Abstract
The transcriptional repressor BLIMP1 is a master regulator of B and T cell differentiation. To examine the role of BLIMP1 in innate immunity we used a conditional knockout (CKO) of Blimp1 in myeloid cells and found that Blimp1 CKO mice were protected from lethal infection induced by Listeria monocytogenes. Transcriptome analysis of Blimp1 CKO macrophages identified the murine chemokine (C-C motif) ligand 8, CCL8 as a direct target of Blimp1-mediated transcriptional repression in these cells. BLIMP1-deficient macrophages expressed elevated levels of Ccl8 and consequently Blimp1 CKO mice had higher levels of circulating CCL8 resulting in increased neutrophils in the peripheral blood, promoting a more aggressive anti-bacterial response. Mice lacking the Ccl8 gene were more susceptible to L. monocytogenes infection than wild type mice. While CCL8 failed to recruit neutrophils directly, it was chemotactic for γ/δ T cells and CCL8-responsive γ/δ T cells were enriched for IL-17F. Finally, CCL8-mediated enhanced clearance of L. monocytogenes was dependent on γ/δ T cells. Collectively, these data reveal an important role for BLIMP1 in modulating host-defenses by suppressing expression of the chemokine CCL8.
Introduction
B-lymphocyte induced maturation protein 1 (BLIMP1) is a transcriptional repressor critical for early embryonic development in multiple species (1–3). BLIMP1 was first identified as a factor, which bound to the positive regulatory domains I and III of the Interferon-β (IFN-β) enhancer, where it was shown to compete with Interferon regulatory factors in order to attenuate IFN-β gene transcription (4, 5). BLIMP1 is best studied, however, in the context of terminal differentiation of B lymphocytes to plasma cells (6–8) and in the differentiation, homeostasis and/or function of both CD4 and CD8 T cells (9, 10). It also plays a central role in the differentiation and function of keratinocytes (11) and sebocytes (12) and in the tolerogenic function of DCs (13). The role of BLIMP1 in macrophages and anti-microbial defenses however has not been examined in detail.
Macrophages are one of the major mediators of inflammation and critical effector cells of innate immunity. They serve as host cells for intracellular pathogens including Listeria monocytogenes (L. monocytogenes) (14). L. monocytogenes is a Gram-positive facultative intracellular bacterium, which causes disease in humans as a result of ingestion of contaminated foods. L. monocytogenes infections are generally limited; however lethal infections can occur in immunocompromised individuals, pregnant women and neonates. Upon ingestion, bacteria invade the intestinal epithelium, enter the draining lymph node and disseminate via the bloodstream to the liver and spleen (15). This rapid clearance of bacteria from the peripheral blood is generally attributed to the resident liver macrophages; the Kupffer cells which line the liver sinusoids, and to the hepatocytes. Within the first 24 hours, a coordinated interaction between neutrophils, γ/δ T cells, monocytes and NK cells is instrumental in preventing the bacterium from spreading and ensuring the survival of the host. L. monocytogenes is an excellent model system to evaluate the role of innate immunity in anti-microbial defenses. Several classes of germ-line encoded pattern recognition receptors (PRRs) have been implicated in the recognition of L. monocytogenes (14, 16). Toll-like Receptor (TLR)-2 plays a key role in bacterial recognition at the cell surface (17–19). Upon escape of intracellular bacteria into the cytosol, L. monocytogenes engages nucleotide-binding oligomerization domain 2 (NOD2), members of the NACHT-, LRR- and pyrin-domain- containing protein family, as well as the cytosolic DNA sensor Absent In Melanoma 2 (AIM2) all of which play a role in the recognition of intracellular L. monocytogenes and release of IL1–β through the activation of caspase-1 (20–23). Additionally, L. monocytogenes triggers type I IFN gene transcription via Stimulator of Interferon Genes (STING) (24–28), TANK-binding kinase 1 (TBK1) and IRF3 (29–31). Bacterial DNA and the second messenger cyclic-di-AMP represent the L. monocytogenes products driving these responses (31–33).
In this study, we examined the role of BLIMP1 in regulating the innate immune response using L. monocytogenes as a model system. We found that Blimp1 gene expression was rapidly induced by this bacterium in macrophages as well as by a range of viral and bacterial pathogens. Since BLIMP1 deficient mice are embryonic lethal (6) we evaluated the role of BLIMP1 in innate immune defenses using a myeloid-specific Blimp1 conditional knockout mouse (Blimp1 CKO). Blimp1 CKO mice were less susceptible to L. monocytogenes infection than littermate controls. We focused this study on the role of BLIMP1 in macrophages. Blimp1 deletion in macrophages had no effect on type I IFN gene transcription but did affect transcription of several other genes including the murine chemokine (C-C motif) ligand 8, CCL8, also called monocyte chemoattractant protein 2 (MCP2). Chromatin Immunoprecipitation (ChIP) assays demonstrated that Ccl8 was a direct BLIMP1 target gene. Increased levels of CCL8 led to higher numbers of circulating neutrophils in the peripheral blood, promoting a more rapid and robust anti-bacterial response resulting in a better clearance of the pathogen. Mobilization of neutrophils was mediated by CCL8-induced recruitment of IL17F-producing γ/δ T cells. Mice lacking Ccl8 were also more sensitive to L. monocytogenes infection than wild type mice.
Our study identifies a new regulatory mechanism in macrophages mediated by BLIMP1 and CCL8 expression in early host defenses against intracellular bacteria. BLIMP1 likely acts as a gatekeeper of initial inflammatory responses following infection to prevent detrimental effects associated with excess inflammation at the level of target tissues.
Materials and Methods
Reagents and bugs
L. monocytogenes (clinical isolate 10403s) and its derivative mutant LLO, S. aureus (ATCC 25923), E. coli (ATCC 27325 (W3110) were used at MOI 5. Sendai virus, (Cantrell strain) was from Charles River Laboratories (Boston, MA) and was used at 300 HAU/ml. Recombinant IL1-β, Murine CCL8, CCL2 and CXCL2 were from Peprotech (Rocky Hill, NJ). IL1-β was used at 100ng/ml. The IL1R antagonist Kineret (or anakinra, Amgen.com) was used at 150μl/ml, a dose, which inhibits 100% of the IL-1 activity when used at 100ng/ml. The inhibitors SB203580 and MG132, both used at 40μM, were from Calbiochem (San Diego, CA).
Mice
Prdm1flox/floxmice were generously provided by K. Calame (Columbia University, NY)[3] and were crossed with LysMCre mice (Jackson Laboratories, Bar Harbor, ME) to generate Prdm1flox/floxLysMCreand Prdm1flox/floxLysMCre+ mice. C57BL/6 mice and IL1R−/− were purchased from Jackson Laboratories. TLR2−/− and MyD88−/− mice were from S. Akira (Osaka University, Osaka, Japan). IRF3−/− mice were from T. Taniguchi, (University of Tokyo, Tokyo, Japan). RIP2−/− mice were from Genentech (San Fransisco, CA). Ccl8/Ccl12 and Ccl12 KO, were from I. Charo (Gladstone Institute of Cardiovascular Disease, University of California, San Francisco, CA). Seven-to-eight-week-old animals were used in all experiments. All mouse strains were bred and maintained under specific pathogen-free conditions in the animal facilities at the University of Massachussetts Medical School. All experiments involving live animals were carried out in accordance with the guidelines set forth by the University of Massachussetts Medical School Department of Animal Medicine and the Institute Animal Care and Use Committee. γ/δ T cell deficient mice were from Jackson laboratories (Bar Harbor, ME) and CCR8−/− mice were from S. Lira (Mount Sinai School of Medicine, New York, NY).
Cell isolation
Bone marrow derived macrophages were generated as described [53]. Peritoneal macrophages were obtained from mice by washing their peritoneal cavities with 5 ml ice-cold PBS after 3 days from i.p. injection with 3 ml of thyoglicolate (Remel, Inc., Lenexa, KS). Splenocytes were isolated by disrupting aseptically harvested spleens in RPMI 10% FCS using a 70-mm pore size mesh cell strainer (BD Biosciences) to obtain single-cell suspensions. Whole blood samples were obtained by cardiac puncture using 1-ml syringes containing EDTA and from tails in sterile 5 ml polypropylene tubes containing PBS with 10mM EDTA. Lysis of red blood cells was performed using Red blood cell lysis buffer (Sigma-Aldrick, St. Louis, MI). Neutrophils were isolated from peritoneal cavity (thioglycollate 4 hrs) or from peripheral blood by negative selection (Miltenyi).
Isolation and chemotaxis of bone marrow neutrophils and γδ T cells
Freshly harvested bone marrow leukocytes from C57Bl/6 mice were first stained with PE-conjugated Ly6G antibody (BD Biosciences), and neutrophils were then immunomagnetically isolated using EasySep® PE selection kits (Stem Cell Technologies). Purity was assessed by flow cytometry (FACSCalibur). Pooled γδ T cells were purified from splenocytes of age matched wild-type C57BL/6 and CCR8−/− mice. Splenocytes were labeled with PE conjugated Hamster Anti-Mouse γδ T cell receptor antibody (UC7-13D5, BD Biosciences) after pre-incubation with anti-mouse CD16/CD32 (BD Pharmingen), and then magnetically purified with anti-PE-antibody (Miltenyi Biotec) coated microbeads. Isolated γδ T cells were rested in 0.5% FCS RPMI overnight prior to assays. Neutrophils (1 × 105 per well) and γδ T cells (2.5 × 104 per well) in RPMI containing BSA were respectively placed on the top of a 3-μm and a 5-μm pore size 96 well ChemoTx chemotaxis apparatus (Neuroprobe) in chemotaxis assays. Neutrophils and γδ T cells were incubated for 1 h and 2h respectively at 37°C. In the neutrophil assays, all non-migrated cells were manually removed from the upper side of the filter, while the migrated cells remained adherent to the lower side of the filter. The filters were stained with Hema 3 (Fisher Scientific) and mounted on glass slides, and cell counts were determined by manual counting using a light microscope. In the γδ T cell assays, cell counts were determined in the lower wells by manual counting using an inverted microscope. Chemokines were placed in the lower wells of the chemotaxis apparatus in triplicate in all assays. The number of cells migrating at each concentration of chemokine was normalized to the number of cells migrating in the presence of media alone (chemokinesis) to calculate chemotactic index.
Flow cytometry analysis
The differentiation state of BMDM and peritoneal macrophages was confirmed by staining for F4/80 and CD11b (BD Pharmingen, San Jose, CA) using monoclonal Abs as direct conjugates and their isotype controls. Splenocytes and peripheral blood cells were stained with indicated combination of the following fluorochrome-conjugated monoclonal antibodies: CD19-PerCP, B220-APC, CD11b-PerCP, F4/80-PE, Ly6G-PE, Ly6C-Fitc, CD11c-FITC, CD4-APC and CD8-PB (BD Pharmingen). Viable cells (2×105) in the lymphocyte gate, as defined according to side and forward scatters, were analyzed. Flow cytometry was performed using a LSR II instrument (BD Biosciences), and the results were analyzed using the FlowJo software (Tree Star, Inc.).
ELISA
Serum samples derived from naïve and L. monocytogenes infected WT or Blimp1 CKO mice and supernatants derived from cultures of untreated or L. monocytegenes treated BMDM were assayed for CCL8 levels by using a specific ELISA kit (ANTIGENIX AMERICA Inc.) according to the manufacturer’s instructions.
In vivo infections
Pre-titered TSB-glycerol stocks of L. monocytogenes strain 10403S were stored at −80C. Prior to infection, 1-ml bacterial aliquots were recovered for 1 h at 37C in 9 ml of TSB (BD Biosciences), washed and resuspended to the desired cfu in PBS. Mice were injected i.v. with 3×104 cfu of L. monocytogenes (strain 10403S) in 0.3 ml of PBS. At defined time points, infected animals were killed by CO2 asphyxiation. Livers and spleens of infected animals were aseptically harvested, weighed, and homogenized in 0.02% Triton X-100. Prior to homogenization, samples of infected livers were taken for slide preparation and staining with Hematoxylin & Eosin or Gram dye. Aliquots of serial 5-fold dilutions in sterile water were plated in duplicate on TSB agar (BD Biosciences) plates containing 10 mg/ml streptomycin. After overnight incubation, the number of bacteria per gram of tissue was determined by counting colonies at the appropriate dilution.
Microarray hibridization and analysis
RNA (5μg) was isolated from WT and Blimp1 CKO BMDM untreated or infected for 2 hours with L. monocytogenes (MOI=5). cDNA and cRNA labeling and hybridization were performed by the Genomic Core Facility (University of Massachusetts Medical School, Worcester, MA) by using kits and GeneChip Scanner 3000 by Affymetrix (Santa Clara, CA). The gene chip mouse genome 430 2.0 Array (Affymetrix) was used. CEL files were normalized in Bioconductor [54] using the GCRMA method [55]. Analyses were restricted to samples where at least 25% of the fluorescence units were greater than log2100 and the interquartile range was at least 0.5 on the log base 2 scale. Differentially regulated genes were identified using a moderated t-test [56]. False discovery rate adjusted P-values were calculated using the method of Benjamini and Hochberg [57]. GO enrichment analysis was performed using the GOstats package [58]. The data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE53145 www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53145.
RNA isolation and Quantitative real-time PCR
DNase I-treated total RNA from BMDM and PEC was extracted with RNeasy kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s instructions. cDNA was synthesized, and quantitative real time RT-PCR analysis was performed on a DNA engine Opticon 2 cycler (MJ Research, Watertown MA) using the SuperScript III Two-Step qRT-PCR Kit with SYBR Green (Invitrogen, Carlsbad, CA). The specificity of amplification was assessed for each sample by melting curve analysis. Relative quantification was performed using standard curve analysis. The quantification data are presented as a ratio to the β-actin level. The standard errors (95% confidence limits) were calculated using the Student’s t test. All gene expression data are presented as a ratio of gene copy number per 100 copies of β-actin ± SD. The following primer pairs have been used: β-Actin_for 5-TTGAACATGGCATTGTTACCAA-3, β-Actin_rev 5- TGGCATAGAGGTCTTTACGGA-3; BLIMP1_for 5-GAGGATCTGACCCGAATCAA-3, BLIMP1_rev 5-GTTGCTGTGAGGCAACTTCA-3; CCL8_for TAAGGCTCCAGTCACCTGCT, CCL8_rev 5-TTCCAGCTTTGGCTGTCTCT-3; Atm_for 5-CCAGGCTTGAAGGACTGAAG-3, Atm_rev 5-TCACCACAATGCCATCTTGT-3; Mpa2l_for 5-TGGAGCAGCTGCATTATGTC-3, Mpa2l_rev 5-GCATTCTGGGTTTGTCACCT-3; Arhgap25_for 5-ATGTCTGGGCTAAGGTGGTG-3, Arhgap25_rev 5-ACCCTCAAGGATCCGTCTTT-3; Arhgap26_for 5-AAGACCATCAGCCCGTACAC-3, Arhgap26_rev 5-CTGGCCTGTCTACTGCTTCC-3; Pldn_for 5-GGACTTCCCTGTGGACGATA-3, Pldn_rev 5-AGCTTGGCGTGATAGTGCTT-3; Rasgrp1_for 5-TGTCACAGCTCCATCTCCAG-3, Rasgrp1_rev 5-TTCACCTTCCCATCTTCCAG-3; Tm7sf4-for 5-CCAAGGAGTCGTCCATGATT-3, Tm7sf4-rev 5-GGCTGCTTTGATCGTTTCTC-3; Tmem87a-for 5-AATGTGGTGCACACTCCAAA-3, Tmem87a-rev 5-AGGGCGAATAATCACAGCAC-3.
Cell extracts
BMDM were infected with L. monocytogenes 10403s at MOI 5. After 1 hour of infection cells were washed once with 10% FCS/DMEM and then treated with 10 mg/ml gentamycin to kill extracellular bacteria. Bone marrow cells were then left for other 2 hours and nuclear lysates prepared as previously described [53].
Western blot analysis
Nuclear cell extracts were separated by 7.5% SDS-PAGE gel and blotted onto nitrocellulose membranes. Blots were incubated with mouse monoclonal Abs against murine BLIMP1 (Novus biological) or β-Actin (Sigma), as indicated, and detected using an ECL system (Amersham, Piscataway, NJ).
ChIP assay
ChIP experiment were performed essentially as described [59]. Briefly, approximately 4×107 BMDM were used to perform immunoprecipitation using rabbit polyclonal recognizing the C-terminus of BLIMP1 (a kind gift of KL Calame, Columbia University, NY) [31]. Quantitative real-time RT-PCR was performed on immunoprecipitated and input fractions from the immunoprecipitation. The following set of primers was used to amplify the different regions of the Ccl8 promoter, as indicated: unr_for 5-CATGTGCAAGGTTCTGGCTA-3, unr_rev 5-GGCCTAAGTCTGGTCCCTTT-3; +141_for 5-ACGCTAGCCTTCACTCCAAA-3, +141_rev 5-GTGCCTGCTAAACCATTTCC-3; −288_for 5-GGGACCAGACTTAGGCCTTT-3, −288_rev 5-GAAGGGCAAGCATCTGAAAC-3; −85_for 5- CTGATCCAGTGGGACTGCTT, −85_rev 5-GAAGGCTAGCGTCCTCTCAA-3.
Transwell chemotaxis and Intracellular cytokine staining of γ/δ T cells
γ/δ T cells were first isolated as described above from pooled lymph nodes and spleens of 10–20 wild-type C57BL/6 mice and rested overnight in 0.5% FCS RPMI supplemented with HEPES, L-glutamate, non-essential amino acids and Na-pyruvate. The next day γ/δT cells (5 × 106 per ml in 100 μl) were harvested and placed in the upper chamber of Transwells (5-μm pore size polycarbonate membrane, 12 mm diameter) (Costar, Fisher) and 600 μl of chemokine or media was placed in the lower chamber. Migration was allowed to proceed for 2–3 h at 37°C and was monitored after 2 h with an inverted microscope and compared to media controls to determine completion of assay. A fraction of input total spleen and LN γ/δ T were set aside and simultaneously incubated at 37°C during the course of the assay. At the end of the assay, total input γ/δ T cells, and CCL8- or CCL2-responsive migrated γ/δ T cells in the transwells were harvested for intracellular staining and activated with 50 ng/ml PMA (Sigma- Aldrich) and 500 ng/ml ionomycin (Sigma-Aldrich) for 2 hours in the presence of Brefeldin A (BD Biosciences). At the end of the activation, cells were fixed and permeabillized with Fix & Perm Medium A & B (Invitrogen), and then stained with anti-IL-17A Alexa Fluor 647 (TC11-18H10.1, Biolegend) and anti-IL-17F Alexa Fluor 488 (eBio18F10, Ebiosciences). Cells were acquired on a Becton-Dickinson FACScalibur and analyzed with FlowJo 8.8 software.
Statistical Analysis
Statistical analysis was calculated using a two-tailed Student’s t-test, if not otherwise stated. A p value ≤ of 0.05 was considered statistically significant.
Results
Regulation of Blimp1 gene expression following L. monocytogenes infection
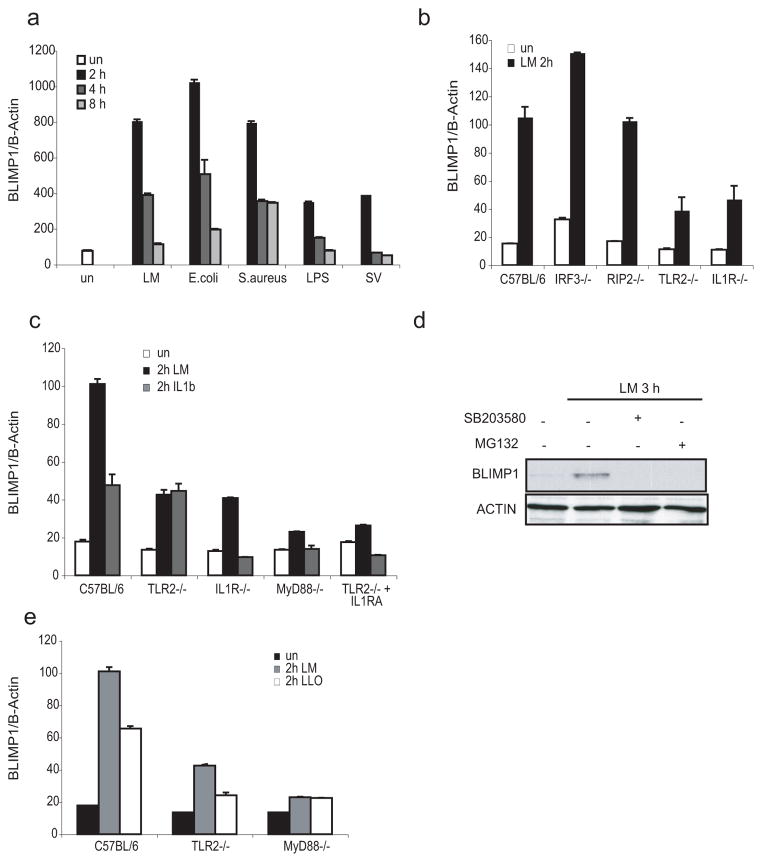
While studying immune response genes in murine bone-marrow derived macrophages (BMDM) infected with bacterial and viral pathogens, we found that expression of the Prdm1 gene (which encodes BLIMP1) was markedly increased upon infection with a variety of pathogens including L. monocytogenes (LM), Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), the paramyxovirus Sendai (SV) and the Gram (-) derived lipopolysaccharide (LPS). In all cases, the induction was strikingly rapid and transient with expression peaking 2 hours post-infection (hpi) (Figure 1a). We also extended this timecourse to later timepoints, which revealed a second wave of inducible BLIMP1 expression at 24 hourstime points (Supplementary Figure 1a). We focused on L. monocytogenes as a model pathogen to understand both the molecular basis for BLIMP1 induction and the role of BLIMP1 in anti-microbial defenses. L. monocytogenes is readily phagocytosed by macrophages and once inside a host vacuole, the essential virulence factor listeriolysin (LLO), a pore forming toxin encoded by the hemolysin (hly) gene, enables the bacterium to permeabilize the vacuolar membrane and enter the cytosol (34). Several classes of pattern recognition receptors can sense L. monocytogenes-derived products at different stages of infection. These include TLR2 (17–19, 35), NOD2 (20), NLRP3 (21), AIM2 (22, 23), STING well as an as yet unidentified DNA sensor that turns on IFN-β gene transcription (28, 31). To understand how Blimp1 gene induction was regulated we monitored L. monocytogenes induced Blimp1 transcript levels in macrophages derived from mice with targeted deletions in components of these signaling pathways. These included macrophages deficient in TLR2; in receptor-interacting serine-threonine kinase 2 (RIP2), the downstream effector of NOD1/2 signaling; in Interleukin-1 receptor chain 1 (IL1R1), which fails to signal in response to IL-1β released upon the activation of caspase-1 and finally IRF3, the key transducer of the cytosolic DNA sensing pathway. Induction of Blimp1 gene transcription upon L. monocytogenes infection was induced normally in macrophages lacking RIP2 and IRF3 but was reduced in both TLR2−/− and IL1R−/− macrophages (Figure 1b). Moreover, this response was completely lost in TLR2−/− macrophages pre-treated with the IL1R antagonist (IL1RA), showing that for a full induction of Blimp1 transcription both these signaling pathways have to be activated by L. monocytogenes (Figure 1c). Indeed, L. monocytogenes infection of BMDM lacking MyD88, a key adaptor downstream of both TLR2 and IL1R intracellular cascades, fails to induce Blimp1 expression. Furthermore, inhibition of p38MAPK and NF-κB signaling using the respective specific inhibitors SB203580 or MG132, prevented L. monocytogenes-induced expression of BLIMP1 protein in macrophages (Figure 1d). These data suggest that cell surface TLR2 and cytosolic PRRs, which regulate IL-1β production, cooperate in control of Blimp1 transcription. In support of this idea, an LLO-deficient strain of L. monocytogenes, which is not capable of entering the cytosol of the host cell and as a consequence is not able to trigger IL-1β or type I IFN production, was less potent than wild-type bacteria in inducing Blimp1 transcription. This response to LLO-deficient bacteria was entirely dependent on TLR2, since macrophages lacking TLR2 failed to up-regulate Blimp1 (Figure 1e).
Figure 1. Blimp1 is rapidly induced by L. monocytogenes via TLR2 and cytosolic sensors.
RNA was isolated from BMDM left untreated (un) or infected with L. monocytogenes (Lm), E. coli, S. aureus (MOI=5), Sendai virus (SV) (300 HAU) or treated with LPS (100ng/ml) for the indicated times and Blimp1 gene expression was quantified by qPCR. Data were normalized with β-Actin and are representative of 3 independent experiments. b, BMDM from C57BL/6, IRF3−/−, RIP2−/−, TLR2−/− and IL1R−/− mice were stimulated with LM for 2 hours. mRNA expression for Blimp1 was measured by qPCR. One of three different experiments conducted singularly is shown. c, RNA was isolated from LM-infected or IL1β treated BMDM from C57BL/6, TLR2−/−, IL1R−/−, MyD88−/− mice or from TLR2−/− pre-incubated with IL1R antagonist (IL1RA, 150μg/ml). Blimp1 gene expression was quantified by real-time RT-PCR. Results are mean ± SD of triplicate values and are representative of at least 4 experiments. d, Lysates were prepared from BMDM treated for 3 hours with LM (MOI=5) either before or after pre-treatment (30 minutes) with an inhibitor for p38/MAPK (SB203580, 40μM) or an inhibitor for NF-κB (MG132, 40μM). BLIMP1 and β-actin expression were detected by Western blotting using the respective specific antibodies. e, Blimp1 gene expression was evaluated by real-time RT-PCR in RNA derived from C57BL/6, TLR2−/−, and MyD88−/− macrophages and treated 2 hours with MOI=5 of either the wild-type or the LLO mutant L. monocytogenes strains.
Generation of a myeloid-specific Blimp1 conditional knockout mouse
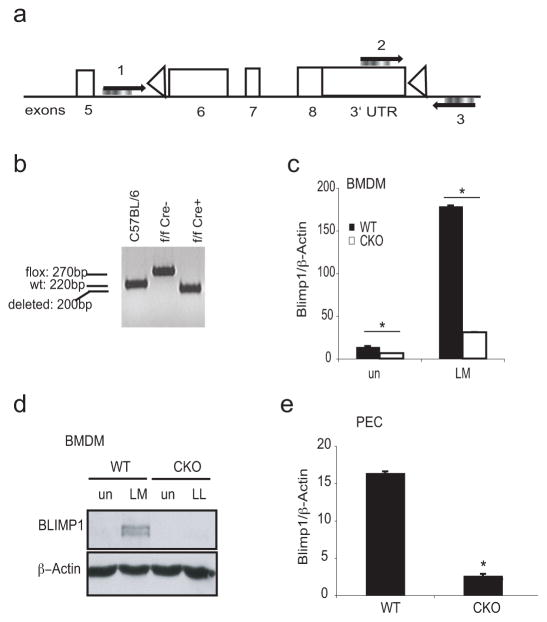
Direct-targeted disruption of the gene encoding BLIMP1 (Prdm1) is lethal for mouse embryos (6). Therefore to define the role of this protein in the host response to microbial challenge, we generated a myeloid-specific Blimp1 conditional knockout mouse via lysozyme M-driven Cre-mediated recombination (36). Prdm1flox/flox M-lysozyme Cre negative (6) and the Prdm1flox/flox M-lysozyme Cre positive mice (hereafter referred to as WT and Blimp1 CKO mice, respectively) were developmentally normal and fertile. Efficient deletion of the floxed Prdm1 sequence in the Blimp1 CKO macrophages was confirmed by PCR analysis of BMDM genomic DNA (Figure 2a and b). Both the mRNA and protein were undetectable in BMDM from Blimp1 CKO mice either before or after infection with L. monocytogenes (Figure 2c and d, respectively). Similar results were obtained when elicited peritoneal macrophages (PEC) from WT and Blimp1 CKO mice were examined ex vivo for the mRNA expression of the Blimp1 gene (Figure 2e). M-lysozyme is expressed in both macrophages and granulocytes so we measured BLIMP-1 in both of these cell types. While BLIMP-1 was expressed in macrophages and dendritic cells its expression was very low by comparison in neutrophils (Supplementary Figure 1b). We did observe efficient deletion of the floxed Prdm1 DNA sequence by PCR in neutrophils isolated from the peritoneal cavity of Blimp1 CKO mice (Supplementary Figure 1c). Although BLIMP-1 was poorly expressed in resting neutrophils its expression was however inducible upon L. monocytogenes infection (Supplementary Figure 1c and d).
Figure 2. Generation of prdm1flox/flox M-lysozime Cre+ mice.
a, Schematic representation of the locus targeted by M-lysozyme Cre within the Prdm1 gene with indicated primer sites used for screening the deletion (1, 2 and 3). b, Genomic DNA was isolated from BMDM from C57BL/6, Prdm1flox/floxCre− (WT) or Prdm1flox/floxCre+ (CKO) mice. PCR analysis for cre-targeted region on the Prdm1 gene was performed using a mix of primer 1, 2 and 3 on genomic DNA. Amplicon sizes are shown. c–e, Lysozyme M-driven cre-mediated deletion of Blimp1 was confirmed in untreated or LM-treated BMDM from WT or Blimp1 CKO mice at mRNA level by qPCR (c), at protein level by Western blotting (d) or at steady state level in peritoneal exudates cells (PEC) from WT or Blimp1 CKO mice by qPCR (e). P-values were calculated by two-tailed Student’s t-test. *p<0.001.
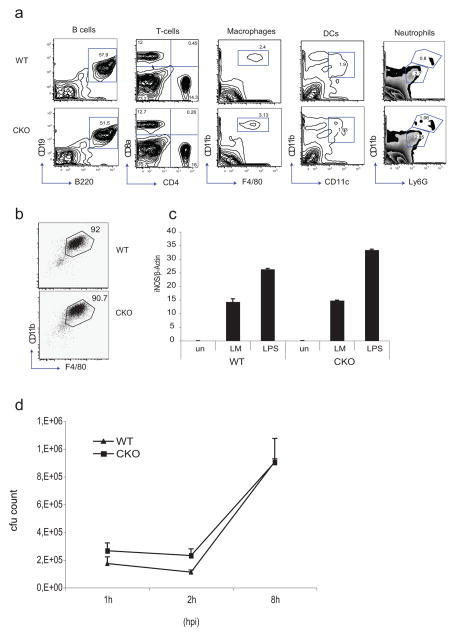
Since BLIMP1 is a key factor controlling the differentiation of many different cell types we examined immune cell development in Blimp1 CKO mice. As expected, no significant differences in the numbers of B cells or T cells was observed when BLIMP1 was deleted only in myeloid cells (Figure 3a). Previous studies using overexpression of a truncated version of BLIMP1 in a human monocytic cell line suggested that BLIMP1 was important in the differentiation of myeloid cells (37). Flow cytometry analysis revealed normal numbers of macrophages, neutrophils and dendritic cells in the spleen of Blimp1 CKO mice (Figure 3a). Similar numbers of elicited peritoneal cells (PEC) were also obtained from WT and Blimp1 CKO mice after thioglycolate injection and expression of CD11b and F4/80 expression on these cells was equivalent between WT and CKO mice (data not shown). We also examined the differentiation of Blimp1 CKO macrophages ex vivo from bone marrow. BMDM from WT and Blimp1 CKOs expressed similar levels of CD11b and F4/80 (Figure 3b) and induced equivalent levels of nitric oxide synthase (iNOS) in response to L. monocytogenes or after treatment with LPS (Figure 3c). Moreover, L. monocytogenes grew at a comparable rate in WT and Blimp1 CKO BMDM (Figure 3d). Collectively, these data demonstrate that BLIMP1 is not required for the development or normal functions of macrophages.
Figure 3. Blimp1 deficiency does not influence normal macrophage function.
a, Flow cytometric analysis of splenocytes in Blimp1 CKO mice. Single cell suspensions were prepared from the spleens of 8-week-old mice and stained as indicated. Viable cells (2×105) in the lymphocyte gate, as defined according to side and forward scatters, were analyzed. The numbers represent the percentage of cells contained in each gated region. b, FACS analysis of CD11b and F4/80 expression in WT or Blimp1 CKO BMDM. A representative experiment is shown. The analysis was repeated four additional times. c, iNOS expression was evaluated by real-time RT-PCR in WT or Blimp1 CKO BMDM treated with LM (MOI=5) or LPS (100ng/ml) for 4 hours. d, L. monocytogenes intracellular growth in WT or Blimp1 CKO BMDM 1, 2 and 8 hours post infection (hpi). In the graph is reported the average ± SD of cfu counts obtained from BMDM derived from 6 mice of each genotype. BMDM lysate dilutions were plated over-night in duplicate on TSB agar plates containing streptomycin before counting. Reported values represent maens of cfu counts obtained in each condition ± SD.
Blimp1 deficiency in myeloid cells protects mice from L. monocytogenes infection
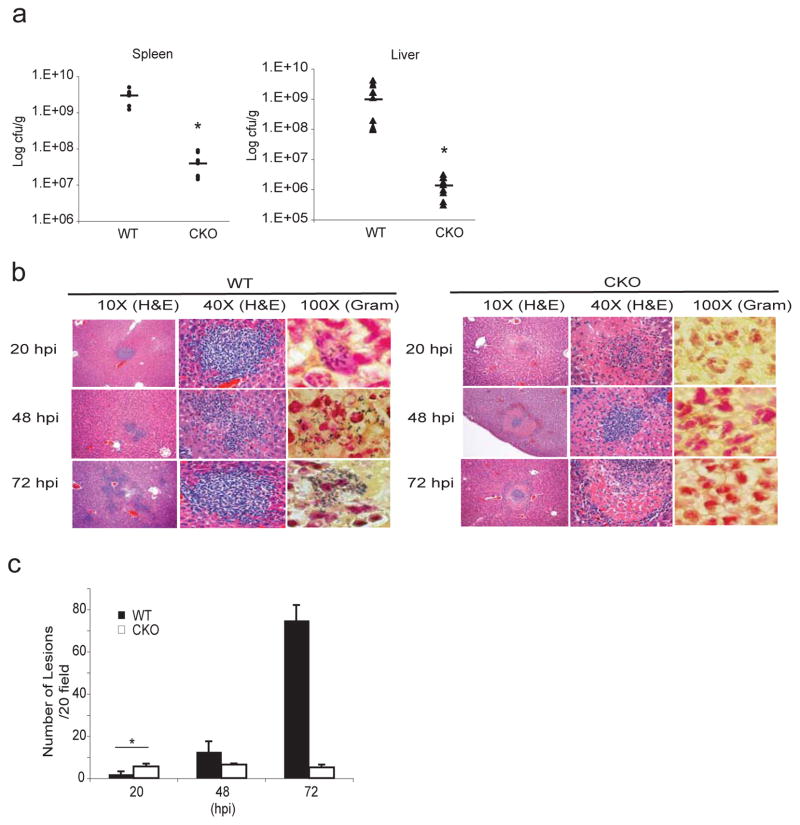
To examine the role of BLIMP1 in innate immunity, we performed in vivo infection experiments using L. monocytogenes. We first monitored bacterial numbers in the spleens and livers 48 hpi (3 × 104 cfu delivered i.v.). Both organs in the Blimp1 CKO mice had lower bacterial loads than WT littermate controls (Figure 4a). While the bacterial load in both organs was comparable between WT and Blimp1 CKO mice 20 hpi, it progressively increased in WT but not in Blimp1 CKO spleens and livers at the later time points examined (Supplementary Figure 1e). Furthermore, Blimp1 CKO mice had a greater rate of survival and quickly recovered from a dose of L. monocytogenes delivered i.v. that was lethal to most control mice (Supplementary Figure 1f). Importantly, we found no differences in bacterial load in liver or spleen when C57BL/6 wild type mice and C57BL/6 mice expressing M-lysozyme Cre only were examined (Supplementary Figure 1g), suggesting that there was no adverse effect of the Cre recombinase in this system.
Figure 4. Myeloid-specific Blimp1 CKO mice are protected from L. monocytogenes infection.
a, WT or Blimp1 CKO mice were infected with 3×104cfu of L. monocytogenes i.v. and bacterial loads were quantified in spleens and livers 48 hpi. Results from 3 different experiments are expressed as mean ± SEM. *p<0.001 (two-tailed Student’s t-test). b, Hematoxylin & Eosin staining (10X and 40X magnification) and Gram staining (100X magnification) of WT or CKO livers 20, 48 and 72 hpi by L. monocytogenes. The experiment shown is representative of 3 experiments conducted separately. c, Numbers of microabscesses per 20 fields of 3 different experiments were quantified and expressed as mean ± SEM. *p<0.05 (two-tailed Student’s t-test).
Histological analysis of liver sections revealed microabscesses containing neutrophils and macrophages by 48 hpi and a further increase in the number of these lesions by 72 hpi in WT animals (Figure 4b [Hematoxylin & Eosin staining] and c) in line with bacterial counting described in Supplementary Figure 1a. In contrast, very few microabsesses were present in the Blimp1 CKO livers. The lesions in the Blimp1 CKO mice while present at higher numbers 20 hpi, did not increase in number during the course of the infection (Figure 4b [Hematoxylin & Eosin staining] and c). Notably, the lesions in Blimp1 CKO livers were morphologically distinct from those found in the WT mice. Neutrophils were present at what was presumably the site of infection, surrounded by a zone rich in the ghost like remnants of dead hepatocytes, characteristic of coagulative necrosis, a pathological finding that results from a highly robust neutrophil response. Gram staining of liver sections revealed large numbers of bacteria in microabsesses of WT livers, but found no bacteria in the Blimp1 CKO lesions (Figure 4b, right panel [Gram staining]). These observations suggested that there was a more robust and rapid clearance of bacteria from the livers of Blimp1 CKO mice. The presence of hepatocytes in a state of coagulative necrosis also supports this idea, since enzymes released from neutrophils are thought to control this process.
The chemokine CCL8 is a target of BLIMP1 mediated transcriptional repression
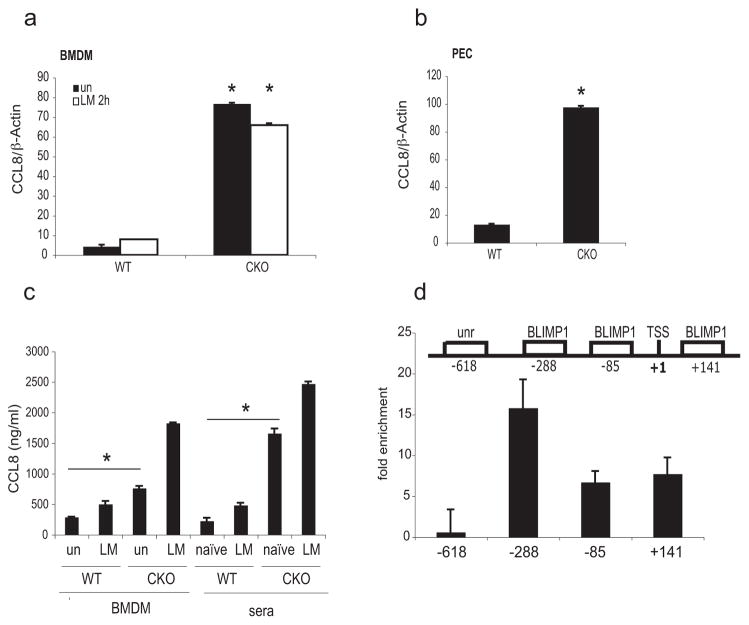
To elucidate the molecular mechanisms responsible for the heightened clearance of L. monocytogenes in Blimp1 CKO mice, we focused on macrophages and compared the gene expression profile of WT and Blimp1 deficient macrophages before and after infection with L. monocytogenes. Supplementary figure 2a and supplementary table 1 shows genes with significantly different expression levels (P-value ≤ 0.05) between WT and Blimp1 CKO BMDM for either untreated or infected cells as evaluated by microarray analysis. The set of genes were significantly enriched in a number of GO terms (Supplementary Table 2) related to macrophage activation and the immune system. Comparison of transcriptomes of untreated WT with untreated CKO macrophages identified a small number of genes that had significantly different expression levels. Importantly, these observations revealed that deletion of Blimp1 had surprisingly little effect on global gene expression in macrophages but rather identified a specific and restricted subset of genes that were differentially expressed in macrophages in the absence of this factor. One gene in particular, Ccl8, encoding the chemokine belonging to the C-C family, was expressed approximately 17 times higher in Blimp1-deficient than in WT macrophages, as measured by quantitative real time RT-PCR (qPCR) (Figure 5a). The expression profile of seven additional genes was also confirmed by qPCR (Supplementary Figure 2b). Ccl8 transcription levels were also significantly elevated in macrophages isolated from the peritoneum (PEC) of Blimp1 CKO mice in the absence of infection, as compared to the levels found in WT mice (Figure 5b). Similarly, CCL8 protein levels were higher in the culture medium of untreated BMDM and uninfected sera from Blimp1 CKO mice as compared to WT littermate controls (Figure 5c).
Figure 5. Ccl8 gene is a direct target of BLIMP1-mediated transcriptional silencing.
a, Ccl8 expression was examined by qPCR in RNA isolated from untreated or 2h LM-treated BMDM from WT or Blimp1 CKO mice. Data were normalized with β-actin and expressed as means ± SD. Data shown are representative of 3 independent experiments. *p=0.001 (two-tailed Student’s t-test). b, Gene expression of Ccl8 was evaluated also in PEC derived from WT or Blimp1 CKO mice in the absence of infection by real-time RT-PCR. *p=0.001 (two-tailed Student’s t-test). c, CCL8 protein level was analyzed by ELISA assay in both culture media and sera derived from uninfected and L. monocytogenes-treated WT or Blimp1 CKO mice. *p<0.05 (two-tailed Student’s t-test). d, Upper panel: Schematic representation and relative position of conserved BLIMP1 binding sites on the murine Ccl8 promoter and of an unrelated (unr) sequence. TSS; transcription starting site. Lower panel: ChIP analysis of BMDM using an antibody to BLIMP1 or pre-immune serum is shown. Analysis of enrichment of selected sequences was performed by qPCR. The results are expressed as fold enrichment of signal in anti-BLIMP1 immunoprecipitates over that of pre-immune serum. The graph shows one representative of two similar experiments.
Sequence analysis of a genomic region spanning 1000bp upstream and 500bp downstream of the Ccl8 transcription start site (TSS) using the software MatInspector (http://www.genomatix.de/matinspector.html) (38) identified three BLIMP1 consensus binding sites, at positions −288, −85 and +141 (Figure 5d, upper panel). Chromatin Immunoprecipitation (ChIP) analysis using an antibody to BLIMP1 (39) in macrophages derived from C57BL/6 mice revealed weak binding of BLIMP1 to both the −85 and +141 sites and a very strong binding to the −288 site (Figure 5d, lower panel). Similar results were obtained when ChIP was performed on infected BMDM (data not shown). No binding was observed when an unrelated (unr) sequence at position −618 of the Ccl8 promoter was examined (Figure 5d, upper and lower panels). These data indicate that BLIMP1 directly binds to the Ccl8 promoter in vivo.
Critical role of CCL8 in antibacterial resistance to L. monocytogenes
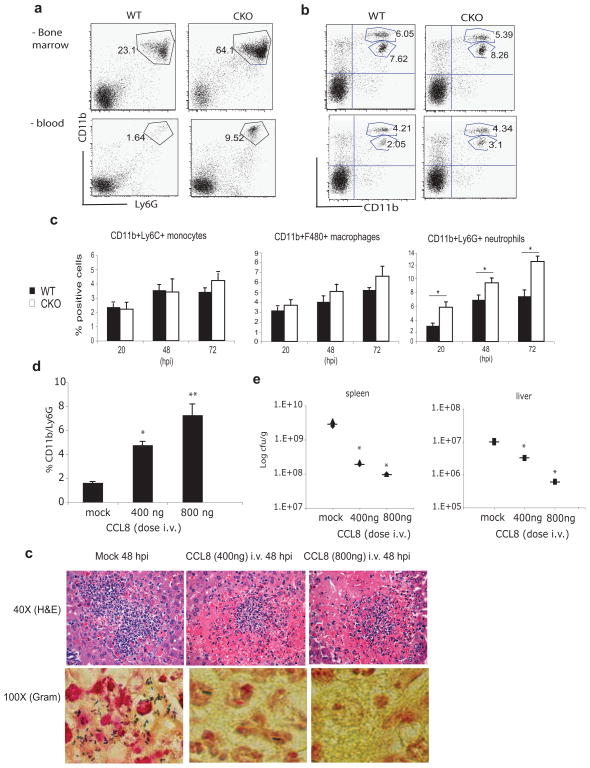
Neutrophils play a key role in the early control of L. monocytogenes growth, appearing in the liver within the first 24–36 hpi (15). First generated in the bone marrow, neutrophils are elicited into the peripheral blood and attracted to the liver where they contribute to antibacterial resistance by killing bacteria, lysing infected host cells, stimulating anti-microbial activity of infected macrophages and inducing hepatocyte apoptosis (40). We speculated that the elevated expression of CCL8 in Blimp1 CKO mice could encourage egress of neutrophils from the bone marrow and promote their infiltration into the livers of L. monocytogenes infected mice. Consistent with this hypothesis, both the bone marrow and peripheral blood of uninfected Blimp1 CKO mice showed a higher percentage of CD11b+/Ly6G+ neutrophils as compared to WT mice, while the percentage of CD11bint/Ly6Chi inflammatory monocytes remained constant in both compartments (Figure 6a and b). Accordingly, in infected spleens of Blimp1 CKO there was a marked and statistically significant increase in the percentage of neutrophils following L. monocytogenes infection as compared to that observed in WT organs, while no differences were observed for monocytes or macrophages (Figure 6c).
Figure 6. Role of CCL8 in antibacterial resistance and host defense against L. monocytogenes.
a–b, Bone marrow and peripheral blood were harvested from naïve WT or Blimp1 CKO mice. Cells were stained for CD11b, Ly6G or Ly6C and their expression evaluated in a large gate drawn in the live granulocytes/ lymphocyte/ monocyte population. Cytometric analysis of CD11b/Ly6G neutrophils (a) and CD11bint/Ly6Chi monocytes (b) from bone marrow (upper panels) or peripheral blood cells (lower panels) is shown. A representative experiments of three conducted separately is depicted. c, CD11bint/Ly6Chi monocytes, CD11b+F480+ macrophages and CD11b+Ly6G+ neutrophils derived from three WT or three CKO infected spleens were analyzed by FACS 20, 48 and 72 hpi. Mean percentages are shown. Error bars represent SEM. * p < 0,001 (two-tailed Student’s t-test). d, Percentages of CD11b/Ly6G positive cells evaluated by facs analysis in peripheral blood from wild-type mice mock treated or injected i.v. with 400 or 800ng of recombinant CCL8 for two consecutive days are shown. Each bar represents the average of three to four mice of two independent experiments. Error bars represent SEM. * p = 0.012; ** p < 0,001 as compared with mock controls (two-tailed Student’s t-test). e, Bacterial burdens from spleens and livers were calculated in mice mock treated or injected with 400 or 800ng of recombinant CCL8 and then infected with L. monocytogenes i.v. (n=3 mice per genotype). *p<0.05 versus mock (two-tailed Student’s t-test). f, Representative Hematoxylin & Eosin staining (40X) and Gram staining (100X) of livers derived from mock treated or CCL8 injected mice subsequently infected with L. monocytogenes i.v. Mice were sacrificed 48hpi.
To test if CCL8 would be responsible for this increased mobilization of neutrophils in Blimp1 CKO mice, we injected C57/Bl6 mice i.v. with two increasing doses of recombinant CCL8 for two consecutive days and monitored CD11b+/Ly6G+ neutrophil numbers in the peripheral blood. The numbers of CD11b+/Ly6G+ cells in mock treated mice was very low, but this percentage significantly increased in a dose-dependent manner with the administration of CCL8 (Figure 6d). Both CCL8- and mock-treated mice were then infected with L. monocytogenes and bacterial loads in the spleen and liver examined 48 hpi. The bacterial burden in both organs analyzed was much lower in the CCL8- treated than mock-treated mice and the numbers of bacteria decreased proportionally with the increase of the chemokine dose (Figure 6e). Histological analysis of liver sections by Gram staining confirmed these findings revealing fewer bacterial laden microabsesses in CCL8-treated mice than mock treated animals (Figure 6f, lower panels). Most interestingly, the lesions present in the livers of CCL8-injected mice looked very similar to those we observed in Blimp1 CKO mice. Zones of hepatocytes in a state of coagulative necrosis surrounded infiltrating neutrophils (Figure 6f, upper panels). We also administered CCL2 (also called MCP1) i.v., a chemokine known to selectively induce migration of monocytes (40–42), and monitored the percentage in the blood of both CD11b+/Ly6G+ neutrophils and CD11bint/Ly6Chi monocytes (Supplementary Figure 2c). As expected, we found a significant increase in the monocyte numbers but no effect on the neutrophil population.
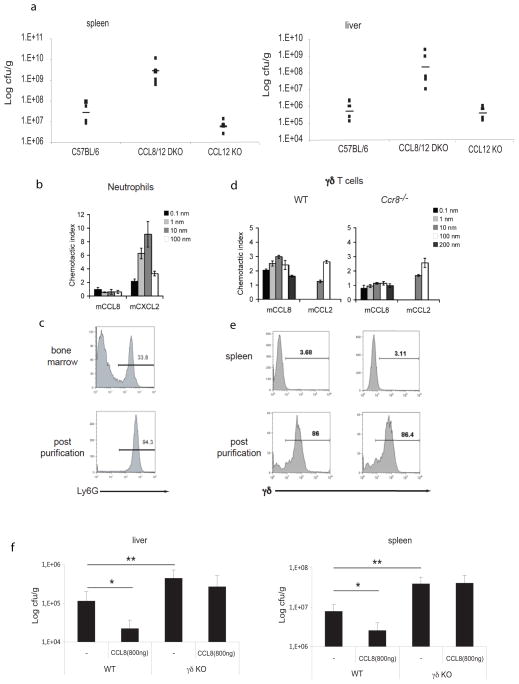
To further define the role of CCL8 in antimicrobial resistance, we infected mice lacking Ccl8 with L. monocytogenes. Ccl8 single KOs for are not available however mice lacking Ccl8 and Ccl12 (also called MCP5) or mice lacking Ccl12 alone were studied (41). Consistent with a role for CCL8 in the clearance of bacteria during infection, the bacterial load was much higher in mice lacking Ccl8/Ccl12 than in Ccl12 single KOs or C57BL/6 WT mice (Figure 7a). CCL8 was recently found to be highly expressed in the skin where it acts via the chemokine receptor CCR8. CCL8 responsiveness defined a population of highly differentiated, CCR8-expressing inflammatory T helper type 2 (TH2) cells enriched for IL-5 (43, 44). The ability of CCL8 to signal via CCR8 rather than CCR2 distinguishes CCL8 from all other MCP chemokines. In an effort to understand the role of CCL8 in innate immune responses to L. monocytogenes, we first examined if CCL8 was chemotactic for neutrophils by monitoring the migration of neutrophils in response to this chemokine using an in vitro chemotaxis assay. Neutrophils were purified from bone marrow and tested for their ability to migrate in response to CCL8 (Figure 7b and c). We found that in contrast to MIP2 (also called CXCL2,- an agonist of CXCR2 expressed on neutrophils), CCL8 was not chemotactic for neutrophils (45)(Figure 7c). Neutrophils can also be mobilized to the site of infection through IL-17-mediated events that stimulate granulopoiesisinducing cytokines and granulocyte-attracting chemokines (46). The major IL-17 producers at the early stage of Listeria infection in the liver are γ/δ T cells (47), which also express CCR8 on their surface (48). We therefore next investigated whether γ/δ T cells were the target of CCL8 action and found that CCL8 was chemotactic for these cells (Figure 7d and e). The migration of γ/δ T cells to CCL8 was similar to that seen with CCL2, a known chemotactic factor for these cells (49). We next wanted to examine if the enhanced clearance of bacteria in animals treated with exogenous CCL8 was dependent on γ/δ T cells. In agreement with published studies, γ/δ T cell–deficient mice had significantly higher bacterial loads in liver and spleen compared to their wild type counterparts following L. monocytogenes infection (Figure 7f). While wild type animals treated with exogenous CCL8 cleared bacteria efficiently, bacterial loads in γ/δ T cell–deficient mice treated with CCL8 were unaltered.
Figure 7. γ/δ T cells are the cellular target of CCL8 chemotactic activity.
a, L. monocytogenes in vivo infection was performed in six different Ccl8/12 DKO or six Ccl12 single KO mice. Six sex and age-matched C57BL/6 mice were used as controls. Infected spleens and livers were harvested 48 hpi and bacterial loads calculated. Data represent means ± SD. *p=0.001 (two-tailed Student’s t-test). b. Neutrophils purified from the bone marrow of WT mice were assayed for migration to mCCL8 and the positive control mCXCL2 in a Neuroprobe chemotaxis chamber. c. Purity of neutrophils determined by Ly6G staining. d. γ/δ T cells isolated from WT and Ccr8 −/− mouse spleens were assayed for migration to mCCL8 and the positive control mCCL2 in a Neuroprobe chemotaxis chamber. e. Purity of WT and Ccr8 −/− γ/δ T cells determined by γ/δ TcR staining. Data in b–e are representative of three independent experiments. f, L. monocytogenes in vivo infection was performed in γ/δ T cell KO or C57BL/6 WT mice that had received CCL8 i. v. Infected spleens and livers were harvested 48 hpi and bacterial loads calculated. Data are from 2 different experiments conducted separately. P-values were calculated by two-tailed Student’s t-test. *p<0.001 untreated versus CCL8-treated WT mice; *p<0.05 untreated WT versus untreated γδ KO mice.
Notably, the IL-17 cytokine family includes different members and three of them, IL-17A, IL-17F and IL-17E (or IL-25) have been best characterized both in vivo and in vitro and have been shown to be pro-inflammatory in nature (50). In particular, while IL-17E induces TH2 cytokine production and eosinophilia, both IL-17A and F are involved in the recruitment of neutrophils. In models of muco-epithelial bacterial infections and autoimmunity IL-17A and F were both shown to play important roles in host defense but to have distinct activity in inflammatory processes and to be produced by different cell types (51). However, γ/δ T cells are able to produce both of these cytokines amplifying Th17 responses (52). Hence, we evaluated in our model which type of IL-17 was produced by CCL8-attracted γ/δ T cells. To this end, we first compared by intracellular staining IL-17A and IL-17F expression in γ/δ T cells that migrate in response to CCL8 after transwell migration assays as compared to the total input population of γ/δ T cells isolated from WT spleen and lymph nodes (Figure 8a and b). Our experimental setting shows that CCL8-responding γ/δ T cells are the main producers of IL-17F, even if IL-17A was also expressed, in contrast to what was observed for γ/δ T cells responding to CCL2, which only weakly induced IL-17F. We conclude from these studies, therefore, that the enhanced anti-bacterial effects associated with CCL8 are dependent on IL-17F producing γ/δ T cells.
Figure 8. mCCL8-responsive γ/δ T cells are enriched for IL-17F.
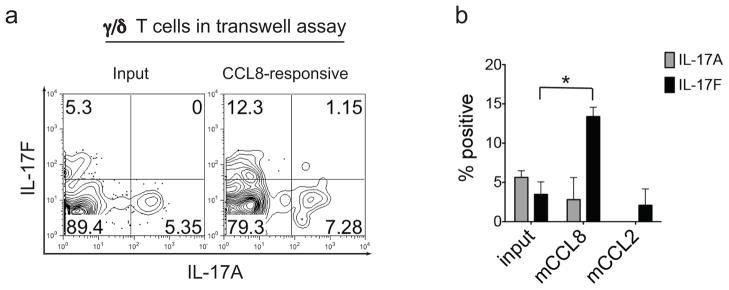
a–b, Intra-cytoplasmic staining was performed after transwell migration assays to compare IL-17A and IL-17F expression by the total input population of γ/δ T cells isolated from WT spleen and lymph nodes and γ/δ T cells that migrate to mCCL8 and mCCL2. a, Representative FACS plots of IL-17A and IL-17F expression by total input γ/δ T cells (left panel) and γ/δ T cells that migrate to mCCL8 (right panel) from three independent experiments is depicted. b. Percentage of IL-17A+ and IL-17F+ cells among input spleen and lymph node γ/δ T cells, and γ/δ T cells that migrate to mCCL8 and mCCL2 expressed as mean + SEM. Data represent a total of three independent experiments for input and mCCL8-responsive γ/δ T cells, and two independent experiments for mCCL2-responsive γ/δ T cells. *p<0.01 (two-tailed Student’s t-test).
Discussion
The role of BLIMP1 as a transcriptional repressor in embryonic development (53) (3) as well as in the differentiation of B and T lymphocytes is well established (9, 10, 54, 55). In this study we have identified a novel role for BLIMP1 in the regulation of early host-defenses in macrophages. Using myeloid-specific Blimp1 CKO mice we found that mice lacking Blimp1 in myeloid cells had greater survival than their wild type counterparts following infection with L. monocytogenes. Analysis of the Blimp1 CKO infected organs revealed little or no bacteria in the liver and spleen of these animals as compared to wild type mice. The livers of CKO animals had lesions with considerable coagulative necrosis, a pathological finding that results from a highly robust neutrophil response. Both the bone marrow and the peripheral blood of uninfected Blimp1 CKO mice had a higher percentage of CD11b+/Ly6G+ neutrophils as compared to WT mice.
Genome wide transcriptional profiling of WT and Blimp1-deficient macrophages identified the gene encoding the chemokine CCL8 as differentially modulated in the presence or absence of BLIMP1. Even in the absence of infection, Ccl8 was expressed at much higher levels in Blimp1 CKO macrophages both at the transcriptional and protein levels. Chromatin immunoprecipitation analysis indicated that BLIMP1 occupied the Ccl8 promoter in wild type cells. Basal expression of BLIMP1 in unstimulated macrophages could bind ISRE sites on target promoters and compete with ISGF3 or IRF transcription factors for these sites resulting in basal repression of these genes. BLIMP1 binding sites present within the CCL8 promoter bound by IRF factors, such as IRF1 or IRF7, would induce its transcription. Since IRF1 and IRF7 are themselves regulated via ISGF3-binding sites, their transcription might also be held in check by BLIMP1. In the context of BLIMP1 CKO mice therefore these transcription factors are likely also expressed at elevated levels which could lead to elevated CCL8 transcription explaining this high expression of CCL8 in uninfected BLIMP1 KO BMDM.
In an effort to link elevated CCL8 levels to the enhanced neutrophil response and heightened ability of Blimp1 CKO mice to clear Listeria, we administered CCL8 in vivo to C57BL/6 mice to mimic the elevated levels of the chemokine observed in the Blimp1 CKO mice. CCL8-treated C57BL/6 mice upon L. monocytogenes infection had significanly lower bacterial loads in the infected organs. The heightened clearance of this bacterium in response to CCL8 correlated with a higher percentage of neutrophils in the peripheral blood. Conversely, Ccl8 KO mice were considerably more susceptible to L. monocytogenes infection having higher bacterial loads in both spleen and liver than their wild type counterparts.
Based on these observations, we hypothesized that under normal circumstances, BLIMP1 acts to suppress the production of CCL8 as a means to limit neutrophil responses and excess inflammation, which can be deleterious to the host. We speculate that the elevated expression of CCL8 in Blimp1 CKO mice could either directly or indirectly promote neutrophils to egress from the bone marrow and infiltration into the livers of L. monocytogenes-infected mice. Our study, however, demonstrates that neutrophils do not directly migrate in response to CCL8. Rather we propose an indirect mechanism involving CCL8 in neutrophil mobilization via an IL-17-mediated feedback loop. IL-17 cytokine family comprises six isoforms, with IL-17A and its closest homolog IL-17F best characterized in vivo and in vitro (50). Cellular sources of IL-17 are mainly the innate immune cells α/β, γ/δ and NK T cells (56) and the primary function of this cytokine is to curb infection by bacterial and fungal pathogens (57, 58). Both IL-17A and IL-17F play a particularly significant role in regulating neutrophil recruitment and granulopoiesis (59–61) by inducing expression of granulocyte-colony stimulating factor and IL-6. In addition, the most strongly induced IL-17 target genes are neutrophil-recruiting chemokines, such as CXCL-1, CXCL-5 and CXCL-2 (62–64).
γ/δ T cells are specialized innate T cells that signal through a TCR formed by TCR-γ and -δ chains (65, 66). These cells are rapidly activated, producing IFN-γ, TNF-α and IL-17 in several mouse 24 models of bacterial infection including Mycobacterium bovis, Salmonella enterica and L. monocytogenes (47, 67, 68). It has been also recently shown that the negative regulation of γ/δ T cell-mediated IL-17 production by type I IFN induced by primary Influenza virus infection is of key importance for the increased susceptibility to secondary Streptococcus pneumoniae superinfection (69). Evidences that all together confirm how the IL-17- γ/δ T cell axis plays a pivotal role in resistance to bacterial infection. In particular, for L. monocytogenes it was reported that IL-17 is expressed in the liver of infected mice from an early stage of infection and that the major IL-17-producing cells were γ/δ T cells expressing TCR Vγ4 or Vγ6 (47). Hamada et al. also stated that IL-17A-deficient mice fail to control bacterial growth in vivo. IL-17 therefore is an effector molecule produced by TCR γ/δ T cells, which is important in innate immunity against L. monocytogenes in the liver. Although murine CCL8, is a member of the MCP family, until recently it has been poorly characterized. Members of the C-C family of chemokines, such as CCL2 (also called MCP1) and CCL7 (also called MCP3), are known to attract monocyte lineage cells bearing CC chemokine receptor 2 (CCR2) on their surfaces (40, 41, 70). Luster and colleagues identified CCR8 rather than CCR2 as the receptor for CCL8 (44). Thus, in our study we finally demonstrated that the real target of CCL8 in the context of L. monocytogenes infection are γ/δ T cells, known to express CCR8 on their surface (48). γ/δ T cells are able to produce IL-17A and IL-17F, both involved in neutrophil recruitment, amplifying Th17 responses (52). Indeed, here we show that γ/δ T cells directly migrate in response to CCL8 and through the release of IL-17, and in particular IL-17F, likely encourage mobilization of neutrophils to the site of infection to promote a rapid innate immune response in situ. Even if the effects of removing γ/δ T cells or injecting CCL8 are milder on CFU counts than those seen in both Ccl8/12 DKOs and Blimp1 CKO, we believe that our findings together support our model of CCL8-driven IL-17- γ/δ T cell dependent resistance to bacterial infection.
The importance of neutrophils in the early resistance of mice to Listeria infection is still somewhat unclear. Early studies based primarily on cell depletions with anti-Gr-1 (clone RB6-8C5), an antibody that binds to the cell surface markers Ly6C and Ly6G suggested that neutrophils played a critical role in reducing bacterial burden in the liver and spleen early during infection. Ly6C is expressed on neutrophils, monocytes, and subsets of CD8+ T cells, while Ly6G is expressed solely on neutrophils. Recent studies using highly selective neutrophil depletion strategies indicated that neutrophils are important for control of L. monocytogenes in the liver but not the spleen during infection (71). Additional studies however indicate that they are dispensable for control of this bacterium and instead proposed that inflammatory monocytes (which were also depleted in the early anti-Gr1 studies) were essential for bacterial clearance (72). These disparate observations may indicate that the role of neutrophils may depend on the dose of bacteria. At lower doses of bacteria where the bacteria are primarily intracellular, neutrophils may be less important, while at higher doses of bacteria which cause host cells heavily infected to lyse and release extracellular bacteria, this scenario may necessitate neutrophil clean up. In the context of Blimp1-deletion, neutrophil-mediated bacterial clearance rather than inflammatory monocyte-dependent mechanisms appear to control enhanced bacterial clearance.
Our results describe a previously unknown mechanism involving BLIMP1 dependent control of CCL8 in macrophages used by the host to control L. monocytogenes infection. Our data demonstrates that BLIMP1 normally acts as a gatekeeper to curb CCL8-induced inflammation. Importantly, since BLIMP1 is also inducible in neutrophils and since LyzM Cre would also modulate BLIMP1 levels in neutrophils, our data cannot exclude additional roles for neutrophil expressed BLIMP1 in this model. In the macrophage scenario however, it is not clear if induction of Blimp1 transcription early during infection acts to minimize the release of CCL8 to control indiscriminant cytolytic acivity of neutrophils and minimize tissue damage or if this is a result of the pathogen’s strategy to defeat host innate defenses. A common feature of pathogens is their ability to evade or suppress innate immunity, in order to avoid being eliminated and establish a successful infection. The ability of multiple pathogens to induce BLIMP1 suggests that this may be a common strategy of immune evasion.
Therapeutic manipulation of Blimp1 gene expression levels or BLIMP1 protein function may therefore represent a novel means to boost protective immunity in an early microbial infection. Furthermore, dysregulation of BLIMP1 could exacerbate autoimmunity by tipping the balance towards uncontrolled inflammation. In this regard, it is noteworthy that SNPs in Blimp1 gene have been linked to Systemic Lupus Erythematosus. Enhancing Blimp1 expression under these circumstances could therefore also represent a novel means of alleviating persistent inflammatory and autoimmune diseases. Our discovery of BLIMP1’s contribution to immune function reveals novel intervention points in the treatment of inflammatory diseases.
Supplementary Material
Acknowledgments
This work was supported by the NIH (AI067497 to K.A.F, R37AI040618 to A.D.L and AI060991 to V.L.B) and by the Italy-USA Program #28C6 (to M.S).
We thank Kathryn Calame for providing Prdm1fl/fl mice and Anna Cerny for backcrossing these animals as well as for animal husbandry and genotyping.
References
- 1.Ng T, Yu F, Roy S. A homologue of the vertebrate SET domain and zinc finger protein Blimp-1 regulates terminal differentiation of the tracheal system in the Drosophila embryo. Development genes and evolution. 2006;216:243–252. doi: 10.1007/s00427-005-0044-5. [DOI] [PubMed] [Google Scholar]
- 2.Roy S, Ng T. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr Biol. 2004;14:1772–1777. doi: 10.1016/j.cub.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 3.Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- 4.Ren B, Chee KJ, Kim TH, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 7.Savitsky D, Cimmino L, Kuo T, Martins GA, Calame K. Multiple roles for Blimp-1 in B and T lymphocytes. Adv Exp Med Biol. 2007;596:9–30. doi: 10.1007/0-387-46530-8_2. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J Exp Med. 2005;202:1471–1476. doi: 10.1084/jem.20051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 10.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 11.Magnusdottir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci U S A. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horsley V, O’Carroll D, Tooze R, Ohinata Y, Saitou M, Obukhanych T, Nussenzweig M, Tarakhovsky A, Fuchs E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SJ, Zou YR, Goldstein J, Reizis B, Diamond B. Tolerogenic function of Blimp-1 in dendritic cells. J Exp Med. 2011;208:2193–2199. doi: 10.1084/jem.20110658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812– 823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 15.Cousens LP, Wing EJ. Innate defenses in the liver during Listeria infection. Immunol Rev. 2000;174:150–159. doi: 10.1034/j.1600-0528.2002.017407.x. [DOI] [PubMed] [Google Scholar]
- 16.Zenewicz LA, Shen H. Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect. 2007;9:1208–1215. doi: 10.10110/2/076/j.micinf.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seki E, Tsutsui H, Tsuji NM, Hayashi N, Adachi K, Nakano H, Futatsugi-Yumikura S, Takeuchi O, Hoshino K, Akira S, Fujimoto J, Nakanishi K. Critical roles of myeloid differentiation factor 88-dependent proinflammatory cytokine release in early phase clearance of Listeria monocytogenes in mice. J Immunol. 2002;169:3863–3868. doi: 10.4049/jimmunol.169.7.3863. [DOI] [PubMed] [Google Scholar]
- 18.Edelson BT, Unanue ER. MyD88-Dependent but Toll-Like Receptor 2- Independent Innate Immunity to Listeria: No Role for Either in Macrophage Listericidal Activity. J Immunol. 2002;169:3869–3875. doi: 10.4049/jimmunol.169.7.3869. [DOI] [PubMed] [Google Scholar]
- 19.Ozoren N, Masumoto J, Franchi L, Kanneganti TD, Body-Malapel M, Erturk I, Jagirdar R, Zhu L, Inohara N, Bertin J, Coyle A, Grant EP, Nunez G. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 21.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 22.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V. Listeria monocytogenes is sensed by the NLRP3 and AIM2 Inflammasome. Eur J Immunol. 2010 doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witte CE, Archer KA, Rae CS, Sauer JD, Woodward JJ, Portnoy DA. Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv Immunol. 2012;113:135–156. doi: 10.1016/B978-0-12-394590-7.00002-6. [DOI] [PubMed] [Google Scholar]
- 27.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-Nnitrosourea- induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, Muller M, Decker T. IFN regulatory factor 3- dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2- independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 30.O’Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, Cheng G. Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TNFR-associated NF kappa B kinasebinding kinase 1. J Immunol. 2005;174:1602–1607. doi: 10.4049/jimmunol.174.3.1602. [DOI] [PubMed] [Google Scholar]
- 31.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3- dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bielecki J, Youngman P, Connelly P, Portnoy DA. Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature. 1990;345:175– 176. doi: 10.1038/345175a0. [DOI] [PubMed] [Google Scholar]
- 35.Flo TH, Halaas O, Lien E, Ryan L, Teti G, Golenbock DT, Sundan A, Espevik T. Human toll-like receptor 2 mediates monocyte activation by Listeria monocytogenes, but not by group B streptococci or lipopolysaccharide. J Immunol. 2000;164:2064–2069. doi: 10.4049/jimmunol.164.4.2064. [DOI] [PubMed] [Google Scholar]
- 36.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 37.Chang DH, Angelin-Duclos C, Calame K. BLIMP-1: trigger for differentiation of myeloid lineage. Nat Immunol. 2000;1:169–176. doi: 10.1038/77861. [DOI] [PubMed] [Google Scholar]
- 38.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173:5556– 5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 40.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 41.Tsou CL, Peters W, Si Y, Slaymaker S, Aslanian AM, Weisberg SP, Mack M, Charo IF. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest. 2007;117:902–909. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurihara T, Warr G, Loy J, Bravo R. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J Exp Med. 1997;186:1757–1762. doi: 10.1084/jem.186.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delano MJ, Kelly-Scumpia KM, Thayer TC, Winfield RD, Scumpia PO, Cuenca AG, Harrington PB, O’Malley KA, Warner E, Gabrilovich S, Mathews CE, Laface D, Heyworth PG, Ramphal R, Strieter RM, Moldawer LL, Efron PA. Neutrophil mobilization from the bone marrow during polymicrobial sepsis is dependent on CXCL12 signaling. J Immunol. 2011;187:911–918. doi: 10.4049/jimmunol.1100588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Islam SA, Chang DS, Colvin RA, Byrne MH, McCully ML, Moser B, Lira SA, Charo IF, Luster AD. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat Immunol. 2011;12:167–177. doi: 10.1038/ni.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Hamada S, Umemura M, Shiono T, Tanaka K, Yahagi A, Begum MD, Oshiro K, Okamoto Y, Watanabe H, Kawakami K, Roark C, Born WK, O’Brien R, Ikuta K, Ishikawa H, Nakae S, Iwakura Y, Ohta T, Matsuzaki G. IL-17A produced by gammadelta T cells plays a critical role in innate immunity against listeria monocytogenes infection in the liver. J Immunol. 2008;181:3456–3463. doi: 10.4049/jimmunol.181.5.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ebert LM, Meuter S, Moser B. Homing and function of human skin gammadelta T cells and NK cells: relevance for tumor surveillance. J Immunol. 2006;176:4331–4336. doi: 10.4049/jimmunol.176.7.4331. [DOI] [PubMed] [Google Scholar]
- 49.Penido C, Costa MF, Souza MC, Costa KA, Candea AL, Benjamim CF, Henriques MG. Involvement of CC chemokines in gammadelta T lymphocyte trafficking during allergic inflammation: the role of CCL2/CCR2 pathway. Int Immunol. 2008;20:129–139. doi: 10.1093/intimm/dxm128. [DOI] [PubMed] [Google Scholar]
- 50.Kawaguchi M, Adachi M, Oda N, Kokubu F, Huang SK. IL-17 cytokine family. J Allergy Clin Immunol. 2004;114:1265–1273. doi: 10.1016/j.jaci.2004.10.019. quiz 1274. [DOI] [PubMed] [Google Scholar]
- 51.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 53.Ng LG, Sutherland AP, Newton R, Qian F, Cachero TG, Scott ML, Thompson JS, Wheway J, Chtanova T, Groom J, Sutton IJ, Xin C, Tangye SG, Kalled SL, Mackay F, Mackay CR. B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J Immunol. 2004;173:807–817. doi: 10.4049/jimmunol.173.2.807. [DOI] [PubMed] [Google Scholar]
- 54.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 55.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 56.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 57.Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2:403–411. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linden A, Laan M, Anderson GP. Neutrophils, interleukin-17A and lung disease. Eur Respir J. 2005;25:159–172. doi: 10.1183/09031936.04.00032904. [DOI] [PubMed] [Google Scholar]
- 60.Forlow SB, Schurr JR, Kolls JK, Bagby GJ, Schwarzenberger PO, Ley K. Increased granulopoiesis through interleukin-17 and granulocyte colony-stimulating factor in leukocyte adhesion molecule-deficient mice. Blood. 2001;98:3309–3314. doi: 10.1182/blood.v98.12.3309. [DOI] [PubMed] [Google Scholar]
- 61.Linden A, Adachi M. Neutrophilic airway inflammation and IL-17. Allergy. 2002;57:769– 775. doi: 10.1034/j.1398-9995.2002.02164.x. [DOI] [PubMed] [Google Scholar]
- 62.Witowski J, Pawlaczyk K, Breborowicz A, Scheuren A, Kuzlan-Pawlaczyk M, Wisniewska J, Polubinska A, Friess H, Gahl GM, Frei U, Jorres A. IL-17 stimulates intraperitoneal neutrophil infiltration through the release of GRO alpha chemokine from mesothelial cells. J Immunol. 2000;165:5814–5821. doi: 10.4049/jimmunol.165.10.5814. [DOI] [PubMed] [Google Scholar]
- 63.Ruddy MJ, Shen F, Smith JB, Sharma A, Gaffen SL. Interleukin-17 regulates expression of the CXC chemokine LIX/CXCL5 in osteoblasts: implications for inflammation and neutrophil recruitment. J Leukoc Biol. 2004;76:135–144. doi: 10.1189/jlb.0204065. [DOI] [PubMed] [Google Scholar]
- 64.Prause O, Laan M, Lotvall J, Linden A. Pharmacological modulation of interleukin- 17-induced GCP-2-, GRO-alpha- and interleukin-8 release in human bronchial epithelial cells. Eur J Pharmacol. 2003;462:193–198. doi: 10.1016/s0014-2999(03)01341-4. [DOI] [PubMed] [Google Scholar]
- 65.O’Brien RL, Roark CL, Jin N, Aydintug MK, French JD, Chain JL, Wands JM, Johnston M, Born WK. gammadelta T-cell receptors: functional correlations. Immunol Rev. 2007;215:77–88. doi: 10.1111/j.1600-065X.2006.00477.x. [DOI] [PubMed] [Google Scholar]
- 66.Thedrez A, Sabourin C, Gertner J, Devilder MC, Allain-Maillet S, Fournie JJ, Scotet E, Bonneville M. Self/non-self discrimination by human gammadelta T cells: simple solutions for a complex issue? Immunol Rev. 2007;215:123–135. doi: 10.1111/j.1600-065X.2006.00468.x. [DOI] [PubMed] [Google Scholar]
- 67.Schulz SM, Kohler G, Holscher C, Iwakura Y, Alber G. IL-17A is produced by Th17, gammadelta T cells and other CD4- lymphocytes during infection with Salmonella enterica serovar Enteritidis and has a mild effect in bacterial clearance. Int Immunol. 2008;20:1129– 1138. doi: 10.1093/intimm/dxn069. [DOI] [PubMed] [Google Scholar]
- 68.Umemura M, Yahagi A, Hamada S, Begum MD, Watanabe H, Kawakami K, Suda T, Sudo K, Nakae S, Iwakura Y, Matsuzaki G. IL-17-mediated regulation of innate and acquired immune response against pulmonary Mycobacterium bovis bacille Calmette-Guerin infection. J Immunol. 2007;178:3786–3796. doi: 10.4049/jimmunol.178.6.3786. [DOI] [PubMed] [Google Scholar]
- 69.Li W, Moltedo B, Moran TM. Type I interferon induction during influenza virus infection increases susceptibility to secondary Streptococcus pneumoniae infection by negative regulation of gammadelta T cells. J Virol. 2012;86:12304–12312. doi: 10.1128/JVI.01269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 71.Carr KD, Sieve AN, Indramohan M, Break TJ, Lee S, Berg RE. Specific depletion reveals a novel role for neutrophil-mediated protection in the liver during Listeria monocytogenes infection. Eur J Immunol. 2011;41:2666–2676. doi: 10.1002/eji.201041363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi C, Hohl TM, Leiner I, Equinda MJ, Fan X, Pamer EG. Ly6G+ neutrophils are dispensable for defense against systemic Listeria monocytogenes infection. J Immunol. 2011;187:5293–5298. doi: 10.4049/jimmunol.1101721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.