Abstract
The mammalian intestine encounters many more microorganisms than any other tissue in the body thus making it the largest and most complex component of the immune system. Indeed, there are greater than 100 trillion (1014) microbes within the healthy human intestine where the total number of genes derived from this diverse microbiome exceeds that of the entire human genome by at least 100-fold. Our coexistence with the gut microbiota represents a dynamic and mutually beneficial relationship that is thought to be a major determinant of health and disease. Because of the potential for intestinal microorganisms to induce local and/or systemic inflammation, the intestinal immune system has developed a number of immune mechanisms to protect the host from pathogenic infections while limiting the inflammatory tissue injury that accompanies these immune responses. Failure to properly regulate intestinal mucosal immunity is thought to be responsible for the inflammatory tissue injury observed in the inflammatory bowel diseases (IBD; Crohn's disease, ulcerative colitis). An accumulating body of experimental and clinical evidence strongly suggest that IBD results from a dysregulated immune response to components of the normal gut flora in genetically-susceptible individuals. The objective of this review is to present our current understanding of the role that enteric microbiota play in intestinal homeostasis and pathogenesis of chronic intestinal inflammation.
Keywords: commensal bacteria, inflammatory bowel disease, Crohn's disease, ulcerative colitis, Th1 effector cells, symbiont, pathobiont, Th17 effector cells, regulatory T cells, dysbiosis, fecal transplant
Introduction
The mammalian intestine encounters many more microorganisms than any other tissue in the body thus making this tissue the largest and most complex component of the immune system. Indeed, the healthy human small and large intestine is home to hundreds of trillions of bacteria, viruses, archaea and fungi[1–5]. It is becoming clear that our coexistence with the gut microbiota represents a dynamic and mutually beneficial relationship that is thought to be a major determinant of health and disease. Because of the potential for intestinal microorganisms to induce local and systemic inflammation, the intestinal mucosal immune system has developed a plethora of mechanisms to protect the host from pathogenic infections while limiting the inflammatory tissue damage that accompanies these innate and adaptive immune responses. Failure to properly regulate these protective immune responses induces a chronic inflammatory response that is thought to be a critical immunopathological mechanism responsible for the development of the inflammatory bowel diseases (IBD; e.g. Crohn's disease and ulcerative colitis). These idiopathic inflammatory diseases affect primarily the small and/or large bowel and are characterized by the infiltration of large numbers of inflammatory leukocytes (e.g. neutrophils, monocytes, and lymphocytes) into the intestinal lamina propria where they directly or indirectly promote tissue injury and dysfunction including edema, loss of goblet cells, fibrosis, erosions and ulcerations. Although the etiology of IBD remains to be definitively defined, it is becoming increasingly appreciated that chronic intestinal inflammation results from a complex interaction among genetic, immune and microbial factors[5–7]. Based upon a large body of experimental and clinical evidence generated over the past 20 years, investigators hypothesize that chronic gut inflammation results from a dysregulated immune response to components of the normal gut flora in genetically-susceptible individuals[8, 9]. The objective of this review is to present our current understanding of the role that commensal enteric microbiota play in intestinal homeostasis and pathogenesis of chronic intestinal inflammation.
Intestinal Microbiota in Health and Disease
Bacterial Colonization and Redox Metabolism
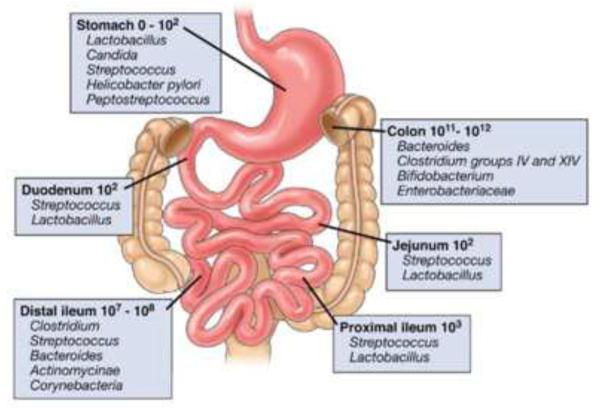
The only time that the human body is devoid of its microbial residents is during gestation. Despite some evidence suggesting that small numbers of bacteria may be present in the amniotic fluid, umbilical cord blood and/or meconium of healthy neonates[10], it is widely accepted that the fetus is maintained essentially in a germ-free state during development[3]. Upon birth, the newborn becomes colonized with commensal microbiota that arise from the mother's vagina, skin, feces and breast milk[11, 12]. The lactate-metabolizing bacteria Bifidobacterium and Lactobacillus derived from the vaginal canal and breast milk are the primary microorganisms that initially colonize in the intestinal tract during the first three months of life[12, 13]. These initial microbial residents lay the foundation for the subsequent colonization of the complex microbial communities that reside within the gut[14]. As the infant grows, phylogenetic diversity of the microbial community increases with bacterial-dependent metabolism becoming more complex and specialized ultimately establishing the adult intestinal microbiota[13]. There is also a distinct distribution of aerobic and anaerobic bacteria along the length of the gastrointestinal (GI) tract. Although the proximal part of the GI tract has many fewer bacteria than the distal portion, it is colonized by a much higher percentage of aerobic and/or facultative anaerobic bacteria than the distal small bowel and colon (Figure 1)[5]. Indeed, the distal portion of the GI tract is dominated almost exclusively by enormous numbers of anaerobic bacteria that are two to three orders of magnitude more prevelant than aerobic bacteria. Interestingly, it has been determined that anaerobic bacteria fail to colonize the newborn gut unless the bowel is first colonized by aerobic and/or facultative anaerobic bacteria[15]. The distribution of oxygen tolerant and intolerant bacteria within the healthy adult gut not only reflects the luminal oxygen gradient along the length of the GI tract but is also responsible for creating the hypoxic/reducing environment withiin the distal bowel lumen[15]. The redox relationship between the intestine and its microbiota is poorly understood at the present time. It is well known that the reductive environment does not prevent the production of reactive nitrogen species such as nitric oxide (NO). In fact, the intestinal lumen contains >200 ppb NO [15]. Although nitric oxide synthase-like enzymes have been identified in a select few enteric bacteria, the vast majority of luminal NO is produced by anaerobic bacteria via the reductive metabolism of nitrate and nitrite[15]. The respiration and growth of certain bacteria have been shown to be reversibly suppressed by NO in the presence of low oxygen tension[15, 16]. Intestinal bacteria also produce large amounts (millimolar levels) of hydrogen sulfide (H2S) which is capable of suppressing mitochondrial metabolism in epithelial cells as well[16, 17].
Figure 1. Composition and luminal concentrations of the major microbial species in different regions of the gastrointestinal tract.
Box panels show the numbers of organisms/gram luminal contents (Reproduced from reference 5, with permission).
Stability and Regulation of Microbial Communities
The healthy adult intestine contains more than 100 trillion (1014) bacteria that has, until recently, been estimated to harbor more than 1,000 different species (Figure 1)[3, 5]. More recent studies, using a newly developed method for low-error amplicon sequencing of bacterial 16S ribosomal RNA genes suggest that the human microbiota actually harbors ~200 strains of bacteria representing slightly more than 100 different bacterial species[18]. Nevertheless, the total number of genes derived from this diverse microbiome exceeds that of the entire human genome by at least 100-fold[2, 3]. Although >90% of all the intestinal bacteria belong to the Bacteroidetes and Firmicutes phyla, smaller but substantial numbers of bacteria associated with the Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia and Cyanobacteria phyla are also found in the human intestine[1–5]. In addition to commensal enteric bacteria, the gut is home to more than a quadrillion (1015) bacteriophages and viruses as well as substantial numbers of archaea and fungi[1, 3, 4, 19, 20]. The viral communities are considered stable, with more than 95% of the virotypes still present in the feces one year after the initial determinations[21]. An emerging area of research is beginning to examine how alterations in bacteriophage populations may alter the numbers and populations of enteric bacteria in the gut[21–24].
Despite the fact that the healthy human gut contains a rather stable community of microrganisms, these different populations may be highly variable between individuals[4, 18, 25]. Factors that promote microbial diversity in healthy adults include genetics, diet, environment and gender, among others[4, 26, 27]. While changes in diet are known to induce significant alterations in the numbers and composition of the intestinal microbiota, this effect as well as that produced by antibiotic treatment are generally thought to be reversible in nature, a term called resilience[14, 28]. The role that the environment and genetics play in shaping our intestinal microbial communities are well known and are currently being actively investigated[25, 29] (see below).
The Gut Microbiota is a “Virtual Organ”
The realization that the human body is actually a “super-organism” that contains 10 times more microbial cells than all cells in the mammalian body[1–5] has revolutionized our thinking about human systems' biology and the management of health and disease at the systemic level[3]. Although the mammalian intestine is a rich environment for supporting microbial growth, it demands that the microbial communities also contribute to the well-being of the host or at least do no harm. In fact, the intestinal bacteria function as a “virtual organ” in that these permanent residents provide essential metabolic functions that the host cannot perform such as degrading complex carbohydrates to produce important energy sources (e.g. short chain fatty acids) as well as synthesizing important vitamins[13, 19]. Another critical function of commensal microorganisms is their ability to promote immune system development and maturation. It has been long-appreciated that animals raised under germ-free conditions have defective T and B cell function, poorly developed lymphoid tissues, lower numbers of circulating CD4+ T cells and decreased antibody production, all of which can be restored upon association with commensal bacteria[30, 31]. Colonization with intestinal microorganisms may also help to suppress inadvertent immune responses to these helpful microbes and the accompanying inflammation that would ensue[5, 32]. The mechanisms by which commensal bacteria induce immune tolerance have not been entirely defined however, it is becoming clear that certain enteric bacteria are capable of inducing the frequency and numbers of protective CD4+ T helper cells within the gut. For example, it has been shown that segmented filamentous bacteria (SFB) are required for the induction of T helper 17 (Th17) cells in the intestine[33, 34]. These gram-positive bacteria induce Th17 cell differentiation in the small intestinal lamina propria where these T effector cells effectively protect the host from fungal and bacterial infections via the secretion of IL-17, IL-22 and IL-21[33–35]. In addition, certain Th17-derived cytokines (IL-22) enhance epithelial cell tight junctions as well as induce mucin and antimicrobial protein production, all of which decrease the likelihood of pathogenic invasion in the gut. However, it should be noted that although Th17 cells are important for protecting against pathogenic infection, they can, under certain situations, promote inflammation as observed in mouse models of psoriasis, rheumatoid arthritis and Type 1 Diabetes[36]. In addition to SFB, certain strains of Clostridia (clusters IV, XIVa and XVIII) and Bacteroidetes have been reported to enhance the abundance of intestinal CD4+Foxp3+ regulatory T cells (Tregs)[37–39]. These immunosuppressive T cells are well-known for their ability to maintain tolerance to commensal bacteria, suppress aggressive immune responses to auto- and bacterial antigens and promote epithelial wound repair via the activation of NFκB-dependent signaling pathways[5, 9, 40, 41].
Based upon the description of host-microbe interactions described above, one could argue that the relationship that exists between host and intestinal microbiota more closely resembles one of mutualism rather than commensalism. Mutualism implies a symbiotic association where both host and microbes benefit whereas commensalism infers that one organism benefits without affecting the other[1, 3, 42]. In reality, both symbiotic and commensal microbes are important residents of the intestinal tract (see below).
Immune Mechanisms that Maintain Intestinal Homeostasis
Minimizing Contact of Intestinal Bacteria with the Epithelium
Given the presence of such enormous numbers of microorganisms that reside in such close proximity to a tissue with the largest surface area in the body (300 m2), it is not surprising that the intestinal immune system possesses multiple layers of protection to limit the ability of resident microbes to gain access to the systemic circulation and thus the systemic immune system. It must be remembered that commensal bacteria are not innocuous and are capable of inducing potentially life threatening inflammatory responses (e.g. sepsis) if they gain access to the systemic immune system. Thus, the mammalian intestine has evolved several protective immune mechanisms that function to: a) minimize contact of the microbes with the epithelium, b) detect and rapidly destroy microbes that penetrate the epithelial barrier and c) confine/compartmentalize invading enteric bacteria to intestinal tissue thereby minimizing their exposure to the systemic immune system[43]. Limiting contact of the intestinal microbiota with the gut epithelium is accomplished by the elaboration and secretion of mucus as well as secretion of a variety of different antimicrobial agents by different intestinal epithelial cells. For example, goblet cells secrete large amounts of mucus, a highly glycosylated protein that is resistant to digestive enzymes. The mammalian small intestine is covered by a loose mucus layer that is not attached to the epithelial surface. In the colon, this loose mucus layer sits atop an inner layer of mucus that is firmly attached to the epithelial cells. Although commensal bacteria may reside within the loose mucus layer, they are not able to penetrate the thicker inner mucus layer thereby minimizing contact of these microbes with the colonic epithelium[44]. Certain pathogenic bacteria have devised ways to penetrate the mucus barrier thereby allowing them to attach to the epithelium and invade the tissue[45, 46]. Mice that lack the gene that encodes for a major mucus protein (MUC2) are largely devoid of the protective mucus layer allowing direct and prolonged contact with the gut epithelium. These mice develop spontaneous intestinal inflammation by 5 weeks of age[47, 48]. As stated above, certain commensal bacteria (e.g. SFB, Clostridia) can penetrate the mucus barrier and bind to the epithelium but do not produce pathological responses as they are required for fine-tuning the intestinal immune system[34].
Production of Antimicrobial Peptides and Antibodies
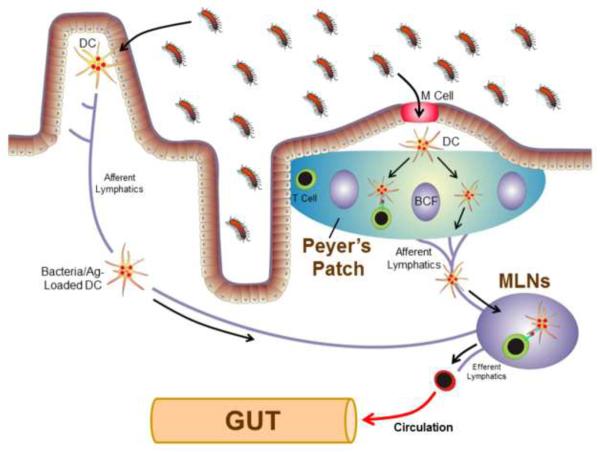
Another mechanism utilized by all intestinal epithelial cells to minimize their contact with gut bacteria is their secretion of potent antimicrobial peptides (AMPs). It is well known that epithelial cells secrete a group of bactericidal lectins such as Reg3 which are capable of directly killing gram-positive bacteria[43, 49]. Although Paneth cells are located within the intestinal crypts in relatively small numbers, they secrete potent AMPs such as defensins and cathelicidins that permeabilize bacterial cell membranes[50, 51]. Other important epithelial cell-derived AMPs secreted into the lumen of the bowel include lipocalin-2 and cathepsin K[52, 53]. A third mechanism utilized by the intestinal immune system for reducing contact between enteric bacteria and epithelial cells is the secretion of immunoglobulin A (IgA)[54]. There is good evidence to suggest that dendritic cells (DCs) within the Peyer's patches (PPs) of the small bowel as well as the intestinal lamina propria continuously sample luminal bacteria via endocytosis of the resident microbiota. Bacteria- and/or antigen (Ag)-loaded DCs within the PPs and those that have migrated to the mesenteric lymph nodes (MLNs) via the lymphatics (Figure 2) induce the activation and differentiation of naive B cells to yield plasma cells that produce IgA that is specific for commensal bacteria[43, 55]. Following their formation, plasma cells exit the PPs and MLNs via the efferent lymphatics, enter the systemic circulation and home to the intestinal lamina propria where they secrete IgA into the gut interstitium. This antimicrobial immunoglobulin is then taken up by the epithelial cells and transported to their apical surface and into the bowel lumen where it binds to commensal bacteria thereby limiting their ability to penetrate the epithelial barrier. Thus, IgA induced immune responses remain restricted to the intestinal tissue[43, 56–59]. Furthermore, it has been shown that the live bacteria transported to the MLNs by DCs never gain access to systemic immunity and thus are not able to induce systemic immune responses and distant organ inflammation providing what has been termed the “mucosal immune firewall”[32, 56–58].
Figure 2. Systemic and intestinal immune responses to enteric bacteria and their antigens.
Luminal intestinal bacteria and/or antigens (Ag) enter the Peyer's patches (PPs) via transport by the M cells where they are endocytosed by dendric cells (DCs) within the sub-epithelial dome region. Bacteria and Ag-loaded DCs may then interact with T and B cells within the PPs to prime the T and B cells or they may migrate from the PPs to the gut-draining mesenteric lymph nodes (MLNs) by way of the afferent lymphatics. Naïve T-cells (colored in green) that enter the MLNs will interact with these DCs resulting in the priming, polarization and expansion of the T-cells to yield effector cells (colored in red). These effector T cells then exit the MLNs via the efferent lymphatics, return to the systemic circulation and home to the gut lamina propria. Enteric bacteria and antigens may also be endocytosed by DCs located within the gut lamina propria and migrate to the MLNs via the afferent lymphatics. BCF refers to B cell follicle (Modified from reference 55, with permission).
Phagocytic Leukocytes, Intraepithelial Lymphocytes and T cells
Given the sheer numbers of enteric bacteria, it is inevitable that some commensal microorganisms will breach the epithelial barrier and gain access to the lamina propria. Fortunately, these opportunistic microbes encounter a second line of defense that greatly limits their ability to gain access to the systemic immune system. It has been demonstrated that those bacteria that do make their way into intestinal tissue are phagocytosed and destroyed by intestinal macrophages[60]. It is important to note that despite their effective antimicrobial activity, intestinal macrophages produce much lower amounts of inflammatory cytokines when compared to peripheral/inflammatory macrophages thereby limiting tissue inflammation[60–62]. Another important function performed by intestinal macrophages is their ability to promote tissue repair. Anti-inflammatory “M2” macrophages in which MyD88 signaling is specifically downregulated, migrate to the region of the gut where a breach has occurred and induce proliferation of progenitor epithelial cells to repopulate the epithelial surface[63].
Another population of immune cells that play an important role in maintaining intestinal homeostasis in the face of extraordinary bacterial and antigenic challenge are the intraepithelial lymphocytes (IELs). A certain subset of IELs termed CD8αα+ γδ IELs has been shown to repair breaches in the intestinal epithelium via their production of keratinocyte growth factor[64]. These IELs express the natural killer cell receptor NKG2D endowing them with the ability to bind to and become activated by intestinal epithelial cells resulting in the generation of the antimicrobial proteins Reg3β and Reg3γ[65]. More recent studies suggest that IL-22-producing innate lymphoid cells (ILCs) that reside within intestinal tissue and MLNs are critical for preventing the systemic dissemination of commensal enteric bacteria[66, 67]. These landmark studies clearly demonstrate that depletion of ILCs in healthy mice results in the systemic dissemination of commensal bacteria and induction of systemic inflammation. Furthermore, the authors convincingly show that both access of the intestinal bacteria to the systemic circulation and the resulting inflammatory tissue injury were largely abrogated by administration of IL-22 suggesting that IL-22 producing ILCs play a critical functional role in the mucosal firewall[67]. Finally, certain subsets of natural killer cells are thought to contain penetrant bacteria via IL-22- and Reg3-dependent mechanisms[66–70].
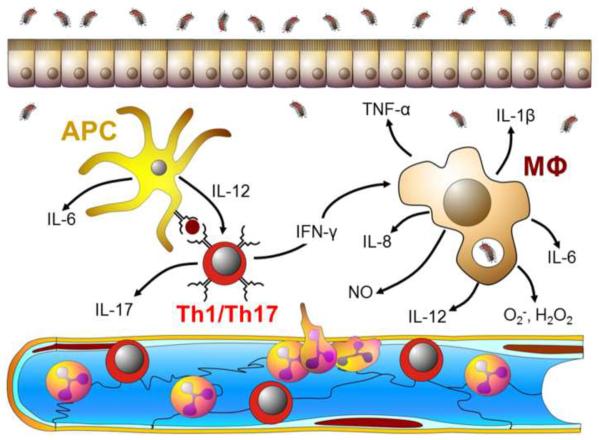
Invasion of the intestinal tissue by pathogenic bacteria may also trigger a well-coordinated and tightly regulated immune response involving a number of different leukocytes including T cells, macrophages and myeloid cells[71]. For example, Ag-loaded DCs that migrate from the gut to the draining MLNs will interact with naïve T-cells resulting in the priming (activation), polarization and expansion of the T-cells to yield Th1 and Th17 effector cells (Figure 2). These effector T cells then exit the MLNs via the efferent lymphatics, return to the systemic circulation and home to the gut lamina propria where the pathogen resides (Figure 3A). Th1 and Th17-derived inflammatory cytokines (e.g. IFN-γ, IL-17 and TNF-α) interact with and activate antigen presenting cells (APCs) and tissue macrophages resulting in the generation of additional inflammatory mediators including IL-1β, IL-6, IL-8, and IL-12 as well as a variety of reactive oxygen species (e.g. superoxide, hydrogen peroxide) and nitric oxide (Figure 3A). Together, these mediators enhance the microbicidal activity of tissue macrophages thereby “helping” these phagocytes destroy the invading microbes. In addition, many of these mediators are known to enhance the expression of different adhesion molecules (e.g. ICAM-1,VCAM-1 and E-selectin) on the surface of the post-capillary endothelial cells thereby facilitating the recruitment of additional leukocytes (e.g. neutrophils, monocytes, macrophages, lymphocytes) into the intestinal tissue to help in host defense. Under normal circumstances, the intestinal immune responses to invading microbes are tightly regulated to suppress or limit the development of inflammatory tissue injury that may accompany these responses. There is good evidence to suggest that intestinal immunity is regulated by the action of CD4+Foxp3+ Tregs[9, 32, 41, 71]. Regulatory T cells have been shown to suppress a wide range of inflammatory responses that are dependent upon the production of IL-10 and/or TGF-β as well as the expression of the transcription factor Foxp3[72]. It is well-recognized that Tregs are critical for regulating immune responses to auto-antigens and enteric bacteria as defects in Treg function associated with loss of expression of Foxp3, IL-10 and/or TGFβ is associated with the development of systemic and/or intestinal inflammation[9, 71, 72]. As described above, the presence of certain microbial populations within the commensal microbiota drives the development of Tregs within the gut and its associated lymphoid tissue (e.g. PPs, MLNs).
Figure 3. Adaptive and innate immune responses to invading bacteria.
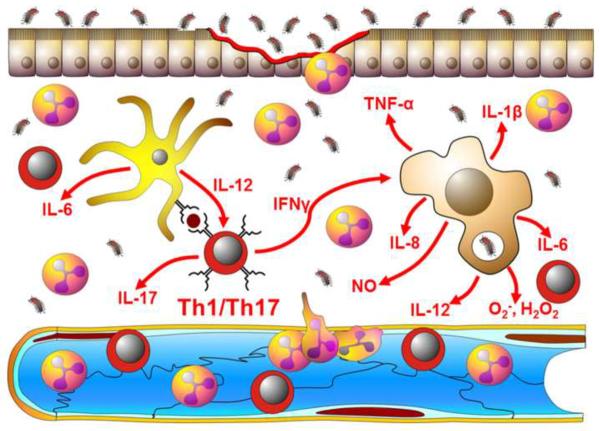
A). Enteric antigen-activated Th1 and Th17 effector T-cells produce large amounts of inflammatory cytokines such as IFN-γ, IL-17 and TNF-α within the intestinal lamina propria. These inflammatory cytokines interact with and activate antigen presenting cells (APC) and tissue macrophages (MΦ) to produce additional inflammatory mediators including IL-1β, IL-6, IL-8, and IL-12 as well as a variety of reactive oxygen and nitrogen species including superoxide (O2−), hydrogen peroxide (H2O2) and nitric oxide (NO). Together, these mediators enhance the microbicidal activity of tissue macrophages thereby “helping” these phagocytes destroy the invading microbes. In addition, many of these mediators are known to enhance the expression of different adhesion molecules (e.g. ICAM-1,VCAM-1 and E-selectin) on the surface of the post-capillary endothelial cells thereby facilitating the recruitment of additional leukocytes (e.g. neutrophils, monocytes, lymphocytes) into the intestinal tissue to help in killing bacteria. Under normal circumstances, these immune responses are tightly regulated to limit the development of inflammatory tissue injury that may accompany these protective responses. B). In the absence of appropriate regulatory mechanisms, the sustained, overproduction of the different inflammatory mediators promote epithelial and endothelial injury and dysfunction leading to erosions, ulcerations, fibrosis and edema.
Induction of Chronic Intestinal Inflammation
A Breakdown in Homeostasis
The ability of most individuals to remain healthy in the presence of such enormous numbers of intestinal microbes is testament to the effectiveness of the intestinal immune system in maintaining homeostasis. If there is a breakdown in homeostasis, chronic intestinal inflammation and tissue injury will ensue[5, 6, 41, 66, 73]. Indeed, there is a growing body of experimental and clinical evidence to suggest that a dysregulated immune response to components of the intestinal flora plays an important role in the pathogenesis of the inflammatory tissue injury observed in Crohn's disease (CD) and ulcerative colitis (UC) (Figure 3B). These chronic relapsing inflammatory disorders affect the small bowel and/or colon in approximately 1.5 million people in the US with a calculated annual cost for both medical expenses and work loss of almost $4 billion dollars[74]. Individuals who suffer from IBD experience bloody diarrhea, abdominal pain, fever and weight loss. In CD patients, inflammation is most often localized to the ileum and colon, however in some individuals, the inflammation may involve any part of the gastrointestinal tract. CD-associated inflammation is discontinuous and transmural in nature and is characterized by the presence of large numbers of myeloid cells as well as Th1 and Th17 effector cells within the gut interstitium (Figure 3B). UC-associated inflammation is, by definition, restricted to the colon where the inflammatory infiltrate is confined to the mucosal portion of the large bowel. It is thought that the lack of appropriate regulation of normal immune responses to enteric bacteria induces the sustained, overproduction of the multiple inflammatory cytokines and reactive oxygen and nitrogen species that may directly or indirectly damage intestinal tissue leading to erosions, ulcerations, fibrosis and edema.
Genetics of IBD
Evidence generated over the past two decades suggests that IBD may result from a complex interaction among genetic, immune and environmental factors[6, 75]. Genome wide association studies have identified a number of genetic variants that are shared by and unique to CD and UC[6, 73, 76]. Of the 163 susceptibility gene loci that have been identified in individuals with IBD, the large majority (110) are associated with both CD and UC whereas only 30 and 23 loci are specific for CD and UC, respectively[76, 77]. Not surprisingly, a significant number of susceptibility genes that are associated with both CD and UC are important in immune function related to the IL-23/Th17 and IL-10 pathways[76, 78]. Despite this large overlap in susceptibility genes for CD and UC, a number of recent studies have identified several CD-specific genes involved in innate immunity and autophagy including the nucleotide oligomerization domain 2 (NOD2), autophagy related protein 16-like 1(ATG16L1) and Immunity-Related GTPase Family M (IRGM) genes[76, 78]. Based upon the known functions of these genes, it has been proposed that CD may arise, in part, from defective processing of intracellular bacteria[76, 78]. Although not as genetically transmissible as CD, a number of UC-specific genes have also been identified that appear to be involved in epithelial barrier function (e.g. HNF4A, CHD1, LAMB1 genes), HLA class II-mediated antigen presentation (e.g. HLA-DQA1) and inflammatory mediator production (e.g. TNFRSF14, TMFRSF9, IL1R2, IL8Ra, IL7R)[76, 78].
Environmental Factors Contribute to IBD
There is no question that genetics play an important role in the pathogenesis of IBD. However, the fact that genetically-identical twins express a relatively low concordance rate for both CD (~30–35%) and UC (~10–15%) coupled to the well-documented increase in the incidence rates and prevalence of IBD over the past 50 years in countries that have adapted a “Westernized” or “Modernized” lifestyle, suggest that environmental factors (e.g. lifestyle, diet, use of antibiotics, hygiene) may be major contributors for the induction of disease in genetically susceptible individuals[76, 78–81]. Furthermore, the rapid increase in the incidence of IBD in nations that have transitioned from undeveloped to developed societies, underscores the fact that alterations in environmental factors may play an important role in the appearance of disease in countries that historically have reported few cases of IBD prior to modernization[79, 81, 82]. Examples of how modernization of society may alter the environment and thus alter the intestinal microbiota are summarized in Table 1 [82]. Many of the changes associated with a modern lifestyle involve major improvements in sanitation and hygiene thereby decreasing exposure of newborns and children to infectious microorganisms. Implementation of these environmental changes has been proposed as the basis for the “hygiene hypothesis” to explain the increased incidence of autoimmune and allergic diseases in developed countries[83]. There is good experimental evidence suggesting that the increased incidence and prevalence of a variety of different autoimmune and allergic diseases in Westernized countries is a direct result of decreased exposure of newborns and children to infectious microorganisms that may help in the development of the immune system[79, 84, 85]. IBD appears to represent one of several immunological disease whose increased incidence and prevalence appear to correspond to major changes in societal lifestyle and environment[79, 81].
Table 1.
Changes in Lifestyle May Alter the Intestinal Microbiota
| Traditional Lifestyle | Modern Lifestyle |
|---|---|
|
| |
| Vaginal Delivery at home | Birth in hospital; increasing rate of caesarean delivery |
| Large family size and crowding | Small family size |
| Tend to live in a rural setting in contact with soil microorganisms | Often live in an urban setting, surrounded by concrete |
| Ancestral colonization of the living environment | Sanitation of living spaces: environment colonized by resistant microorganisms (including resistant bacteria, fungi and acari) |
| Lack of antibiotics in infant life | Antibiotic usage early in life |
| Limited access to hot water and soap | Daily body wash with hot water and soap |
| High rate of Helicobacter pylori colonization | Low rate of Helicobacter pylori colonization |
| Parasitic worm infection | Decline in endemic parasitism |
| Food conserved by microbial fermentation | Food conserved by refrigeration |
| Consumption of natural foods | Consumption of processed foods |
Table modified from reference 82, with permission.
Role of the Intestinal Microbiota in the Pathogenesis of IBD
Despite the growing evidence that enteric microbes may be involved in the pathogenesis of IBD in genetically susceptible individuals, there has been no convincing evidence reported to suggest that CD or UC is caused by a pathogenic microorganism. Furthermore, neither CD nor UC are considered classical autoimmune diseases as no clear evidence has been reported to suggest that these diseases develop as a result of an unrestrained immune response to tissue self-antigens. If however, we define “self” to include those microbial cells that dramatically outnumber our own and reside within the intestinal tract for the entire lifetime of the individual, IBD may need to be reclassified as an autoimmune disease [86]. There is a large body of clinical and experimental data implicating the commensal microbiota as a major “driver” of the unrestrained inflammatory responses observed in human and experimental IBD [5, 73]. The first evidence that intestinal microbes play a pathogenetic role in IBD came from clinical studies demonstrating that diversion of the fecal stream from a segment of inflamed small bowel attenuated intestinal inflammation in patients with CD[87]. Furthermore, restoration of the fecal stream to a segment of surgically resected bowel that was judged to be histologically normal, resulted in the induction of intestinal inflammation suggesting that components in the fecal stream are capable of inducing IBD[87, 88]. In addition, a variety of studies have demonstrated dramatic alterations in the luminal composition of the microbiota (termed dysbiosis) in patients with IBD[5, 25]. The majority of these studies have demonstrated decreased microbial diversity, especially in members within the Firmicutes and Bacteroidetes phyla [5, 73, 82, 89–92](Figure 4). Several groups of investigators have confirmed significant reductions in the butyrate-producing bacterium Faecalibacterium prausnitzii, a member of the Firmicutes phyla [73, 93]. This bacterium has well-documented anti-inflammatory activity [93]. As described in a later section, butyrate produced by these types of commensals has been shown to exert a protective effect on the gut by enhancing mucosal barrier function, increasing intestinal mucous production, stimulating the production of immunosuppressive cytokines (e.g. IL-10) and decreasing the generation of pro-inflammatory mediators[94]. In addition, the production of butyrate by fermentation of fiber enhances the generation of immunosuppressive Tregs[95]. Reductions in certain members of Bacteroidetes such as Bacteroides fragilis, may also contribute to the induction and/or perpetuation of chronic gut inflammation by virtue of its protective/antiinflammatory effects as well[39, 73, 96].
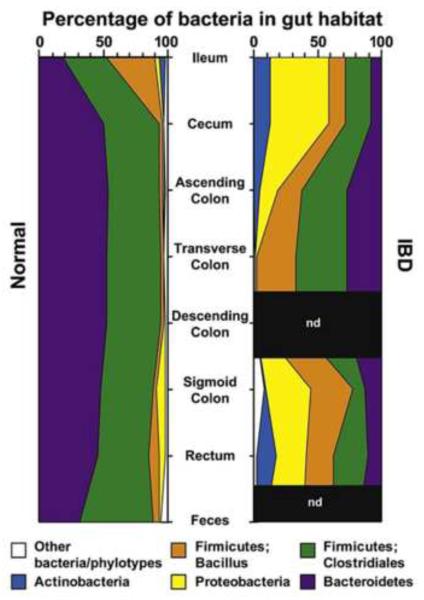
Figure 4. Distribution of predominant bacterial phylotypes in the human intestinal tract.
These figures illustrate the relative abundance as a function of location along the healthy and inflamed small bowel (ileum) and colon. (Reproduced from reference 92, with permission).
In contrast to the observed decreases in Firmicutes and Bacteroidetes, investigators have reported increases in members associated with the Proteobacteria and Actinobacteria in patients with active IBD[5, 73, 82, 89–92, 97, 98]. For example, it has been consistently reported that the numbers of certain strains of E. coli are increased in patients with CD and are associated with intestinal epithelial cells[73, 82, 91, 99]. Isolates of these adherent-invasive E. coli (AIEC) have been shown to not only adhere to epithelial cells but they also invade and replicate within these cells as well as macrophages[100]. It may be that the presence of increased numbers of AIEC in individuals with one or more genetic defects in their ability to kill intracellular bacteria[76, 78], may result in the induction and/or progression of chronic intestinal inflammation. The adhesion and invasion of AIEC may be facilitated by the increased numbers of mucolytic bacteria (Ruminococcus gnavus and Ruminococcus torques) in patients with IBD[101]. Although Clostrium boltae and Clostrium symbiosum have also been shown to be increased in stool samples collected with patients with IBD, the significance of these findings is not clear at the present time[102].
In addition to increased numbers of mucosal- and epithelial-associated bacteria in biopsies obtained from patients with IBD, there is also ample evidence demonstrating enhanced bacterial translocation in patients with active disease[73, 82, 91, 99, 100]. The fact that patients with IBD have antibodies directed against a variety of microbial antigens derived from intestinal bacteria such as Escherichia coli and Pseudomonas fluorescens as well as yeast (e.g. Saccharomyces) suggest that invading microorganisms may induce intestinal immune responses that trigger the induction and/or progression of IBD[73]. As mentioned above, the recent identification of polymorphisms in genes that are involved in intracellular processing and killing of bacteria in patients with CD(NOD2, ATG16L1 and IRGM) suggests that inappropriate innate immune responses to luminal bacteria may promote chronic gut inflammation in genetically-susceptible individuals[73, 76, 78]. Thus, it is not surprisingly that therapeutic administration of certain antibiotics have proven effective in attenuating distal bowel inflammation in patients with IBD[103].
Although a great deal of excitement has been generated over the past several years regarding the potential role that dysbiosis and enteric bacteria play in the pathogenesis of IBD, it is not clear whether alterations in the gut microbiota represent a cause and/or consequence of chronic intestinal inflammation. Joossens and coworkers have reported that asymptomatic, first degree relatives of patients with IBD exhibit dysbiosis when compared to unrelated healthy controls suggesting a “predysbiosis” may precede the development of IBD[104]. However, it must be remembered that inflammation, in general, is capable of creating dysbiosis[105]. Thus, despite these intriguing associations between the microbiota and IBD, many of these observations in humans remain indirect in nature.
The first direct evidence demonstrating that commensal enteric bacteria play an important role in the induction of chronic gut inflammation was reported in 1993 with the simultaneous publication of three different animal studies demonstrating that chronic colitis developed in genetically-engineered mice that were rendered devoid of the IL-10, IL-2 or T-cell receptor gene[106–108]. Within one month following these publications, Powrie and coworkers reported that adoptive transfer of antigen-inexperienced (naïve) T cells into lymphopenic mice induced a chronic and unrelenting colitis in these immunodeficient recipients[109]. Subsequent studies using these models, as well as others, revealed that the development of colonic inflammation required the presence of commensal enteric bacteria in genetically-susceptible hosts. Animals raised in a germ-free environment or treated with broad spectrum antibiotics failed to develop chronic disease[5, 6, 73]. In more recent studies using genetically engineered mice with genetic defects in genes known to be important in adaptive immunity, Garrett et. al. found that Rag2−/−/Tbet−/− mice developed chronic and unrelenting colitis that appeared histopathologically similar to human UC when raised under specific pathogen free (SPF) conditions[110, 111]. Furthermore, these investigators showed that healthy wild type (WT) mice developed chronic colitis when co-housed with colitic mice or when WT mice were gavaged with feces from colitic animals. Subsequent studies revealed that deletion of both the Rag2 and Tbet genes created a dysbiosis characterized by large and significant increases in two proteobacteria populations (e.g. Proteus mirabilis and Klebsiella pneumonia) that appeared to be responsible for induction of disease[111]. Another example of how the intestinal microbiota drives intestinal inflammation in mice with genetic deficiencies in systemic immunity was recently reported by Kang and coworkers using mice with defective IL-10 and TGFβ signaling pathways (i.e. IL10R2−/−/TGFβR2−/− mice)[112]. These mice develop a severe and fulminant ulcerative colitis that could be completely suppressed by treatment with broad spectrum antibiotics. In addition, Bloom et. al. showed that transfer of the gut microbiota from colitic mice induced colonic inflammation in antibiotic-treated IL10R2−/−/TGFβR2−/− but not in antibiotic treated mice that were deficient in only one of the two genes. Taken together, these studies demonstrated that colitogenic bacteria were present in IL10R2−/−/TGFβR2−/− mice and disease could only be transferred to those mice with defects in both IL-10 and TGFβ signaling[113]. A third example of how genetics, environment and the immune system may interact to promote chronic gut inflammation can be found in a recent study by Devkota et. al.[114]. These investigators demonstrate that the ingestion of saturated (but not polyunsaturated) fat by IL-10−/− mice induces a more severe form of chronic colitis than normally develops in these genetically-engineered mice. Devkota and coworkers show that saturated fat induces the formation of taurocholine-conjugated bile acids that creates intestinal dysbosis characterized by the overgrowth of a rare sulfate-reducing bacteria called Bilophilia wadsworthia[114]. These investigators demonstrated that the fat-induced bloom of B. wadsworthia was responsible for the more severe disease in IL-10−/− mice. These types of studies underscore the importance of understanding the complex interactions among diet (environment), genes, intestinal microbiota and the immune system.
Role of the Intestinal Microbiota in Other Autoimmune and Chronic Inflammatory Diseases
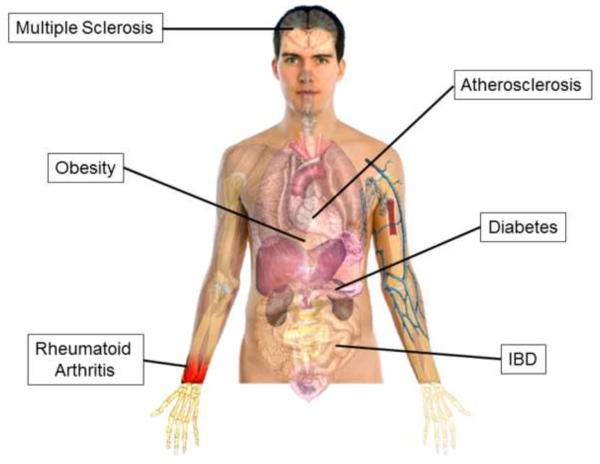
In addition to IBD, the intestinal microbiota has been implicated in the pathogenesis of several autoimmune and chronic inflammatory diseases (Figure 5). Wu and coworkers have demonstrated that the spontaneous arthritis observed in offspring produced by crossing KRN T cell receptor (TCR) transgenic mice with nonobese diabetic (NOD) mice is remarkably attenuated when mice are raised under germ-free conditions[115]. The KRN TCR recognizes the 42–56 peptide of bovine pancreatic ribonuclease bound to I-Ak. Furthermore, they show that the monocolonization of these germ free mice with a gut-associated bacterial species (termed segmented filamentous bacteria) induces the formation of lamina propria Th17 cells and autoantibody production as well as induces robust arthritis. A recent study also suggests that certain hormones may play a major role in establishing gender-specific microbiomes. In an attempt to explain why female nonobese diabetic (NOD) mice develop more rapid and severe Type1 diabetes (T1D) when compared to their male counterparts, Markle et al. recently reported that the microbiota obtained from sexually mature male and female mice were signficantly different[27]. In addition, they found that colonization of female NOD mice with male microbiota prior to the onset of sexual maturity blunted the development of diabetes in mature females[27]. Suppression of disease in females was associated with increased production of testosterone and was abrogated by administration of the androgen receptor antagonist flutamide (Eulexin). Taken together, these data suggest that the intestinal microbiota may regulate sex hormone generation and may influence an individual's susceptibility to different types of autoimmune diseases such as the well-established female gender bias in T1D and rheumatoid arthritis.
Figure 5.
Autoimmune and chronic inflammatory diseases in which the intestinal microbiota has been implicated.
The intestinal microbiota has also been implicated in the pathogenesis of the autoimmune, neurodegenerative disease multiple sclerosis (MS). Studies using transgenic mice that spontaneously develop autoimmune encephalomyelitis (a model of MS) demonstrated that commensal enteric bacteria were required for induction of disease as mice raised under germ-free conditions developed little or no neuroinflammation[116]. In addition to classic autoimmune disorders, the enteric microbiota has been suggested to play an important role in the development of the chronic inflammatory disease atherosclerosis. Two recent studies have shown that intestinal microbiota metabolism of certain dietary constituents (L-carnitine, choline and phosphotidylcholine) found in foods high in fat and cholesterol (red meat, etc) results ultimately in the generation of the proatherogenic metabolite trimethylamine-N-oxide (TMAO) in both mice and humans[117, 118]. Production of this reactive metabolite accelerates the development of atherosclerosis in mice that can be abrogated via the use of antibiotics[117, 118]. These data suggest that targeting the intestinal microbiota and/or their biochemical pathways may prove to be reasonable therapeutic strategies to reduce the formation of proatherogenic metabolites.
Finally, recent studies suggest that the gut microbiota may represent a major environmental factor that contributes to the development of obesity. Turbaugh et. al. have shown that the composition of intestinal bacteria of genetically obese mice (or obese human volunteers) is associated with significant alterations in Bacteroidetes and Firmicutes when compared to their lean counterparts[119]. Using bomb calorimetry to quantify total energy content in feces of lean vs. obese mice, these investigators demonstrate that the obese microbiota harvests more energy from the diet than the lean commensal flora. In addition, they found that colonization of germ-free mice with microbiota obtained from obese but not lean mice results in the generation of mice with a significantly greater amount of total body fat. Taken together, these data suggest that the obesity phenotype is transmissible. In a more recent study, Ridaura and coworkers show that colonization of adult germ free mice with feces obtained from an obese twin results in increased body weight and adiposity when compared to mice that received feces from the discordant lean twin[120]. Surprisingly, these investigators reported that co-housing of mice that were colonized with microbiota obtained from the lean twin with mice that received the obese twin's microbiota abrogated the increase in body weight and adiposity in mice colonized with the obese bacteria. Ridaura and coworkers determined that the protective effect was diet dependent and was due to the invasion of certain subgroups of Bacteroidetes residing within the lean twin mice into the obese twin animals. These remarkable results further suggest that an obesity phenotype may be transmissible.
A New Paradigm for Understanding the Role of Intestinal Microbiota in Health and Disease
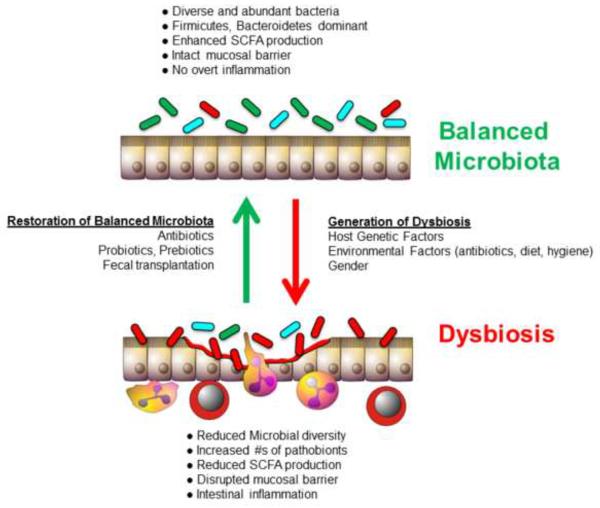
A conceptual framework that helps explain how a life-long residential population of nonpathogenic bacteria may, under certain situations, promote local (i.e.gut) and/or distant organ inflammation has recently been proposed by Mazmanian and coworkers. These investigators suggest that the healthy gut microbiota is composed of a well-balanced community of three highly evolved groups of bacteria termed symbionts, commensals and pathobionts (Figure 6)[39, 96, 121]. Symbionts are the resident bacteria that possess health-promoting effects whereas commensal bacteria thrive within the gut but have no positive or negative effect on the host (Figure 6). Pathobionts are also constituents of the healthy microbiota however these bacteria possess the potential to induce pathology (i.e. chronic inflammation) under certain situations (Figure 6). There is good evidence to suggest that alterations in the genetics, immune system and/or environment may induce a dysbiosis that facilitates the expansion of certain pathobionts that induce intestinal inflammation and/or may enter the systemic circulation where they initiate distant organ disease (Figure 5)[39, 96, 121]. It should be noted that pathobionts are not classical pathogens as they are permanent residents of the microbiota and are innocuous to the healthy host. Pathogens on the other hand, gain access to the intestine from the external environment and induce acute infections and inflammation in healthy individuals. Indeed, this paradigm may help to explain the complex immunopathological mechanisms responsible for induction of disease in different mouse models of IBD.
Figure 6. Immunological dysregulation associated with dysbiosis of the microbiota.
A) A healthy microbiota contains a balanced composition of different classes of bacteria. Symbionts are organisms with known health-promoting functions. Commensals are permanent residents of this complex ecosystem and provide no benefit or detriment to the host. Pathobionts are also permanent residents of the microbiota and have the potential to induce pathology. B) Dysbiosis is created by an alteration in the composition of the microbiota resulting in either a reduction in the numbers of symbionts and commensal and/or an increase in the numbers of pathobionts. This may result in the induction of inflammation in genetically susceptible individuals (Modified from reference 121, with permission).
Targeting the Intestinal Micriobiota as a Therapeutic Strategy for Treating IBD
Currently, there is no medical cure for either CD or UC. There are only a handful of pharmacologic and biologic agents available to treat these debilitating inflammatory disorders however many of the current and future therapies are immunosuppressive agents that may have significant side-effects not to mention their expense or cumbersome delivery system (i.e. intravenous infusion). While many of the new immunosuppressive therapeutics and biologics target specific inflammatory mediators or pathways that are thought to contribute to the overly aggressive immune responses associated with active disease, few if any target the immunologic “trigger” (i.e. the dysbiosis) that is thought to initiate and perpetuate IBD in susceptible individuals. Thus, there is a clear need for the development of nontoxic and inexpensive therapies to treat patients with IBD. The use of antibiotics, probiotics, prebiotics and/or fecal transplant to restore a healthy microbial community may offer a simple and inexpensive adjunct and/or alternative therapeutic strategy to treat patients with IBD (Figure 7). Although the clinical efficacy of antibiotics and probiotics in the treatment of IBD remains to be confirmed in rigorously-controlled, multicenter studies, there is good evidence to suggest that administration of certain antibiotics (e.g. metronidazole and/or ciprofloxin) may be effective in treating Crohn's colitis and ileocolitis while administration of specific probiotics may be useful in suppressing relapse of ulcerative colitis[5, 94, 122, 123]. A more recent approach to correct the intestinal dysbiosis in patients with IBD involves dietary administration of non-digestible, oligosaccharides termed prebiotics[94, 122]. The most commonly used prebiotics are beta-fructan oligosaccharides such as inulin and oligofructose that are found associated with plant-based fiber. Prebiotics are neither digested nor absorbed by the upper GI tract and thus will enter the colon unaltered. Once in the colon, specific anaerobic bacteria will metabolize these oligosaccharides to yield large concentrations of short chain fatty acids (SCFA; 50–100 mM) such as acetate, propionate and butyrate. There is very good evidence to suggest that increasing the concentrations of the SCFA within the intestinal lumen via ingestion of prebiotics enhances the growth of protective bacteria (symbionts) while limiting the growth of pathobionts[94]. SCFA have been shown to enhance mucosal barrier function, increase intestinal mucus production, stimulate the production of immunosuppressive cytokines (e.g. IL-10), decrease the generation of pro-inflammatory mediators and increase the generation of immunosuppressive Tregs [94, 95]. In addition, several preclinical animal studies have demonstrated protective or anti-inflammatory effects of prebiotic administration in different models of experimental IBD[94].
Figure 7. Potential therapeutic strategies for modulating intestinal dysbiosis.
(Modified from reference 123, with permission).
A recent report provides evidence that feeding mice the three SCFA individually or as a mixture of all three increases the frequency and numbers of Tregs as well as their expression of Foxp3 and the immunosuppressive cytokine IL-10 within the colonic interstitium[95]. In addition, Smith et. al. showed that SCFA administration attenuated the development of chronic inflammation in two different mouse models of IBD[95]. Data presented in this recent study showed that SCFA (especially propionate) appear to mediate their effects by inhibiting histone deacetylases (HDACs) 6 and 9 in a G protein-coupled via a receptor 43 (GPR43)-mediated process[95]. These data complement and extend previous studies by Tao et. al. who showed that pharmacological inhibition of HDAC enhances Treg function in vitro and in vivo[124]. Taken together, these studies appear to provide a mechanistic rationale for the well-known association between reduced consumption of plant-based fiber and increased intestinal disease. Despite these exciting preclinical results, the efficacy of prebiotics in the limited clinical studies performed on patients with IBD have been somewhat disappointing. Based upon the results of a promising, open-labeled pilot study using small numbers of IBD patients (N=10)[125], Benjamin et. al. undertook large and adequately powered (N=103), randomized, double-blind, placebo-controlled trial to assess the efficacy of fructo-oligosaccharide (FOS) administration to patients with CD[126]. They found that FOS administration did not significantly reduce inflammation in patients with CD when compared to placebo controls.
A novel and potentially powerful therapeutic method to manipulate the intestinal microbiota is through fecal microbiota transplantation (FMT)[127]. Based upon the early anecdotal accounts as well as work by Eiseman and coworkers[128], van Nood et. al. initiated a randomized, controlled clinical study that compared FMT with antibiotic treatment (vancomycin) in patients with recurring Clostridium difficile infection (CDI)[129]. In many but not all cases, the symptoms of CDI may occur following treatment with certain broad spectrum antibotics that disturb the intestinal flora thereby allowing overgrowth of the C.difficile. The resulting expansion of this pathobiont results in severe diahrrea that may progress to an inflammatory condition known as pseudomembranous colitis. Over the past decade, the number of CDI cases and fatality rate have more than doubled making this bacterial infection one of the major causes of bacterial-induced diarrhea in the US. Although antibiotic therapy is reasonably effective for the treatment of CDI, recurrent CDI is very difficult to treat with standard antibiotics. Van Nood and coworkers found that 94% of the recurring CDI patients infused with FMT had complete resolution of diarrhea whereas only 31% and 23% of the patients given vancomycin alone or vancomycin with bowel lavage, respectively showed resolution of diarrhea. Because of the dramatic protective effects of FMT, the study was terminated after treating only 43 of the 120 patients enrolled as it was deemed to be unethical to continue this study. Retrospective and meta analyses studies of small numbers of IBD patients treated with FMT show promising results[130]. There are currently six ongoing clinical studies to test the efficacy of FMT in patients with IBD (www.clinicaltrials.gov).
Summary and Conclusions
The mammalian intestine encounters and responds to more microbial antigens than any other tissue in the body making this tissue the largest and most complex component of the immune system. Our peaceful coexistence with these “residents for life” represents a dynamic and mutually beneficial relationship that is thought to be a major determinant of health and disease. Because of the potential for intestinal microorganisms to induce local and systemic inflammation and tissue injury, the intestinal immune system has developed a plethora of mechanisms to protect the host from pathogenic infections while limiting the inflammatory tissue injury that accompanies these immune responses. New and exciting research suggests that alterations in intestinal microbiota induced by genetic, dietary and/or environmental factors may play an important role in the pathogenesis of IBD as well as other autoimmune diseases.
Highlights
The healthy human intestine is home to more than 100 trillion (1014) bacteria.
The intestine is the largest and most complex component of the immune system.
Failure to regulate immune responses to enteric microbiota induces chronic intestinal inflammation.
Targeting intestinal micriobiota is a therapuetic strategy for treating inflammatory bowel disease.
Acknowledgements
Some of the work reported in this manuscript was supported by grants from the NIH (DK091269)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- [1].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol. 2011;17:557–66. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- [4].Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- [6].Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- [8].Coombes JL, Robinson NJ, Maloy KJ, Uhlig HH, Powrie F. Regulatory T cells and intestinal homeostasis. Immunol Rev. 2005;204:184–94. doi: 10.1111/j.0105-2896.2005.00250.x. [DOI] [PubMed] [Google Scholar]
- [9].Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–38. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- [10].Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–93. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- [11].Mandar R, Mikelsaar M. Transmission of mother's microflora to the newborn at birth. Biol Neonate. 1996;69:30–5. doi: 10.1159/000244275. [DOI] [PubMed] [Google Scholar]
- [12].Solis G, de los Reyes-Gavilan CG, Fernandez N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16:307–10. doi: 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- [13].Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Espey MG. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med. 2013;55:130–40. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- [16].Yu H, Sato EF, Nagata K, Nishikawa M, Kashiba M, Arakawa T, et al. Oxygen-dependent regulation of the respiration and growth of Escherichia coli by nitric oxide. FEBS Lett. 1997;409:161–5. doi: 10.1016/s0014-5793(97)00494-8. [DOI] [PubMed] [Google Scholar]
- [17].Circu ML, Aw TY. Intestinal redox biology and oxidative stress. Semin Cell Dev Biol. 2012;23:729–37. doi: 10.1016/j.semcdb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haynes M, Rohwer F. The human virome. In: Nelson KE, editor. Metagenomics of the Human Body. Springer; New York: 2011. pp. 63–78. [Google Scholar]
- [21].Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–8. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4:4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–17. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–22. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- [26].Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21:517–23. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- [27].Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–8. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- [28].de La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–92. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mathis D, Benoist C. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev. 2012;245:239–49. doi: 10.1111/j.1600-065X.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- [32].Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–69. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- [33].Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ivanov II, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol. 2011;14:106–14. doi: 10.1016/j.mib.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377–82. doi: 10.1016/j.smim.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Waite JC, Skokos D. Th17 response and inflammatory autoimmune diseases. Int J Inflam. 2012;2012:819467. doi: 10.1155/2012/819467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- [38].Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- [40].Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–9. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- [41].Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- [42].Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- [43].Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013 doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A. 2009;106:14321–6. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kawai F, Paek S, Choi KJ, Prouty M, Kanipes MI, Guerry P, et al. Crystal structure of JlpA, a surface-exposed lipoprotein adhesin of Campylobacter jejuni. J Struct Biol. 2012;177:583–8. doi: 10.1016/j.jsb.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].van der Sluis M, de Koning BA, De Bruijn AC, Velcich A, Meijerink JP, van Goudoever JB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–29. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- [48].van der Sluis M, Bouma J, Vincent A, Velcich A, Carraway KL, Buller HA, et al. Combined defects in epithelial and immunoregulatory factors exacerbate the pathogenesis of inflammation: mucin 2-interleukin 10-deficient mice. Lab Invest. 2008;88:634–42. doi: 10.1038/labinvest.2008.28. [DOI] [PubMed] [Google Scholar]
- [49].Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–71. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- [50].Biswas A, Petnicki-Ocwieja T, Kobayashi KS. Nod2: a key regulator linking microbiota to intestinal mucosal immunity. J Mol Med (Berl) 2012;90:15–24. doi: 10.1007/s00109-011-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pamer EG. Immune responses to commensal and environmental microbes. Nat Immunol. 2007;8:1173–8. doi: 10.1038/ni1526. [DOI] [PubMed] [Google Scholar]
- [52].Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–21. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- [53].Sina C, Lipinski S, Gavrilova O, Aden K, Rehman A, Till A, et al. Extracellular cathepsin K exerts antimicrobial activity and is protective against chronic intestinal inflammation in mice. Gut. 2013;62:520–30. doi: 10.1136/gutjnl-2011-300076. [DOI] [PubMed] [Google Scholar]
- [54].Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–6. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- [55].Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3:331–41. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- [56].Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E, et al. Luminal Bacteria Recruit CD103(+) Dendritic Cells into the Intestinal Epithelium to Sample Bacterial Antigens for Presentation. Immunity. 2013;38:581–95. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–5. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- [58].Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–8. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- [59].Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–7. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- [60].Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011;4:31–42. doi: 10.1038/mi.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Smythies LE, Shen R, Bimczok D, Novak L, Clements RH, Eckhoff DE, et al. Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation. J Biol Chem. 2010;285:19593–604. doi: 10.1074/jbc.M109.069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, Stappenbeck TS. Efficient colonic mucosal wound repair requires Trem2 signaling. Proc Natl Acad Sci U S A. 2009;106:256–61. doi: 10.1073/pnas.0803343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci U S A. 2002;99:14338–43. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A. 2011;108:8743–8. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–7. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–5. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sanos SL, Diefenbach A. Innate lymphoid cells: from border protection to the initiation of inflammatory diseases. Immunol Cell Biol. 2013;91:215–24. doi: 10.1038/icb.2013.3. [DOI] [PubMed] [Google Scholar]
- [70].Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–9. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- [71].Shale M, Schiering C, Powrie F. CD4(+) T-cell subsets in intestinal inflammation. Immunol Rev. 2013;252:164–82. doi: 10.1111/imr.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rudensky AY. Regulatory T cells and Foxp3. Immunol Rev. 2011;241:260–8. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–8. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- [74].Longobardi T, Jacobs P, Bernstein CN. Work losses related to inflammatory bowel disease in the United States: results from the National Health Interview Survey. Am J Gastroenterol. 2003;98:1064–72. doi: 10.1111/j.1572-0241.2003.07285.x. [DOI] [PubMed] [Google Scholar]
- [75].Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–21. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond Gene Discovery in Inflammatory Bowel Disease: The Emerging Role of Epigenetics. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Bernstein CN, Shanahan F. Disorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseases. Gut. 2008;57:1185–91. doi: 10.1136/gut.2007.122143. [DOI] [PubMed] [Google Scholar]
- [80].Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–94. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- [81].Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- [82].Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- [83].Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–60. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Okada H, Kuhn C, Feillet H, Bach JF. The `hygiene hypothesis' for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shanahan F, Bernstein CN. The evolving epidemiology of inflammatory bowel disease. Curr Opin Gastroenterol. 2009;25:301–5. doi: 10.1097/MOG.0b013e32832b12ef. [DOI] [PubMed] [Google Scholar]
- [86].Mathis D, Benoist C. Microbiota and autoimmune disease: the hosted self. Cell Host Microbe. 2011;10:297–301. doi: 10.1016/j.chom.2011.09.007. [DOI] [PubMed] [Google Scholar]
- [87].Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet. 1991;338:771–4. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- [88].D'Haens GR, Geboes K, Peeters M, Baert F, Penninckx F, Rutgeerts P. Early lesions of recurrent Crohn's disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114:262–7. doi: 10.1016/s0016-5085(98)70476-7. [DOI] [PubMed] [Google Scholar]
- [89].Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Rehman A, Lepage P, Nolte A, Hellmig S, Schreiber S, Ott SJ. Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J Med Microbiol. 2010;59:1114–22. doi: 10.1099/jmm.0.021170-0. [DOI] [PubMed] [Google Scholar]
- [91].Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–27. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Looijer-van Langen MA, Dieleman LA. Prebiotics in chronic intestinal inflammation. Inflamm Bowel Dis. 2009;15:454–62. doi: 10.1002/ibd.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly Y, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- [97].Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–54. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- [98].Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–30. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- [99].Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009;15:872–82. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- [100].Chassaing B, Rolhion N, de VA, Salim SY, Prorok-Hamon M, Neut C, et al. Crohn disease--associated adherent-invasive E. coli bacteria target mouse and human Peyer's patches via long polar fimbriae. J Clin Invest. 2011;121:966–75. doi: 10.1172/JCI44632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–8. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- [102].Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, Zaneveld J, et al. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 2012;22:1974–84. doi: 10.1101/gr.138198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Feller M, Huwiler K, Schoepfer A, Shang A, Furrer H, Egger M. Long-term antibiotic treatment for Crohn's disease: systematic review and meta-analysis of placebo-controlled trials. Clin Infect Dis. 2010;50:473–80. doi: 10.1086/649923. [DOI] [PubMed] [Google Scholar]
- [104].Joossens M, Huys G, Cnockaert M, De P, V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–7. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- [105].Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- [106].Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- [107].Mombaerts P, Mizoguchi E, Grusby MJ, Glimcher LH, Bhan AK, Tonegawa S. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:274–82. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- [108].Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–61. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- [109].Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 1993;5:1461–71. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- [110].Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, et al. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–41. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- [117].Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–84. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- [120].Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–31. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res. 2013;69:75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- [124].Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- [125].Lindsay JO, Whelan K, Stagg AJ, Gobin P, Al-Hassi HO, Rayment N, et al. Clinical, microbiological, and immunological effects of fructo-oligosaccharide in patients with Crohn's disease. Gut. 2006;55:348–55. doi: 10.1136/gut.2005.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Benjamin JL, Hedin CR, Koutsoumpas A, Ng SC, McCarthy NE, Hart AL, et al. Randomised, double-blind, placebo-controlled trial of fructooligosaccharides in active Crohn's disease. Gut. 2011;60:923–9. doi: 10.1136/gut.2010.232025. [DOI] [PubMed] [Google Scholar]
- [127].Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic Potential of Fecal Microbiota Transplantation. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.08.058. [DOI] [PubMed] [Google Scholar]
- [128].EISEMAN B, SILEN W, BASCOM GS, KAUVAR AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958;44:854–9. [PubMed] [Google Scholar]
- [129].van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- [130].Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic Potential of Fecal Microbiota Transplantation. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.08.058. [DOI] [PubMed] [Google Scholar]