Abstract
NLR (nucleotide-binding domain, leucine-rich repeat containing) proteins have rapidly emerged as central regulators of immunity and inflammation with demonstrated relevance to human diseases. Much attention has focused on the ability of several NLRs to activate the inflammasome complex and drive proteolytic processing of inflammatory cytokines; however, NLRs also regulate important inflammasome-independent functions in the immune system. In this review, we will discuss several of these functions, including the regulation of canonical and non-canonical NF-κB activation, MAP kinase activation, cytokine and chemokine production, antimicrobial reactive oxygen species production, type I interferon production, and RNase L activity. We will also explore the mechanistic basis of these functions and present current challenges in the field.
The genomic mining of evolutionarily conserved gene families with structural similarity has led to the discovery of a large gene family (NLRs) encoding proteins with a characteristic arrangement of nucleotide binding domain (NBD) and leucine rich region (LRR) in both plants and animals. NLRs share structural similarity with a subgroup of plants disease resistance (R) genes, which confer resistance to infection caused by fungal, viral, parasitic and insect pathogens by inducing cell death of infected cells (1). Among animals, NLR proteins are found in species ranging from sea urchin to human (2). The most prominent function of NLRs is the intracellular sensing of structures shared by classes of microbes and endogenous molecules associated with inflammation, known as pathogen-associated molecular patterns (PAMPs) and damaged-associated molecular patterns (DAMPs), respectively. The precise mechanism by which this “sensing” occurs, however, remains a major challenge in the field (2). NLR proteins also have functions outside of the innate immune system, such as the regulation of major histocompatibility complex (MHC) genes to affect adaptive immunity, or the regulation of cell death. This review will primarily focus on the role of non-inflammasome NLRs in innate immunity because data are converging to indicate that these NLRs can be categorized into functional subgroups which regulate other crucial innate immune pathways such as canonical and noncanonical NF-κB, MAPK, type I interferon, cytokines and chemokines.
An underlying reason for the intense attention paid to NLRs is their association with genetic immunologic disorders in humans. For example, mutations in the class II, major histocompatability complex, transactivator (CIITA), the master activator of MHC class II gene transcription, result in immunodeficiency. Mutations in NOD2 are associated with susceptibility to Crohn's disease (a type of inflammatory bowel disease) and Blau syndrome (a granulomatous inflammatory disorder). Mutations in the NLRP3 gene predispose patients to a variety of autoinflammatory disorders. Association of NLRs with asthma, vitiligo (disease characterized by patchy depigmentation of skin) and urticaria skin rash has also been shown. Thus, NLRs are important determinants of human inflammatory disorders and an in depth understanding of their molecular mechanisms of action is crucial to understand their basic biology and also to develop targeted therapies.
How does activation of NOD1 and NOD2 regulate immunity in the host?
NOD1 and NOD2 were two of the first characterized members of the NLR family. Shortly after their identification, it was recognized that several polymorphisms encoding non-conservative amino acids or frameshift mutations in the leucine rich repeat of NOD2 were found in some familial cases of Crohn's disease. A different polymorphism in the nucleotide binding domain of NOD2 was found to associate Blau syndrome (3). Despite years of intensive research, however, the mechanism by which variant NOD2 proteins lead to the enhanced inflammation associated with Crohn's diseases remains enigmatic. There are at least three working models. The first posits that NOD2 is a positive regulator of immune defense and defective NOD2 cannot contain pathogen infection (4). Indeed, several Crohn's disease-associated NOD2 mutations confer impaired activation of the transcription factor nuclear factor kappa B (NF-κB), whereas Blau syndrome-associated mutations lead to constitutive NF-κB activation (5). Nod2-deficient mice show a reduction in the level of antimicrobial α-defensins in Paneth cells of the intestine (4), consistent with samples from ileal Crohn's disease patients indicating an association between the NOD2 variant genotype and reduced α-defensin (6). However the association of NOD2 mutation with α-defensin was not found by all (7). The second model suggests that NOD2 is protective against Crohn's disease because it negatively regulates Toll like receptor (TLR)-mediated responses to the intestinal bacterial flora. This model is supported by the analysis of a second Nod2-deficient mouse, which showed increased T helper 1-associated cytokine production and NF-κB activation in response to a TLR2 agonist (8). In support of this, TLR2 ligand administration in control but not Nod2-deficient mice greatly reduced TLR2-induced inflammatory responses. TLR2 ligand delivery also reduced chemically-induced colitis in a NOD-dependent fashion. Remarkably, reintroduction of wildtype NOD2 into Nod2-deficient mice led to resistance to colitis (9). A third model hypothesizes that Crohn's-associated NOD2 variants cause increased inflammatory response, because the replacement of Nod2 with a disease variant form resulted in an elevated inflammatory responses, including enhanced interleukin (IL)-1β secretion and NF-κB activation in mice (10). Monocytes from Crohn's disease patients homozygous for this allele, however, actually demonstrate impaired IL-1β secretion (11), suggesting there may be context-dependent, species-specific differences. Finally, recent work suggests that a different Crohn's disease associated mutation in NOD2 leads to a novel interaction between the mutant NOD2 and heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). This interaction results in inhibition of hnRNP A1-mediated production of the anti-inflammatory cytokine, IL-10 (12). Interestingly, the interaction between mutant NOD2 and hnRNP A1 was observed only with human NOD2 and the human IL-10 promoter but not the murine counterparts. The contribution of this novel inhibitory effect of disease associated NOD2 to Crohn's disease pathogenesis remains to be determined.
Early studies sought to identify the PAMPs responsible for activating NOD1 and NOD2. NOD1 and NOD2 respond to bacterial peptidoglycan-derived molecules meso-diaminopimelic acid (DAP) and muramyl dipeptide (MDP), respectively (3, 13). NOD1 can also be activated by meso-lanthionine, another peptidoglycan-associated diamino-amino acid. N-glycolylated MDP, which is made by mycobacteria and actinomycetes, is substantially more potent in its ability to elicit NOD2-dependent activation of NF-κB than N-acylated MDP, generated by typical gram-positive and gram-negative bacteria (14). Thus, NOD1 and NOD2 are activated in response to a number of peptidoglycan-derived moieties stemming from a broad range of bacterial sources. Although NLR proteins are now frequently referred to as receptors, it is important to note that neither NOD1 nor NOD2 has been shown to directly interact with their activating peptidoglycans in a manner consistent with a pattern recognition receptor. The LRR domains of these proteins are required to confer responsiveness to their respective stimuli, leading some to suggest that these domains either bind directly to the cognate peptidoglycan components or to other intracellular protein(s) that act as an intermediate between these bacterial products and NOD1 or NOD2.
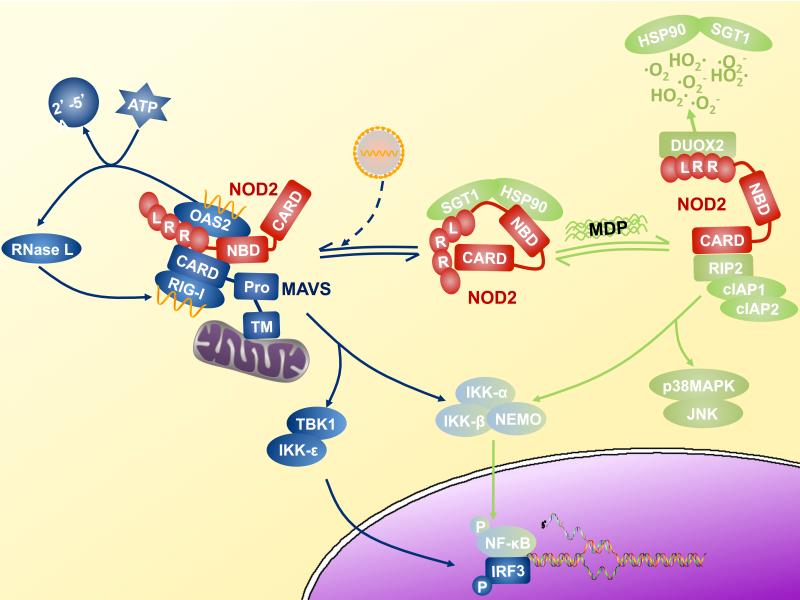
Similar to other NLR molecules, NOD2 signals by acting as a scaffold for the assembly of large multicomponent signaling complexes (Fig. 1). NOD2 induces multiple effector pathways that are involved in the host response to pathogenic bacteria. The best characterized effector signaling pathway of NOD2 leads to activation of NF-κB through interactions with the receptor interacting protein-2 (RIP2, also known as RICK or CARDIAK), a serine/threonine kinase (3). After MDP is internalized by phagocytosis or bacterial invasion of the cytoplasm, intracellular NOD2 translocates to the plasma membrane (15). There, it associates with RIP2, through the homotypic interactions of caspase activation and recruitment domains (CARD), thereby allowing membrane translocation of RIP2. Rather than inducing NF-kB activation through RIP2 mediated phosphorylation of IκB kinase, the NOD2-RIP2 complex activates the IκB kinase complex through K63-linked polyubiquitination of its γ subunit (also known as NEMO). This is achieved through the recruitment of the ubiquitin ligases cIAP1 and 2 to the signaling complex (16). In parallel to the activation of NF-κB, the NOD2-RIP2 complex also stimulates the mitogen activated protein kinases (MAPK) p38 and JNK. The molecular mechanisms that regulate the formation of a membrane-bound, RIP2-NOD2 complex are not fully determined, however numerous cellular proteins have been implicated in NOD2 signaling . The mechanism by which these multiple factors interact to regulate NOD2 signaling during physiologic responses to infection and under pathologic conditions such as Crohn's disease require further investigation (17).
Fig. 1.
NOD2 signaling bifurcates into antibacterial and antiviral effector arms. A model of NOD2 signaling is presented in which NOD2 is bound to the chaperonin and ubiquitin ligase pair, HSP90 and SGT in the basal state (Complex A). This is thought to hold the inactive NOD2 in a signaling competent form (48). Upon stimulation with MDP, NOD2 binds to RIP2, and activates NF-κB and MAPK (p38 and JNK) through recruitment of several intracellular proteins, including cIAP1 and cIAP2 (Complex B). This leads to in the induction of chemokines, cytokines and defensins, which mediate the antimicrobial responses. Similarly, viral infection can activate NOD2, leading to its translocation to the mitochondria, association with MAVS, and the induction of the antiviral cytokine, type I interferon (Complex C). Double stranded RNA, and possibly single stranded RNA, induces OAS-2 to interact with NOD2. NOD2 then stimulates the production of -5’ oligoadenylate synthase type 2, which activates antiviral responses including RNaseL activation (Complex C). RIG-I is depicted because it is the PRR that binds to viral nucleic acids, while TBK and IKK-e lies downstream of MAVS to activate IRF-3.
MDP-mediated activation of NOD2-RIP2 complex is known to modulate both innate and adaptive immune responses by activating expression of numerous cytokines and chemokines. NOD2 control of cytokine expression is mediated largely through its ability to activate NF-κB and p38 MAPK-dependent signaling. Several studies, however, suggest that NOD2 has a role in caspase-1 activation and subsequent IL-1α processing in response to MDP. One study carried out in cells isolated from NOD2-deficient mice suggests that MDP induces caspase-1 activation and IL-1α processing through the NOD2 and ASC/NLRP3 inflammasome (18) , although another found MDP activation of caspase-1 is independent of NOD2 (19). These discrepancies are likely due to the distinct inflammasome activation protocols used. There is also evidence that indicates NLRP1 activates caspase-1 in an MDP responsive fashion (20, 21). NOD2 has been reported to associate with NLRP1 and cooperate in caspase-1 activation in response to MDP (21). This heterotypic NLR:NLR interaction is interesting because in vitro cell-free reconstitution of MDP-stimulated NLRP1 inflammasome formation represents the only evidence that any NLR protein actually directly senses a PAMP. Verification of such a finding in the presence or absence of purified NOD2 would be crucial in assessing if NLRP1 might be the (or one of the) cellular proteins required to confer MDP- responsiveness on NOD2 because direct binding of MBP by NOD2 has yet to be observed.
Besides regulating cytokine production, signaling through NOD2 also induces host antimicrobial responses in both immune and epithelial cells. Most of the NOD2-induced antimicrobial responses require NF-κB and MAPK activation, which regulates expression of antimicrobial peptides including α defensins, β defensins, and cryptidins (4). NOD2 also associates with the NAD(P)H oxidase family member DUOX2 (22). Microbicidal reactive oxygen species (ROS) production by DUOX2 appears to be regulated through MDP-NOD2 signaling. These results indicate that recognition of MDP by NOD2 induces multiple effector responses to enhance intercellular communication through cytokine, chemokine and defensin production, and antimicrobial function by ROS production. These new observations present the pressing challenge of defining if these disparate actions of NOD2 are mediated by a single multifunctional complex or by distinct biochemical complexes
How do NLRs affect the mito-signalosome?
Besides the biochemical complexes defined above, recent reports have found a new linkage between NLRs and a newly defined multimeric complex that is located in the mitochondria, which together regulates the production of antiviral type I interferon (IFN) and inflammatory cytokines. The mitochondria have been typically associated with pivotal roles in oxidative phosphorylation, ATP generation, ROS production, cell survival, programmed cell death and autophagy; however, recent evidence suggests that mitochondria can act as central platforms for innate antiviral responses. This function centers on the MAVS/IPS-1/VISA/Cardif protein, which is an immune-activating adaptor for type I IFN production that is located in the mitochondria (23-26). Functional and physical associations of MAVS with NLR proteins have been demonstrated, and are proposed to regulate type I IFN and other inflammatory cytokines. This complex, which also includes helicases that directly bind to viral single stranded RNA, double stranded viral RNA intermediates, RNaseL-cleaved host RNA and other regulatory proteins is referred to in this review as the mito-signalosome.
MAVS is ubiquitously expressed and its elimination dramatically ablates type I IFN production in response to specific signals (Fig. 2). Some cell types are heavily dependent on MAVS for the type I IFN response caused by RNA viruses, yet MAVS-independent, Toll like receptor-dependent machinery is important for type I IFN production in other cell types (25, 27). MAVS activates the transcription factor interferon response factor (IRF)-3 and NF-κB. NF-κB, in turn, drives increased production of type I IFN and pro-inflammatory cytokines like IL-6. MAVS can also induce apoptosis in response to some viral infections; however, certain viral proteins such as those from SARS-coronavirus and hepatitis C virus can antagonize this function (28).
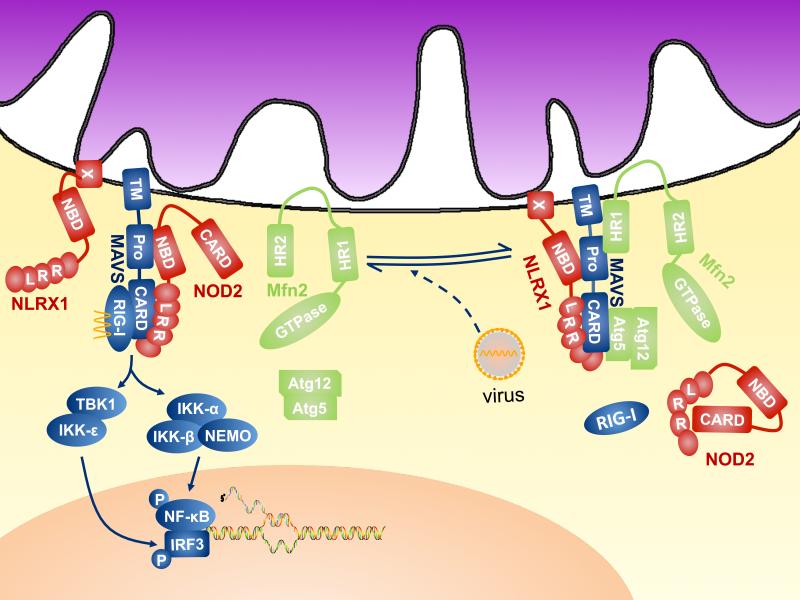
Fig. 2.
NLR proteins and regulation of the mito-signalosome. In the quiescent state (Complex A), the CARD:CARD homotypic interaction between MAVS and RIG-I is prevented by MAVS association with NLRX1, Atg5-Atg12 conjugate, perhaps by steric hindrance. Mfn2 interacts with the C-terminus and the transmembrane region of MAVS to abolish its immune-activating function. The three regulatory proteins target different regions of MAVS to execute the “molecular brake”. In the presence of cytosolic 5'-triphosphate single stranded RNA or double stranded RNA, these brakes are released, which renders the assembly of the active form of mitosignalosome (Complex B), in which MAVS interacts with NOD2 and RLRs, such as RIG-I, which directly interact with viral nucleic acid to trigger type I IFN production.
A key pathway by which MAVS regulates inflammation is through its interaction with the RIGI-like family of pathogen recognition receptors (RLRs). Two of the RLR family members, retinoic-acid-inducible protein I (RIG-I) and melanoma-differentiation-associated gene 5 (MDA-5) share a conserved domain structure consisting of two N-terminal CARD domains and a DExD/H-box helicase domain; however, they exhibit distinct preferences for the molecular features of RNA ligands and RNA viruses (29). RIG-I binds to short dsRNA and 5’-triphosphate-bearing ssRNA whereas MDA5 recognizes long dsRNA (30)
Given the substantial impact of type I IFN production, it is not surprising that MAVS is subjected to meticulous checks-and-balances by negative regulators. One such negative regulator is the NLR protein, NLRX1 (31). NLRX1 is an unusual NLR member in that it is located at the mitochondria and contains a mitochondrial targeting sequence at its amino terminus; however its precise mitochondrial location is in dispute (31, 32). Furthermore, it contains an N-terminal effector domain that bears similarity to both Pyrin and CARD domains but cannot be categorized as either. Finally, the sequences encoding the central nucleotide binding domain is split into two exons at the genomic level and the encoded domain lacks a Walker A motif that is required for ATP binding found in other NLRs. Instead of direct microbial sensing, NLRX1 interacts with MAVS to prevent its binding to RIG-I, thus compromising the activation of NF-κB and IRF3 in response to cytosolic RNA and leading to the inhibition of type I IFN and proinflammatory cytokine production (31). Overexpression studies also suggest that NLRX1 might positively regulate the production of ROS from mitochondria in response to an intracellular bacterial pathogen, although verification with a non-overexpression system is necessary (33). Further delineation of the physiologic function of NLRX1 would be aided by generation of Nlrx1-deficient mice.
Similar to NLRX1, five other proteins, including Atg5-Atg12 conjugate, gC1qR, Mfn2, PSMA7 and PCBP2, have recently been identified in the mito-signalosome negative regulatory module (34-38). Reduction of protein expression by RNA interference results in enhanced type I IFN production in response to certain RNA viruses. These proteins, however, are unlikely to be functionally redundant because they regulate mito-signalosome activation via distinct mechanisms, including molecular steric hindrance, autophagy and post-transcriptional destabilization of MAVS. Interestingly, MAVS resides in a high-molecular weight complex in the quiescent state and a substantial fraction of MAVS migrates into lower molecular weight fractions after stimulation, suggesting it is released from a negative regulatory complex which precludes signal transduction at baseline (37). NLRX1 and several negative regulators of MAVS are proposed to work by steric hindrance. For example, NLRX1 inhibits Sendai virus-induced homotypic CARD:CARD interactions between RIG-I and MAVS (31). Atg5-Atg12 conjugate-mediated inhibition of MAVS signaling acts by a similar mechanism (35). In contrast, Mfn2 interacts with the C-terminal and transmembrane region of MAVS rather than the N-terminal CARD domain (37), although it is unclear whether Mfn2 can sterically preclude upstream RLR engagement by MAVS. These findings are also compatible with the possibility that binding of inhibitory factors induces conformational changes in MAVS that reduce its affinity for RLRs. Furthermore, MAVS activity is also attenuated by its association with the proteosome subunit, PMSA7 (34), and the ubiquitin ligase, AIP4, via PCBP2 (38). Whether these are connected to the NLR pathway has not been investigated.
Besides NLRX1, a recent report demonstrated that interaction between MAVS and NOD2 regulates type I IFN (Fig. 2). Over-expression of NOD2 but not other NLRs such as NOD1, NLRC4, NAIP or NLRC3 provides HEK293 cells the capability to activate IRF3 in response to single stranded RNA or infection with the single stranded RNA virus, respiratory syncytial virus (RSV) (39). Interestingly, endogenous expression of NOD2 was also shown to be induced by single stranded RNA treatment or RSV infection. Depletion of NOD2 ablated type I interferon production in these cells. Furthermore, immunoprecipitation of NOD2 led to the recovery of RSV-specific RNA, which was substantiated in a cell-free system. Thus this study suggests that NOD2 can positively regulate type I IFN by direct or indirect association with viral single stranded RNA. Patients with NOD2 polymorphisms that lead to altered NOD2 function, however, are not known to have difficulties with viral infection. This could be a species-specific difference in that the anti-viral role of NOD2 is specific to mice or that the disease-associated NOD2 variants are not affected in their anti-viral function. As with MDP, however, it is still premature to define NOD2 as a bona fide pattern recognition receptor for single stranded RNA because immunoprecipitation is likely to pull down NOD2-interacting proteins. MAVS associates with both NOD2 and RIG-I/MDA-5, thus it is possible that the recovery of RSV-specific RNA is the result of co-immunoprecipitation of RIG-I.
NOD2 also has been found to interact with 2’-5’ oligoadenylate synthase type 2 (OAS2), a known RNA binding protein (40). Binding of double stranded RNA to OAS2 results in the generation of 2’-5’- adenosine oligomers, which activate intracellular RNase L. RNase L then destroys viral RNA and impairs further viral production in the infected cell by degrading cellular RNAs (41). RNase L-generated RNA species can also activate RLRs, thus endowing the mitosignalosome with a positive feedback mechanism. These data suggest that the association with OAS2 links NOD2-mediated type I IFN production to RNase L activation, which synergistically amplify host antiviral responses (40). It is intriguing to consider that the induction of type I IFN and RNase L activation by NOD2 in response to viral infection might parallel the bifurcated signaling and antimicrobial response seen during NOD2 activation by bacterially-derived MDP (Fig. 1). It will be of great interest to assess if NOD2 can bind RNA in the absence of accessory molecules such as RIG-I or OAS2 because evidence of direct binding would establish the structural basis for the classification of NOD2 as a pattern recognition receptor.
How does NLR affect the non-canonical NF-κB pathway?
One of the first pyrin-encoding NLR genes to be identified was NLRP12 (also known as Monarch-1 or Pypaf7) (42-44). The expression of this gene is restricted to the myeloid-monocytic compartment, thus implicating a role in immunity. Early work using over-expression of NLRP12 demonstrated it can activate NF-κB, and furthermore, the introduction of NLRP12, pro-IL1β, pro-caspase-1 and the common inflammasome adaptor, ASC/Pycard into 293T cells can activate caspase-1 with a corresponding increase in IL-1β production (43). Thus, NLRP12 exhibits properties of an inflammasome NLR; however, gene silencing by short hairpin RNAs or gene deletion has not verified an affect of NLRP12 on IL-1β production. One possible explanation for these opposing results is that the specific activator of the NLRP12-dependent inflammasome was not used in these experiments.
In contrast to its proinflammatory role in inflammasome activation, NLRP12 has also been shown to impede activation of the non-canonical NF-κB pathway downstream of the tumor necrosis factor receptor (TNFR) pathway (42). Activation of NF-κB can occur through two distinct mechanisms, referred to as the canonical and non-canonical pathways (42, 45). Whereas the canonical pathway is triggered rapidly after cell stimulation, the non-canonical pathway exhibits much slower kinetics and is entirely dependent upon the NF-κB-inducing kinase (NIK), which is not required for canonical NF-κB activation. NIK associates with p100 subunit of NF-κB and induces its cleavage to its active form, p52, which causes the expression of a distinct subset of inflammatory genes. NLRP12 downregulates the function of several important molecules involved in TNFR signaling, including NIK, RIP1, TNF Receptor Associated Factor (TRAF) 2 and TRAF6 in human cell lines. Furthermore, both over-expression and small interfering RNA approaches show that NLRP12 reduces the level of NIK by a proteosome-dependent pathway, although the precise mechanism is unknown. Besides NLRP12, NOD2 may also regulate NIK function (46, 47). This is supported by several observations: when over-expressed, NIK can associate with NOD2, NOD2 and MDP can enhance p100 processing, and NIK is required for MDP-induced transcription of the chemokine, CXCL13. NIK function is only partially attenuated in macrophages lacking NOD2, suggesting that there might be redundant regulation of NIK by other proteins, including other NLRs.
What are the major challenges in the NLR field?
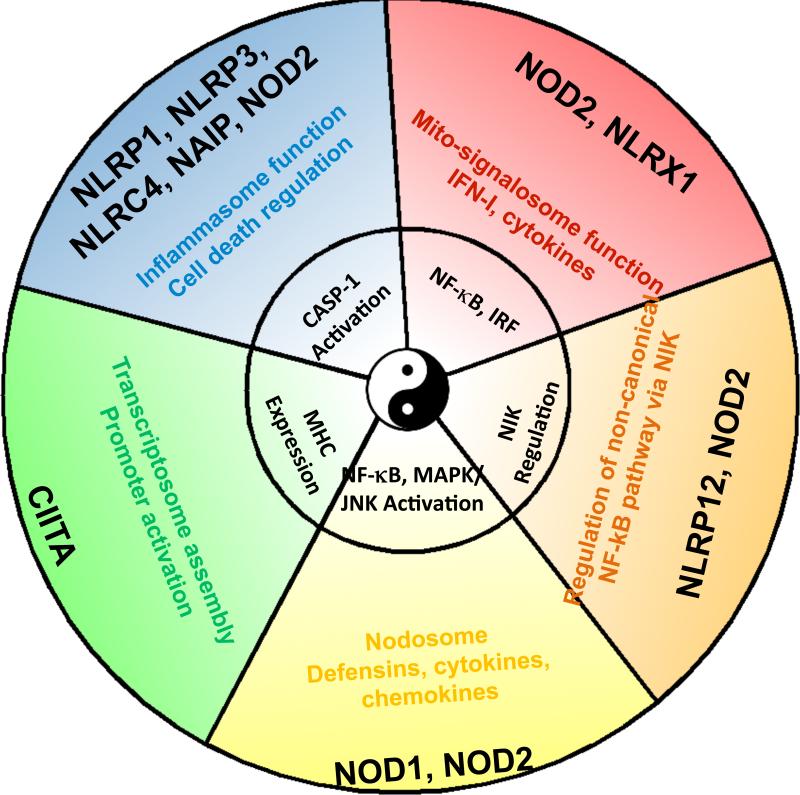
This review summarizes the noninflammasome immune functions of NLRs. In total, studies of NLRs show that they interface with major regulatory pathways that impact the immune system, including caspase activation, cytokine, chemokine and defensin production, canonical and non-canonical NF-κB pathways, MAPK, antimicrobial ROS, interferon production, RNase L activity, MHC expression and various forms of cell death (Fig. 3). A major challenge is to assess how NLRs function outside of the immune system because many NLRs have broad tissue distribution and they participate in signaling pathways that are broadly employed throughout the organism. Such fields of research include developmental and reproductive biology, metabolism and cancer, where connections with NLRs are less-developed but ripe for exploration.
Fig. 3.
NLR proteins signal through different multi-component signalosomes. NLR signaling modules include the CIITA transcriptosome, the caspase-1 activating inflammasomes, the interferon/cytokine inducing mito-signalosome, the NF-kB/MAPK activating NOD1/2 complex (referred to as the NODosome), and the NIK pathway. This figure depicts the concept that one NLR can serve multiple functions, while multiple NLRs can also serve similar functions.
Another challenge is to further understand the in vivo function of both well-studied and understudied NLRs in immune and non-immune systems. This will require that we understand the cellular and tissue distribution of the endogenous, not just over-expressed, NLRs. It also requires that we understand dynamic changes in their sub-cellular localization upon activation with the appropriate agonists. This has not been addressed for most NLRs because of the lack of good antibodies specific for endogenous NLRs,.
A third challenge is to determine which NLRs can undergo homo- and hetero-typic association with other NLRs. Such interactions have long been proposed (2) but their significance might be unappreciated. The functional and physical interactions of NOD2 with NLRP1 or NLRP3 are prime examples where MDP stimulation of the inflammasome is achieved (18, 21). Thus the potential heterotypic association of 22 NLRs should exponentially expand repertoire of biologic functions ascribed to NLRs not only in the immune system, but more broadly in all organs and tissues. A natural extension of such a model is that each NLR is not in a tidy functional group, but rather has multiple functions. Multiple functions could be executed by partnering with other NLRs and participating in large functional complexes, which depending on the stimulatory signal, would be distinct in their composition and function (Fig. 3).
This leads to a fourth challenge, which is to decipher how a single NLR mediates distinct functions. For example, several NLRs participate in both caspase-1 activation and the induction of cell death. NOD2 affects not only RIP2, NF-κB, MAPK and NIK, but also the mitosignalosome. Are these distinct functions achieved by its participation in the same or different signalosomes? Does NOD2 partner with different adaptors and NLR proteins?
A fifth challenge, which is fundamental to our understanding of NLRs, is: are NLRs simply components of large signaling complexes, or are they co-receptors for pattern recognition receptors or even true pattern recognition receptors? Or is it combination of these possibilities? As presented in this review, there is abundant evidence that each NLR is a component of a large signaling complex. In the case of NLRP3, an inflammasome-activating NLR, it is difficult to envision how a single molecule might be the receptor for the multitude of PAMPs with divergent chemical compositions that can activate this inflammasome. In other cases where the function appears highly restricted, however, a receptor-ligand function seems to be a greater possibility.
This leads to sixth challenge that when met, will help resolve the mechanism of action of NLRs: what is the structure of NLR proteins? A resolution of the protein structure of each NLR protein will help to resolve the mechanism by which NLRs interact with other proteins within a signalosome and also whether NLRs directly bind to PAMPs. NLRs are notoriously difficult to purify, and thus resolving their full-length structure will be a major technical hurdle, whereas solving the structure of individual domains is more feasible. Although these are challenging issues to resolve, considering the rapid pace of NLR research, much progress is likely to be accomplished in the near future.
Acknowledgments
We would like to acknowledge the very large number of researchers who have contributed to this field whose work was not cited or was cited through other's review articles due to space limitations. This work is supported by AI063031, DE016326, U19AI077437, DK38108 and SERCEB. JAD is supported by NIH CA131645, AI057157, AI031496 and by the Burroughs Wellcome Fund Career Award for Medical Scientists.
References and Notes
- 1.Ausubel FM. Nat Immunol. 2005 Oct;6:973. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 2.Ting JP, Davis BK. Annu Rev Immunol. 2005;23:387. doi: 10.1146/annurev.immunol.23.021704.115616. [DOI] [PubMed] [Google Scholar]
- 3.Kanneganti TD, Lamkanfi M, Nunez G. Immunity. 2007 Oct;27:549. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi KS, et al. Science. 2005 Feb 4;307:731. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 5.Chamaillard M, et al. Proc Natl Acad Sci U S A. 2003 Apr 18;100:3455. doi: 10.1073/pnas.0530276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wehkamp J, et al. Gut. 2004 Nov;53:1658. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simms LA, et al. Gut. 2008 Jul;57:903. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe T, Kitani A, Murray PJ, Strober W. Nat Immunol. 2004 Aug;5:800. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe T, et al. J Clin Invest. 2008 Feb;118:545. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda S, et al. Science. 2005 Feb 4;307:734. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 11.Li J, et al. Hum Mol Genet. 2004 Aug 15;13:1715. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 12.Noguchi E, Homma Y, Kang X, Netea MG, Ma X. Nat Immunol. 2009 May;10:471. doi: 10.1038/ni.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritz JH, Le Bourhis L, Magalhaes JG, Philpott DJ. Trends Immunol. 2008 Jan;29:41. doi: 10.1016/j.it.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Coulombe F, et al. J Exp Med. 2009 Aug 3;206:1709. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecine P, et al. J Biol Chem. 2007 May 18;282:15197. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand MJ, et al. Immunity. 2009 Jun 19;30:789. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Tattoli I, Travassos LH, Carneiro LA, Magalhaes JG, Girardin SE. Semin Immunopathol. 2007 Sep;29:289. doi: 10.1007/s00281-007-0083-2. [DOI] [PubMed] [Google Scholar]
- 18.Pan Q, et al. J Leukoc Biol. 2007 Jul;82:177. doi: 10.1189/jlb.1006627. [DOI] [PubMed] [Google Scholar]
- 19.Kanneganti TD, et al. Immunity. 2007 Apr;26:433. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Faustin B, et al. Mol Cell. 2007 Mar 9;25:713. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 21.Hsu LC, et al. Proc Natl Acad Sci U S A. 2008 Jun 3;105:7803. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipinski S, et al. J Cell Sci. 2009 Oct 1;122:3522. doi: 10.1242/jcs.050690. [DOI] [PubMed] [Google Scholar]
- 23.Kawai T, et al. Nat Immunol. 2005 Oct;6:981. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 24.Meylan E, et al. Nature. 2005 Oct 20;437:1167. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 25.Seth RB, Sun L, Ea CK, Chen ZJ. Cell. 2005 Sep 9;122:669. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Xu LG, et al. Mol Cell. 2005 Sep 16;19:727. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Kumar H, et al. J Exp Med. 2006 Jul 10;203:1795. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei Y, et al. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, et al. Nature. 2006 May 4;441:101. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 30.Yoneyama M, Fujita T. Immunity. 2008 Aug 15;29:178. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Moore CB, et al. Nature. 2008 Jan 31;451:573. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 32.Arnoult D, et al. J Cell Sci. 2009 Sep 1;122:3161. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tattoli I, et al. EMBO Rep. 2008 Mar;9:293. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia Y, et al. J Immunol. 2009 Oct 1;183:4241. doi: 10.4049/jimmunol.0901646. [DOI] [PubMed] [Google Scholar]
- 35.Jounai N, et al. Proc Natl Acad Sci U S A. 2007 Aug 28;104:14050. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Xiao N, Liu F, Ren H, Gu J. Proc Natl Acad Sci U S A. 2009 Feb 3;106:1530. doi: 10.1073/pnas.0811029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasukawa K, et al. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 38.You F, et al. Nat Immunol. 2009 Nov;1 [Google Scholar]
- 39.Sabbah A, et al. Nat Immunol. 2009 Oct;10:1073. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dugan JW, et al. Mol Immunol. 2009 Oct;22 [Google Scholar]
- 41.Malathi K, et al. Proc Natl Acad Sci U S A. 2005 Oct 11;102:14533. doi: 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lich JD, et al. J Immunol. 2007 Feb 1;178:1256. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, et al. J Biol Chem. 2002 Aug 16;277:29874. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 44.Williams KL, Taxman DJ, Linhoff MW, Reed W, Ting JP. J Immunol. 2003 Jun 1;170:5354. doi: 10.4049/jimmunol.170.11.5354. [DOI] [PubMed] [Google Scholar]
- 45.Bonizzi G, Karin M. Trends Immunol. 2004 Jun;25:280. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Ogura Y, et al. J Biol Chem. 2001 Mar 16;276:4812. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 47.Pan Q, et al. Infect Immun. 2006 Apr;74:2121. doi: 10.1128/IAI.74.4.2121-2127.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J. Nat Immunol. 2007 May;8:497. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]