Abstract
High-throughput and unbiased binding assays have proven useful in probe discovery for a myriad of biomolecules, including targets of unknown structure or function and historically challenging target classes. Over the past decade, a number of novel formats for executing large-scale binding assays have been developed and used successfully in probe discovery campaigns. Here we review the use of one such format, the small-molecule microarray (SMM), as a tool for discovering protein-small molecule interactions. This review will briefly highlight selected recent probe discoveries using SMMs as well as novel uses of SMMs in profiling applications.
Introduction
Small molecules are essential components of a growing toolbox used to study cellular processes and develop effective therapies. Advances in genomics and proteomics, have led to the identification of a vast number of biomolecules implicated in human disease. Our understanding the function of these new targets will benefit from small molecule probes that can bind directly to them and modulate their activity.
Since the introduction of DNA microarrays, which allowed scientists to rapidly assess the expression of thousands of genes, the microarray platform has played a pivotal role in understanding complex biological systems [1]. By immobilizing small molecules onto microarray slides, Schreiber and coworkers realized they could discover protein-small molecule interactions using this format [2]. Over a decade after the creation of the first small-molecule microarray (SMM), this unbiased approach to detecting ligand-protein interactions has become commonplace in both academia and industry [3-6].
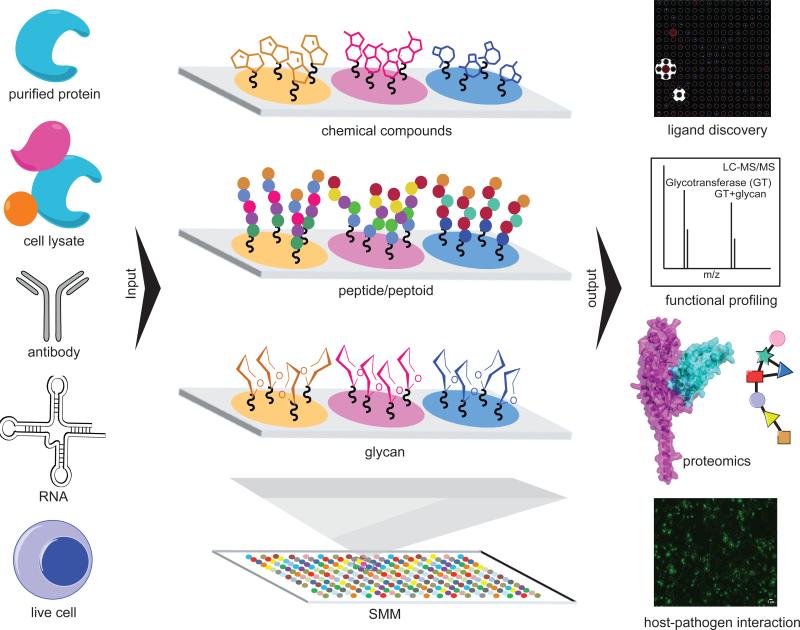
In recent years, SMMs have also moved from simple ligand discovery applications to include new uses in functional proteomics. Organic compounds, natural products, peptides and carbohydrates have all been immobilized on SMM slides using a range of surface chemistries. In addition, a variety of biomolecules can be incubated with SMMs, including purified proteins, cell lysates, antibodies, RNAs and even living cells (Figure 1). We refer the reader to several excellent and thorough reviews and volumes published previously that cover approaches to SMM manufacture in great detail [7-11]. This review will focus on selected SMM-facilitated probe discovery stories and functional proteomics applications from the last five years.
Figure 1.
Schematic representation of screens involving small molecule microarrays and various applications. Inputs ranging from purified protein to live cells can be incubated with functionalized microarrays containing immobilized small molecules, peptides or glycans. The SMM is an established tool for small molecule probe discovery, and is fast becoming a powerful platform for high throughput proteomic profiling. Binding is most commonly detected using readouts such as fluorescence, surface plasmon resonance, or mass spectrometry (right).
Probe discovery
Over the past decade, small-molecule microarrays (SMMs) have emerged as engines for probe discovery. Novel SMM-facilitated screens are reliably yielding ligands that bind several different functional classes of proteins, allowing researchers to develop probes for previously elusive protein targets [9].
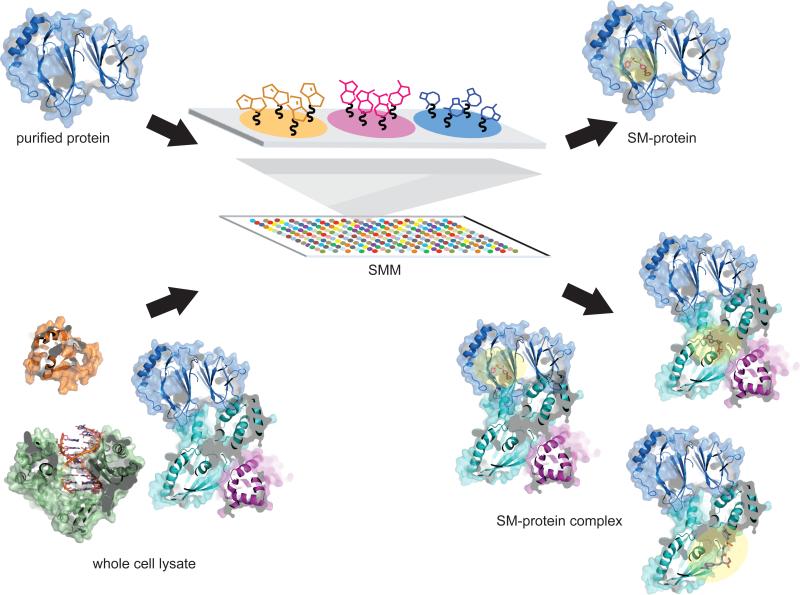
SMM screening strategies can be separated into two broad categories: those that use purified protein, and those that use cellular lysate (Figure 2). Traditionally, SMMs have been incubated with purified target protein and binding is detected using readouts such as surface plasmon resonance (SPR) or fluorescently labeled antibodies against the protein or an epitope tag on the protein [12,13]. In cases where the protein of interest has no known binding partners or specified function, the use of purified proteins in SMM screens is a good starting starting point for probe discovery. However, this approach does not guarantee that the compounds will bind the target in a relevant cellular context. More recently, applying cell lysates to SMMs has become more common [12,14,15]. This format allows for the detection of probes that can bind targets with relevant post-translational modifications, as well as interact with protein complexes. This format also enables probe discovery for proteins that elude purification. Since SMM assay positives may directly or indirectly bind the target, lysate screens require secondary assays to pinpoint where in the protein complex small molecules are binding. Classical secondary binding assays (e.g. SPR, thermal shifts or isothermal calorimetry) and target identification methods are used to validate binders using both purified and lysate approaches to screening. Both strategies have been used to target a range of functional classes of proteins, and each has yielded effective probes and inhibitors. Selected discovery stories for both approaches are outlined herein.
Figure 2.
Small molecule microarrays can be incubated with either purified proteins or cellular lysates. Purified protein SMMs favor discovery of probes that bind directly to the target protein (highlighted as yellow), while lysate screening allows for detection of compounds that can also indirectly interact with the protein of interest by binding its partner proteins.
Purified protein screening
SMMs have been used to identify direct probes of various types of proteins, including enzymes such as kinases [16,17], proteases [18,19], and deacetylases [20,21] as well as classically challenging proteins such as transcriptional regulators [22,23] and growth factors [24,25], among others [11,26-28]. In one recent example that highlights the discovery of a novel enzyme inhibitor, Lee and Park developed Jeffamine-coated slides with the aim of reducing nonspecific interactions and enhance fluorescent detection of small molecules that bind target proteins [29]. These arrays were used to identify 2,4,4’-trihydroxychalcone as a potent binder of tyrosinase, a major regulator of melanin production (Kd = 0.4 μM, Table 1). Skin disorders such as melasma and hyperpigmentation result from abnormally high levels of melanin, and can be treated using tyrosinase inhibitors [30]. 2,4,4’-trihydroxychalcone's activity was confirmed by an inhibition assay that measured the rate of the tyrosinase-catalyzed conversion of L-DOPA to DOPAchrome (IC50 = 0.76 μM).
Table 1.
Summary of recently discovered ligands from SMM
| Ligand | Structure | Target | Kd [μM] | Screening strategy | Surface chemistry |
|---|---|---|---|---|---|
| 2,4,4'- trihydroxychal cone [29] |
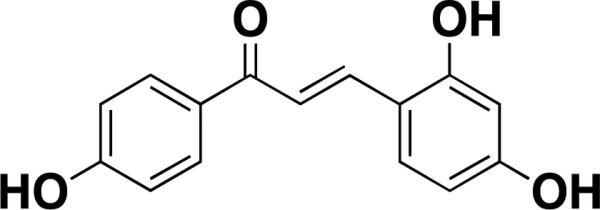
|
tyrosinase | 0.4[a] | Cy5-purified protein | isocyanate |
| NSC143101 [24] |
|
VEGF-KDR | 0.33[b] | Label-free purified protein mixture | isocyanate |
| Robotnikinin [25] |
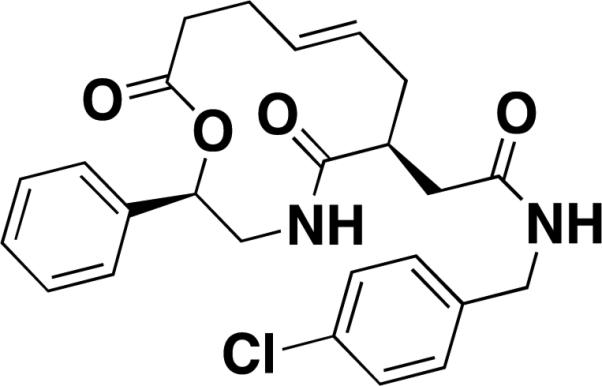
|
sonic hedgehog | 3.1[c] | epitope-purified protein | isocyanate |
| 2002-H20 [34] |
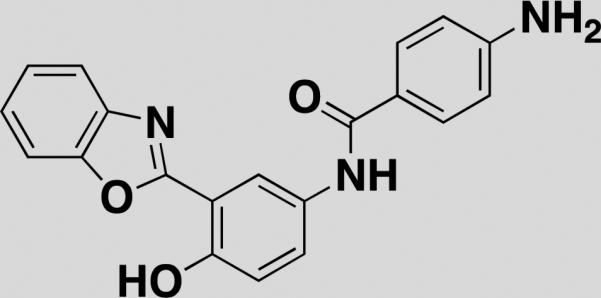
|
amyloid β-peptide | - | fluorescent-peptide | isocyanate |
| TPh A [15] |
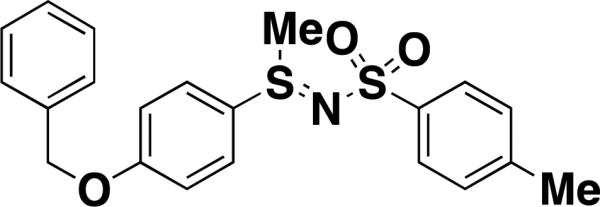
|
Pirin | 0.6[d] | HEK293T cell lysates (DsRed-fused pirin) | photo affinity linker |
| 4-4-3-12 [37] |
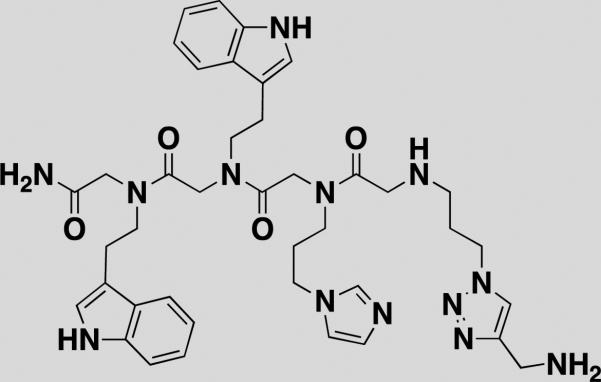
|
C. albicans ribozyme | 158±14[e] | 32P-labeled ribozyme | alkyne-agarose |
| 15-AB-21 [38] |
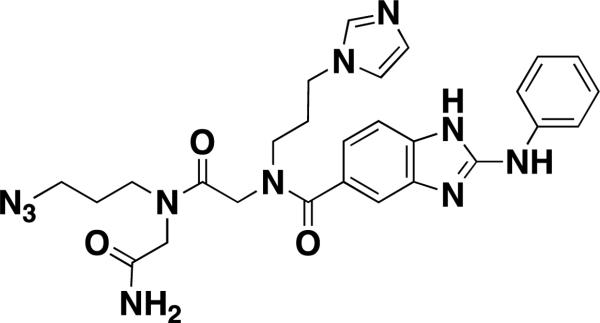
|
secondary structures of RNAs | 154±22[f] | 32P-labeled nucleotide internal loop | alkyne-agarose |
| Tyr-NH2 [40] |
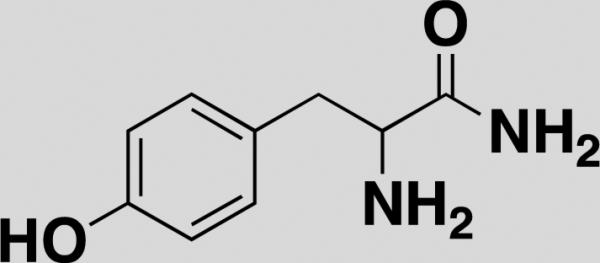
|
S. aureus, B. subtilis, P. aeruginosa, E.coli | - | Fluorophore stained bacteria strain | amine |
| c-c-5 [46] |
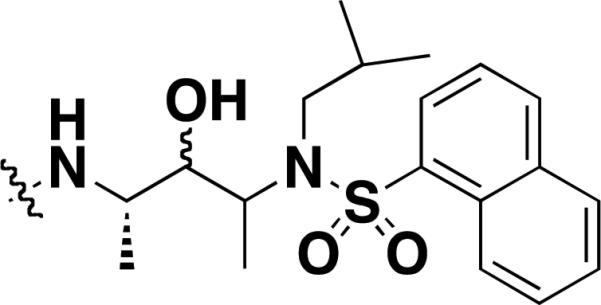
|
γ-secretase | - | Fluorescent r30 cell lysate | biotin tag |
The binding affinity of compound 1 with tyrosinase was measured by SPR spectroscopy.
The microarray-based IC50 of NSC143101 was calculated using the OI-RD signal. The cell-based IC50 was 0.09 μM from nonlinear curve fitting to the chemiluminescence data.
Robonikinin was measured by SPR spectroscopy.
The binding affinity of TPh A for purified His6-pirin was measured by ITC.
The lC50 for peptoids that bound the ribozyme were calculated for their ability to inhibit self-splicing of the C. albicans group I intron precursor.
The binding affinity of 15-AB-21 with selected internal RNA loop was measured by fluorescence-based assay.
Extracellular proteins that bind membrane receptors often play pivotal roles in tumorigenesis. For example, overexpression of vascular endothelial growth factor (VEGF) is known to drive tumor proliferation and survival, and disruption of the VEGF-kinase domain receptor (KDR) interaction, is an established cancer intervention strategy [31]. Yet, small molecule inhibition of proteins of this class has historically proven exceedingly difficult. Landry and coworkers executed various types of SMM screens to identify inhibitors of VEGF-KDR binding [24]. In the first assay, the team identified ligands of VEGF by incubating the protein with an SMM containing 7,916 small molecules immobilized on the glass slide via an isocyanate attachment strategy. The second assay exposed KDR to an identical SMM slide, followed by subsequent incubation with VEGF in order to find compounds that bind KDR but do not interrupt formation of the VEGF-KDR complex. A final assay involved capture of KDR on a glass slide using the compounds identified in the second assay, followed by incubation with VEGF and the VEGF binders identified in the first screen. This elegant approach yielded 12 inhibitors that disrupted VEGF-KDR formation, with half-maximal inhibitory concentrations (IC50s) ranging from 0.3 to 60 μM (Table 1). Inhibitors were validated in a cell-based assay that measured levels of phospho-KDR (proxy for VEGF-KDR binding) in response to different concentrations of inhibitor.
In another example, Stanton et al. used SMMs to identify a small-molecule inhibitor of the extracellular signaling protein Sonic hedgehog (Shh) [25]. Proper Shh signaling is essential for embryonic development and dysregulation of this pathway has implicated in cancer [32,33]. An SMM screen involving bacterially expressed ShhN against a library of ~10,000 diversity-oriented synthesis (DOS) compounds yielded a small subset of related macrocycles as assay positives. SPR was used to evaluate binding of the positives in a quantitative assay and one compound was prioritized for phenotypic studies. The compound showed moderate pathway inhibition in luciferase reporter assays in Shh-LIGHT2 cells. Structure-activity relationship data lead to the development of robotnikinin, an improved 12-membered macrocycle that demonstrated ShhN binding with a Kd of 3.1 μM (Table 1). Real-time PCR studies in human keratinocytes showed that robotnikin decreased transcription of downstream targets in the Shh signaling pathway. The compound also blocks Shh signaling in a synthetic model of human skin and cell lines, suggesting potential efficacy as a therapeutic agent in basal cell carcinoma, where the Shh pathway is known to be abnormally active.
SMMs are also yielding probes for proteins that have significant regions of disorder in their structure. In one example, Chen et al. developed a novel SMM assay to detect binders of amyloid-β (Aβ) [34]. Aβ proteins form toxic aggregates with a complex structure and are hallmarks of Alzheimer's disease (AD) [35]. Compounds that interfere directly with complex Aβ aggregates are desirable because they have the potential to halt AD pathogenesis. 17,904 natural products, commercial synthetic compounds, and DOS compounds were screened for binding to fluorescently labeled Aβ-40 monomers. Primary SMM positives were then tested in an MTT-based viability rescue assay involving PC12 cells treated with exogenous Aβ-42; fifteen of these compounds increased cell viability of PC12 cells by >30%. Compound 2002-H20 (Table 1) was selected for follow up studies as it increased cell survival by 41%, reproducibly bound Aβ-40 in the SMM, and was commercially available. 2002-H20 was found to structurally resemble known Aβ-binders, and caused a dose-dependent increase in amount of fibril formation, as measured by Congo red spectral shift assays and transmission electron microscopy. This suggests that 2002-H20 inhibits cytotoxicity by accelerating AB-42 aggregation toward fibril formation and away from neurotoxic tangle formation, a rational therapeutic approach to stemming the development of AD. This compound would not have been identified in a traditional ThT assay focused on identifying inhibitors of fibrillar aggregates and highlights how unbiased binding assays may identify compounds with novel mechanisms.
Cellular lysate screening
An impressive example of probe development for a protein of unknown function using SMMs involves the nuclear protein pirin. Pirin has been shown to interact with the nuclear factor I/CCAAT box transcription factor and is highly conserved in both eukaryotes and prokaryotes. Pirin also binds Bcl3, a protein whose overexpression enhances proliferation, survival and migration of many human tumors. Osada and coworkers sought to identify small molecule probes of pirin in an effort to further characterize pirin's cellular roles. Using SMMs, the team screened >20,000 compounds from the RIKEN Natural Products Repository for binding to pirin residing in cell lysates [15]. Slides were incubated with HEK293T cell lysates that overexpressed DsRed or DsRed-fused pirin and binders were detected fluorescent readout. An SMM positive, Triphenyl compound A (TPh A) was evaluated by isothermal calorimetry (ITC) and was found to bind with high-affinity to pirin (Kd = 0.6 μM, Table 1). A crystal structure of pirin complexed with TPh A revealed that the compound binds in a cavity between two beta-barrel domains, next to a Fe2+ -binding site. Subsequent GST pull-down assays using GST-Bcl3 and His6-pirin showed that TPh A reduced the amount of pirin-bound Bcl3. Thus TPh A inhibits binding of pirin to Bcl3. The authors next showed that treatment of melanoma cells with TPh A inhibited cell migration, and also caused a downregulation SNA12 expression, a gene implicated in cancer cell motility. Identifying a high-affinity inhibitor of pirin enabled study of the protein's role in cell signaling and melanoma cell migration.
Novel screening strategies
Ribonucleic acids (RNAs) play vital roles in transcription, protein synthesis and biocatalysis. The discovery of small-molecule probes that inhibit specific RNA function has wide implications for understanding fundamental biology or validating RNAs as therapeutic targets. Disney and coworkers developed a novel SMM-based screen that enables targeting RNAs [36]. A designed peptoid library was screened for binding to the C. albicans group I ribozyme [37]. The group I intron is essential for the assembly of active ribosomes since the intron is embedded in the large subunit ribosomal RNA precursor. They immobilized 109 peptoids on an agarose-coated microarray and incubated it with a library of RNA internal loops, representing the RNA secondary structure. Twelve hits identified from the SMM inhibited self-splicing and subsequently led to rational inhibitor design. Velagapudi et al. used SMMs to demonstrate interactions between series of benzimidazole compounds and RNA internal loops. Benzimidazole was previously identified as a pharmacophore that competes with aminoglycosides for binding in the bacterial rRNA A-site. These studies identified diverse elements that impart affinity and specificity for binding RNA internal loops [38,39]. 2-aminobenzimidazole I (15-AB-21) displayed mid-micromolar Kds using a fluorescence-based assay against single RNA motifs in solution (Table 1). Understanding which small molecules bind RNA secondary structure using data generated through simple binding assays will advance the future of rational design.
Finally, Lee et al. developed an SMM platform to identify small molecules that bind to the surface of live pathogenic bacteria [40]. Small molecules were first immobilized on amine-reactive functionalized microarray slide with PEG-linker. After incubation with fluorophore-stained live bacterial cells, they scored the affinity and specificity of the small molecules to 4 different bacterial stains with different wavelength fluorophores for 4-channel imaging. This study describes a novel platform to develop new classes of antibiotics and provides proof-of-principle for applying live cells to SMMs.
Functional proteomics
Human proteomic projects have identified over 200,000 proteins and those proteins participate in virtually all cell functions through biomolecular interactions [41]. Traditionally, proteomics has relied on electrophoresis [42], liquid chromatography [43] and mass spectrometry [44], which allow quantitative identification and characterization of protein structure and function. However, proteins are regulated at many different levels besides expression, including posttranslational modifications, protein-protein interactions and cellular networking. This complexity requires analysis of a large number of samples to detect patterns that are consistently associated with a specific biological state. Thus it is increasingly essential for proteomic experiments to be carried out in a high-throughput fashion. The SMM format minimizes reagent consumption and enable highly parallel experiments for various proteomic applications, including protein expression profiling, molecular interaction mapping, biomarker and drug discovery [45]. A subset of recent proteomic examples is covered herein.
Reactivity profiling
Activity-based protein profiling (ABPP) utilizes active site-directed probes to determine the functional state of enzymes in complex proteomes. Shi and coworkers created an SMM platform that was used to identify affinity-based probes (AfBPs) against γ-secretase, an aspartic protease implicated in the pathology of Alzheimer's disease [46]. Fluorescently labeled cell lysates overexpressing γ-secretase were applied to an SMM containing 198 hydroxylethylene-based small molecule inhibitors. Several of the strongest binders were converted to AfBPs using Cu-catalyzed “click chemistry” between azido intermediates and TER-BP or biotin alkynes. AfBPs of γ-secretase were subsequently confirmed by in-gel fluorescence labeling and pull-down assays using cell lysates (Table 1). The team added 86 compounds to their library of hydroxyethylamine-derived inhibitors, and screened them against 8 different lysates from human cell lines [47]. Eleven binders were converted into fluorescent AfBPs and protein targets were detected using pull-down assays coupled to mass spectrometric analysis. Putative protein targets included well-known aspartic proteases implicated in disease, such as cathepsin D. These experiments enabled quick identification of AfBPs appropriate for profiling of aspartic proteases and other potential biomarkers in mammalian proteomes. AfBPs have the potential to bind previously undiscovered proteins and point researchers toward novel disease biomarkers. More recently, they have successfully demonstrated large-scale functional profiling of cysteine proteases present in native apoptotic biological samples, including fluorescence-labeled cellular lysates and parasite-infected red blood cells (RBCs) [48].
In another example involving chemically labeled peptides, Gurard-Levin et al. measured endogenous cellular lysine deacetylase (KDAC) activity and substrate specificity [49]. A library of 361 hexapeptides was synthesized with acetylated lysine and various amino acids at the X and Z positions for the consensus peptide GRKACXZC. The peptides were immobilized onto a maleimide-terminated monolayer on a gold planar substrate then treated with either purified KDAC enzymes or nuclear extracts. Deacetylation was detected using SAMDI (self-assembled monolayers for matrix assisted laser desorption/ionization time-offlight mass spectrometry) with the appearance of peak corresponding to a 42 m/z shift. From the various lysine deacetylases, KDAC3 activity corresponded to the altered acetylation state of histone H4. An SMM followed by SAMDI was also used to characterize novel glycosyltransferase activity. These SMM screens were performed by applying glycosyltransferase and sugar donors onto the microarrays presenting carbohydrate acceptors. From the 57,120 reactions tested in the screen, 44 had new glycosylation products. This method provided a label-free method for the rapid functional annotation of putative enzymes.
Biomarker discovery
The availability of systematic and general high-throughput platforms for measuring changes across cellular states is critical to understanding unique disease characteristics at the proteomic level. Reddy et al. compared serum samples from six patients with Alzheimer's disease (AD) [50]. The samples were incubated on microarrays comprised of 15,000 structurally novel peptoids, and IgG binding patterns were visualized using secondary antibody. The AD peptoids 1-3 were identified from the screen, and further characterization revealed at least two candidate auto-antibody biomarkers for AD. In another example involving peptides, Dai and colleagues developed a new peptide-antigen microarray format on a plasmonic gold substrate capable of enhanced NIR florescence and background minimization [51]. A model histone peptide microarray afforded ~ 100-fold enhancement in NIR fluorescence and exhibited three orders of magnitude increased sensitivity relative to streptavidin-glass beaded arrays. The gold-coated platform provides a novel method of profiling antibodies in human samples and identifying peptides or peptoids against low abundance or affinity antibodies. This platform may be compatible with small molecules in the future.
Conclusion
This review covers only a subset of recent probe discovery efforts and novel applications involving SMMs. Based on the results from the last decade, SMMs and other unbiased binding assay format should prove useful in probe discovery for new proteins and other types of biomolecules associated with disease-states. This is particularly the case for targets of unknown function and are not compatible with functional assay development or proteins that lack structure or function in isolation and may require screens from cellular lysates. The SMM format goes beyond simple ligand discovery applications as demonstrated by arrays used for reactivity profiling. Finally, many of the exciting new profiling applications in development or use with other types of arrays (e.g. peptide and antibody arrays) may be translatable to arrays printed with small molecule content in the future.
Highlights.
We review selected recent probe discoveries and novel applications using SMMs.
SMM screening can use purified protein and proteins from cellular lysates.
Both strategies have yielded effective probes and inhibitors for a wide variety of targets.
SMMs enables functional proteomics- biomarker discovery and reactivity profiling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacBeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. Journal of the American Chemical Society. 1999;121:7967–7968. [Google Scholar]
- 3.Foong YM, Fu J, Yao SQ, Uttamchandani M. Current advances in peptide and small molecule microarray technologies. Current opinion in chemical biology. 2012;16:234–242. doi: 10.1016/j.cbpa.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Shi H, Uttamchandani M, Yao SQ. A method for small molecule microarray-based screening for the rapid discovery of affinity-based probes. Methods in molecular biology. 2010;669:57–68. doi: 10.1007/978-1-60761-845-4_5. [DOI] [PubMed] [Google Scholar]
- 5.Uttamchandani M, Yao SQ. The expanding world of small molecule microarrays. Methods in molecular biology. 2010;669:1–15. doi: 10.1007/978-1-60761-845-4_1. [DOI] [PubMed] [Google Scholar]
- 6.Walsh DP, Chang YT. Recent advances in small molecule microarrays: applications and technology. Combinatorial chemistry & high throughput screening. 2004;7:557–564. doi: 10.2174/1386207043328427. [DOI] [PubMed] [Google Scholar]
- 7.Casalena DE, Wassaf D, Koehler AN. Ligand discovery using small-molecule microarrays. Methods in molecular biology. 2012;803:249–263. doi: 10.1007/978-1-61779-364-6_17. [DOI] [PubMed] [Google Scholar]
- 8.Lee HY, Park SB. Small molecule microarray: functional-group specific immobilization of small molecules. Methods in molecular biology. 2010;669:23–42. doi: 10.1007/978-1-60761-845-4_3. [DOI] [PubMed] [Google Scholar]
- 9.Vegas AJ, Fuller JH, Koehler AN. Small-molecule microarrays as tools in ligand discovery. Chemical Society reviews. 2008;37:1385–1394. doi: 10.1039/b703568n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H, Ge J, Uttamchandani M, Yao SQ. Small molecule microarrays: the first decade and beyond. Chemical communications. 2011;47:5664–5670. doi: 10.1039/c1cc11464f. [DOI] [PubMed] [Google Scholar]
- 11.Duffner JL, Clemons PA, Koehler AN. A pipeline for ligand discovery using small-molecule microarrays. Current opinion in chemical biology. 2007;11:74–82. doi: 10.1016/j.cbpa.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 12.Bradner JE, McPherson OM, Koehler AN. A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature protocols. 2006;1:2344–2352. doi: 10.1038/nprot.2006.282. [DOI] [PubMed] [Google Scholar]
- 13.Kanoh N, Kyo M, Inamori K, Ando A, Asami A, Nakao A, Osada H. SPR imaging of photo-cross-linked small-molecule arrays on gold. Analytical chemistry. 2006;78:2226–2230. doi: 10.1021/ac051777j. [DOI] [PubMed] [Google Scholar]
- 14.Bradner JE, McPherson OM, Mazitschek R, Barnes-Seeman D, Shen JP, Dhaliwal J, Stevenson KE, Duffner JL, Park SB, Neuberg DS, et al. A robust small-molecule microarray platform for screening cell lysates. Chemistry & biology. 2006;13:493–504. doi: 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 15••.Miyazaki I, Simizu S, Okumura H, Takagi S, Osada H. A small-molecule inhibitor shows that pirin regulates migration of melanoma cells. Nature chemical biology. 2010;6:667–673. doi: 10.1038/nchembio.423. [This is the first example of a cellular lysate SMM being used to profile a protein target with unknown function, culminating in a crystal structure and new knowledge about the protein's cellular role.] [DOI] [PubMed] [Google Scholar]
- 16.Miao H, Tallarico JA, Hayakawa H, Munger K, Duffner JL, Koehler AN, Schreiber SL, Lewis TA. Ring-opening and ring-closing reactions of a shikimic acid-derived substrate leading to diverse small molecules. Journal of combinatorial chemistry. 2007;9:245–253. doi: 10.1021/cc060135m. [DOI] [PubMed] [Google Scholar]
- 17.Kwon SJ, Lee MY, Ku B, Sherman DH, Dordick JS. High-throughput, microarray-based synthesis of natural product analogues via in vitro metabolic pathway construction. ACS chemical biology. 2007;2:419–425. doi: 10.1021/cb700033s. [DOI] [PubMed] [Google Scholar]
- 18.Harris J, Mason DE, Li J, Burdick KW, Backes BJ, Chen T, Shipway A, Van Heeke G, Gough L, Ghaemmaghami A, et al. Activity profile of dust mite allergen extract using substrate libraries and functional proteomic microarrays. Chemistry & biology. 2004;11:1361–1372. doi: 10.1016/j.chembiol.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Urbina HD, Debaene F, Jost B, Bole-Feysot C, Mason DE, Kuzmic P, Harris JL, Winssinger N. Self-assembled small-molecule microarrays for protease screening and profiling. Chembiochem : a European journal of chemical biology. 2006;7:1790–1797. doi: 10.1002/cbic.200600242. [DOI] [PubMed] [Google Scholar]
- 20.Vegas AJ, Bradner JE, Tang W, McPherson OM, Greenberg EF, Koehler AN, Schreiber SL. Fluorous-based small-molecule microarrays for the discovery of histone deacetylase inhibitors. Angewandte Chemie. 2007;46:7960–7964. doi: 10.1002/anie.200703198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemp MM, Weiwer M, Koehler AN. Unbiased binding assays for discovering small-molecule probes and drugs. Bioorganic & medicinal chemistry. 2012;20:1979–1989. doi: 10.1016/j.bmc.2011.11.071. [DOI] [PubMed] [Google Scholar]
- 22.Koehler AN, Shamji AF, Schreiber SL. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. Journal of the American Chemical Society. 2003;125:8420–8421. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 23.Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 24••.Landry JP, Fei Y, Zhu X, Ke Y, Yu G, Lee P. Discovering small molecule ligands of vascular endothelial growth factor that block VEGF-KDR binding using label-free microarray-based assays. Assay and drug development technologies. 2013;11:326–332. doi: 10.1089/adt.2012.485. [The team developed a three part SMM-based screen to detect compounds that interrupt the interaction between an extracellular protein (VEGF) and its receptor (KDR).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Stanton BZ, Peng LF, Maloof N, Nakai K, Wang X, Duffner JL, Taveras KM, Hyman JM, Lee SW, Koehler AN, et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nature chemical biology. 2009;5:154–156. doi: 10.1038/nchembio.142. [The authors used a purified protein SMM to develop robotnikinin, a small molecule that binds Shh and decreases downstream activity in a synthetic model of human skin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clemons PA, Bodycombe NE, Carrinski HA, Wilson JA, Shamji AF, Wagner BK, Koehler AN, Schreiber SL. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemons PA, Wilson JA, Dancik V, Muller S, Carrinski HA, Wagner BK, Koehler AN, Schreiber SL. Quantifying structure and performance diversity for sets of small molecules comprising small-molecule screening collections. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6817–6822. doi: 10.1073/pnas.1015024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes-Seeman D, Park SB, Koehler AN, Schreiber SL. Expanding the functional group compatibility of small-molecule microarrays: discovery of novel calmodulin ligands. Angewandte Chemie. 2003;42:2376–2379. doi: 10.1002/anie.200351043. [DOI] [PubMed] [Google Scholar]
- 29.Lee HY, Park SB. Surface modification for small-molecule microarrays and its application to the discovery of a tyrosinase inhibitor. Molecular bioSystems. 2011;7:304–310. doi: 10.1039/c0mb00122h. [DOI] [PubMed] [Google Scholar]
- 30.Seo SY, Sharma VK, Sharma N. Mushroom tyrosinase: recent prospects. Journal of agricultural and food chemistry. 2003;51:2837–2853. doi: 10.1021/jf020826f. [DOI] [PubMed] [Google Scholar]
- 31.Muller YA, Li B, Christinger HW, Wells JA, Cunningham BC, de Vos AM. Vascular endothelial growth factor: crystal structure and functional mapping of the kinase domain receptor binding site. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7192–7197. doi: 10.1073/pnas.94.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & development. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 33.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nature reviews. Drug discovery. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 34•.Chen J, Armstrong AH, Koehler AN, Hecht MH. Small molecule microarrays enable the discovery of compounds that bind the Alzheimer's Abeta peptide and reduce its cytotoxicity. Journal of the American Chemical Society. 2010;132:17015–17022. doi: 10.1021/ja107552s. [The authors used a purified protein SMM to identify binders of Aβ monomer. These compounds are potentially effective therapies for Alzheimer's disease.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakob-Roetne R, Jacobsen H. Alzheimer's disease: from pathology to therapeutic approaches. Angewandte Chemie. 2009;48:3030–3059. doi: 10.1002/anie.200802808. [DOI] [PubMed] [Google Scholar]
- 36.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS chemical biology. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 37.Labuda LP, Pushechnikov A, Disney MD. Small molecule microarrays of RNA-focused peptoids help identify inhibitors of a pathogenic group I intron. ACS chemical biology. 2009;4:299–307. doi: 10.1021/cb800313m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velagapudi SP, Pushechnikov A, Labuda LP, French JM, Disney MD. Probing a 2-aminobenzimidazole library for binding to RNA internal loops via two-dimensional combinatorial screening. ACS chemical biology. 2012;7:1902–1909. doi: 10.1021/cb300213g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velagapudi SP, Seedhouse SJ, French J, Disney MD. Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. Journal of the American Chemical Society. 2011;133:10111–10118. doi: 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, Park S, Hyun H, Bordo MW, Oketokoun R, Nasr KA, Frangioni JV, Choi HS. High-throughput screening of small molecule ligands targeted to live bacteria surface. Analytical chemistry. 2013;85:3508–3514. doi: 10.1021/ac303199x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES. Distinguishing protein-coding and noncoding genes in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3–10. [PubMed] [Google Scholar]
- 43.Xie F, Smith RD, Shen Y. Advanced proteomic liquid chromatography. Journal of chromatography. A. 2012;1261:78–90. doi: 10.1016/j.chroma.2012.06.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annual review of biochemistry. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 45.Sun H, Chen GY, Yao SQ. Recent advances in microarray technologies for proteomics. Chemistry & biology. 2013;20:685–699. doi: 10.1016/j.chembiol.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Shi H, Liu K, Xu A, Yao SQ. Small molecule microarray-facilitated screening of affinity-based probes (AfBPs) for gamma-secretase. Chemical communications. 2009:5030–5032. doi: 10.1039/b910611a. [DOI] [PubMed] [Google Scholar]
- 47.Shi H, Uttamchandani M, Yao SQ. Applying small molecule microarrays and resulting affinity probe cocktails for proteome profiling of mammalian cell lysates. Chemistry, an Asian journal. 2011;6:2803–2815. doi: 10.1002/asia.201100523. [DOI] [PubMed] [Google Scholar]
- 48.Wu H, Ge J, Yang PY, Wang J, Uttamchandani M, Yao SQ. A peptide aldehyde microarray for high-throughput profiling of cellular events. Journal of the American Chemical Society. 2011;133:1946–1954. doi: 10.1021/ja109597v. [DOI] [PubMed] [Google Scholar]
- 49.Gurard-Levin ZA, Kilian KA, Kim J, Bahr K, Mrksich M. Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity. ACS chemical biology. 2010;5:863–873. doi: 10.1021/cb100088g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, German D, Kodadek T. Identification of candidate IgG biomarkers for Alzheimer's disease via combinatorial library screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [The team discloses a method for profiling synthetic molecules against serum samples from case samples and controls to identify candidate biomarkers for Alzheimer's disease. This approach should prove general for biomarker discovery campaigns involving other diseases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang B, Jarrell JA, Price JV, Tabakman SM, Li Y, Gong M, Hong G, Feng J, Utz PJ, Dai H. An integrated Peptide-antigen microarray on plasmonic gold films for sensitive human antibody profiling. PloS one. 2013;8:e71043. doi: 10.1371/journal.pone.0071043. [DOI] [PMC free article] [PubMed] [Google Scholar]