5.95±0.06 | 0.390 | ||
| Recessive | 164 | 8.76±0.30 | 0.871 | Recessive | 159 | 5.95±0.06 | <0.0001 | ||
CEU: 90 Utah residents with ancestry from northern and western Europe; YRI: 90 Yoruba in Ibadan, Nigeria; Asian: 45 unrelated Han Chinese in Beijing and 45 unrelated Japanese in Tokyo.
Genotyping data and mRNA expression levels for Fas and FasL by genotypes were obtained from the HapMap phase II release 23 data from EBV-transformed lymphoblastoid cell lines from 270 individuals.
Two-side Student's t test within the stratum was used.
There were missing data for unavailable genotyping data.
Statistically significant results were in bold.
Table 6. Fas and FasL mRNA expression by the ethnicity, using data from the HapMap1.
| Fas -670A>G | FasL -844C>T | ||||||
| Ethnicity | No. | Mean ± SD | P 2 | Ethnicity | No. | Mean ± SD | P 2 |
| CEU3 | 81 | 8.83±0.31 | CEU3 | 81 | 5.94±0.07 | ||
| YRI3 | 84 | 8.67±0.30 | 0.002 | YRI3 | 81 | 5.95±0.06 | 0.120 |
| Asian3 | 85 | 8.79±0.30 | 0.391 | Asian3 | 83 | 5.94±0.06 | 0.398 |
CEU: 90 Utah residents with ancestry from northern and western Europe; YRI: 90 Yoruba in Ibadan, Nigeria; Asian: 45 unrelated Han Chinese in Beijing and 45 unrelated Japanese in Tokyo.
Genotyping data and mRNA expression levels for Fas and FasL by genotypes were obtained from the HapMap phase II release 23 data from EBV-transformed lymphoblastoid cell lines from 270 individuals.
Two-side Student's t test within the stratum was used.
There were missing data for unavailable genotyping data.
Statistically significant results were in bold.
Test of heterogeneity
There was significant heterogeneity across the studies focused on these three polymorphisms as evaluated by Q-test. Then, we evaluated the heterogeneity for dominant model comparison by subgroups (cancer type, ethnicity, source of controls and genotyping method). As a result, ethnicity (χ2 = 13.44, degree of freedom = 3, Ph = 0.004) and cancer type (χ2 = 22.26, degree of freedom = 11, Ph = 0.022), but not source of controls (χ2 = 1.49, degree of freedom = 1, Ph = 0.222) or genotyping method (χ2 = 1.48, degree of freedom = 4, Ph = 0.830) contributed to substantial heterogeneity of the Fas -670A>G polymorphism. For the Fas -1377G>A polymorphism, the test revealed cancer type (χ2 = 22.60, degree of freedom = 8, Ph = 0.004), but not ethnicity (χ2 = 4.81, degree of freedom = 3, Ph = 0.187), source of controls (χ2 = 0.42, degree of freedom = 1, Ph = 0.518), or genotyping method (χ2 = 0.51, degree of freedom = 3, Ph = 0.917) contributed to substantial heterogeneity. For the FasL -844C>T polymorphism, genotyping method (χ2 = 9.21, degree of freedom = 3, Ph = 0.027), but not cancer type (χ2 = 4.33, degree of freedom = 7, Ph = 0.741), ethnicity (χ2 = 5.64, degree of freedom = 3, Ph = 0.131), or source of controls (χ2 = 0.08, degree of freedom = 1, Ph = 0.777) contributed to substantial heterogeneity.
Sensitivity analyses
To assess the stability of the results and the source of the heterogeneity, sensitivity analysis was performed by sequential removal of each individual eligible study. For Fas -670A>G and FasL -844C>T polymorphisms, statistically similar results were observed after sequential removal of individual study in dominant and homozygote model, respectively, and the summary ORs in the other genetic models were not materially altered, suggesting that the results were stable. For the Fas -1377G>A polymorphism, sensitivity analysis indicated that study by Shao et al [38] was responsible for heterogeneity. The heterogeneity was decreased when this study was removed (AA+GA vs GG: Ph = 0.075, I2 = 26.5). Although the genotype distribution in 11 studies (listed in Table 1) didn't follow HWE, the corresponding summary ORs were not materially altered with or without including these studies for the three polymorphisms. In addition, no other single study altered the pooled ORs by sensitivity analysis.
Publication bias
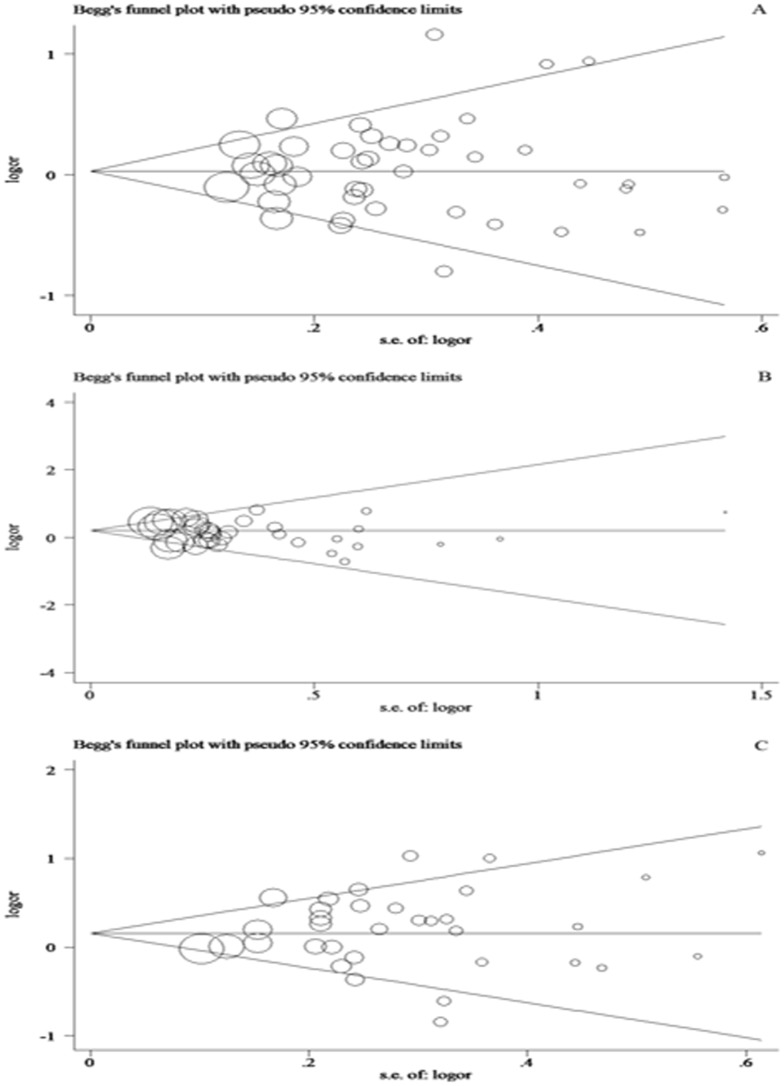
To assess the publication bias, Begg's funnel plot and Egger's test were performed and the shapes of funnel plots didn't show any obvious asymmetry in all genetic models of the three polymorphisms (Figure 4A–C). Therefore, to provide statistical evidence of funnel plot symmetry, Egger's test was performed for each of these polymorphisms and the results confirmed the absence of publication bias (P>0.05).
Figure 4. Begg's funnel plot of Egger's test for publication bias for three polymorphisms.
Each circle represents as an independent study for the indicated association. Log[OR], natural logarithm of OR. Horizontal lines mean effect size. A: Begg's funnel plot of publication bias test for Fas -670A>G polymorphism. B: Begg's funnel plot of publication bias test for Fas -1377G>A polymorphism. C: Begg's funnel plot of publication bias test for FasL -844C>T polymorphism.
Discussion
Fas, a potent member of the death receptor family, plays a crucial role in apoptotic signaling in many cell types [40]. Meanwhile, interactions between Fas and its receptor FasL trigger the death signal cascade, and subsequently induce apoptotic cell death [41]. Previous studies have indicated that down-regulation of Fas expression and/or up-regulation of FasL expression could be detected in many types of human tumors [42], [43]. The reason may be that down-regulation of Fas could protect tumor cells from elimination by anti-tumor immune responses, whereas up-regulation of FasL could increase the ability of tumor cells to counterattack the immune system by inducing apoptosis [44], [45], [46]. Therefore, it is believed that Fas and FasL play a crucial role in carcinogenesis. Given the important roles of Fas and FasL in carcinogenesis process, it is biologically plausible that Fas and FasL polymorphisms that possess the potential to influence the expression of Fas and/or FasL may be associated with cancer risk. Therefore, associations between the Fas -670A>G, Fas -1377G>A and FasL -844C>T polymorphisms and cancer risk were determined in this meta-analysis.
In this meta-analysis, 52 published studies were enrolled to determine the association between the three potentially functional polymorphisms within the Fas and FasL and cancer risk. This study revealed that the Fas -1377G>A and FasL -844C>T, but not the Fas -670A>G polymorphisms were associated with significantly increased overall cancer risk. Previous studies have identified that the -1377A allele had markedly reduced ability to bind transcription factor stimulatory protein 1 as compared with the -1377G allele, whereas the -670A and G alleles had similar ability to bind transcription factor signal transducers and activators of transcription 1 (STAT1)[47]. As the Fas -1377A allele reduced the ability to bind transcription factor stimulatory protein 1 that is a crucial transcriptional activator, the expression of Fas was decreased in carriers of the Fas -1377AA genotype as expected, but the Fas -670G allele didn't influence the expression of Fas [47], [48]. Therefore, it is reasonable that the Fas -1377A allele increased the overall cancer risk, and that the Fas -670G allele had no marked effect on overall cancer risk, which was consistent with our results. For the FasL -844T>C polymorphism, which is located in a binding motif for transcription factor CAAT/enhancer binding protein β, could influence the promoter activity of the FasL gene [49]. Additionally, it has been proposed that compared with the -844T allele, -844C allele strongly increased the expression of FasL on T cells and was associated with an enhanced rate of activation-induced cell death of T cells, which may lead to less powerful immune surveillance and increase the susceptibility to cancer [6].
The Fas -670GG genotype was associated with decreased risk of prostate cancer and melanoma according to the cancer type subgroup analysis. It was suggested that Fas -670A>G polymorphism might have the same effect on these two cancers. However, these results were based on 44 studies, which could affect the results owing to small amount of studies. Therefore, to draw a more precise conclusion, more related studies are needed.
For the Fas -1377G>A polymorphism, this study revealed that those who carried the -1377AA genotype had an increased risk for breast cancer, gastric cancer and esophageal cancer, while the melanoma risk was decreased. As described above, the different risk factors could contribute to the discrepancies. Also other unidentified causal genes would influence the effect of this polymorphism on different cancers.
For the FasL -844C>T polymorphism, the -844CC associated with increased cancer risk was observed in gastric cancer, esophageal cancer, and ovarian cancer among the previous studies, indicating that this polymorphism had similar effect on these three cancers. Although these cancers had different mechanisms of carcinogenesis, small amount of studies, publication bias, and other unidentified causal genes would be the result of the discrepancies, which contributed to the similar association between the FasL -844C>T polymorphism and three cancers.
In the subgroup analysis by ethnicity, an increased cancer risk in carriers of the Fas -670GG genotype was found in African, while the result of mRNA expression showed that GG genotype expressed higher levels of Fas in Asian populations. Meanwhile, the previous studies showed increased cancer risk in carriers of the Fas -1377AA and FasL -844CC genotype were found in Asian subjects, which was evidenced in mRNA expression by genotypes in Asian populations. However, this association was not proved in other ethnicities. The discrepancies in racial backgrounds and environment they lived in would lead to the differences. In addition, these polymorphisms might be masked by the presence of other unidentified causal genes involved in carcinogenesis. Due to the small size of population for the ethnicities, well-designed, large randomized case-control studies should be performed.
The pooled results of this study may be affected by polymorphism genotyping methods applied in the enrolled studies. Previous studies revealed that the pooled results of the Fas -670A>G polymorphism were not affected by the studies with genotyping methods of both PCR-RFLP and TaqMan. While Fas -1377AA genotype carriers increased cancer risk in the studies using PCR-RFLP but not TaqMan, and similar result was found in the FasL -844CC genotype carrier. The discrepancy across the studies applied different polymorphism genotyping methods may result from the different sensitivity and accuracy of genotyping methods. Meanwhile, the quality control is crucial to cause discrepancy as well. In general, studies [12], [17], [50] selected 10% repeated, random sample of subjects to test twice by standard genotyping method or different investigator, which was used to confirm the accuracy of results, while Mandal et al [51] and Ter-Minassian et al [22] tested 5% repeated samples. As a result, the consistency rate of quality control was 100% in almost all studies. However, the study by Crew et al [19] showed that the consistency rate was 100% for Fas -1377G>A, 94% for Fas -670G>A and 96% for FasL -844C>T. Therefore, the results of further studies should be confirmed by a standardized genotyping method. In addition, the limited amount of studies would also contribute to the discrepancy.
Heterogeneity is an important factor which can interpret the results of the meta-analysis. Therefore, we stratified the studies by cancer type, ethnicity, source of controls and genotyping method, respectively. The results showed that the main heterogeneity existed for cancer type and ethnicity. The reason might be that different cancers have different mechanisms of carcinogenesis. Virus infections, hormone levels, smoking, drinking, family history all could contribute to the different cancers. Meanwhile, different genetic backgrounds and different environmental factors among different ethnicities were the main factor of heterogeneity as well. Geographic differences, exposure of the Sun, eating habits, and environmental pollutes could exist in different ethnicities, which contributed to the heterogeneity.
Some limitations of the meta-analysis should be addressed. First, only studies in English were enrolled in this meta-analysis, which might miss some studies in other languages consistent with inclusion criteria. Second, some eligible studies included in the meta-analysis were hospital-based controls, which could generate the selection bias. Third, only a limited amount of studies was included, which might limit the strength of the associations. Finally, some suspected factors such as drinking, smoking, age, sex, and living habits were not considered in the meta-analysis. Regardless of such limitations, this meta-analysis still had some strengths. We investigated heterogeneity that may result from ethnicity of subjects, the types of cancer, the source of control subjects, and various genotyping methods. In addition, we analyzed the relationship between the mRNA expressions and genotypes, which partly supported the results of this meta-analysis.
In summary, this meta-analysis indicates that the Fas-1377G>A and FasL -844T/C polymorphisms are associated with increased cancer risk, but that no significant association is observed for the Fas -670A>G polymorphism and cancer risk. A definite conclusion should be made in the future through well-designed, unbiased, powered, population-based case–control association studies.
Supporting Information
PRISMA Checklist.
(DOC)
Funding Statement
This work was supported by Program of Healthy Talents' Cultivation for Nanjing City, and Social Development Technology Projects of Nanjing City, China (QYK11175). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. (2000) Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343: 78–85. [DOI] [PubMed] [Google Scholar]
- 2. Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, et al. (1991) The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell 66: 233–243. [DOI] [PubMed] [Google Scholar]
- 3. Oehm A, Behrmann I, Falk W, Pawlita M, Maier G, et al. (1992) Purification and molecular cloning of the APO-1 cell surface antigen, a member of the tumor necrosis factor/nerve growth factor receptor superfamily. Sequence identity with the Fas antigen. J Biol Chem 267: 10709–10715. [PubMed] [Google Scholar]
- 4. Kang S, Dong SM, Seo SS, Kim JW, Park SY (2008) FAS -1377 G/A polymorphism and the risk of lymph node metastasis in cervical cancer. Cancer Genet Cytogenet 180: 1–5. [DOI] [PubMed] [Google Scholar]
- 5. Zoodsma M, Nolte IM, Schipper M, Oosterom E, van der Steege G, et al. (2005) Interleukin-10 and Fas polymorphisms and susceptibility for (pre)neoplastic cervical disease. Int J Gynecol Cancer 15 Suppl 3282–290. [DOI] [PubMed] [Google Scholar]
- 6. Sun T, Zhou Y, Li H, Han X, Shi Y, et al. (2005) FASL -844C polymorphism is associated with increased activation-induced T cell death and risk of cervical cancer. J Exp Med 202: 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lai HC, Lin WY, Lin YW, Chang CC, Yu MH, et al. (2005) Genetic polymorphisms of FAS and FASL (CD95/CD95L) genes in cervical carcinogenesis: An analysis of haplotype and gene-gene interaction. Gynecol Oncol 99: 113–118. [DOI] [PubMed] [Google Scholar]
- 8. Dybikowska A, Sliwinski W, Emerich J, Podhajska AJ (2004) Evaluation of Fas gene promoter polymorphism in cervical cancer patients. Int J Mol Med 14: 475–478. [PubMed] [Google Scholar]
- 9. Lai HC, Sytwu HK, Sun CA, Yu MH, Yu CP, et al. (2003) Single nucleotide polymorphism at Fas promoter is associated with cervical carcinogenesis. Int J Cancer 103: 221–225. [DOI] [PubMed] [Google Scholar]
- 10. Zhang W, Li C, Wang J, He C (2012) Functional polymorphisms in FAS/FASL system contribute to the risk of occurrence but not progression of gastric cardiac adenocarcinoma. Hepatogastroenterology 59: 141–146. [DOI] [PubMed] [Google Scholar]
- 11. Kupcinskas J, Wex T, Bornschein J, Selgrad M, Leja M, et al. (2011) Lack of association between gene polymorphisms of Angiotensin converting enzyme, Nod-like receptor 1, Toll-like receptor 4, FAS/FASL and the presence of Helicobacter pylori-induced premalignant gastric lesions and gastric cancer in Caucasians. BMC Med Genet 12: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu L, Wu C, Wang Y, Zhong R, Wang F, et al. (2011) Association of candidate genetic variations with gastric cardia adenocarcinoma in Chinese population: a multiple interaction analysis. Carcinogenesis 32: 336–342. [DOI] [PubMed] [Google Scholar]
- 13. Zhou RM, Wang N, Chen ZF, Duan YN, Sun DL, et al. (2010) Polymorphisms in promoter region of FAS and FASL gene and risk of cardia gastric adenocarcinoma. J Gastroenterol Hepatol 25: 555–561. [DOI] [PubMed] [Google Scholar]
- 14. Wang M, Wu D, Tan M, Gong W, Xue H, et al. (2009) FAS and FAS ligand polymorphisms in the promoter regions and risk of gastric cancer in Southern China. Biochem Genet 47: 559–568. [DOI] [PubMed] [Google Scholar]
- 15. Hsu PI, Lu PJ, Wang EM, Ger LP, Lo GH, et al. (2008) Polymorphisms of death pathway genes FAS and FASL and risk of premalignant gastric lesions. Anticancer Res 28: 97–103. [PubMed] [Google Scholar]
- 16. Hashemi M, Fazaeli A, Ghavami S, Eskandari-Nasab E, Arbabi F, et al. (2013) Functional polymorphisms of FAS and FASL gene and risk of breast cancer - pilot study of 134 cases. PLoS One 8: e53075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang W, Zheng Z, Yu W, Lin H, Cui B, et al. (2012) Polymorphisms of the FAS and FASL genes and risk of breast cancer. Oncol Lett 3: 625–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahfoudh W, Bouaouina N, Gabbouj S, Chouchane L (2012) FASL-844 T/C polymorphism: a biomarker of good prognosis of breast cancer in the Tunisian population. Hum Immunol 73: 932–938. [DOI] [PubMed] [Google Scholar]
- 19. Crew KD, Gammon MD, Terry MB, Zhang FF, Agrawal M, et al. (2007) Genetic polymorphisms in the apoptosis-associated genes FAS and FASL and breast cancer risk. Carcinogenesis 28: 2548–2551. [DOI] [PubMed] [Google Scholar]
- 20. Zhang B, Sun T, Xue L, Han X, Lu N, et al. (2007) Functional polymorphisms in FAS and FASL contribute to increased apoptosis of tumor infiltration lymphocytes and risk of breast cancer. Carcinogenesis 28: 1067–1073. [DOI] [PubMed] [Google Scholar]
- 21.Krippl P, Langsenlehner U, Renner W, Koppel H, Samonigg H (2004) Re: Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96: : 1478–1479; author reply 1479. [DOI] [PubMed] [Google Scholar]
- 22. Ter-Minassian M, Zhai R, Asomaning K, Su L, Zhou W, et al. (2008) Apoptosis gene polymorphisms, age, smoking and the risk of non-small cell lung cancer. Carcinogenesis 29: 2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gormus U, Ergen A, Yaylim-Eraltan I, Yilmaz H, Turna A, et al. (2007) Fas-1377 A/G polymorphism in lung cancer. In Vivo 21: 663–666. [PubMed] [Google Scholar]
- 24. Park SH, Choi JE, Kim EJ, Jang JS, Lee WK, et al. (2006) Polymorphisms in the FAS and FASL genes and risk of lung cancer in a Korean population. Lung Cancer 54: 303–308. [DOI] [PubMed] [Google Scholar]
- 25. Zhang X, Miao X, Sun T, Tan W, Qu S, et al. (2005) Functional polymorphisms in cell death pathway genes FAS and FASL contribute to risk of lung cancer. J Med Genet 42: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holm K, Melum E, Franke A, Karlsen TH (2010) SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics 11: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He J, Qiu LX, Wang MY, Hua RX, Zhang RX, et al. (2012) Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet 131: 1235–1244. [DOI] [PubMed] [Google Scholar]
- 28. The International HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 29. Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 31. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 32. Bye H, Prescott NJ, Matejcic M, Rose E, Lewis CM, et al. (2011) Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis 32: 1855–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ho T, Li G, Zhao C, Zheng R, Wei Q, et al. (2008) Fas single nucleotide polymorphisms and risk of thyroid and salivary gland carcinomas: a case-control analysis. Head Neck 30: 297–305. [DOI] [PubMed] [Google Scholar]
- 34. Ueda M, Terai Y, Kanda K, Kanemura M, Takehara M, et al. (2006) Fas gene promoter -670 polymorphism in gynecological cancer. Int J Gynecol Cancer 16 Suppl 1179–182. [DOI] [PubMed] [Google Scholar]
- 35. Kordi Tamandani DM, Sobti RC, Shekari M (2008) Association of Fas-670 gene polymorphism with risk of cervical cancer in North Indian population. Clin Exp Obstet Gynecol 35: 183–186. [PubMed] [Google Scholar]
- 36. Zhang Z, Wang LE, Sturgis EM, El-Naggar AK, Hong WK, et al. (2006) Polymorphisms of FAS and FAS ligand genes involved in the death pathway and risk and progression of squamous cell carcinoma of the head and neck. Clin Cancer Res 12: 5596–5602. [DOI] [PubMed] [Google Scholar]
- 37. Bel Hadj Jrad B, Mahfouth W, Bouaouina N, Gabbouj S, Ahmed SB, et al. (2006) A polymorphism in FAS gene promoter associated with increased risk of nasopharyngeal carcinoma and correlated with anti-nuclear autoantibodies induction. Cancer Lett 233: 21–27. [DOI] [PubMed] [Google Scholar]
- 38. Shao P, Ding Q, Qin C, Wang M, Tang J, et al. (2011) Functional polymorphisms in cell death pathway genes FAS and FAS ligand and risk of prostate cancer in a Chinese population. Prostate 71: 1122–1130. [DOI] [PubMed] [Google Scholar]
- 39. Yang M, Sun T, Wang L, Yu D, Zhang X, et al. (2008) Functional variants in cell death pathway genes and risk of pancreatic cancer. Clin Cancer Res 14: 3230–3236. [DOI] [PubMed] [Google Scholar]
- 40. Andera L (2009) Signaling activated by the death receptors of the TNFR family. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 153: 173–180. [DOI] [PubMed] [Google Scholar]
- 41. Kim R, Emi M, Tanabe K, Uchida Y, Toge T (2004) The role of Fas ligand and transforming growth factor beta in tumor progression: molecular mechanisms of immune privilege via Fas-mediated apoptosis and potential targets for cancer therapy. Cancer 100: 2281–2291. [DOI] [PubMed] [Google Scholar]
- 42. Rabinowich H, Reichert TE, Kashii Y, Gastman BR, Bell MC, et al. (1998) Lymphocyte apoptosis induced by Fas ligand- expressing ovarian carcinoma cells. Implications for altered expression of T cell receptor in tumor-associated lymphocytes. J Clin Invest 101: 2579–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, et al. (2001) Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene 20: 7437–7446. [DOI] [PubMed] [Google Scholar]
- 44. Griffith TS, Brunner T, Fletcher SM, Green DR, Ferguson TA (1995) Fas ligand-induced apoptosis as a mechanism of immune privilege. Science 270: 1189–1192. [DOI] [PubMed] [Google Scholar]
- 45. Strand S, Hofmann WJ, Hug H, Muller M, Otto G, et al. (1996) Lymphocyte apoptosis induced by CD95 (APO-1/Fas) ligand-expressing tumor cells–a mechanism of immune evasion? Nat Med 2: 1361–1366. [DOI] [PubMed] [Google Scholar]
- 46. Reichmann E (2002) The biological role of the Fas/FasL system during tumor formation and progression. Semin Cancer Biol 12: 309–315. [DOI] [PubMed] [Google Scholar]
- 47. Sibley K, Rollinson S, Allan JM, Smith AG, Law GR, et al. (2003) Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res 63: 4327–4330. [PubMed] [Google Scholar]
- 48. Huang QR, Morris D, Manolios N (1997) Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol 34: 577–582. [DOI] [PubMed] [Google Scholar]
- 49. Wu J, Metz C, Xu X, Abe R, Gibson AW, et al. (2003) A novel polymorphic CAAT/enhancer-binding protein beta element in the FasL gene promoter alters Fas ligand expression: a candidate background gene in African American systemic lupus erythematosus patients. J Immunol 170: 132–138. [DOI] [PubMed] [Google Scholar]
- 50.Shao P, Ding Q, Qin C, Wang M, Tang J, et al. (2011) Functional polymorphisms in cell death pathway genes FAS and FAS ligand and risk of prostate cancer in a Chinese population. Prostate. [DOI] [PubMed]
- 51. Mandal RK, Mittal RD (2012) Are cell cycle and apoptosis genes associated with prostate cancer risk in North Indian population? Urol Oncol 30: 555–561. [DOI] [PubMed] [Google Scholar]
- 52. Zucchi F, da Silva ID, Ribalta JC, de Souza NC, Speck NM, et al. (2009) Fas/CD95 promoter polymorphism gene and its relationship with cervical carcinoma. Eur J Gynaecol Oncol 30: 142–144. [PubMed] [Google Scholar]
- 53. Ivansson EL, Gustavsson IM, Magnusson JJ, Steiner LL, Magnusson PK, et al. (2007) Variants of chemokine receptor 2 and interleukin 4 receptor, but not interleukin 10 or Fas ligand, increase risk of cervical cancer. Int J Cancer 121: 2451–2457. [DOI] [PubMed] [Google Scholar]
- 54. Ikehara SK, Ikehara Y, Matsuo K, Hirose K, Niwa T, et al. (2006) A polymorphism of C-to-T substitution at -31 IL1B is associated with the risk of advanced gastric adenocarcinoma in a Japanese population. J Hum Genet 51: 927–933. [DOI] [PubMed] [Google Scholar]
- 55. Wang LE, Cheng L, Spitz MR, Wei Q (2003) Fas A670G polymorphism, apoptotic capacity in lymphocyte cultures, and risk of lung cancer. Lung Cancer 42: 1–8. [DOI] [PubMed] [Google Scholar]
- 56. Jain M, Kumar S, Lal P, Tiwari A, Ghoshal UC, et al. (2007) Role of BCL2 (ala43thr), CCND1 (G870A) and FAS (A-670G) polymorphisms in modulating the risk of developing esophageal cancer. Cancer Detect Prev 31: 225–232. [DOI] [PubMed] [Google Scholar]
- 57. Sun T, Miao X, Zhang X, Tan W, Xiong P, et al. (2004) Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96: 1030–1036. [DOI] [PubMed] [Google Scholar]
- 58. Qureshi A, Nan H, Dyer M, Han J (2010) Polymorphisms of FAS and FAS ligand genes and risk of skin cancer. J Dermatol Sci 58: 78–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang H, Sun XF, Synnerstad I, Rosdahl I (2007) Importance of FAS-1377, FAS-670, and FASL-844 polymorphisms in tumor onset, progression, and pigment phenotypes of Swedish patients with melanoma: a case-control analysis. Cancer J 13: 233–237. [DOI] [PubMed] [Google Scholar]
- 60. Li C, Larson D, Zhang Z, Liu Z, Strom SS, et al. (2006) Polymorphisms of the FAS and FAS ligand genes associated with risk of cutaneous malignant melanoma. Pharmacogenet Genomics 16: 253–263. [DOI] [PubMed] [Google Scholar]
- 61. Nelson HH, Kelsey KT, Bronson MH, Mott LA, Karagas MR (2001) Fas/APO-1 promoter polymorphism is not associated with non-melanoma skin cancer. Cancer Epidemiol Biomarkers Prev 10: 809–810. [PubMed] [Google Scholar]
- 62. Li Y, Hao YL, Kang S, Zhou RM, Wang N, et al. (2013) Genetic polymorphisms in the Fas and FasL genes are associated with epithelial ovarian cancer risk and clinical outcomes. Gynecol Oncol 128: 584–589. [DOI] [PubMed] [Google Scholar]
- 63. Gormus U, Ergen A, Yilmaz H, Dalan B, Berkman S, et al. (2007) Fas-1377A/G and FasL-844 T/C gene polymorphisms and epithelial ovarian cancer. Anticancer Res 27: 991–994. [PubMed] [Google Scholar]
- 64. Lima L, Morais A, Lobo F, Calais-da-Silva FM, Calais-da-Silva FE, et al. (2008) Association between FAS polymorphism and prostate cancer development. Prostate Cancer Prostatic Dis 11: 94–98. [DOI] [PubMed] [Google Scholar]
- 65. Cao Y, Miao XP, Huang MY, Deng L, Lin DX, et al. (2010) Polymorphisms of death pathway genes FAS and FASL and risk of nasopharyngeal carcinoma. Mol Carcinog 49: 944–950. [DOI] [PubMed] [Google Scholar]
- 66. Zhu Q, Wang T, Ren J, Hu K, Liu W, et al. (2010) FAS-670A/G polymorphism: A biomarker for the metastasis of nasopharyngeal carcinoma in a Chinese population. Clin Chim Acta 411: 179–183. [DOI] [PubMed] [Google Scholar]
- 67. Gangwar R, Mittal RD (2010) Association of selected variants in genes involved in cell cycle and apoptosis with bladder cancer risk in North Indian population. DNA Cell Biol 29: 349–356. [DOI] [PubMed] [Google Scholar]
- 68. Li C, Wu W, Liu J, Qian L, Li A, et al. (2006) Functional polymorphisms in the promoter regions of the FAS and FAS ligand genes and risk of bladder cancer in south China: a case-control analysis. Pharmacogenet Genomics 16: 245–251. [DOI] [PubMed] [Google Scholar]
- 69. Zhu J, Qin C, Wang M, Yan F, Ju X, et al. (2010) Functional polymorphisms in cell death pathway genes and risk of renal cell carcinoma. Mol Carcinog 49: 810–817. [DOI] [PubMed] [Google Scholar]
- 70. Wang LH, Ting SC, Chen CH, Tsai CC, Lung O, et al. (2010) Polymorphisms in the apoptosis-associated genes FAS and FASL and risk of oral cancer and malignant potential of oral premalignant lesions in a Taiwanese population. J Oral Pathol Med 39: 155–161. [DOI] [PubMed] [Google Scholar]
- 71. Koshkina NV, Kleinerman ES, Li G, Zhao CC, Wei Q, et al. (2007) Exploratory analysis of Fas gene polymorphisms in pediatric osteosarcoma patients. J Pediatr Hematol Oncol 29: 815–821. [DOI] [PubMed] [Google Scholar]
- 72. Erdogan M, Karadeniz M, Berdeli A, Tamsel S, Ertan Y, et al. (2007) Fas/Fas ligand gene polymorphism in patients with papillary thyroid cancer in the Turkish population. J Endocrinol Invest 30: 411–416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOC)