Abstract
Development of high-throughput approaches to map the RNA interaction sites of individual RNA binding proteins (RBPs) transcriptome-wide is rapidly transforming our understanding of post-transcriptional gene regulatory mechanisms. Here we describe a ribonucleoprotein (RNP) footprinting approach we recently developed for identifying occupancy sites of both individual RBPs and multi-subunit RNP complexes. RNA:protein immunoprecipitation in tandem (RIPiT) yields highly specific RNA footprints of cellular RNPs isolated via two sequential purifications; the resulting RNA footprints can then be identified by high-throughput sequencing (Seq). RIPiT-Seq is broadly applicable to all RBPs regardless of their RNA binding mode and thus provides a means to map the RNA binding sites of RBPs with poor inherent ultraviolet (UV) crosslinkability. Further, among current high-throughput approaches, RIPiT has the unique capacity to differentiate binding sites of RNPs with overlapping protein composition. It is therefore particularly suited for studying dynamic RNP assemblages whose composition evolves as gene expression proceeds.
Keywords: RNA binding proteins, immunoprecipitation, RIP-Seq, RIPiT, formaldehyde crosslinking, high-throughput sequencing
1. Introduction
Nucleic acid-protein interactions govern all aspects of gene expression in every organism. Therefore, elucidating precisely when and where proteins bind to DNA and RNA is central to our understanding of the exquisite intricacies of how genetic information is decoded and regulated within cells. To execute RNA-mediated control, the human genome encodes >1000 RNA binding proteins (RBPs) as per current estimates [1–3]. A large fraction of these engage with RNA polymerase II transcripts (precursors to messenger RNAs; pre-mRNAs) to form ribonucleoprotein particles (RNPs) and exert control over post-transcriptional events such as pre-mRNA processing (splicing and polyadenylation) and intracellular localization, translation and decay of product mRNAs [1, 2, 4–6]. Importantly, most RBPs function within multi-protein complexes that also consist of non-RBPs. Further, whereas some RBPs are restricted to just a few transcripts, others act on tens of thousands of distinct species. The diverse features recognized by RBPs include common structural elements (e.g., the 7-methyl-G cap and polyA tail), short sequence motifs (e.g., exonic splicing enhancers and silencers; ESEs and ESSs), particular secondary structures (e.g., double-stranded regions; dsRNA) and modified nucleotides (e.g., inosine, 6-methyl-adenosine). To recognize these features, RBPs utilize a variety RNA recognition modes, with >50 different types of RNA-binding domains having been identified to date [4, 7]. Among RNPs, messenger RNPs (mRNPs) are particularly dynamic, shedding proteins and acquiring others as they move from one cellular compartment to another and/or are acted upon by numerous macromolecular machines (e.g., the spliceosome, nuclear pore complex, and ribosome). A major goal in understanding post-transcriptional control of gene expression is therefore to identify the RNA targets directly bound to individual RBPs and their partner proteins in cells, as well as the precise binding sites of these proteins on their target RNAs.
The post-genomic era has seen rapid advances in technologies to study nucleic acid-protein interactions on an unprecedented scale. The panoply of RBP targets within cells has recently been illuminated by approaches that generally combine enrichment of RBP-bound RNAs via immunoprecipitation (IP) with high-throughput transcript profiling methods. The first such transcriptome-wide approach, dubbed RIP-chip [8, 9], combined RNA IP (RIP) with microarray identification of precipitated RNAs. With the advent of next generation sequencing, RIP-Seq has also become a viable alternative [10–13].
A new dimension to the analysis of RBP-RNA target interactions in vivo came with the confluence of ultraviolet (UV) cross-linking and IP (CLIP) and next generation sequencing (CLIP-seq or HITS-CLIP; [14, 15]). By inducing covalent protein-RNA linkages in living cells, CLIP allows identification of the sites of direct contact between an RBP and its RNA target at single nucleotide resolution. Crosslinking efficiencies can be boosted through intracellular incorporation of photoreactive ribonucleosides (photoactivable ribonucleoside-enhanced CLIP; PAR-CLIP; [16]). In both methods, after UV-irradiation, cell lysis and partial RNase digestion, the RBP of interest and the RNA fragments crosslinked to it are immunoprecipitated (IPed) with a specific antibody. To reveal the identity of the crosslinked RNAs, the latter are converted into cDNA libraries for sequencing on massively parallel next generation DNA sequencing platforms.
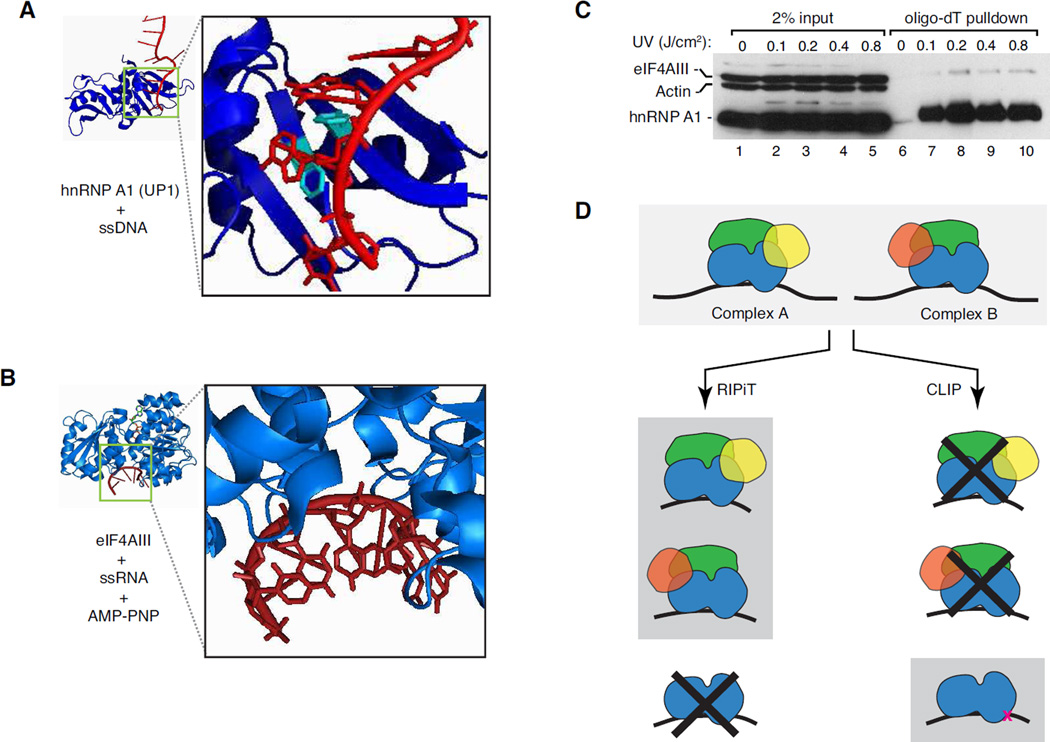
At present, CLIP-based approaches have been the most widely used to map transcriptome-wide RBP binding sites, having been now applied to several dozen RBPs [17]. Despite this resounding success, however, the varying degrees of UV-crosslinkability inherent to different RBP families limits CLIP’s applicability to all RBPs. Formation of RNA-protein crosslinks upon UV irradiation requires close juxtaposition of photo-reactive groups (i.e., nucleic acid bases and aromatic amino acid side chains) at the RNA-protein interface. Such intimate association between bases and aromatic amino acids underlies sequence-specific RNA recognition by many RBP families (e.g. hnRNP and SR proteins) [18, 19], and these proteins readily UV-crosslink to RNA (Figures 1A and 1C; [20–22]). However, other RBP families that primarily interact with other features (e.g., the sugar-phosphate backbone or double stranded RNA, dsRNA) often lack the appropriate spatial arrangements between RNA bases and aromatic amino acids required for efficient UV-crosslinking. This is exemplified by the crystal structure of the DEAD-box protein eIF4AIII bound to RNA. Binding of eIF4AIII is sequence independent – it interacts with RNA via the sugar-phosphate backbone (Figure 1B; [23, 24]). Consistently, when exposed to short-wave UV light we found that eIF4AIII crosslinked with much reduced efficiency to polyA-RNA than did hnRNPA1 (Figure 1C). Further, among proteins containing various classical RNA binding domains (RBDs), those with DEAD-box motifs or double-stranded RBDs (dsRBDs) are most under-represented in the fraction of the human proteome directly UV-crosslinked to mRNAs (See Figure 5B in [2]). Even in the case of readily UV-crosslinkable proteins, RNA sequence-driven differences can be observed between CLIP and PAR-CLIP approaches [2]. Thus, the biggest pitfall of the CLIP approach is the likely bias towards identification of sites that are most efficiently cross-linkable, which may not represent the complete RNA binding landscape for any single RBP. Such biases are likely to be even more pronounced in the case of proteins that inherently crosslink poorly to RNA.
Figure 1. UV-crosslinking bias of different RBPs dictates the suitability of different approaches for identifying binding sites.
A. hnRNPA1:ssDNA interface. Crystal structure of two-RRM-containing UP1 domain of hnRNP A1 (PDB ID: 2UP1; blue) in complex with a target containing its AGGG preferred recognition motif (in this case, within single-stranded DNA, ssDNA; red). Black box. Enlarged view of the DNA:protein interface. Aromatic residues (Phe-17 and Phe-59 from RRM1) that stack with the nucleobases are shown in cyan.
B. eIF4AIII:RNA interface. Crystal structure of eIF4AIII (PDB ID: 2J0S; blue) complexed with RNA (red) and AMP-PNP. Black box. Enlarged view of the RNA:protein interface. Note that the RNA bases are pointing away from the bound protein.
C. Comparison of UV-crosslinkability of RBPs to polyA+ RNA. Levels of proteins detected by western blots in input (lanes 1–5) or in oligo-dT pulldown fractions from cells irradiated with the dosage of short-wave UV light indicated above each lane (lanes 6–10). Oligo-dT pulldowns were performed as in [30].
D. RIPiT and CLIP yield different types of information. Top: Two similar yet compositionally distinct hypothetical multi-subunit RNPs. RBPs (blue), non-RBPs (green) and proteins unique to each complex are shown (complex A: yellow; complex B: red). Left: RIPiT can reveal the binding sites of an intact multi-subunit RNP, and can also distinguish between footprints of two compositionally similar complexes (schematics on gray background). However, RIPiT does not conclusively define direct RBP-RNA interactions (crossed-out schematic). Right: On the contrary, while CLIP reveals no information regarding the complexes an RNA-bound RBP is part of (crossed-out schematics), it can unveil the sites of direct contact between an RBP and RNA (bottom schematic).
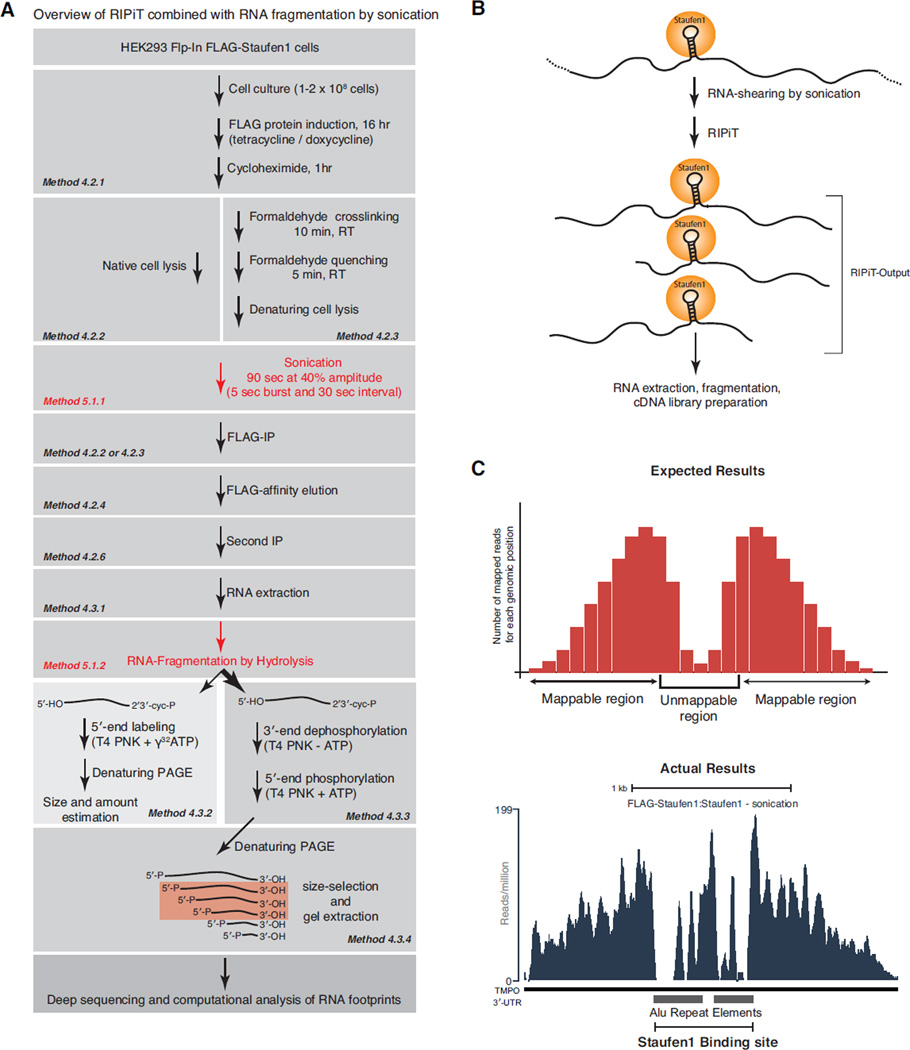
Figure 5. Schematic of the major steps for the RIPiT approach coupled to random target-RNA fragmentation by sonication.
A. Step-by-step description of the overall RIPiT methodology combined with fragmentation of target RNAs by ultrasound sonication.
B. Scheme of the outcome of ultrasound sonication followed by RIPiT of Staufen1. Sonication leads to random RNA cleavage and RIPiT enriches for RNA fragments bound to the RBP of interest. This results in large RNA fragments that are randomly distributed around the RBP binding site. After RIPiT, obtained RNAs are further fragmented for preparation of cDNA libraries suitable for high-throughput sequencing.
C. Upper panel: Histogram of expected results for the number of sequencing reads mapping at a paired Alu element Staufen1 binding site based on the RIPiT approach coupled to sonication. The x-axis corresponds to the genomic position while the y-axis corresponds to the cumulative number of reads mapping across a given genomic position. Notice that signal intensity is correlated to the distance from the binding site but strongly decreases at the binding site because of poor mappability of Alu elements in the human genome. Lower panel: Histogram of observed sequencing reads at a Staufen1 binding site consisting of two Alu elements in the opposite orientation located in the 3’UTR of the human TMPO transcript.
An alternate approach to UV-crosslinking is to map RBP (binding sites by nuclease protection of the sequences directly occupied by a single protein or multiprotein complex (RNA footprinting; Figure 1D). Various footprinting approaches have been used extensively to map RNA binding sites of proteins within homogenous RNA:protein complexes [25]. More recently, the ribosome profiling approach based on isolation and high-throughput sequencing of 80S ribosome-protected mRNA fragments has revealed the transcriptome-wide positions of translating ribosomes [26, 27]. In the case of ribosome profiling, the footprints can be isolated simply based on the size of the complex containing them (i.e., they co-sediment with 80S monosomes). However, in other instances it is desirable to isolate footprints of multi-protein complexes dependent on the exact protein composition of that complex. For example, the exon junction complex (EJC) is a multi-protein complex deposited upstream of exon-exon junctions by the spliceosome during the process of pre-mRNA splicing. The tetrameric EJC core, comprised of eIF4AIII, the Y14-Magoh heterodimer and MLN51, along with various peripheral proteins that assemble onto this core, participate in pre-mRNA splicing and mRNA export, localization, translation and degradation [28]. Very likely, EJC composition dynamically evolves throughout the post-transcriptional lifetime of an mRNP [29]. Thus, to understand the multitude of EJC functions, it was highly desirable to obtain footprints of the core complex and compare them to footprints of the core bound to different peripheral proteins.
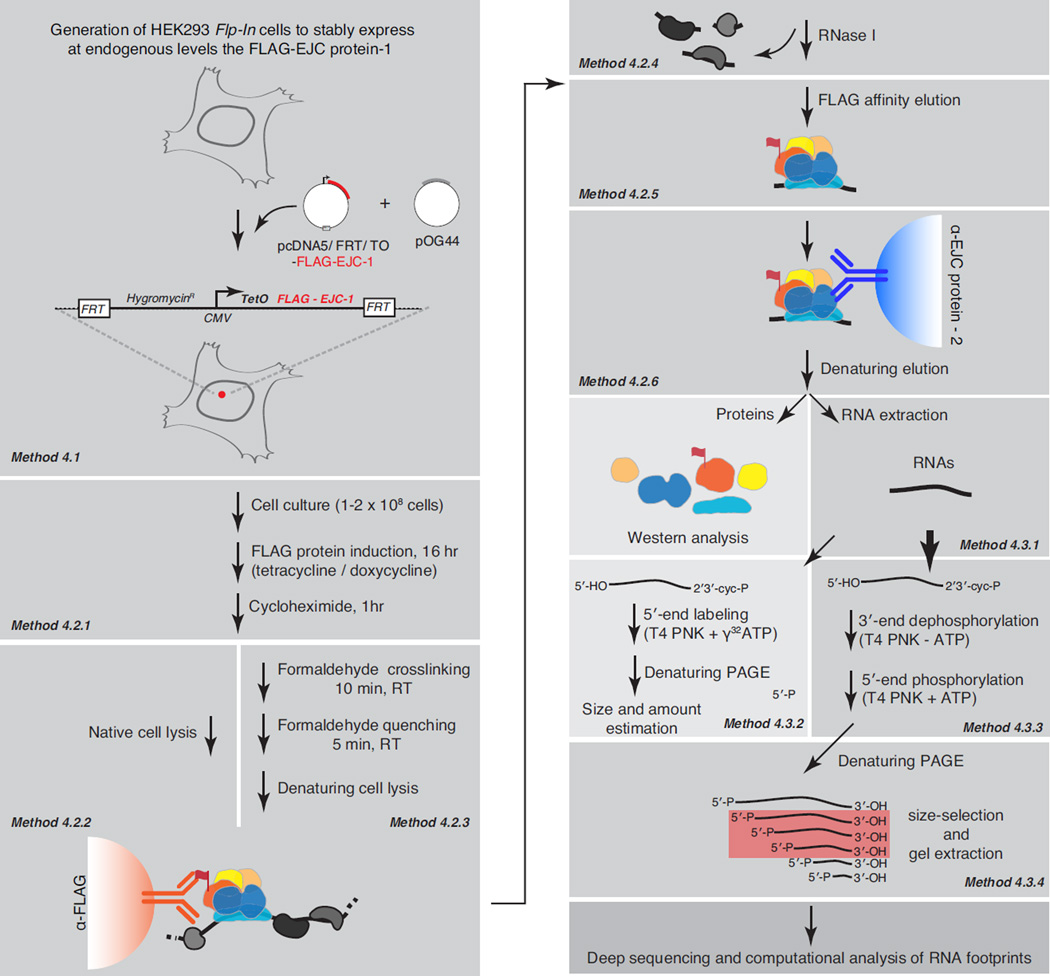
To reveal the in vivo binding landscape of the EJC, we recently developed a high-throughput RNP footprinting approach termed RNA:protein immunoprecipitation in tandem coupled to high-throughput sequencing (RIPiT-seq) (Figure 2; [30]). Subsequently, we have used RIPiT to characterize the binding sites of human Staufen1 [31], a dsRBP that also shows poor UV-crosslinking to its target RNAs [32]. RIPiT, an extension of RIP, involves two sequential IPs. It is therefore also analogous to the tandem affinity purification (TAP) approach widely employed to isolate macromolecular complexes at high purity [33]. The first IP within RIPiT can be performed in lysates either from untreated cells or from cells treated with a protein-protein crosslinking reagent prior to cell lysis to faithfully capture dynamic and unstable RNPs. Between the two IPs, while still bound to the solid support, RNPs enriched from the first IP are treated with a ribonuclease to digest away the unprotected RNA and generate RNA footprints for the complex of interest. These footprinted RNPs are gently eluted under native conditions and subsequently subjected to a second IP using an antibody recognizing a different component of the complex (Figure 2). When main interaction partners of an RBP are unknown, the second IP can be performed with an antibody recognizing a different site on the same protein as in the first IP. Proteins and RNA footprints from the resulting immunoprecipitates can be used for biochemical analyses using standard molecular biology methods (e.g., western blots for proteins or 5’-end labeling to assess footprint length and quantity), or for detailed compositional analyses via proteomic and high-throughput sequencing methods.
Figure 2. A schematic of the major steps involved in EJC RIPiT: FLAG-tag protein expression, RIPiT purification and footprint isolation.
Each major method presented in the text is shown as an individual grey rectangle with reference to the corresponding method section indicated in italics. Terminal steps in the procedure are shown on a lighter background, while downstream steps not discussed in detail are on a darker background. The EJC core proteins are depicted as colored shapes; RNA as a black wavy line; RNA-dependent interactors as grey shapes.
RIPiT combines the complementary strengths of both RIP and CLIP. Most prominently, like RIP, due to its independence from UV-crosslinking, RIPiT is broadly applicable to all RBPs irrespective of their RNA recognition modes. On the other hand, like CLIP, it allows identification of precise RBP binding sites within individual RNA targets. Additionally, sequential IPs within RIPiT offer multiple advantages over both RIP and CLIP. Two opportunities to remove nonspecific interactions lead to a superior signal-to-noise ratio of isolated footprints, even if the same protein is IPed during both steps. Even more importantly, by choosing to IP different and unique pairs of proteins that comprise a dynamic multi-protein complex, RIPiT provides a unique ability to distinguish footprints of RNPs with overlapping protein composition. It is therefore particularly suited to investigate how dynamic RNP assemblies evolve compositionally over time.
2. Materials and Reagents
2.1 Generation, propagation and induction of stable cell lines
Cell line
HEK293 Flp-In TREx cells (Life technologies, R780-07)
Transfection reagent
HEK293TransIT reagent (Mirus, MIR 2704)
Plasmids
pcDNA5/FRT/TO (Life technologies, V6520-20)
pOG44 (Life technologies, V6005-20)
Note: Any immortalized cell line can be converted into Flp-In host cells by sequential stable transfections of plasmids pFRT/lacZeo and pcDNA6-TR that are available as part of Flp-In T-Rex core kit (Life technologies, K6500-01).
Antibiotics
Blasticidin (Life technologies, A11139-02)
Hygromycin (Life technologies, 10687-010)
Zeocin (Life technologies, R250-01)
Tetracycline (1 mg/ml stock prepared fresh every month in 100% ethanol)
Cell growth and propagation reagents
Standard growth medium: Dulbecco’s modified eagle medium (DMEM; Life technologies, 11965-092) supplemented with 10% fetal bovine serum (FBS; Sigma, F2442-500ML) and 1% penicillin/streptomycin (Life technologies, 15140-122)
Flp-In host cell growth medium: Standard growth medium supplemented with 400 µg/ml Zeocin
Flp-In stable cell selection medium: Standard growth medium supplemented with 15 µg/ml Blasticidin and 100 µg/ml Hygromycin
Trypsin-EDTA (Life technologies, 25300-062)
Phosphate buffered saline (PBS)
2.2. Cell lysis and FLAG IP
Hypotonic Lysis Buffer (HLB): 20 mM Tris-HCl pH 7.5, 15 mM NaCl, 10 mM EDTA, 0.5% NP-40, 0.1% Triton X-100, 10 µg/ml Aprotinin (Sigma, A1153-10MG), 1 µg/ml Leupeptin (Sigma, L9783-5MG), 1 µM Pepstatin (Sigma, L4265-5MG), 1 mM PMSF (Sigma, P7626-5G), 100 µg/ml cycloheximide (Sigma, C1988-1G)
Denaturing lysis buffer (DLB): HLB supplemented with 0.1 % SDS and 0.1% sodium deoxycholate
Formaldehyde 37% stock solution (Fisher, F79-1)
Quenching buffer (QB): 2.5 M Glycine, 25 mM Tris-base
Branson Digital Sonifier-250 with microtip (Fisher, 15-338-125)
Anti-FLAG agarose (Sigma, A2220)
Isotonic wash buffer (IsoWB): 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% NP-40
Wash buffer 300 (WB300): 20 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.1% NP-40
Denaturing wash buffer (DWB): IsoWB supplemented with 0.1 % SDS and 0.1% sodium deoxycholate
2.3. RNase I and FLAG elution
RNase I (Life technologies, AM2294)
Thermomixer (Eppendorf, 5355 000.011)
FLAG peptide (Sigma, F3290) – prepare 5 mg/ml stock in Tris-buffered saline (TBS) and freeze aliquots at −20°C.
2.4. Second IP
2X IP2 buffer: 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 20 mM EDTA, 1 % NP-40, 0.2% Triton X-100, 20 µg/ml Aprotinin, 2 µg/ml Leupeptin, 2 µM Pepstatin, 2 mM PMSF, 200 µg/ml bovine serum albumin (BSA; NEB B9001S)
Protein-A Dynabeads (Life technologies, 10001D)
Protein-G Dynabeads (Life technologies, 10003D)
Clear sample buffer (CSB): 100 mM Tris-HCl pH 6.8, 4% SDS, 10 mM EDTA, 100 mM DTT
2.5. RNA extraction
Phenol:Chloroform:Iso-amyl alcohol, pH 4.5 (Life technologies, AM9720)
RNA precipitation stocks: 5 mg/ml Glycogen (Life technologies, AM9510), 3M Sodium acetate, pH 5.2, 2M MgCl2, 200 proof (100%) ethanol
2.6. Estimation of footprint size and amount
5′-end labeling: T4 Polynucleotide Kinase (NEB, M0201S), γ32P-ATP, 20mM DTT, 1mM ATP
2X formamide loading buffer (FLB): 95% Formamide (deionized; Life technologies, AM9342), 10 mM EDTA, 0.025% Xylene cyanol, 0.025% bromophenol blue; Alternative: Gel load buffer II (Life technologies, AM8547)
Gel electrophoresis equipment: 20 × 27 cm Glass plates, Vertical gel electrophoresis apparatus, Heat shield
-
Denaturing PAGE stocks:
-
-
Acrylamide mix: 20% acrylamide, 6M urea, 0.5X TBE
-
-
Dilution mix: 6M urea, 0.5X TBE
-
-
TEMED
-
-
10% APS – Dissolve 1 g in 10 ml sterile water. Store at 4°C.
-
-
Oligos and ladders: Low molecular weight ssDNA ladder (USB, 76410), 100bp ladder (NEB, N3231S)
Gel drying and imaging: 3M whatman filter paper, Saran wrap, Gel dryer, Phosphorimager screens, Typhoon scanner
2.7. RNA size-selection and gel extraction
SYBR-Gold (Life technologies, S-11494)
Blue-light LED transilluminator, ultrabright (New EnglandBio Group)
RNA elution buffer (REB): 0.3M Sodium acetate pH 5.2, 0.1 mM EDTA
Spin-X column (Corning, 8161)
2.8. RNA fragmentation via sonication and hydrolysis
RNA fragmentation reagents (Life technologies, AM8740): Contains 10X RNA fragmentation buffer, Stop solution
3. The RIPiT strategy
As summarized in Section 1, RIPiT involves two sequential IPs separated by an RNase treatment step to generate RNP footprints. A key requirement of this procedure is expression of an epitope-tagged RNP component so that an RNP can be IPed via this epitope, footprinted while still bound to the affinity matrix and then gently eluted for input into the second IP. Because RBPs often exhibit promiscuous binding when expressed at high levels (i.e., are overexpressed), it is also essential that the tagged protein be expressed at near endogenous levels. For purification of endogenous EJC's and their RNA footprints via RIPiT, we expressed FLAG-tagged EJC core proteins from a Tetracycline (Tet)-inducible promoter in human embryonic kidney (HEK) 293 cells. We begin the following “Methods” section by describing the use of a site-specific recombination system to generate stable cell lines capable of Tet-inducible expression of FLAG-tagged proteins close to their endogenous levels (Method 4.1).
RIPiT should be broadly applicable to enrich any RNP from a biological system of choice where an epitope-tagged protein can be stably expressed. It can be particularly straightforward to apply to genetically tractable model organisms such as yeasts or flies, where epitope tags can readily be inserted into the genome via homologous recombination to drive tagged protein expression from its endogenous promoter. In fact, we tagged different spliceosome proteins at their genomic loci in Schizosaccharomyces pombe with a split TAP-tag (protein A tag and calmodulin binding peptide tag on two separate proteins) to reveal footprints of a stable spliceosomal complex via a RIPiT-like strategy [34]. In the case of mammalian systems, where genetic tagging is still a time- and resource-intensive exercise, a cultured cell line-based approach – such as the one we describe for expressing FLAG-tagged EJC proteins – presents a more rapid alternative. When proteins are exogenously expressed using such a system, the use of an inducible promoter is extremely important to express the tagged protein at endogenous levels (Figure 3A) to obtain physiologically relevant targets and binding sites for the RBP or multiprotein complex of interest.
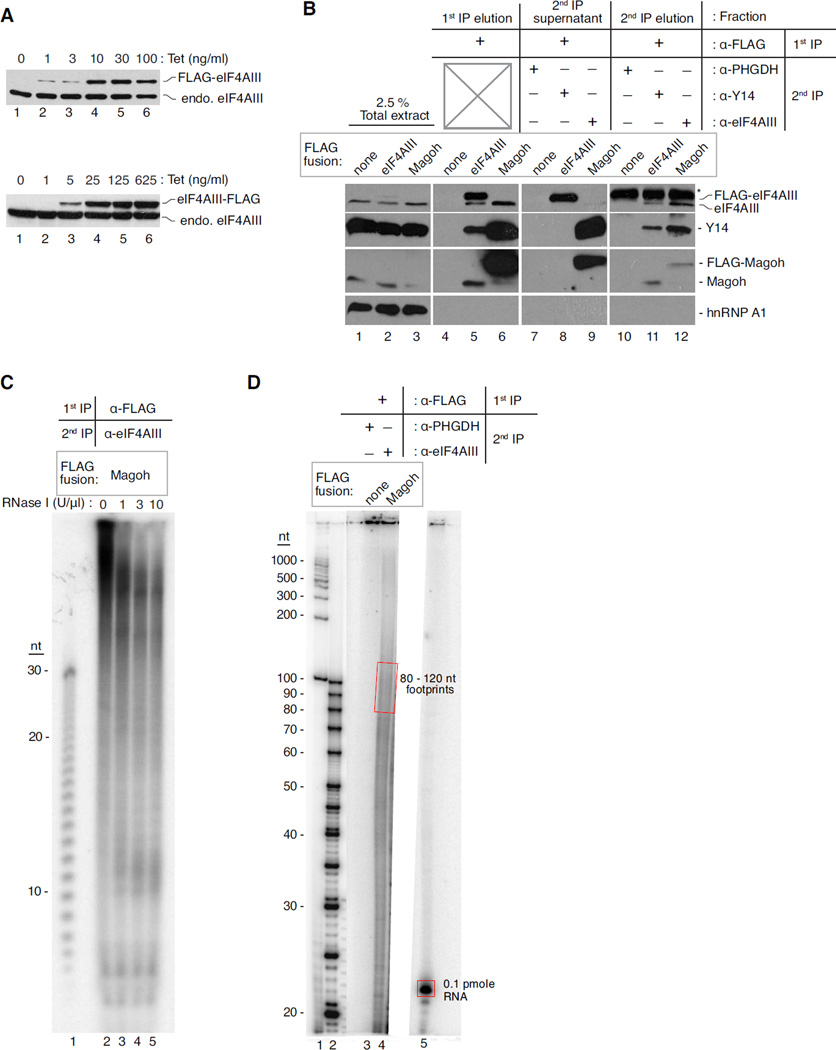
Figure 3. Biochemical analysis of proteins and RNAs from the RIPiT procedure.
A. Western blots showing tetracycline (Tet)-mediated induction of eIF4AIII protein with the FLAG tag at its N- or C-terminus (top and bottom panels, respectively). The Tet concentration used for induction is indicated at the top of each lane; protein identities are indicated to the right.
B. Levels of proteins detected by western blots in different fractions during EJC RIPiT. The table on the top indicates the different fractions from the RIPiT procedure and the antibodies used for 1st and 2nd IPs. The stably expressed FLAG-tag fusion protein used in each sample is indicated directly above lane. Proteins detected by western blot are indicated to the right.
C. Size distribution of EJC footprints upon RNase I titration. An autoradiogram of 26% denaturing PAGE with 5′ [32P]-labeled RNA fragments from base-hydrolysis of poly U30 oligonucleotide (lane 1) or FLAG-Magoh:eIF4AIII RIPiT (lanes 2–5). RNase I concentrations used are indicated at top of each lane; nucleotide (nt) lengths are to the left.
D. Quantification of desired size RNA footprints in RIPiT elution. An autoradiogram of 20% denaturing PAGE with 5′ [32P]-labeled 100 bp NEB DNA ladder (lane 1), low molecular weight ssDNA ladder (lane 2), footprints of RIPiTs indicated on top (lanes 3 and 4) and a 21 nt ssRNA oligo of known specific activity (lane 5). The signal from red rectangles is quantified in comparison to that of the 21 nt oligo to estimate the amount of RIPiT footprints in the indicated size range.
Like any affinity purification scheme, the choice of epitope-tag for RIPiT requires careful consideration regarding its size, nature and position (i.e., fusion to N- or C-terminus of a protein). We chose to express the EJC proteins with the FLAG-tag, a short peptide tag that binds with high affinity to anti-FLAG antibody. Importantly, following IP, FLAG-fusion proteins can be eluted from the FLAG affinity matrix under native conditions with a competing FLAG peptide. Due to its popularity for affinity purification, the FLAG affinity matrix and competing FLAG peptide are readily available from multiple commercial sources. Any other affinity tag that rivals the critical features of the FLAG-tag (i.e., high affinity binding and efficient elution under native conditions) can be used in its place. However, on a cautionary note based on our experience (A. Butterworth, N. Rozovsky and M.J.Moore, unpublished data), fusion of an RBP with a His6-tag can dramatically alter its innate RNA binding properties due to the strong positive charge of the His-tag at physiological pH. Thus, unless there is a way to confirm that a His-tag has no effect on RNA binding or protein function, its use is highly discouraged. The size and location of any epitope tag can also interfere with a protein's physiological function by occluding its functional domains. For example, fusion of the GST-tag to the N-terminus of eIF4AIII leads to its engagement with abnormal protein partners [35] whereas fusion of the same tag to the C-terminus of the protein allows its normal protein-protein interactions [36]. In contrast to the relatively larger GST-tag, we have not observed any such differences in eIF4AIII's interactions with other EJC proteins when the much smaller FLAG-tag was placed at either the N- or C-terminus (G. Singh and M. J. Moore, unpublished data).
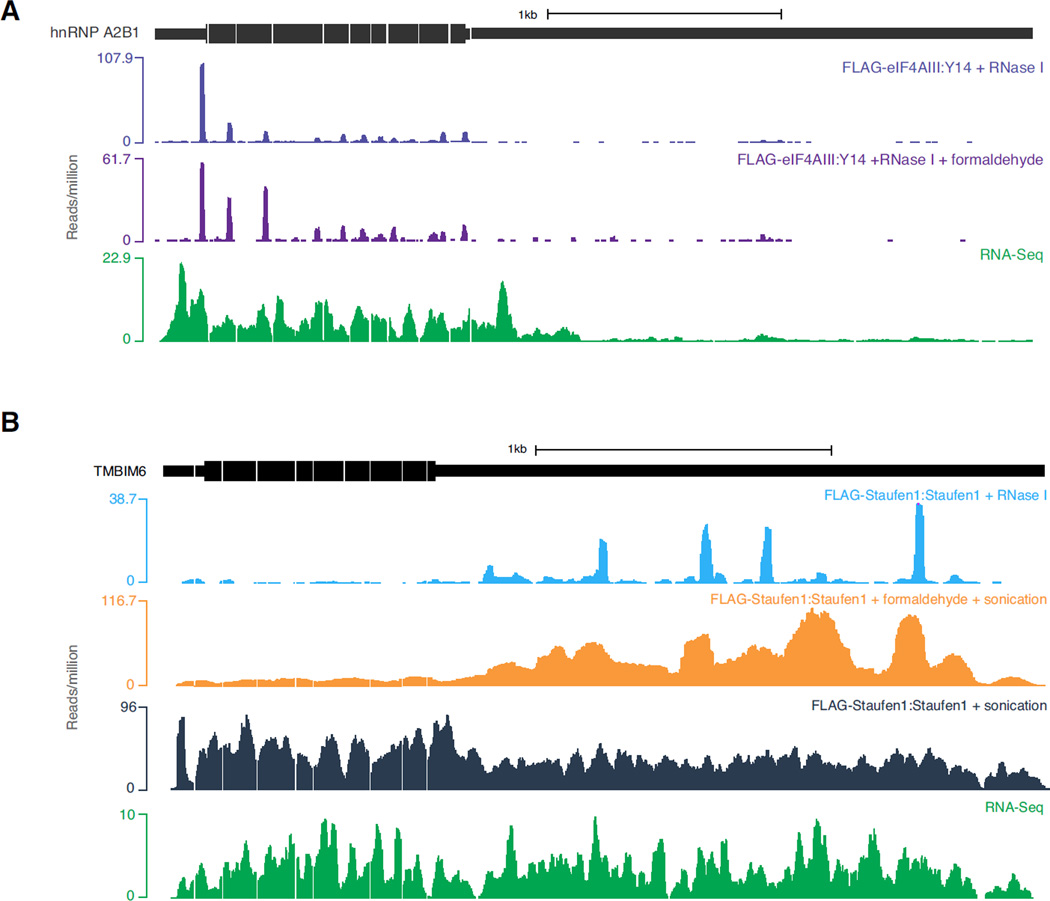
Next in the RIPiT procedure, we describe steps for growth and induction of FLAG-protein expressing cell lines (Method 4.2.1). Following this, we normally IP FLAG-EJC proteins under native conditions (Method 4.2.2) to obtain EJC footprints via RIPiT. Once formed, the EJC maintains an extremely stable grip on spliced mRNAs; the EJC thus represents an extreme case of a stable mRNP component. Most other proteins/protein complexes are likely to be less stably bound to their RNA targets; such proteins can easily dissociate from their natural targets upon cell lysis and subsequently engage in artificial interactions [37]. Native IP of such less stably associated proteins thus carries a heightened risk of both false negative and false positive identification of target RNA binding sites. To overcome such issues, formaldehyde crosslinking has previously been shown to preserve native RNA-protein interactions [38]. To study more dynamic RNA-protein interactions, we have successfully incorporated formaldehyde crosslinking in our RIPiT procedure [30], and we describe an alternate protocol below to perform FLAG IP from formaldehyde crosslinked cells (Figure 2; Method 4.2.3). Because formaldehyde crosslinking and IPs under denaturing conditions do adversely affect overall yields, it is advisable to use more starting material for crosslinked samples. While EJC footprint profiles obtained with or without formaldehyde crosslinking were largely indistinguishable (Figure 4A; compare top two tracks), this treatment vastly improved our ability to map the endogenous binding sites for Staufen1 [Figure 4B; compare the orange track (+ formaldehyde) to the dark blue track (no formaldehyde)], a double-stranded RBP that binds RNA with nanomolar affinity [39].
Figure 4. Observed EJC and Staufen1 occupancies in native and formadehyde crosslinked samples.
A. Distributions of EJC footprinting reads on the hnRNPA2B1 transcript from uncrosslinked (FLAG-eIF4AIII:Y14; top) and formaldehyde crosslinked RIPiT (FLAG-eIF4AIII:Y14 + formaldehyde; middle). Bottom: polyA+ RNA-Seq (RNA-Seq). Note the high similarity in observed EJC footprints between the uncrosslinked and crosslinked libraries.
B. Read distribution on the TMBIM6 transcript from the Staufen1 footprints or from polyA+ RNA-Seq as indicated above each track on the right. Notice the enrichment of Staufen-specific signal in the 3’UTR for the crosslinked sonication library (FLAG-Staufen 1:Staufen1 + formaldehyde + sonication; orange) contrary to the native sonication RIPiT library (FLAG-Staufen1:Staufen1 + sonication; dark blue). It also important to notice the difference between the RNase footprinting (light blue) and sonication output (orange). While the sonication shows a wide-peak around the Staufen1 binding site, the RNase footprinting is able to precisely map the interaction at almost single nucleotide precision.
After FLAG IP under native or denaturing conditions, RNPs captured on the solid support via the anti-FLAG antibody are treated with RNase I to digest away RNAs not protected by bound proteins (Method 4.2.4). In our experience, the most reproducible RNA fragment lengths are obtained when RNase-treatment is performed on the beads following the first IP instead of adding the RNase to the lysate prior to the IP (as is commonly advised in other protocols). Performing digestions on the beads allows for the RNase to be washed away at a specific time, whereas performing the RNase treatment in lysates leads to more variability, both because the digestion continues during the first IP and because cell lysates are much more heterogeneous than are selected RNPs bound to beads. After washing away the RNase I, the bound protein is then eluted with FLAG peptide (Method 4.2.5).
RNPs eluted from the anti-FLAG beads are next input into the second IP (Method 4.2.6). When this second IP utilizes an antibody against a different RNP constitutent, this key step both enriches the desired subcomplex and purifies away any excess, uncomplexed FLAG-tagged protein from the first IP. As can be seen in Figure 3B, the EJC-free FLAG-eIF4AIII (lane 8) and FLAG-Magoh:Y14 heterodimer (lane 9) from the anti-FLAG IP remain in the supernatant upon second IP with antibody against a different EJC protein. The removal of such uncomplexed proteins as well as depletion of non-specific interactors remaining after first IP serves to further enhance the specificity of the footprints isolated by RIPiT. To complete the RIPiT procedure, RNPs captured during the second IP are eluted in a denaturing buffer from which both proteins and RNAs can be recovered for compositional analysis.
RNase I cleavage generates a wide size range of RNA fragments bearing a 5’-OH and a 2’, 3’ cyclic phosphate at their ends. Following RNA extraction (Method 4.3.1), first a small fraction of the RNA footprints are 5′-[32P]-phosphorylated and resolved on denaturing PAGE to reveal their size distribution (Figure 3C; Method 4.3.2). Next, the 3′-phosphatase and 5′-kinase activities of T4 polynucleotide kinase (PNK; Method 4.3.3) are exploited to turn the ends of the remaining bulk RNA fragments into 3′-OH and 5′-P, respectively; this is necessary for adapter ligation during downstream cDNA library preparation. Finally, these cured RNAs are resolved on a denaturing PAGE and fragments of the desired length are excised from the gel and extracted (Figure 3D; Method 4.3.4). Preparation of cDNA libraries for high-throughput sequencing and subsequent data analysis have been extensively described elsewhere [40–42]. Therefore, we include only a short discussion highlighting major issues to consider for library preparation, and direct readers to suitable resources for further details (see Section 6).
4. Methods
4.1. Generation of stable cell lines and conditions for optimal expression of FLAG-tagged bait protein
For controlled expression of FLAG-tagged EJC proteins in human cells, we chose HEK293 TREx cells that allow expression of epitope-tagged protein near its endogenous levels in every cell in the population via a Tet-inducible promoter. To achieve this, a plasmid vector carrying a Tet-inducible cDNA expression cassette is stably integrated into these cells via site-specific recombination between two FRT sites, one in the plasmid and another at a transcriptionally active locus in the host cells. The Tet-inducible cassette expresses the protein of interest fused to an N-terminal FLAG tag. Below we describe the step-wise procedure we used to obtain cell lines with stably integrated pcDNA5/FRT/TO plasmid vector carrying FLAG-EJC cDNA fusions generated by standard molecular biology techniques. As a negative control, we also generated a cell line expressing only the FLAG tag. Alternatively, an RNA binding mutant of the RBP of interest or an unrelated control protein (e.g. green fluorescent protein) can be stably integrated for use in control RIPiT.
Seed 5 × 106 HEK293 TREx cells on 10-cm plates in standard growth media.
Next day (after ~16 hrs), transfect 10 µg plasmid mix containing 1 µg of a pcDNA5/FRT/TO-FLAG construct and 9 µg of pOG44 using 30 µl of HEK293TransIT reagent following manufacturer’s instructions (no need to change growth medium prior to transfection).
After an overnight incubation, trypsinize the transfected cells and seed them at 1:10 dilution on a new 10-cm plate.
After yet another overnight incubation to allow cells to adhere to the surface, replace the growth medium with Flp-In stable cell selection.
Replace the selection medium every 3–4 days. Within a week, most untransfected cells die and are washed away with every medium change, while individual transfected cells begin to grow into colonies.
When colonies become readily visible to the naked eye (~5 mm diameter, 10–14 days of selection), remove medium and rinse the plate once with PBS. Trypsinize the cells with 1 ml of Trypsin-EDTA for 2–3 min (individual colonies should dissociate upon tilting the plate multiple times).
Resuspend cells in 9 ml of Flp-In selection medium and re-plate the cell suspension on a new 10-cm plate. Cells can be expanded from this culture for frozen stocks (in FBS + 5% DMSO) and further experiments.
Seed 5 × 105 stably transfected cells per well in a 12-well tissue culture dish.
After overnight growth, titrate Tet from 0–500 ng/ml in each well.
Incubate cells for 16–24 hrs to induce FLAG protein expression.
Remove medium and resuspend cells by scraping into 100 µl PBS.
Transfer to a 1.5 ml tube and add 100 µl of 2X Lammelli sample buffer.
Heat samples at 95°C for 10 min.
Resolve 5 µl on a SDS-PAGE gel and western blot with an antibody against the untagged endogenous protein.
Notes
We induce expression of FLAG-tagged EJC proteins (or FLAG-Staufen) by adding 10–25 ng/ml tetracycline for 16–18 hrs. As the average half-life of a human mRNA (and hence an mRNP) is about 12 hrs, this time period is sufficient for incorporation of FLAG-tagged EJC proteins and most other mRNP components into the vast majority of newly synthesized mRNPs. However, in cases where the tagged protein is part of a more stable RNP, (e.g. the ribosome, snRNPs, etc.), longer induction times will be needed depending on specific RNP's assembly rate and lifetime.
For inductions longer than 24 hrs, doxycycline (a semi-synthetic analog of tetracycline) should be used because of its greater stability. When using doxycycline, however, it is important to independently determine the optimal concentration for achieving the desired expression level, as this optimal concentration may be different from that of tetracycline.
We generally are able to find SDS-PAGE conditions (gel % and/or pH) that separate the endogenous and FLAG-tagged proteins so optimal tetracycline/doxycycline concentrations are easily determined (see Figure 3A).
4.2. RIPiT Purification
The protocol below describes RIPiT from three (for native purification) or six (for denaturing purification) 150-mm plates of HEK cells. These inputs yield ~1 pmol of RNA footprints (~1015 RNA molecules) in the case of EJC purification through FLAG-eIF4AIII:Y14 or FLAG-Magoh:eIF4AIII IPs. In our hands, this amount is sufficient to generate complex high-throughput sequencing libraries that yield tens of millions of uniquely mapping read species. All steps should also be carried out in parallel starting with negative control cells described in method 4.1. During the entire procedure, all buffers should be pre-chilled on ice and all steps subsequent to cell growth should to be carried out at 4°C unless specified otherwise.
4.2.1. Cell growth and induction
Grow Flp-In TREx-HEK293 cells stably expressing the FLAG-tagged protein in 150-mm plates to ~60% confluency.
Use the time and tetracycline/doxycycline concentration pre-determined in Method 4.1 to induce expression of the FLAG-tagged protein (~16 hrs in the case of EJC proteins).
One hour prior to harvesting cells, add 100 µg/ml cycloheximide to cell growth medium for 1 hr. This treatment leads to translation arrest and limits stripping of proteins from newly made mRNPs by translating ribosomes.
4.2.2. Native lysis and FLAG IP
Rinse the cell monolayer once with ice-cold PBS.
Harvest cells by scraping all plates into a total of 25 ml PBS.
Pellet cells in a 50 ml conical tube at 400 × g for 10 min.
Lyse cells in 3 ml HLB for 10 min on ice and transfer to a 5 ml tube.
Place the tube with cell lysate in a firm holder in an ice bath. Sonicate at 40% amplitude using a Microtip for a total of 16 sec (in 2 sec bursts with 10 sec intervals).
Add NaCl to 300 mM final concentration.
Clear the lysate by centrifugation at 15,000 × g for 10 min.
Dilute the cleared lysate to 10 ml with HLB with a final NaCl concentration of 300 mM.
Aliquot 750 µl of anti-FLAG agarose bead slurry (50%) with a wide-bore 1 ml pipet tip (cut ~5mm from the tip) into a 15 ml tube. The amount of anti-FLAG slurry needed for efficient IP of FLAG-tagged protein will have to be empirically determined depending on the protein expression levels.
To wash beads, add 10 ml IsoWB, mix by inversion and centrifuge at 400 × g for 1 min. Repeat once.
Incubate the clarified, diluted lysate with washed anti-FLAG beads with gentle mixing for 2 hrs.
Pellet beads at 400 × g for 1 min and discard supernatant.
Wash beads (with captured RNPs) as above, twice with 10 ml ice-cold WB300 and then twice with 10 ml IsoWB.
Notes
To increase specificity, we carry out native FLAG-EJC protein IPs from lysates containing 300 mM NaCl. Protein-protein interactions between the EJC core and peripheral proteins remain largely unperturbed under these conditions [30]. For specific enrichment of other protein complexes, the optimal salt concentration should be determined empirically.
To further improve the specificity, cell extracts can be fractionated into nuclear, cytoplasmic or another appropriate fraction where the RNP of interest partitions.
Performing IPs under dilute conditions is key, as it significantly increases specificity. If the total protein concentration is too high, non-specific interactions with the beads and antibodies will prevail, limiting both yields and purity of the desired complexes.
4.2.3. Denaturing lysis and FLAG IP
Harvest and pellet cells as in steps 1 – 3 above in method 4.2.1.
Resuspend cells in 30 ml room temperature (RT) PBS.
Add 81 µl of 37% formaldehyde to obtain a final concentration of 0.1 %.
Incubate the tube with gentle mixing at RT for 10 min.
Add 3 ml of QB and continue incubation at RT for another 5 min.
Pellet cells at 400 × g for 10 min.
Lyse cells in 3 ml DLB on ice.
Transfer to a 5 ml tube and sonicate at 40% amplitude using a Microtip for a total of 30 sec (in 2 sec bursts with 10 sec intervals).
Clear the lysate by centrifugation at 15,000 × g for 10 min.
Dilute the cleared lysate to 10 ml with DLB.
Incubate the clarified, diluted lysate with washed anti-FLAG beads (prepared as in steps 11 and 12 in Method 4.2.2 above) with gentle mixing for 2 hrs.
Discard supernatant. Wash beads twice with 10 ml DWB and then twice with 10 ml IsoWB.
Notes
We determined that crosslinking cells with 0.1 % formaldehyde provided an optimal balance between crosslinking, macromolecular solubilization and RNA fragmentation. The amount of crosslinking agent should be empirically determined for other RNA-protein complexes. If higher amounts of formaldehyde are used, the sonication time should be increased to improve solubility.
In the denaturing lysis and IP buffer described above, FLAG antigen-antibody interactions persist but at a reduced level. We therefore advocate starting with twice the amount of input as recommended for native IPs in order to obtain comparable RNA footprint yields.
4.2.4 RNase I digestion
Transfer the washed beads from native or denaturing IP to a 1.5 ml tube. This is efficiently achieved by resuspending beads in 1 ml IsoWB, pipeting the slurry into the 1.5 ml tube and pelleting beads at 400 × g for 1 min.
Add one bed volume (375 µl) of IsoWB containing 1 U/µl of RNase I. Incubate for 10 min at 37°C with intermittent shaking on a Thermomixer.
Notes
For generating EJC and Staufen footprints, we have used RNase I, which cleaves after every RNA nucleotide without discrimination. RNase I can be substituted by another nuclease (e.g., RNase T1) more compatible with the nature of the RNP footprint sequences desired or depending on the RNP under investigation.
The uncrosslinked EJC interactome survives the 10-minute incubation at 37°C for RNase I treatment. In the case where the RNP of interest is labile at this temperature, this treatment can be performed at lower temperatures with an appropriate increase in RNase concentration.
When RIPiT is performed for the first time, an RNase titration experiment should be performed to identify the conditions yielding the appropriate size RNA footprints. The minimum size of footprint depends on the organism: a minimum of 15–16 nt sequence length is required to map uniquely to the human whereas a minimum of 12 nt long sequence is needed for yeast. We generally aim for footprints that are 2–3 times this minimum length, as longer fragments are always polluted with shorter ones after gel purification. Because the RNase digestion should occur randomly around the actual site of binding, even these longer fragments can reveal the exact binding site when they are piled together on individual genes. To perform the RNase titrations, we divide the FLAG beads post-IP and washes into four equal parts and perform RNase I titrations ranging from 1 to 10 U/µl in 3-fold steps in one bed-volume of IsoWB (Figure 3C). Subsequently, all four reactions are individually processed through all downstream steps, albeit at one-fourth. scale.
4.2.5. FLAG elution
Transfer the RNase I-treated beads to 15 ml conical tube. Wash beads 4 times with ice-cold 10 ml IsoWB.
Transfer washed beads to 1.5 ml tube. Add one bed volume (375 µl) of IsoWB containing 250 µg/ml FLAG peptide.
Shake gently at 4°C for 2 hrs to specifically elute the FLAG-epitope-containing RNPs from the beads.
To recover the eluted material, pellet beads at 400 × g for 1 minute in a microfuge and withdraw 350 µl of the liquid from the top.
4.2.6. Second immunoprecipitation
Aliquot 100 µl of ProteinG-Dyna-beads into a 1.5 ml tube. Following manufacturer’s instructions, couple 1–4 µg of affinity purified antibody that recognizes a different protein of the complex being immunopurified, and for the control RIPiT, same amount of an antibody against an unrelated protein [e.g. 3-phophoglycerate dehydrogenase (3-PHGDH; Figures 3B and D)]. The amount of antibody will have to be empirically determined based upon its efficacy and the fraction of bait protein co-immunopurified during the FLAG IP.
To prepare the material eluted from the anti-FLAG IP for input into the seconds IP, dilute it with 1 volume of 2X IP2 buffer.
Add this diluted elution to antibody-coupled magnetic beads previously washed with IP2 buffer.
Incubate with gentle mixing at 4°C for 2 hr.
Capture beads on a magnet and discard supernatant.
Wash beads six times with 1 ml ice-cold IsoWB.
Discard supernatant. Elute RNA and proteins in 40 µl CSB at 25°C for 5 min. To minimize the amount of released IgG, this elution can be carried out on ice for 20 min in a buffer without DTT. A small fraction (~ 1/10th) of elution can then be used for protein analysis by western blotting (as in Figure 2).
Notes
As compared to protein concentration of cell extract input into the first IP, overall protein concentration is much lower in the FLAG IP elution input into the second IP. In such a case, a significant amount of proteins may be lost on the tube and bead surfaces. To reduce this loss, we include BSA as a carrier in the second IP reaction. However, an appreciable amount of BSA carries over into the final elution and may interfere with detection of proteins similar to its size (~66.5 kDa) during western blotting. A different size carrier protein may be used in such a scenario.
The two sequential IPs during RIPiT lead to dramatic depletion of abundant stable RNA species (e.g. rRNA fragments, tRNAs, etc.) thereby increasing the signal-to-noise ratio by saving the valuable sequence space in the downstream high-throughput sequencing datasets. This is a major advantage of the RIPiT approach. We have achieved RIPiT-like superior signal-to-noise ratios in RNP footprints isolated by single FLAG-eIF4AIII IP followed by fractionation of these eIF4AIII-containing RNPs based on their molecular weight on a gel filtration column [30]. In certain cases two sequential IPs may not be possible, for example due to unavailability of antibodies to endogenous proteins or due to lack of information on RNP composition. In such instances, a combination of single IP and biochemical fractionation (e.g., gel filtration, sucrose or glycerol density gradients) can be used to obtain more pure RNP footprints.
4.3. Extraction, estimation and size-selection of RNA footprints
4.3.1. RNA extraction
Dilute the RIPiT elution to 475 µl with water.
Extract twice with an equal volume of Phenol (pH 4.5):Chloroform:Iso-amyl alcohol (25:24:1) and once with Chloroform:Iso-amyl alcohol (24:1). Leave behind 25 µl of aqueous phase between each extraction. Note: the organic phase in the first and/or second extractions may appear milky white due to extraction of SDS in the CSB.
To precipitate RNA from 400 µl of the recovered aqueous phase, add 10 µg of glycogen, 40 µl 3M sodium acetate pH 5.2 (300 mM final), 2 µl 2M MgCl2 (10 mM final) and 1 ml ice-cold 100% ethanol (2.5 volumes, 70% final). Addition of MgCl2 aids in precipitation of small RNAs. Incubate at 4°C for one hour or overnight at −20°C (for convenience).
Pellet RNA by centrifugation at 12,000 × g for 30 min at 4°C.
Discard the supernatant and wash the pellet with 1 ml 70% ethanol. Centrifuge at 12,000 × g for 5 min at 4°C.
Discard the wash and briefly air-dry RNA.
Resuspend RNA in 5 µl of water and store at −80°C.
Notes
In case of RIPiT from formaldehyde crosslinked cells, the denaturing eluate in CSB should be heated at 75°C for 40 min to reverse the protein crosslinks PRIOR to RNA extraction.
4.3.2. Estimation of footprint size and amount
For visualizing RNA footprints, we label footprinted RNA with 5′-32P using γ32P-ATP and T4 polynucleotide kinase (PNK). We use one-tenth of the precipitated RNA from the undivided second IP (RNA from method 4.3.1, step 7) or RNA from one-half of each of the four reactions that were divided to perform RNase I titrations (Method 4.2.4, final note). To estimate the size-range and molar amounts of the purified RNA, we also 5′-end label oligonucleotide of known size and concentration and run them on the same gel.
- Set-up the following 5’ end-labeling reaction:
10X PNK Buffer 1.0 µl 1mM ATP 0.5 µl γ32P-ATP 10–20 µCi RNA 0.5 µl (or another volume) T4 PNK 0.5 µl Water to 10 µl Incubate at 37°C for 30 min.
Bring up volume of each reaction to 200 µl with water. Precipitate RNA/DNA as described above (Method 4.3.1, steps 3–6).
Resuspend RNAs in 6 µl 2X FLB and DNA ladders in 40 µl 2X FLB.
Assemble gel plates to pour a thin (0.35-0.75 mm) 20 × 27 cm gel.
To 30 ml of Acrylamide mix, add 30 µl of TEMED and 90 µl of 10% APS to begin acrylamide polymerization and quickly pour the gel. Leave at room temperature for at least one hour to allow maximal polymerization.
Pre-run gel at 35W (constant power setting) for at least 20 min.
Load in separate wells all of the labeled immunopurified RNAs and RNA oligonucleotide, and 1/20th of the radiolabeled DNA ladders. Run the gel at 35W for about 90 min.
Stop electrophoresis when the faster migrating bromophenol blue reaches near the bottom of the gel.
Remove gel onto 3M Whatman filter paper and cover with saran wrap.
Dry gel for one hour at 80°C on a gel dryer.
Expose gel to phosphorimager screen overnight to visualize 32P-labeled RNA.
Quantify the amount of immunopurified footprints in a desired size range optimal for sequencing.
Obtain molar estimates of the footprints in this range by comparing their signal to that of the known RNA oligonucleotide (Figure 3D).
Notes
For the ladder, one can either use DNA or RNA. In the case of a DNA ladder, it is important to be aware that ssDNA tends to migrate faster than RNA in denaturing PAGE.
Although it may be tempting to decrease or eliminate the cold ATP from the kinase reaction in order to increase specific activity (and therefore detectability) of the footprints, as well as minimize the amount of radioactivity entering the lower buffer chamber, DO NOT DO THIS. In general, shorter oligos are better substrates for T4 polynucleotide kinase than longer ones; therefore, if ATP is limiting, shorter fragments will seem to be more abundant than longer fragments. In order to ensure equal distribution of the 32P label among all RNA sizes, the total ATP concentration must be at least 4-fold greater than the total concentration of 5′-ends.
4.3.3. RNA end curing
- Set up the following reaction for RNA 3′-end dephosphorylation:
10X PNK Buffer 2.0 µl RNA 4.5 µl (or another volume) T4 PNK 1.0 µl Water to 20 µl Incubate at 37°C for 30 min.
- Add the following mix to the reaction to kinase RNA 5’-end:
10X PNK Buffer 1.0 µl 1 mM ATP 4.0 µl T4 PNK 0.5 µl Water 4.5 µl Incubate again at 37°C for 30 min.
Add 20 µl of 2X FLB and heat denature at 65°C for 2 min.
4.3.4. RNA size selection and gel extraction
Pour and pre-run a preparative (1.0–1.5 mm thick) 12% acrylamide, 6M urea, 0.5X TBE gel (3:2 ratio of Acrylamide mix:Dilution mix) as above.
Load denatured RNA sample flanked on either side by 2 µl each of low molecular weight ssDNA ladder and NEB 100 bp DNA ladder (unlabeled).
Run gel at 35W until the faster migrating bromophenol blue dye has reached the middle.
Remove the gel and stain with 1X SYBR-Gold in 0.5X TBE for 5 min with gentle shaking.
Visualize the gel using blue-light LED transilluminator and cut the desired size range of RNA fragments.
Crush the gel slices by extruding through a 3 ml syringe.
Add 800 µl of REB and nutate overnight at RT (or at 37°C for 1–2 hrs).
Load the slurry onto an empty Spin-X column and centrifuge in a bench-top microfuge at top-speed for 2–5 min until most of the liquid comes out into the collection tube.
Precipitate, pellet and wash RNA as above (Method 4.3.1, steps 3–6).
Resuspend RNA in 5 µl of water and store at −80°C.
5. Alternate methods
In some instances, a large fraction of the binding sites for a particular RBP can occur within repeat sequences or low complexity regions. In this case, identifying the genomic locations of such sites is not possible using a footprinting strategy that generates only short RNA fragments, as the obtained reads will not map uniquely to the genome. If the repeat sequence is a SINE or LINE element (~300 and >500 nts, respectively), even relatively long RNase-digested fragments will fail to map to the genome. To circumvent this, we describe an alternate approach to generate RNA fragments during RIPiT (Figure 5A) where samples can be extensively sonicated before proceeding to the first IP. This harsh sonication results in random fragmentation of RNAs to sizes ranging from 200 to 900 nt depending on the total sonication time. The remaining steps of the RIPiT procedure are identical to those described above with only the RNase treatment between the two IPs being omitted. This strategy can also be used for crosslinked samples, noting that the sonication conditions needed to obtain desired fragment lengths will differ between crosslinked and uncrosslinked samples.
Sonication-induced RNA fragmentation occurs at random places along transcripts, while the RIPiT approach specifically pulls down target RNAs through the RBP of interest (Figure 5B). The expected output is therefore an enrichment of reads at the RBP binding site with a gradual decrease in coverage correlated to distance both upstream and downstream of the binding site. However, if the binding site occurs within a region of poor mappability (e.g., repeat or low complexity region), a dramatic loss of signal at the actual binding will be observed. In this case, the binding site location can be estimated by extrapolating the signal in the adjacent regions (see Figure 5C).
5.1.1. Generation of RNA fragments using sonication
Harvest and lyse cells as previously described in Method 4.2.2.
Place the tube with cell lysate in a firm holder in an ice bath. Sonicate at 40% amplitude using a Microtip for a total of 90 sec (in 5 sec bursts with 30 sec intervals). This will yield ~800–900 nt RNA fragments for uncrosslinked samples and ~200–300 nt fragments for crosslinked samples. Longer sonication times will result in shorter fragments.
Continue with the RIPiT procedure but omit the RNase treatment described in Method 4.2.4.
5.1.2. RNA fragmentation by hydrolysis
Because current high-throughput sequencing platforms are not able to sequence inserts longer than 400–500 nt, it is essential to further fragment the RNA footprints obtained via RIPiT from sonicated extracts before proceeding to library preparation. For our studies, we usually fragment RNAs to a size of ~100 nt that allows an efficient library preparation. It is important to note that this fragmentation step will not affect the output of the data generated by the sonication approach.
Once the RIPiT procedure has been performed, resuspend phenol-chloroform extracted RNAs in 9 µl of water.
To fragment RNAs to a size of 100 nt, add 1 µl of RNA fragmentation buffer and incubate at 70°C for 4 min and 30 sec.
Add 1 µl of the Stop solution and incubate on ice for 1 minute.
Precipitate, pellet and wash RNA as above (steps 3–6, Method 4.3.1).
Resuspend RNA in 5 µl of water and store at −80°C.
Perform steps in Methods 4.3.2, 4.3.3 and 4.3.4 to prepare RNA samples for generation of high-throughput sequencing libraries.
6. Guidelines for high-throughput sequencing and computational analysis of RNA footprinting libraries
High-throughput RNA sequencing methods have quickly become a routine laboratory practice to analyze RNA populations from diverse biological samples. Several different options to generate strand-specific libraries from RNA fragments could be applicable at this stage. Because of their small size, however, the short RNA fragments generated by RIPiT are most suited for a library construction method designed for small RNAs. Multiple commercial sources [e.g., Tru-Seq small RNA kit (Illumina, RS-200-0012), ScriptMiner small RNA-Seq library prep kit (Epicenter, SMMP101212), and NEBNext small RNA library prep set (New England Biolabs, E7330S)] now provide complete kits for generating next generation cDNA libraries from short RNAs. Numerous suitable non-commercial protocols have also been published [41–43]. The most popular methods generally involve either: (i) ligation of adapters to both 5′ and 3′ ends of RNA fragments followed by reverse transcription through both the captured RNA and the 5′-adaptor; or (ii) ligation of an adaptor to fragment 3′ ends followed by reverse transcription and circularization of the cDNA. Because RIPiT footprints contain no residual crosslinked amino acid that can act as a roadblock for reverse transcriptase (as is the case with CLIP), both library prep approaches work equally well on RIPiT-isolated footprints. We have generally found that ~1 pmol of isolated RNA footprints (~35 ng of 80–120 nt fragments) is sufficient to generate libraries yielding 10's of millions of unique species.
Once reads have been generated and mapped to the genome, it can be advantageous to identify peaks (genomic regions where RNA footprints are enriched over background) for further analysis. Because the copy number of individual mRNAs within a cell spans nearly four orders of magnitude [44], it is crucial to use an abundance-sensitive peak-calling algorithm such as ASPeak [45]. For this purpose, we also strongly advocate preparation of an mRNA-Seq [e.g., TruSeq stranded mRNA sample prep kit (Illumina, RS-122–2101)] or a total RNA-Seq [e.g., TruSeq stranded total RNA sample prep kit (Illumina, RS-122–2201)] library from the same cells that were used for RIPiT.
7. Concluding remarks
Here we described a novel approach to isolate and identify RNA footprints of an RBP or an RBP-containing complex from any biological source of choice. Being UV-crosslinking independent, RIPiT should be applicable to any RBP of interest regardless of its RNA recognition mode. Native RIPiT should only be applied to extremely stable RNP complexes, whereas RIPiT of formaldehyde-crosslinked samples can reveal in situ RNA:protein interactions and limit both false positive and false negative results for less stable complexes. A unique advantage of RIPiT over CLIP is its ability to specifically enrich RNA footprints of multi-subunit RNPs via two sequential purification steps. As RBPs often operate within such multi-component structures that are also dynamic in nature, RIPiT is particularly attractive for revealing targets and binding sites of compositionally dynamic RNPs. Parallel RIPiTs, where antibodies against different proteins are used in the second IP, can uncover where the same protein binds when in complex with different interaction partners. While offering many advantages, RIPiT does come with some limitations. Most prominently, RIPiT on its own does not provide conclusive evidence of direct RNA binding by any individual protein, as RBPs often exist in multi-subunit complexes where more than one protein may be in direct contact with RNA. Another technical limitation of RIPiT in its current incarnation is the need to exogenously express a tagged copy of the protein of interest to allow gentle affinity elution of RNPs after the first IP. This limits the use of RIPiT to enrich native endogenous RNPs, particularly from mammalian tissues and primary cells. However, multiple alternatives can be considered in such scenarios: (i) Animal models or primary cells can be engineered using traditional transgenic technologies or more recent ZFN-, TALEN-, or CRISPR-based genome editing approaches to epitope tag an endogenous allele of an RBP of interest; (ii) An antibody against a peptide epitope in an endogenous RNP component can be immobilized on a solid support via protein A for the first IP and then the antibody-bound RNP conjugates eluted using that peptide; or (iii) An alternate initial purification method (e.g., gel filtration or density sedimentation) could be performed prior to IP of the endogenous RBP of interest.
In summary, RIPiT is a powerful and highly adaptable addition to the arsenal of techniques available to RNA biologists to study RBP function in the context of multi-subunit complexes and further uncover the intricacies of the RNP world.
Highlights.
RIPiT-Seq, a novel RNP footprinting approach based on two sequential purifications, can reveal binding sites of single RBPs or multi-subunit complexes transcriptome-wide
RIPiT-Seq can distinguish between footprints of RNPs with overlapping yet distinct composition
When combined with protein-protein crosslinking agents, RIPiT-Seq can be used to study dynamic RNPs
Being independent of UV-crosslinking, RIPiT-Seq is broadly applicable to all RBPs regardless of their RNA recognition mode
Acknowledgements
We thank Alper Kucukural and Can Cenik for bioinformatics analyses of RIPiT-Seq datasets in our laboratory. Erin Heyer and Blandine Mercier are thanked for their critical feedback on the manuscript. This work was funded in part by NIH grant RO1-GM53007 (M.J.M.) and a Charles A. King Trust post-doctoral fellowship (G.S.). M.J.M is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 3.Keene JD. Proc Natl Acad Sci U S A. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore MJ. Science (New York, N.Y. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell SF, Jain S, She M, Parker R. Nature structural & molecular biology. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anantharaman V, Koonin EV, Aravind L. Nucleic acids research. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keene JD, Komisarow JM, Friedersdorf MB. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 9.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Proc Natl Acad Sci U S A. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han YH, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G. The Journal of biological chemistry. 2010 doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Molecular cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salton M, Elkon R, Borodina T, Davydov A, Yaspo ML, Halperin E, Shiloh Y. PloS one. 2011;6:e23882. doi: 10.1371/journal.pone.0023882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brazao TF, Demmers J, van IW, Strouboulis J, Fornerod M, Romao L, Grosveld FG. FEBS Lett. 2012;586:1101–1110. doi: 10.1016/j.febslet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. Science (New York, N.Y. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 16.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konig J, Zarnack K, Luscombe NM, Ule J. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 18.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Genes & development. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. EMBO J. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayrand S, Setyono B, Greenberg JR, Pederson T. The Journal of cell biology. 1981;90:380–384. doi: 10.1083/jcb.90.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dreyfuss G, Choi YD, Adam SA. Molecular and cellular biology. 1984;4:1104–1114. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Steitz JA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 23.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Science (New York, N.Y. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 24.Bono F, Ebert J, Lorentzen E, Conti E. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Draper DE. In: RNA-Protein Interactions. Nagai K, Mattaj IW, editors. New York: Oxford University Press; 1994. pp. 82–102. [Google Scholar]
- 26.Ingolia NT, Lareau LF, Weissman JS. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Science (New York, N.Y. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tange TO, Nott A, Moore MJ. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Bono F, Gehring NH. RNA Biol. 2011;8:24–29. doi: 10.4161/rna.8.1.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer E, Ashar A, Peng LT, Moore MJ. Submitted [Google Scholar]
- 32.Liu ZR, Wilkie AM, Clemens MJ, Smith CW. RNA. 1996;2:611–621. [PMC free article] [PubMed] [Google Scholar]
- 33.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nature biotechnology. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 34.Chen W, Shulha HP, Ashar AJ, Yan J, Query CQ, Rhind N, Weng Z, Moore MJ. In preparation [Google Scholar]
- 35.Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibuya T, Tange TO, Sonenberg N, Moore MJ. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 37.Mili S, Steitz JA. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Methods (San Diego, Calif. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 39.Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. Molecular and cellular biology. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hafner M, Lianoglou S, Tuschl T, Betel D. Methods (San Diego, Calif. 2012;58:94–105. doi: 10.1016/j.ymeth.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langevin SA, Bent ZW, Solberg OD, Curtis DJ, Lane PD, Williams KP, Schoeniger JS, Sinha A, Lane TW, Branda SS. RNA biology. 2013;10:502–515. doi: 10.4161/rna.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafner M, Renwick N, Farazi TA, Mihailovic A, Pena JT, Tuschl T. Methods (San Diego, Calif. 2012;58:164–170. doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 45.Kucukural A, Ozadam H, Singh G, Moore MJ, Cenik C. Bioinformatics (Oxford, England) 2013 doi: 10.1093/bioinformatics/btt428. [DOI] [PMC free article] [PubMed] [Google Scholar]