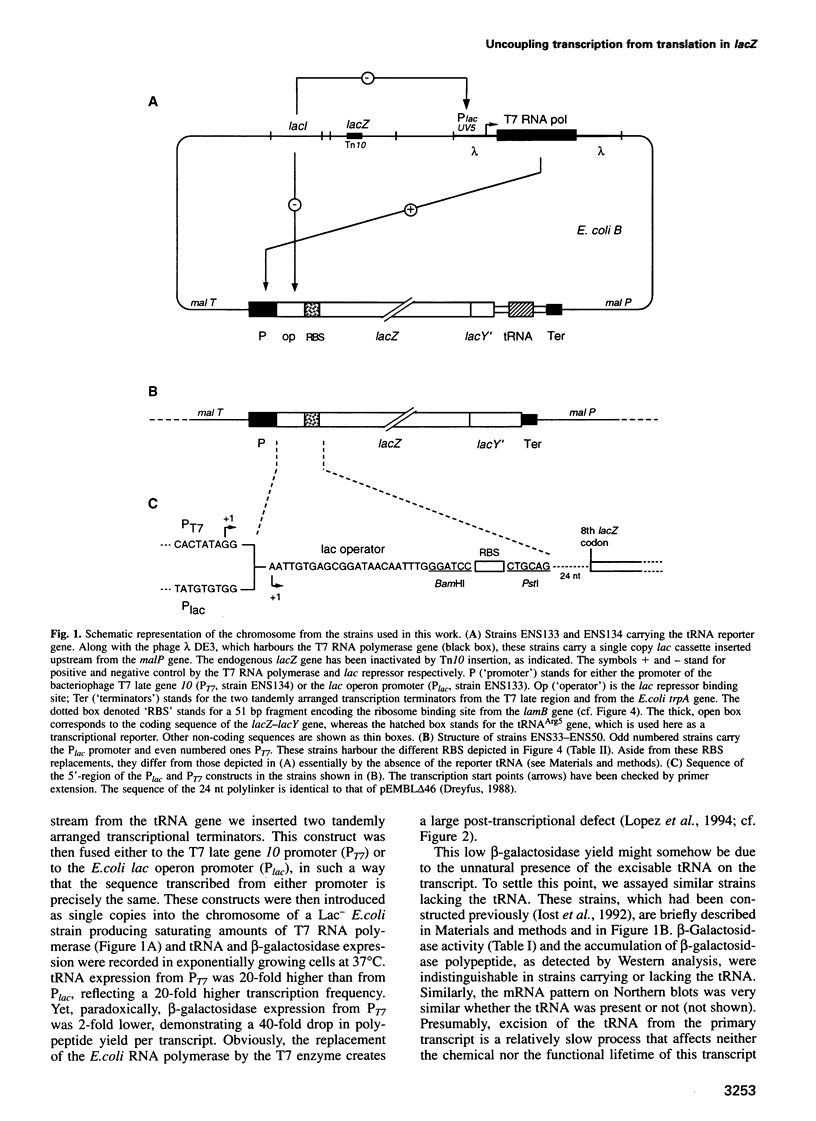
Abstract
We have used either Escherichia coli or T7 RNA polymerase to transcribe in E. coli a series of lacZ genes that differ in the nature of their ribosome binding sites (RBS). Each T7 RNA polymerase transcript yields from 15- to 450-fold less beta-galactosidase than its E. coli polymerase counterpart, the ratio being larger when weaker RBS are used. The low beta-galactosidase yield from T7 transcripts reflects their low stability: the ams-1/rne-50 mutation, which inactivates RNase E, nearly equalizes the beta-galactosidase yields from T7 and E. coli RNA polymerase transcripts. T7 RNA polymerase transcribes the lacZ gene approximately 8-fold faster than the E. coli enzyme. We propose that this higher speed unmasks an RNase E cleavage site which is normally shielded by ribosomes soon after its synthesis when the slower E. coli enzyme is used. This leads to degradation of the T7 transcript, unless the leading ribosome comes in time to shield the cleavage site: the weaker the RBS, the lower this probability and the more severe the inability of T7 RNA polymerase transcripts for beta-galactosidase synthesis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Babitzke P., Kushner S. R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Dubnau D. Induced mRNA stability in Bacillus subtilis. Proc Natl Acad Sci U S A. 1987 Jan;84(2):498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Zen K. H. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):5803–5811. doi: 10.1128/jb.171.11.5803-5811.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Bouvet P., Belasco J. G. Control of RNase E-mediated RNA degradation by 5'-terminal base pairing in E. coli. Nature. 1992 Dec 3;360(6403):488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994 Mar 11;76(5):889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Chevrier-Miller M., Jacques N., Raibaud O., Dreyfus M. Transcription of single-copy hybrid lacZ genes by T7 RNA polymerase in Escherichia coli: mRNA synthesis and degradation can be uncoupled from translation. Nucleic Acids Res. 1990 Oct 11;18(19):5787–5792. doi: 10.1093/nar/18.19.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G. E., Farnham P. J., Platt T. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4180–4184. doi: 10.1073/pnas.78.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R., Nomura M. Changes in the half-life of ribosomal protein messenger RNA caused by translational repression. J Mol Biol. 1986 Apr 5;188(3):383–392. doi: 10.1016/0022-2836(86)90162-2. [DOI] [PubMed] [Google Scholar]
- Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol. 1988 Nov 5;204(1):79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. Purification, structure, and properties of hybrid beta-galactosidase proteins. J Biol Chem. 1983 Dec 10;258(23):14354–14358. [PubMed] [Google Scholar]
- Guillerez J., Gazeau M., Dreyfus M. In the Escherichia coli lacZ gene the spacing between the translating ribosomes is insensitive to the efficiency of translation initiation. Nucleic Acids Res. 1991 Dec 25;19(24):6743–6750. doi: 10.1093/nar/19.24.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S., Schlessinger D. Coupling of rates of transcription, translation, and messenger ribonucleic acid degradation in streptomycin-dependent mutants of Escherichia coli. J Bacteriol. 1976 Jan;125(1):84–93. doi: 10.1128/jb.125.1.84-93.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Iost I., Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994 Nov 10;372(6502):193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- Iost I., Guillerez J., Dreyfus M. Bacteriophage T7 RNA polymerase travels far ahead of ribosomes in vivo. J Bacteriol. 1992 Jan;174(2):619–622. doi: 10.1128/jb.174.2.619-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques N., Guillerez J., Dreyfus M. Culture conditions differentially affect the translation of individual Escherichia coli mRNAs. J Mol Biol. 1992 Aug 5;226(3):597–608. doi: 10.1016/0022-2836(92)90618-t. [DOI] [PubMed] [Google Scholar]
- Jacquet M., Kepes A. Initiation, elongation and inactivation of lac messenger RNA in Escherichia coli studied studied by measurement of its beta-galactosidase synthesizing capacity in vivo. J Mol Biol. 1971 Sep 28;60(3):453–472. doi: 10.1016/0022-2836(71)90181-1. [DOI] [PubMed] [Google Scholar]
- Jensen K. F., Pedersen S. Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol Rev. 1990 Jun;54(2):89–100. doi: 10.1128/mr.54.2.89-100.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug G., Cohen S. N. Effects of translation on degradation of mRNA segments transcribed from the polycistronic puf operon of Rhodobacter capsulatus. J Bacteriol. 1991 Feb;173(4):1478–1484. doi: 10.1128/jb.173.4.1478-1484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landick R., Carey J., Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicki B. T., Margus T., Remme J., Nierhaus K. H. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993 Jun 5;231(3):581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- Lopez P. J., Iost I., Dreyfus M. The use of a tRNA as a transcriptional reporter: the T7 late promoter is extremely efficient in Escherichia coli but its transcripts are poorly expressed. Nucleic Acids Res. 1994 Apr 11;22(7):1186–1193. doi: 10.1093/nar/22.7.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J. R., Zengel J. M., Lindahl L. Intermediates in the degradation of mRNA from the lactose operon of Escherichia coli. Nucleic Acids Res. 1991 May 25;19(10):2767–2776. doi: 10.1093/nar/19.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol Microbiol. 1991 Apr;5(4):857–864. doi: 10.1111/j.1365-2958.1991.tb00759.x. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr, Hamkalo B. A., Thomas C. A., Jr Visualization of bacterial genes in action. Science. 1970 Jul 24;169(3943):392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Yanofsky C. Polarity and the degradation of mRNA. Nature. 1969 Oct 25;224(5217):329–331. doi: 10.1038/224329a0. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Krisch H. M., Higgins C. F. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990 Dec;4(12):2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Murakawa G. J., Kwan C., Yamashita J., Nierlich D. P. Transcription and decay of the lac messenger: role of an intergenic terminator. J Bacteriol. 1991 Jan;173(1):28–36. doi: 10.1128/jb.173.1.28-36.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins P. O., Rangwala S. H. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. J Biol Chem. 1989 Oct 15;264(29):16973–16976. [PubMed] [Google Scholar]
- Ono M., Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J Mol Biol. 1979 Apr 15;129(3):343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991 Mar 22;64(6):1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- Robertson E. S., Aggison L. A., Nicholson A. W. Phosphorylation of elongation factor G and ribosomal protein S6 in bacteriophage T7-infected Escherichia coli. Mol Microbiol. 1994 Mar;11(6):1045–1057. doi: 10.1111/j.1365-2958.1994.tb00382.x. [DOI] [PubMed] [Google Scholar]
- Robertson E. S., Nicholson A. W. Phosphorylation of Escherichia coli translation initiation factors by the bacteriophage T7 protein kinase. Biochemistry. 1992 May 26;31(20):4822–4827. doi: 10.1021/bi00135a012. [DOI] [PubMed] [Google Scholar]
- Rosa M. D. Structure analysis of three T7 late mRNA ribosome binding sites. J Mol Biol. 1981 Mar 25;147(1):55–71. doi: 10.1016/0022-2836(81)90079-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Stanssens P., Remaut E., Fiers W. Inefficient translation initiation causes premature transcription termination in the lacZ gene. Cell. 1986 Mar 14;44(5):711–718. doi: 10.1016/0092-8674(86)90837-8. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Stent G. S. Genetic transcription. Proc R Soc Lond B Biol Sci. 1966 Mar 22;164(995):181–197. doi: 10.1098/rspb.1966.0022. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Bacteriophage T7. Science. 1972 Apr 28;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Summers W. C. The process of infection with coliphage T7. IV. Stability of RNA in bacteriophage-infected cells. J Mol Biol. 1970 Aug;51(3):671–678. doi: 10.1016/0022-2836(70)90015-x. [DOI] [PubMed] [Google Scholar]
- Sørensen M. A., Jensen K. F., Pedersen S. High concentrations of ppGpp decrease the RNA chain growth rate. Implications for protein synthesis and translational fidelity during amino acid starvation in Escherichia coli. J Mol Biol. 1994 Feb 18;236(2):441–454. doi: 10.1006/jmbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraseviciene L., Miczak A., Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991 Apr;5(4):851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- Uzan M., Favre R., Brody E. A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8895–8899. doi: 10.1073/pnas.85.23.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U., Jensen K. F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol. 1994 May;176(10):2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U., Sørensen M., Pedersen S., Jensen K. F., Kilstrup M. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol Microbiol. 1992 Aug;6(15):2191–2200. doi: 10.1111/j.1365-2958.1992.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Wagner L. A., Gesteland R. F., Dayhuff T. J., Weiss R. B. An efficient Shine-Dalgarno sequence but not translation is necessary for lacZ mRNA stability in Escherichia coli. J Bacteriol. 1994 Mar;176(6):1683–1688. doi: 10.1128/jb.176.6.1683-1688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchuk O., Jacques N., Guillerez J., Dreyfus M. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J Mol Biol. 1992 Aug 5;226(3):581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]