Abstract
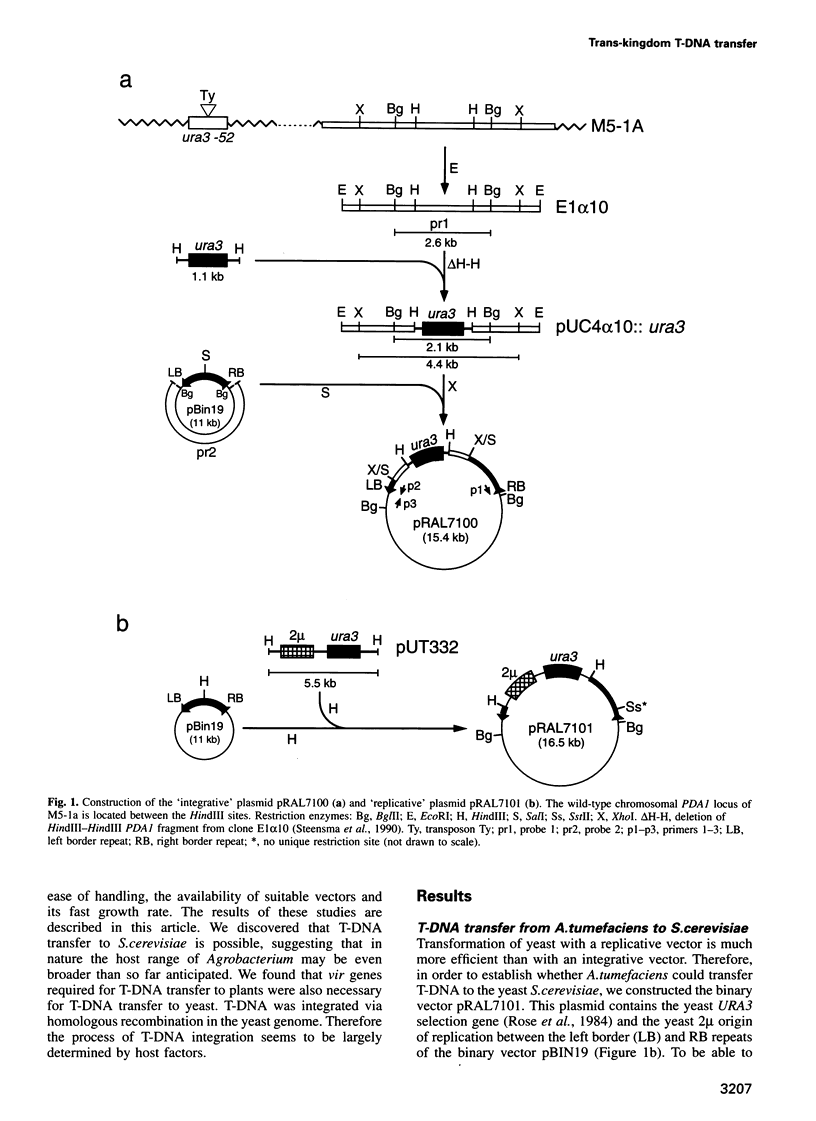
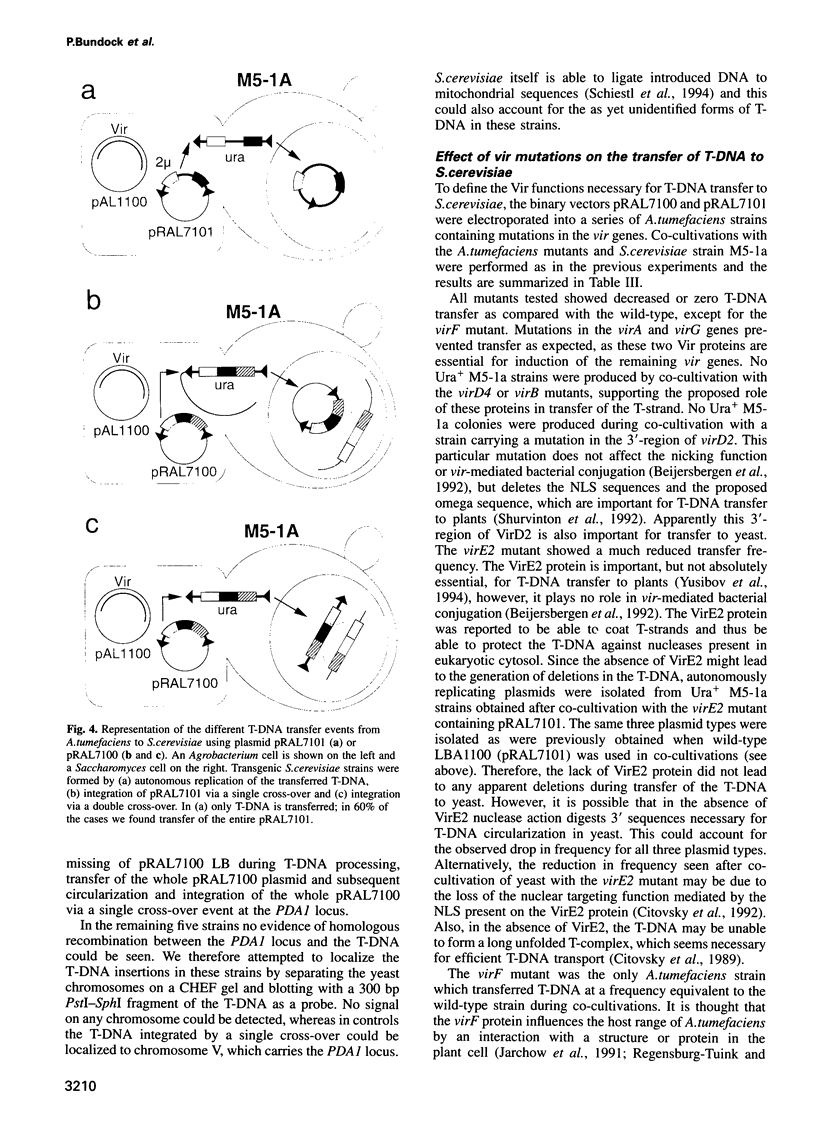
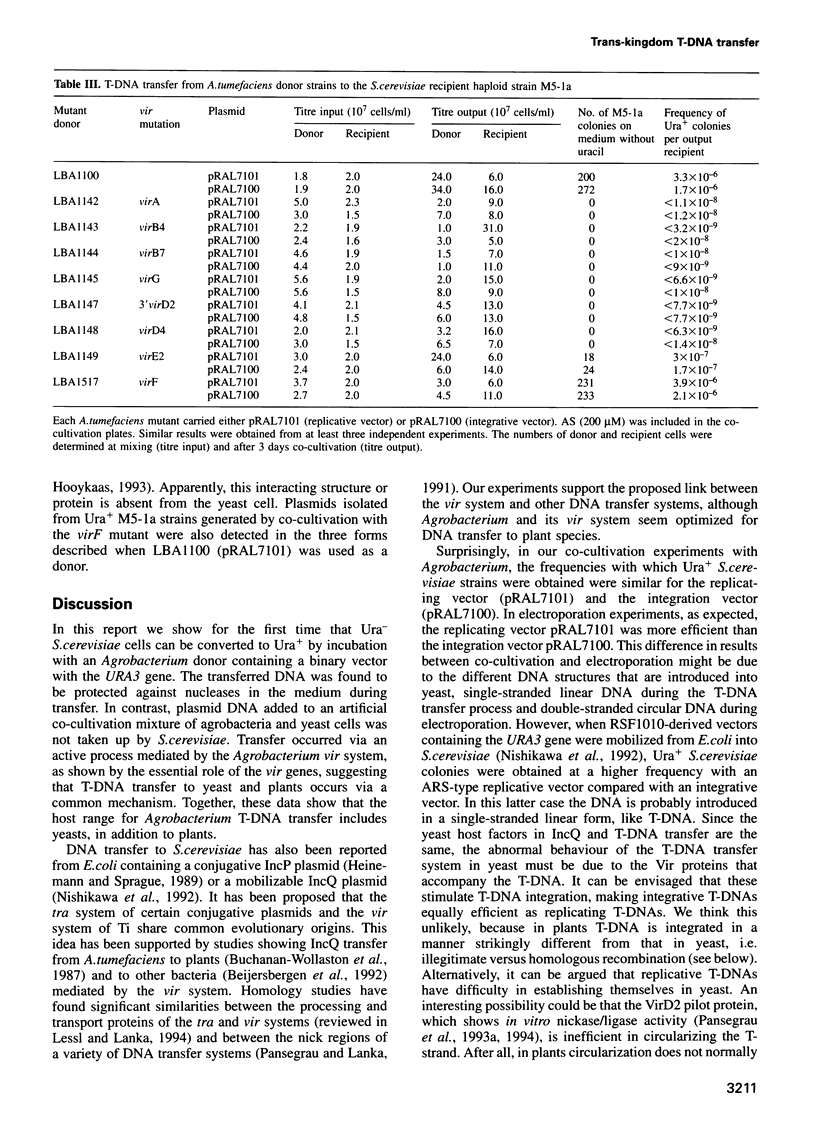
Agrobacterium tumefaciens transfers part of its tumour-inducing (Ti) plasmid, the transferred or T-DNA, to plants during tumourigenesis. This represents the only example of naturally occurring trans-kingdom transfer of genetic material. Here we report that A.tumefaciens can transfer its T-DNA not only to plant cells, but also to another eukaryote, namely the yeast Saccharomyces cerevisiae. The Ti plasmid virulence (vir) genes that mediate T-DNA transfer to plants were found to be essential for transfer to yeast as well. Transgenic S.cerevisiae strains were analysed for their T-DNA content. Results showed that T-DNA circles were formed in yeast with precise fusions between the left and right borders. Such T-DNA circles were stably maintained by the yeast if the replicator from the yeast 2 mu plasmid was present in the T-DNA. Integration of T-DNA in the S.cerevisiae genome was found to occur via homologous recombination. This contrasts with integration in the plant genome, where T-DNA integrates preferentially via illegitimate recombination. Our results thus suggest that the process of T-DNA integration is predominantly determined by host factors.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright L. M., Yanofsky M. F., Leroux B., Ma D. Q., Nester E. W. Processing of the T-DNA of Agrobacterium tumefaciens generates border nicks and linear, single-stranded T-DNA. J Bacteriol. 1987 Mar;169(3):1046–1055. doi: 10.1128/jb.169.3.1046-1055.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkeren G., Koukolíková-Nicola Z., Grimsley N., Hohn B. Recovery of Agrobacterium tumefaciens T-DNA molecules from whole plants early after transfer. Cell. 1989 Jun 2;57(5):847–857. doi: 10.1016/0092-8674(89)90799-x. [DOI] [PubMed] [Google Scholar]
- Beijersbergen A., Dulk-Ras A. D., Schilperoort R. A., Hooykaas P. J. Conjugative Transfer by the Virulence System of Agrobacterium tumefaciens. Science. 1992 May 29;256(5061):1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- Beijersbergen A., Smith S. J., Hooykaas P. J. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid. 1994 Sep;32(2):212–218. doi: 10.1006/plas.1994.1057. [DOI] [PubMed] [Google Scholar]
- Berger B. R., Christie P. J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994 Jun;176(12):3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984 Nov 26;12(22):8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Wong M. L., Zambryski P. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Zupan J., Warnick D., Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992 Jun 26;256(5065):1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- Gatignol A., Dassain M., Tiraby G. Cloning of Saccharomyces cerevisiae promoters using a probe vector based on phleomycin resistance. Gene. 1990 Jul 2;91(1):35–41. doi: 10.1016/0378-1119(90)90159-o. [DOI] [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989 Jul 20;340(6230):205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994 Aug;6(2):271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Hooykaas P. J., Schilperoort R. A. Agrobacterium and plant genetic engineering. Plant Mol Biol. 1992 May;19(1):15–38. doi: 10.1007/BF00015604. [DOI] [PubMed] [Google Scholar]
- Jarchow E., Grimsley N. H., Hohn B. virF, the host-range-determining virulence gene of Agrobacterium tumefaciens, affects T-DNA transfer to Zea mays. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10426–10430. doi: 10.1073/pnas.88.23.10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Michelitch M., Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jul;13(7):3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemoto R. H., Powell A. T., Akiyoshi D. E., Regier D. A., Kerstetter R. A., Nester E. W., Hawes M. C., Gordon M. P. Nucleotide sequence and analysis of the plant-inducible locus pinF from Agrobacterium tumefaciens. J Bacteriol. 1989 May;171(5):2506–2512. doi: 10.1128/jb.171.5.2506-2512.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessl M., Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994 May 6;77(3):321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Martineau B., Voelker T. A., Sanders R. A. On Defining T-DNA. Plant Cell. 1994 Aug;6(8):1032–1033. doi: 10.1105/tpc.6.8.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer R., Koncz-Kalman Z., Nawrath C., Bakkeren G., Crameri A., Angelis K., Redei G. P., Schell J., Hohn B., Koncz C. T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 1991 Mar;10(3):697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazodier P., Cossart P., Giraud E., Gasser F. Completion of the nucleotide sequence of the central region of Tn5 confirms the presence of three resistance genes. Nucleic Acids Res. 1985 Jan 11;13(1):195–205. doi: 10.1093/nar/13.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozo T., Hooykaas P. J. Electroporation of megaplasmids into Agrobacterium. Plant Mol Biol. 1991 May;16(5):917–918. doi: 10.1007/BF00015085. [DOI] [PubMed] [Google Scholar]
- Nishikawa M., Suzuki K., Yoshida K. DNA integration into recipient yeast chromosomes by trans-kingdom conjugation between Escherichia coli and Saccharomyces cerevisiae. Curr Genet. 1992 Feb;21(2):101–108. doi: 10.1007/BF00318467. [DOI] [PubMed] [Google Scholar]
- Offringa R., de Groot M. J., Haagsman H. J., Does M. P., van den Elzen P. J., Hooykaas P. J. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J. 1990 Oct;9(10):3077–3084. doi: 10.1002/j.1460-2075.1990.tb07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991 Jun 25;19(12):3455–3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Schoumacher F., Hohn B., Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Schröder W., Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J Biol Chem. 1994 Jan 28;269(4):2782–2789. [PubMed] [Google Scholar]
- Pansegrau W., Schröder W., Lanka E. Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regensburg-Tuïnk A. J., Hooykaas P. J. Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature. 1993 May 6;363(6424):69–71. doi: 10.1038/363069a0. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Rossi L., Hohn B., Tinland B. The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plant. Mol Gen Genet. 1993 Jun;239(3):345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- Scheeren-Groot E. P., Rodenburg K. W., den Dulk-Ras A., Turk S. C., Hooykaas P. J. Mutational analysis of the transcriptional activator VirG of Agrobacterium tumefaciens. J Bacteriol. 1994 Nov;176(21):6418–6426. doi: 10.1128/jb.176.21.6418-6426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Otto S., Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992 Jan 11;20(1):41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Zhu J., Petes T. D. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Jul;14(7):4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C. E., Hodges L., Ream W. A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11837–11841. doi: 10.1073/pnas.89.24.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Timmerman B., Zambryski P. Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5' virD gene products. EMBO J. 1987 Apr;6(4):857–863. doi: 10.1002/j.1460-2075.1987.tb04831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma H. Y., Holterman L., Dekker I., van Sluis C. A., Wenzel T. J. Molecular cloning of the gene for the E1 alpha subunit of the pyruvate dehydrogenase complex from Saccharomyces cerevisiae. Eur J Biochem. 1990 Aug 17;191(3):769–774. doi: 10.1111/j.1432-1033.1990.tb19186.x. [DOI] [PubMed] [Google Scholar]
- Timmerman B., Van Montagu M., Zambryski P. vir-induced recombination in Agrobacterium. Physical characterization of precise and imprecise T-circle formation. J Mol Biol. 1988 Sep 20;203(2):373–384. doi: 10.1016/0022-2836(88)90005-8. [DOI] [PubMed] [Google Scholar]
- Tinland B., Hohn B., Puchta H. Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8000–8004. doi: 10.1073/pnas.91.17.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Koukolíková-Nicola Z., Hall M. N., Hohn B. The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7442–7446. doi: 10.1073/pnas.89.16.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N., Datta A., Yanofsky M., Nester E. Role of the overdrive sequence in T-DNA border cleavage in Agrobacterium. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8558–8562. doi: 10.1073/pnas.85.22.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Stachel S. E., Timmerman B., VAN Montagu M., Zambryski P. C. Site-Specific Nick in the T-DNA Border Sequence as a Result of Agrobacterium vir Gene Expression. Science. 1987 Jan 30;235(4788):587–591. doi: 10.1126/science.235.4788.587. [DOI] [PubMed] [Google Scholar]
- Winans S. C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992 Mar;56(1):12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusibov V. M., Steck T. R., Gupta V., Gelvin S. B. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):2994–2998. doi: 10.1073/pnas.91.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]






