SUMMARY
Mitochondrial pyruvate dehydrogenase complex (PDC) is crucial for glucose homoeostasis in mammalian cells. The current understanding of PDC regulation involves inhibitory serine phosphorylation of pyruvate dehydrogenase (PDH) by PDH kinase (PDK), whereas dephosphorylation of PDH by PDH phosphatase (PDP) activates PDC. Here we report that lysine acetylation of PDHA1 and PDP1 is common in EGF-stimulated cells and diverse human cancer cells. K321 acetylation inhibits PDHA1 by recruiting PDK1 and K202 acetylation inhibits PDP1 by dissociating its substrate PDHA1, both of which are important to promote glycolysis in cancer cells and consequent tumor growth. Moreover, we identified mitochondrial ACAT1 and SIRT3 as the upstream acetyltransferase and deacetylase, respectively, of PDHA1 and PDP1, while knockdown of ACAT1 attenuates tumor growth. Furthermore, Y381 phosphorylation of PDP1 dissociates SIRT3 and recruits ACAT1 to PDC. Together, hierarchical, distinct post-translational modifications act in concert to control molecular composition of PDC and contribute to the Warburg effect.
INTRODUCTION
Mammalian cells use glucose to generate energy. Normal cells produce ATP in the mitochondria through oxidative phosphorylation (OXPHOS), whereas under hypoxia, glucose is converted to lactate through glycolysis to produce ATP (Cairns et al., 2011; Kroemer and Pouyssegur, 2008). Glucose oxidation starts from the irreversible decarboxylation of glycolytic intermediate pyruvate to acetyl-CoA in mitochondria by pyruvate dehydrogenase complex (PDC), a large complex of three functional enzymes: E1, E2 and E3. PDC is organized around a 60-meric dodecahedral core formed by dihydrolipoyl transacetylase (E2) and E3-binding protein (E3BP) (Hiromasa et al., 2004), which binds pyruvate dehydrogenase (PDH; E1), dihydrolipoamide dehydrogenase (E3) as well as pyruvate dehydrogenase kinase (PDK) and pyruvate dehydrogenase phosphatase (PDP) (Read, 2001). PDH is the first and most important enzyme component of PDC that converts pyruvate to acetyl-CoA, which, along with the acetyl-CoA from the fatty acid β-oxidation, enters the Krebs cycle to produce ATP and electron donors including NADH. Thus, PDC links glycolysis to the Krebs cycle and thus plays a central role in glucose homeostasis in mammals (Harris et al., 2002).
Since PDH catalyzes the rate-limiting step during the pyruvate decarboxylation, activity of PDH determines the rate of PDC flux. The current understanding of PDC regulation involves the cyclic phosphorylation/dephosphorylation of PDH catalyzed by specific PDKs and PDPs, respectively (Holness and Sugden, 2003). PDK1 is a Ser/Thr kinase that inactivates PDC by phosphorylating at least one of three specific serine residues (Sites 1, 2 and 3 are S293, S300, and S232, respectively) of PDHA1 while dephosphorylation of PDHA1 by PDP1 restores PDHA1 and subsequently PDC activity (Roche et al., 2001). The Warburg effect describes the observation that cancer cells take up more glucose than normal tissue and favor aerobic glycolysis more than mitochondrial oxidation of pyruvate (Kroemer and Pouyssegur, 2008; Vander Heiden et al., 2009; Warburg, 1956). An emerging concept suggests that the metabolic change in cancer cells to reply more on glycolysis may be due in part to attenuated mitochondrial function through inhibition of PDC. In consonance with this concept, gene expression of PDK1, in addition to diverse glycolytic enzymes, is upregulated by Myc and HIF-1α in cancer cells (Kim et al., 2007; Kim et al., 2006a; Papandreou et al., 2006). Moreover, we recently also reported that diverse oncogenic tyrosine kinases (TKs), including FGFR1, are localized to different mitochondrial compartments in cancer cells, where they phosphorylate and activate PDK1 to inhibit PDH and consequently PDC, providing a metabolic advantage to tumor growth (Hitosugi et al., 2011).
Here we report a mechanism where lysine acetylation of PDHA1 and PDP1 contributes to inhibitory regulation of PDC, providing complementary insight into the current understanding of PDHA1 regulation through the phosphorylation/dephosphorylation cycle.
RESULTS
K321 and K202 acetylation inhibits PDHA1 and PDP1, respectively
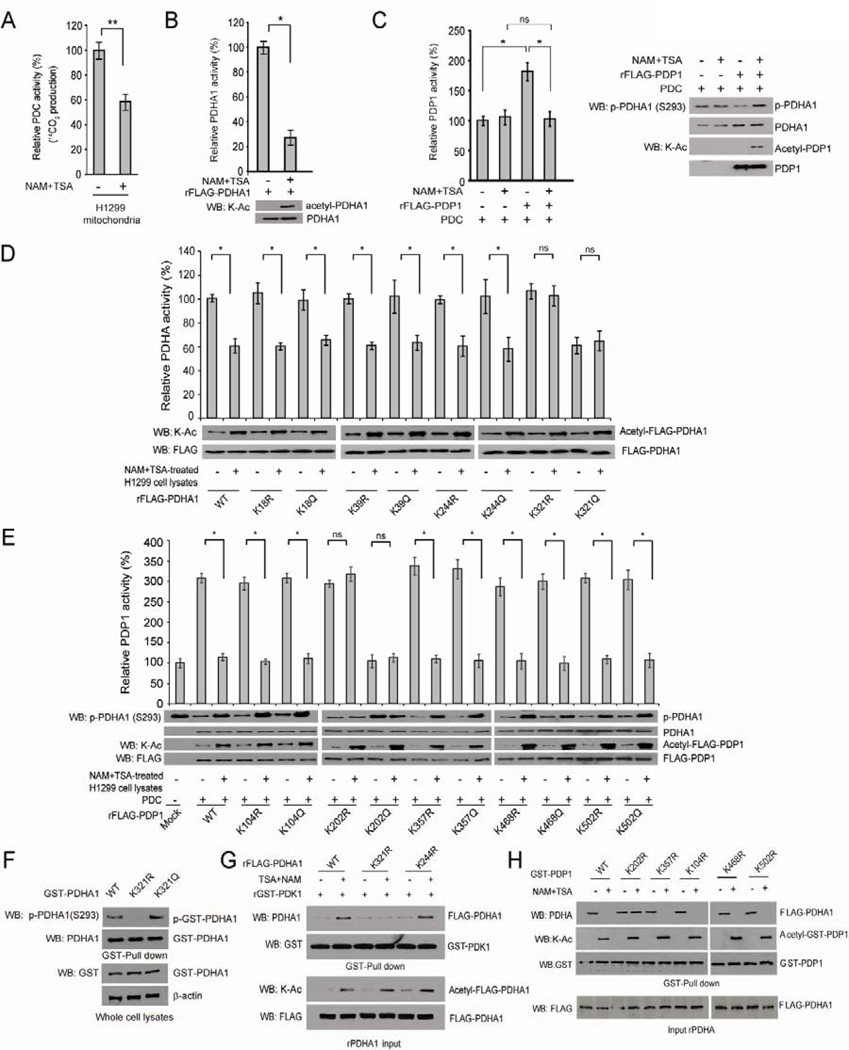
Our recent finding that tyrosine phosphorylation activates PDK1 (Hitosugi et al., 2011) suggests an important role for post-translational modifications in PDC regulation. To examine the potential effect of lysine acetylation on PDC activity, we treated lung cancer H1299 cells that overexpress FGFR1 (Marek et al., 2009) with deacetylase inhibitors nicotinamide (NAM) and Trichostatin A (TSA) for 16 hours, which led to increased global lysine acetylation in cells without affecting cell viability (Figure S1A). NAM+TSA treatment resulted in decreased PDC flux rate in isolated mitochondria from H1299 cells (Figure 1A), suggesting alteration of global lysine acetylation levels leads to PDC inhibition in human cancer cells. Interestingly, multiple proteomics-based studies performed by our collaborators at Cell Signaling Technology (CST) identified key components of PDC including PDHA1 (http://www.phosphosite.org/proteinAction.do?id=1271&showAllSites=true) and PDP1 (http://www.phosphosite.org/proteinAction.do?id=19516&showAllSites=true), but not PDK1 (http://www.phosphosite.org/proteinAction.do?id=2352&showAllSites=true), as acetylated at a group of lysine residues in human cancer cells. To test the hypothesis that lysine acetylation might directly affect PDHA1 and PDP1 activity, we incubated recombinant FLAG-tagged PDHA1 and PDP1 with cell lysates from NAM+TSA treated H1299 cells. Such treatment results in increased lysine acetylation of PDHA1 (Figure 1B; lower) and PDP1 (Figure 1C; right), accompanied by reduced PDHA1 enzyme activity (Figure 1B; upper) and PDP1 phosphatase activity, as assessed by the ability of PDP1 to dephosphorylate S293 of substrate PDHA1 (Figure 1C; left), respectively. These results suggest that lysine acetylation inhibits PDHA1 and PDP1, and consequently PDC flux.
Figure 1. Lysine acetylation inhibits PDHA1 and PDP1.
(A) PDC flux rate was measured using isolated mitochondria from H1299 cells incubated with 14C-labeled pyruvate and treated with NAM (10mM) + TSA (5µM) for 16 hours.
(B) Purified recombinant FLAG-PDHA1 (rFLAG-PDHA) was incubated with lysates of NAM+TSA-treated H1299 cells, followed by in vitro PDHA1 enzyme assay and Western blot using a pan acetyl-Lys antibody (K-Ac).
(C) Purified rFLAG-PDP1 was incubated with purified PDC proteins in the presence or absence NAM+TSA-treated H1299 cell lysates, followed by in vitro PDP1 assay (left). Decreased PDP1 activity was assessed by the increase in phosphor-S293 levels of PDHA1 using Western blot (right).
(D–E) Purified rFLAG-PDHA1 (D) and rFLAG-PDP1 (E) variants were incubated with or without NAM+TSA-treated H1299 cell lysates, followed by in vitro PDHA1 and PDP1 assays, respectively.
(F) Immunoblotting of GST-pull down samples using cell lysates from H1299 cells transiently transfected with distinct GST-PDHA1 variants.
(G) Purified rFLAG-PDHA1 variants were incubated with NAM+TSA treated H1299 cell lysates, followed by incubation with purified GST-PDK1. GST-pull down was performed and bound FLAG-PDHA1 to GST-PDK1 was determined by Western blot.
(H) Purified GST-PDP1 variants were incubated with NAM+TSA treated H1299 cell lysates, followed by incubation with purified FLAG-PDHA1. GST-pull down was performed and bound FLAG-PDHA1 to GST-PDP1 was determined by Western blot. The error bars represent mean values +/− SD.
Also see Figure S1.
To identify the lysine sites that mediate acetylation dependent inhibition of PDHA1 and PDP1 upon treatment with NAM+TSA, we performed mass spectrometry based experiments to examine GST-PDHA1 and GST-PDP1 proteins incubated with NAM+TSA-treated H1299 cell lysates. We identified K18 of PDHA1 and K104, K357, K468 and K502 of PDP1 as acetylated (Figure S1B), in addition to K39, K244 and K321 of PDHA1 and K202 of PDP1 that were posted on the CST website (www.phosphosite.org). We next performed mutational analysis and generated diverse acetyl-deficient K→R and acetyl-mimetic K→Q mutants of PDHA1 and PDP1 to replace each of the lysine residues that were identified as acetylated. We found that only substitutions of PDHA1 K321 and PDP1 K202 conferred resistance to the enzymes in terms of NAM+TSA treatment-induced inhibition of PDHA1 enzyme activity (Figure 1D) and PDP1 phosphatase activity (Figure 1E), respectively. We performed an isoelectric focusing (IEF) experiment and estimated the stoichiometry of lysine acetylation of recombinant PDHA1 (~70%) and PDP1 (~50%) upon treatment with cell lysates from H1299 cells treated with NAM+TSA (Figure S1C). This is consistent with our previous observations that treatment with NAM+TSA-treated cell lysates resulted in a ~70% decrease in enzyme activity of recombinant PDHA1 (Figure 1B) and a ~50% decrease in phosphatase activity of recombinant PDP1 (Figure 1C). Note in Figures 1B and 1C, both the acetyl-deficient K321R and acetyl-mimetic K321Q mutants of PDHA1 as well as K202R and K202Q mutants of PDP1 showed resistance to NAM+TSA treatment, where K→R mutants were catalytically more active while K→Q mutants were catalytically less active, suggesting that the enzyme activity change of PDHA1 and PDP1 was most likely due to lysine acetylation.
We next examined whether K321 acetylation-dependent inhibition of PDHA1 is dependent or independent of serine phosphorylation-dependent inhibition of PDHA1. We found that acetyl-deficient K321R and acetyl-mimetic K321Q variants of GST-PDHA1 that were transiently transfected in H1299 cells demonstrate decreased and increased S293 phosphorylation, respectively, compared to control GST-PDHA1 WT (Figure 1F). These results suggest that K321 acetylation of PDHA1 functions as an upstream event that promotes PDK1-dependent phosphorylation and inhibition of PDHA1. We hypothesized that K321 acetylation may potentiate recruitment of PDK1 to PDHA1 and thus performed a GST-pull down assay. As shown in Figures 1G and S1D, FLAG-PDHA1 WT and control K18R, K29R or K244R mutants incubated with NAM+TSA-treated H1299 cell lysates show increased binding of PDHA1 protein to beads-bound purified GST-PDK1. In contrast, substitution at K321 of PDHA1 abolishes the increased PDK1-binding induced by NAM+TSA treatment. We next tested whether K202 acetylation similarly affects PDP1-PDHA1 association. We found that incubation of purified GST-PDP1 WT and control K104R, K357R, K468 or K502 mutants, but not K202R mutant, with NAM+TSA treated cell lysates abolishes binding of purified FLAG-PDHA1 to beads-bound GST-PDP1 protein (Figure 1H). Together these results suggest that lysine acetylation of PDHA1 and PDP1 regulates the molecular composition of PDC, which facilitates S293 phosphorylation of PDHA1, leading to inhibition of PDHA1 and subsequently PDC.
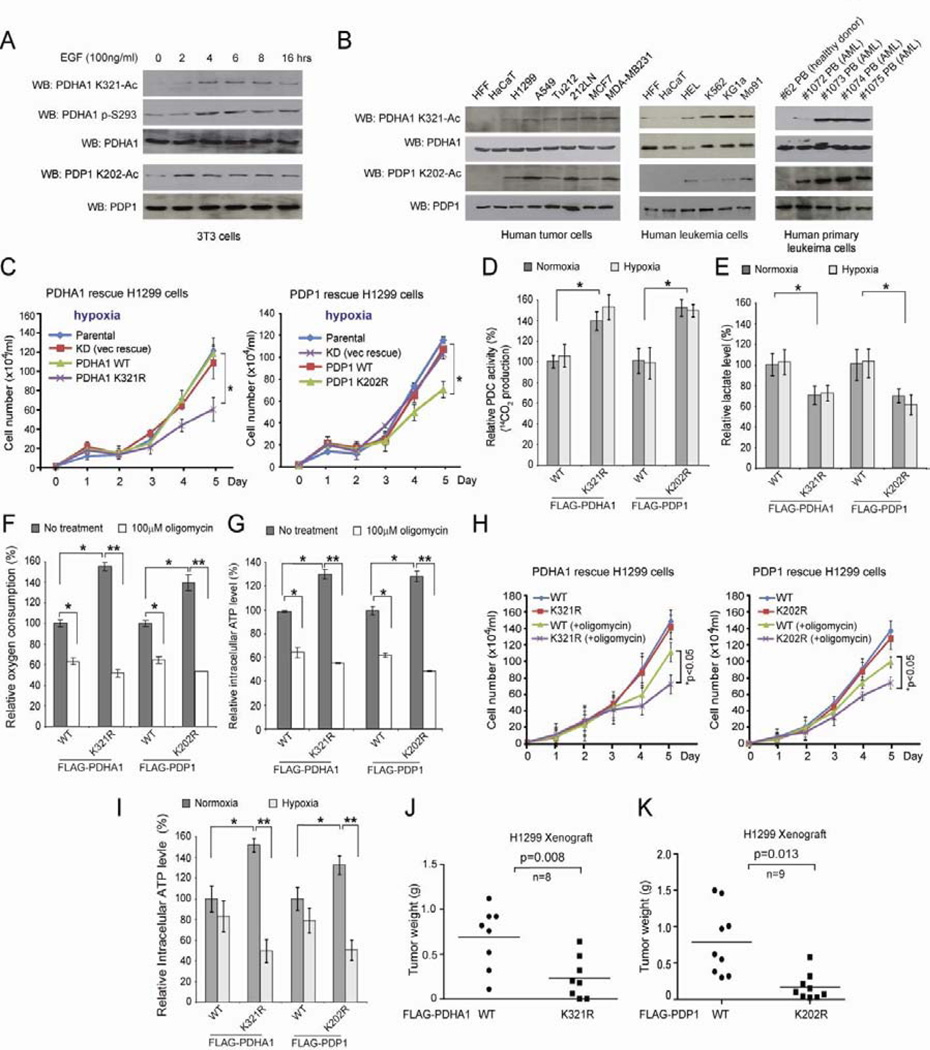
Lysine acetylation of PDHA1 and PDP1 is induced by EGF-stimulation and is common in diverse human cancer cells, which is important to promote glycolysis and tumor growth
We next generated two specific acetyl-PDHA1 and acetyl-PDP1 antibodies and detected NAM+TSA treatment-dependent acetylation at K321 of PDHA1 and K202 of PDP1 proteins, respectively (Figure S1E). Using these acetyl-specific antibodies, we found that K321 acetylation and S293 phosphorylation of PDHA1 as well as K202 acetylation of PDP1 are increased in EGF-treated human 3T3 and 293T cells (Figures 2A and S1F, respectively). Moreover, acetylation of PDHA1 K321 and PDP1 K202 is common in diverse human tumor cells including H1299 and A549 lung cancer, Tu212 and 212LN head and neck cancer, and MCF7 and MDA-MB-231 breast cancer cells (Figure 2B; left), as well as a group of leukemia cells associated with distinct leukemogenic tyrosine kinases including HEL (JAK2 V617F), K562 (BCR-ABL), KG1a (FOP2-FGFR1) and Mo91 (TEL-TrkC) cells (Figure 2B; middle). Furthermore, we observed that acetylation levels of PDHA1 K321 and PDP1 K202 are increased in primary leukemia cells from four acute myeloid leukemia (AML) patients compared to peripheral blood cells from a representative healthy donor (Figure 2B; right).
Figure 2. Lysine acetylation of PDHA1 and PDP1 is important to promote glycolysis as well as consequent cancer cell proliferation under hypoxia and tumor growth.
(A) Immunoblotting of lysates of 3T3 cells treated with EGF stimulation for different time as indicated using specific acetyl- or phospho-PDHA1 and acetyl-PDP1 antibodies.
(B) Immunoblotting to detect acetylation levels of PDHA1 K321 and PDP1 K202 in diverse human tumor (left) and leukemia (middle) cells as well as human primary leukemia cells isolated from peripheral blood (PB) samples from representative AML patients (right). Normal proliferating human foreskin fibroblasts (HFF), HaCaT keratinocyte cells and PB cells from a healthy human donor were included as controls.
(C) Distinct PDHA1 (left) and PDP1 (right) rescue H1299 cells as well as parental H1299 and control knockdown (KD; empty vector rescue) cells were tested for cell proliferation rate under hypoxia. Cell proliferation was determined based on cell numbers counted daily.
(D–H) Distinct PDHA1 and PDP1 WT and K→R mutant rescue H1299 cells were tested for PDC flux rate (D) and lactate production (E) under both normoxia and hypoxia, as well as oxygen consumption (F), intracellular ATP level (G) and cell proliferation rate (H) in the presence and absence of ATP synthase inhibitor oligomycin under normoxia.
(I) Distinct PDHA1 and PDP1 WT and K→R mutant rescue H1299 cells were tested for intracellular ATP level under both normoxia and hypoxia.
(J–K) Tumor masses in xenograft nude mice injected with PDHA1 K321R rescue cells compared to mice injected with control PDHA1 WT rescue cells (J) or mice injected with PDP1 K202R rescue cells compared to mice injected with control PDP1 WT rescue cells (K) are shown. p values were determined by a two-tailed paired Student’s t test. The error bars represent mean values +/− SD.
Also see Figures S1–S2.
Since EGF-stimulation induces glycolysis and promotes cell proliferation while most of cancer cells are highly proliferative and glycolytic (Kroemer and Pouyssegur, 2008; Sumi et al., 1984), we next tested whether K321 and K202 acetylation-dependent inhibition of PDHA1 and PDP1, respectively, is important for glycolysis and cancer cell proliferation. We generated “rescue” H1299 cells with stable knockdown of endogenous PDHA1 or PDP1, followed by rescue expression of shRNA-resistant FLAG-PDHA1 WT and K321R or PDP1 WT and K202R that harbor silent mutations in the target regions of shRNA, respectively (Figure S1G). As shown in Figure 2C and S1H, knockdown or rescue expression of PDHA1 WT and PDP1 WT does not affect H1299 cell proliferation under normoxic or hypoxic conditions. In contrast, rescue expression of catalytically active, acetyl-deficient PDHA1 K321R or PDP1 K202R significantly attenuates cell proliferation under hypoxia but not normoxia. Note lysine acetylation levels of PDP1 and PDHA1 in cells were comparable under normoxia and hypoxia (Figure S1I). In consonance with these findings, PDHA1 K321R and PDP1 K202R cells demonstrate increased PDC flux rate (Figure 2D), decreased glycolytic rate (Figure S1J) and lactate production (Figure 2E) under both normoxic and hypoxic conditions, as well as increased oxygen consumption rate (Figure 2F) with elevated intracellular ATP (Figure 2G) and ROS (Figure S1K) levels under normoxia, compared to corresponding PDHA1 and PDP1 WT cells, respectively. In addition, PDHA1 K321R and PDP1 K202R cells are more sensitive to treatment with ATP synthase inhibitor oligomycin in terms of inhibition of oxygen consumption, ATP production and cell proliferation (Figures 2F–2H, respectively), compared to corresponding control PDHA1 and PDP1 WT cells, respectively. Consistent with these findings, PDHA1 K321R and PDP1 K202R cells are more sensitive to hypoxia in terms of inhibition of ATP production, compared to corresponding control WT cells (Figure 2I). These data together suggest that abolishment of lysine acetylation of PDHA1 and PDP1 results in a metabolic change to allow cells rely more on OXPHOS, providing a metabolic disadvantage to cell proliferation under hypoxia.
We next performed xenograft experiments and found that the growth rate (Figure S2A) and masses of tumors (Figure 2J) derived from PDHA1 K321R rescue H1299 cells were significantly reduced with decreased K321 acetylation and decreased expression of proliferation marker Ki67 (Figure S2B; left and right, respectively), as well as increased PDC flux, decreased lactate production and increased oxygen consumption rate (Figure S2C) in tumor cells, compared to those of tumors formed by the control PDHA1 WT rescue cells. Similar results were obtained in xenograft mice using PDP1 WT and K202R cells (Figures 2K, S2C–S2F). Together these data demonstrate an important role for lysine acetylation of PDHA1 and PDP1 in tumor growth.
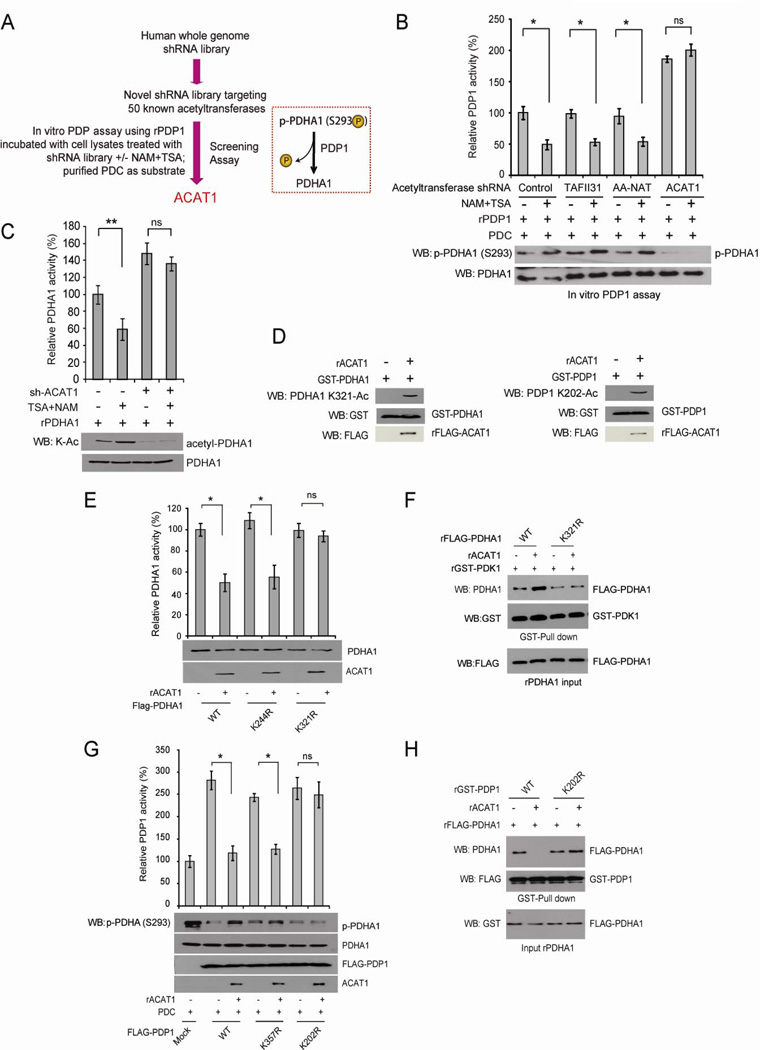
Mitochondrial ACAT1 is the upstream acetyltransferase of PDHA1 and PDP1
To identify the upstream acetyltransferase(s) of PDHA1 and PDP1, we constructed a “targeted” lentiviral shRNA library that target 50 out of 71 known acetyltransferases in the human genome, which are available in the shRNA library targeting the whole human genome (OpenBioSystems) (Figure 3A). Using this library, we performed PDP1 phosphatase assay-based screening studies using recombinant PDP1 incubated with cell lysates from H1299 cells with knockdown of particular acetyltransferases by lentiviral transduction in the presence or absence of NAM+TSA (Figure 3A). We found that lentiviral shRNA-mediated knockdown of acetyl-CoA acetyltransferase 1 (ACAT1) results in increased PDP1 activity with decreased lysine acetylation that are resistant to NAM+TSA treatment, whereas knockdown of other acetyltransferases (TAFII31 and AA-NAT as representatives) does not affect NAM+TSA induced lysine acetylation and inhibition of PDP1 (Figure 3B). Similarly, treatment with cell lysates from H1299 cells with knockdown of ACAT1 results in decreased lysine acetylation and increased enzyme activity of PDHA1 that are resistant to NAM+TSA treatment (Figure 3C). We next confirmed that ACAT1 directly acetylates PDHA1 and PDP1 in an in vitro lysine acetylation assay using purified recombinant ACAT1 incubated with purified GST-tagged PDHA1 and PDP1 where acetylated PDHA1 and PDP1 were detected by pan acetyl-Lys and specific acetyl-PDHA1/PDP1 antibodies (Figures S3A and 3D, respectively).
Figure 3. Identification of ACAT1 as the upstream acetyltransferase for PDHA1 and PDP1.
(A) Schematic representation of a PDP1 activity assay-based screening strategy to identify the upstream acetyltransferase that mediates NAM+TSA treatment-dependent inhibition of PDP1. Purified PDP1 was incubated with cell lysates from H1299 cells that were infected with lentiviruses targeting each acetyltransferase in the presence or absence of NAM+TSA treatment, followed by PDP1 assay.
(B) Purified PDP1 was incubated with cell lysates from H1299 cells that were infected with lentiviruses targeting ACAT1, or control acetyltransferases TAFII31 and AA-NAT in the presence or absence of NAM+TSA treatment, followed by PDP1 activity assay.
(C) Recombinant PDHA1 was incubated with cell lysates from H1299 cells with ACAT1 knockdown treated with or without NAM+TSA, followed by PDHA1 activity assay (top) and Western blot (bottom).
(D) Immunoblotting of recombinant PDHA1 (left) and PDP1 (right) treated with recombinant ACAT1 (rACAT1) using specific acetyl-PDHA1 and acetyl-PDP1 antibodies, respectively.
(E–F) Purified FLAG-PDHA1 variants were incubated with recombinant ACAT1 (rACAT1), followed by in vitro PDHA1 enzyme assay (E) or incubation with purified GST-PDK1 and PDHA1/PDK1 binding assays as described in Figure 1G.
(G–H) Purified FLAG-PDP1 variants were incubated with recombinant ACAT1 (rACAT1), followed by in vitro PDP1 assay (G) or incubation with purified FLAG-PDHA1 and PDP1/PDHA1 binding assay as described in Figure 1H. The error bars represent mean values +/− SD.
Also see Figure S3.
Moreover, incubation with ACAT1 significantly inhibits enzyme activity of PDHA1 WT and control K244R mutant but not K321R mutant (Figure 3E). In consonance with our findings in Figure 1G, ACAT1 incubation promotes PDK1 binding to PDHA1, whereas K321R mutant has decreased ability to bind PDK1 compared to PDHA1 WT control that is resistant to ACAT1 treatment (Figure 3F). In addition, ACAT1 treatment significantly inhibits phosphatase activity of PDP1 WT and control K357R mutant but not K202R mutant (Figure 3G). Consistent with results shown in Figure 1H, ACAT1 treatment abolishes the ability of PDP1 WT but not K202R mutant to bind PDHA1 (Figure 3H). Thus, these results together demonstrate ACAT1 as the upstream acetyltransferase of PDHA1 and PDP1.
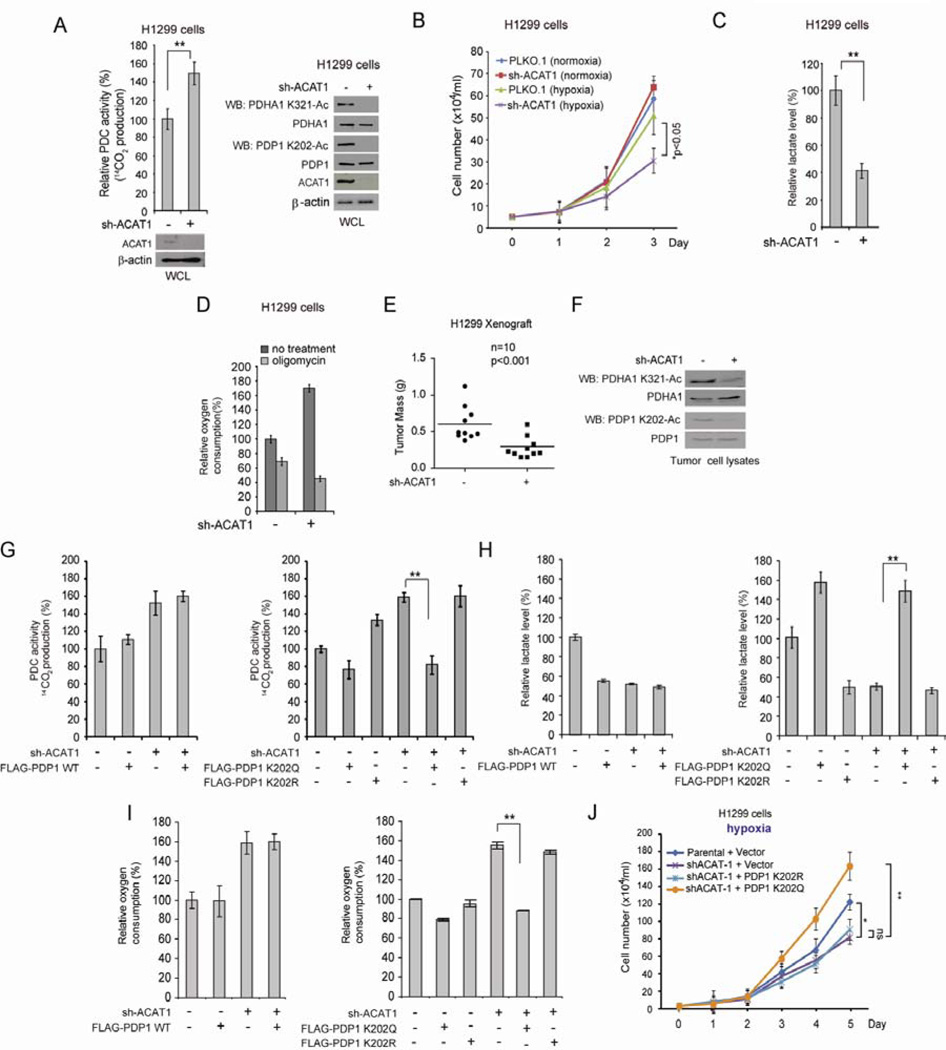
ACAT1 is important for cancer cell proliferation and tumor growth, which signals through the PDP-PDHA1 axis to promote glycolysis
We reasoned that if ACAT1 is responsible for lysine acetylation and subsequently inhibition of PDHA1 and PDP1, ACAT1 is important for the metabolic alteration in cancer cells to rely more on glycolysis. Indeed, we found that stable knockdown of ACAT1 in H1299 cells results in increased PDC flux rate with decreased acetylation levels of PDHA1 K321 and PDP1 K202 (Figure 4A). In addition, similar to PDHA1 K321R and PDP1 K202R cells described in Figures 2 and S1, ACAT1 knockdown cells show decreased cell proliferation rate under hypoxia but not normoxia (Figure 4B) with decreased glycolytic rate (Figure S3B) and lactate production (Figure 4C), as well as increased oxygen consumption rate (Figure 4D) with elevated intracellular ATP (Figure S3C) and ROS (Figure S3D) levels, compared to control cells harboring an empty vector. Moreover, ACAT1 cells are more sensitive to treatment with ATP synthase inhibitor oligomycin in terms of inhibition of oxygen consumption, ATP production and cell proliferation (Figures 4D, S3C and S3E, respectively), compared to control vector cells. Consistent with these findings, we found that expression of PDP1 K202R or PDHA1 K321R, or stable knockdown of ACAT1, did not induce apoptosis, compared to control WT rescue cells (Figure S3F). In contrast, H1299 cells expressing K→R mutants or with ACAT1 knockdown demonstrated a decreased cell proliferation index (PI) with an increased percentage of cells at G0/G1 phase but decreased cell percentages at S and G2/M phases (Figure S3G).
Figure 4. ACAT1 signals through inhibition of PDC by acetylating PDHA1 and PDP1 to promote glycolysis and tumor growth.
(A) H1299 cells with stable knockdown of ACAT1 by shRNA and control cells harboring an empty vector were tested for PDC flux rate (left) and Western blot (right).
(B–D) ACAT1 KD cells were tested for cell proliferation rate under normoxia and hypoxia (B), lactate production (C) and oxygen consumption in the presence and absence of ATP synthase inhibitor oligomycin (D).
(E) Tumor masses in xenograft nude mice injected with ACAT1 knockdown cells compared to mice injected with control vector cells are shown. p values were determined by a two-tailed paired Student’s t test.
(F) Western blot results show K321 acetylation levels of PDHA1 (upper panels) and K202 acetylation levels of PDP1 (lower panels) in tumor lysates.
(G–J) Stable ACAT1 knockdown cells with stable expression of FLAG-PDP1 WT (left panels), or acetyl-mimetic form K202Q or acetyl-deficient form K202R of FLAG-PDP1 (right panels) were tested for PDC flux (G), lactate production (H), oxygen consumption (I) and cell proliferation rate under hypoxia (J). The error bars represent mean values +/− SD.
Also see Figures S3–S4.
Furthermore, in a xenograft experiment, the growth rate (Figure S3F) and masses (Figures 4E and S3G, left) of tumors derived from ACAT1 knockdown cells were significantly reduced, likely due to decreased cell proliferation assessed by decreased IHC staining of proliferation marker Ki67 (Figure S3G, right), compared to those of tumors formed by control cells. In addition, acetylation levels of PDHA1 K321 and PDP1 K202 are markedly reduced in tumors derived from ACAT1 knockdown cells compared to those formed by control vector cells (Figure 4F). Consistent with these findings, we found increased IHC staining of HIF-1a and its target GLUT1 (Figure S3J) and increased mRNA levels of a group of markers of hypoxia including VEGF, GLUT1, CA9 and BHLHB2 that are transcription targets of HIF-1α (Figure S3K), in tumors from mice injected with ACAT1 knockdown cells compared to tumors derived from control vector cells, or PDHA1 K321R or PDP1 K202R rescue cells compared to tumors derived from corresponding control cells. These data together suggest that tumor derived from ACAT1 KD cells, or PDHA1 K321R or PDP1 K202R rescue cells are more hypoxic compared to tumors derived from corresponding control cells.
We acknowledge that PDHA1 and PDP1 might not be the only substrates of ACAT1. To determine whether the effect of ACAT1 knockdown on cancer cell metabolism and cell proliferation is predominantly mediated through activation of the PDP1-PDHA1 axis and subsequently PDC, we next examined the effect of expressing distinct acetyl-mimetic K→Q and acetyl-deficient K→R mutants of PDHA1 and PDP1 on ACAT1 knockdown cells (Figures S4A and S4B). We found that expression of catalytically less active PDP1 K202Q mutant in ACAT1 knockdown cells reverses the ACAT1-deficiency results increased PDC flux rate (Figure 4G), decreased lactate production (Figure 4H), and increased oxygen consumption (Figure 4I), compared to control vector cells. Moreover, expression of PDP1 K202Q not only completely rescued the reduced cell proliferation rate of ACAT1 knockdown cells under hypoxia but also conferred a cell proliferation rate to ACAT1 knockdown cells that exceeds that of control vector cells (Figure 4J). In contrast, H1299 cells with expression of FLAG-PDP1 WT or catalytically more active PDP1 K202R show phenotypes similar to ACAT1 knockdown cells, and PDP1 WT or K202R expression in ACAT1 knockdown cells cannot rescue the aforementioned phenotypes due to loss of ACAT1 as PDP1 K202Q does (Figure 4G–4J). Similar results were obtained using ACAT1 knockdown cells with expression of PDHA1 K321Q and K321R cells (Figures S4C–S4F). These data together demonstrate that ACAT1 predominantly signals through the PDP1-PDHA1 axis to promote glycolysis and confer a proliferative advantage to cancer cells.
Mitochondrial SIRT3 is the upstream deacetylase of PDHA1 and PDP1
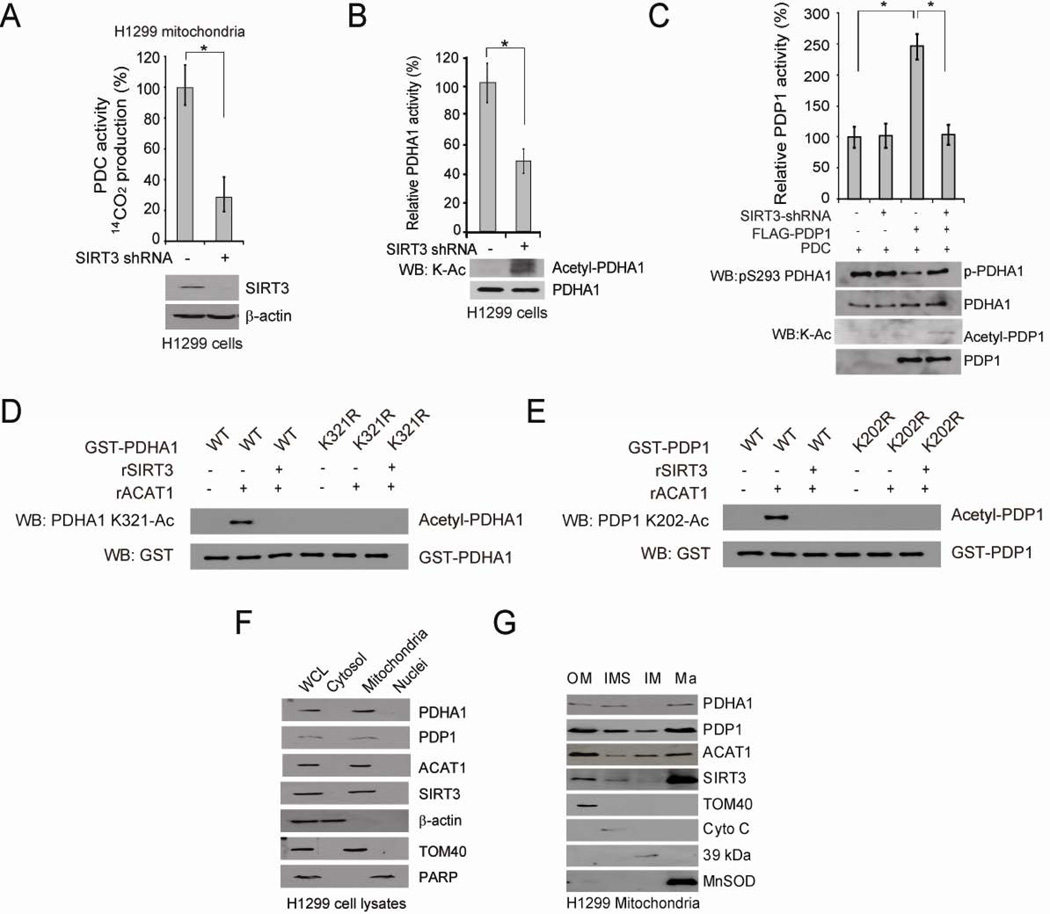
We also sought to identify the mitochondrial deacetylase that acts in concert with ACAT1. We tested NAD+-dependent deacetylase SIRT3, which was reported to regulate global lysine acetylation in mitochondria (Lombard et al., 2007). H1299 cells with stable knockdown of SIRT3 (Figure 5A), but not the control mitochondrial deacetylase SIRT5 (Figure S5A), show decreased PDC flux rate in isolated mitochondria compared to control vector cells. In addition, incubation with cell lysates from SIRT3 knockdown cells results in increased lysine acetylation and inhibition of recombinant PDHA1 and PDP1 (Figures 5B and 5C, respectively). Moreover, as shown in Figures 5D and 5E, treatment with recombinant ACAT1 results in increased K321 and K202 acetylation of PDHA1 and PDP1, respectively, which is abolished by sequential treatment with recombinant SIRT3. Conversely, treatment with recombinant SIRT3 results in decreased lysine acetylation of PDHA1 and PDP1 (Figures S5B and S5C, respectively) and in particular, decreased K321 and K202 acetylation of SIRT3-treated PDHA1 and PDP1, respectively (Figures S5D and S5E, respectively), which is restored by sequential ACAT1 treatment. Moreover, we detected ACAT1 and SIRT3 in mitochondria along with mitochondrial PDHA1 and PDP1 in H1299 cells (Figure 5F). We further localize these proteins within mitochondria biochemically by performing sub-fractionation of highly purified mitochondria from H1299 cells. As shown in Figure 5G, we found that PDHA1, PDP1, ACAT1 and SIRT3 co-localize in not only the mitochondrial matrix (Ma) but also the mitochondrial outer membrane (Om) and the intermembrane space (IMS), while a fraction of PDP1 and ACAT1 but not PDHA1 and SIRT3 were also detected in the mitochondrial inner membrane (IM). These data together demonstrate that SIRT3 directly deacetylates PDHA1 and PDP1 and acts in concert with ACAT1 to regulate PDHA1 and PDP1 by cyclic deacetylation/acetylation in mitochondria.
Figure 5. Mitochondrial SIRT3 is the upstream deacetylase of PDHA1 and PDP1.
(A) Control vector and stable SIRT3 knockdown cells were tested for PDC flux (upper). SIRT3 protein level was detected by Western blot (lower).
(B–C) Purified PDHA1 (B) and PDP1 (C) were incubated with cell lysates from H1299 cells with stable knockdown of SIRT3 or an empty vector, followed by PDHA1 and PDP1 activity assays (upper panels) and Western blot (lower panels).
(D–E) Purified PDHA1 WT and K321R mutant (D) or PDP1 WT and K202R mutant (E) were treated with rACAT1, followed by incubation with recombinant SIRT3 and Western blot.
(F) Western blot results show co-localization of PDHA1, PDP1, ACAT1 and SIRT3 in mitochondria of H1299 cells. Cytosolic β-actin, mitochondrial TOM 40 and nuclear PARP were included as control markers.
(G) Western blot results show co-localization of PDHA1, PDP1, ACAT1 and SIRT3 in the outer membrane (Om), inter-membrane space (IMS) and matrix (Ma) of mitochondria in H1299 cells. TOM40, Cyto C, complex I 39-kDa protein, and MnSOD are markers for Om, IMS, inner membrane (Im), and Ma, respectively. The error bars represent mean values +/− SD.
Also see Figure S5.
Y381 phosphorylation of PDP1 recruits ACAT1 and dissociates SIRT3 to promote lysine acetylation of PDP1 and PDHA1 and the Warburg effect
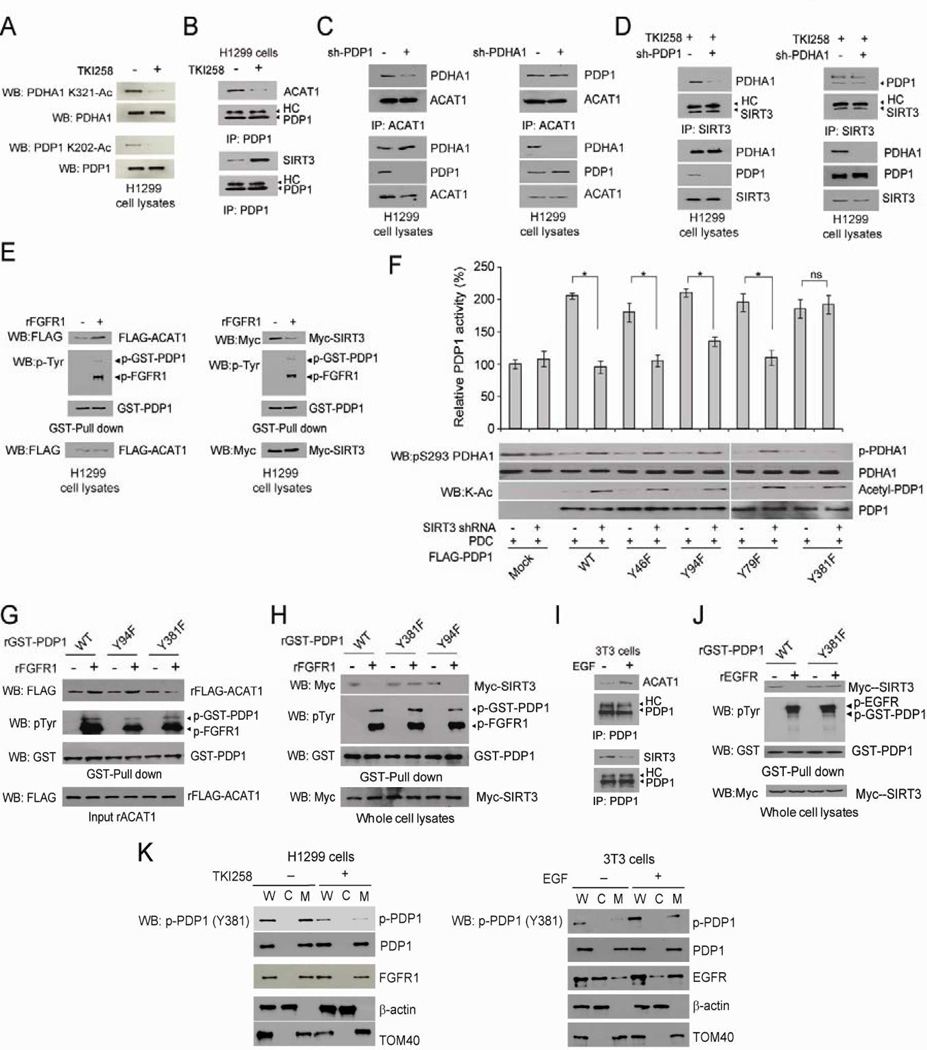
To better understand how PDHA and PDP1 are regulated, we tested whether tyrosine kinase signaling, commonly activated in proliferating cells and cancer cells, regulates lysine acetylation of PDHA and PDP1. We found that treatment with FGFR1 inhibitor TKI258 in lung cancer H1299 cells that overexpress FGFR1 (Marek et al., 2009) results in decreased K321 and K202 acetylation of PDHA1 and PDP1, respectively (Figure 6A). Since we observed that, in a GST-pull down assay, FLAG-ACAT1 and Myc-SIRT3 interact with GST-tagged PDHA1 and PDP1 (Figures S6A and S6B), we next determined whether FGFR1 regulates recruitment of ACAT1 and/or SIRT3 to PDP1. As shown in Figure 6B, treatment with TKI258 results in decreased endogenous ACAT1 but increased endogenous SIRT3 binding to endogenous PDP1 in H1299 cells. Because both PDP1 and PDHA1 are part of PDC that is a huge complex, we next determined which component, PDP1 or PDHA1, is important to recruit ACAT1 and SIRT3 to PDC. We found that stable knockdown of PDP1 results in decreased PDHA1-ACAT1 and PDHA1-SIRT3 association (Figures 6C and 6D, left panels, respectively). In contrast, knockdown of PDHA1 does not affect PDP1-ACAT1 and PDP1-SIRT3 association (Figures 6C and 6D, right panels, respectively). These results together suggest that tyrosine kinase signaling regulates PDP1 but not PDHA1 to control accessibility of ACAT1 and SIRT3 to PDC.
Figure 6. Y381 phosphorylation of PDP1 recruits ACAT1 but dissociates SIRT3 to acetylate PDP1 and PDHA1.
(A) Cell lysates from H1299 cells treated with or without FGFR1 inhibitor TKI258 (1µM) for 4 hours were applied to Western blot to detect acetylation of PDHA1 K202 (upper) and PDP1 K321 (lower).
(B) Endogenous PDP1 was immunoprecipitated from H1299 cells treated with or without FGFR1 inhibitor TKI258. Co-immunoprecipitated endogenous ACAT1 (upper) and SIRT3 (lower) were detected using Western blot. HC: heavy chain.
(C–D) Endogenous ACAT1 (C) or SIRT3 (D) were immunoprecipitated from H1299 cells with stable knockdown of PDP1 (left panels) or PDHA1 (right panels), and co-immunoprecipitated endogenous PDHA1 or PDP1 were detected using Western blot, respectively. SIRT3 was immunoprecipitated from TKI258-treated cells.
(E) Purified GST-PDP1 was incubated with recombinant FGFR1 (rFGFR1), followed by incubation with cell lysates from H1299 cells transiently transfected with FLAG-ACAT1 (left) or Myc-SIRT3 (right). GST-pull-down was performed and FLAG-ACAT1 (left) or Myc-SIRT3 (right) bound to GST-PDP1 were detected by Western blot.
(F) Purified FLAG-PDP1 variants were incubated with purified PDC proteins in the presence or absence of cell lysates from SIRT3 KD cells, followed by in vitro PDP1 assay.
(G) Purified GST-PDP1 WT, Y94F or Y381F were incubated with rFGFR1, followed by incubation with recombinant FLAG-ACAT1. GST-pull-down was performed and FLAG-ACAT1 bound to GST-PDP1 were detected by Western blot.
(H) Purified GST-PDP1 variants were incubated with rFGFR1, followed by incubation with cell lysates from H1299 cells transiently transfected with Myc-SIRT3. GST-pull-down was performed and Myc-SIRT3 bound to GST-PDP1 were detected by Western blot.
(I) 3T3 cells were treated with EGF (100ng/ml) for 4 hours. Endogenous PDP1 was immunoprecipitated and co-immunoprecipitated endogenous ACAT1 (upper) and SIRT3 (lower) were detected using Western blot. HC: heavy chain.
(J) GST-PDP1 variants were incubated with rEGFR, followed by incubation with lysates from H1299 cells expressing Myc-SIRT3. GST-pull-down was performed to detect Myc-SIRT3/GST-PDP1 association.
(K) Immunoblots of FGFR1 (left) or EGFR (right), PDP1 and phospho-PDP1 (Y381) in whole cell lysates (W), cytosolic fraction (C) and mitochondrial fraction (M) of H1299 cells treated with FGFR1 inhibitor TKI258 (left) or 3T3 cells stimulated with EGF (right), respectively.. Cytosolic β-actin and mitochondrial TOM40 were included as controls. The error bars represent mean values +/− SD.
Also see Figure S6.
Moreover, we found that recombinant FGFR1 directly phosphorylates GST-PDP1 in an in vitro FGFR1 kinase assay, leading to increased FLAG-ACAT1 but decreased Myc-SIRT3 binding to tyrosine-phosphorylated GST-PDP1 in the following GST-pull down assay (Figure 6E). To identify the tyrosine site(s) of PDP1 that mediates phosphorylation dependent recruitment of ACAT1 and/or dissociation of SIRT3, we performed mass spectrometry based experiments to examine purified PDP1 protein incubated with recombinant FGFR1. We identified Y79, Y94 and Y381 of PDP1 as phosphorylated (Figure S6C), in addition to Y46 of PDP1 posted on the CST website (www.phosphosite.org). We next performed mutational analysis and generated diverse phosphorylation-deficient Y→F mutants of PDP1 to replace each of the identified phospho-tyrosine residues. As shown in Figure 6F, we found that substitution of PDP1 Y381 with phenylalanine abolishes the inhibition of PDP1 phosphatase activity induced by treatment with SIRT3 knockdown cell lysates. In addition, treatment with FGFR1 results in increased ACAT1 (Figure 6G) but decreased SIRT3 (Figure 6H) binding to PDP1 WT and control Y94F mutant. In contrast, PDP1 Y381F mutant is resistant to FGFR1 treatment, which maintains basal level binding to ACAT1 (Figure 6G) but binds constitutively to SIRT3 (Figure 6H) in the presence of FGFR1.
In addition, EGF treatment results in increased ACAT1 binding but decreased SIRT3 association to PDP1 in 3T3 cells (Figure 6I), and recombinant EGFR (Figure 6J) and several other tyrosine kinases including ABL, FLT3 and JAK2 (Figures S6D–S6F, respectively), which are frequently dysregulated in diverse human cancers, also directly phosphorylate PDP1 WT, leading to decreased SIRT3 binding, whereas Y381F mutation abolishes tyrosine phosphorylation-dependent disruption of PDP1-SIRT3 association. We also generated an antibody that specifically recognizes PDP1 phospho-Y381. Using this antibody, we found that active recombinant FGFR1 and EGFR directly phosphorylate purified recombinant PDP1 at Y381 (Figures S6G). In addition, we found that treatment with FGFR1 inhibitor TKI258 resulted in a decreased Y381 phosphorylation level of mitochondrial PDP1 in H1299 cells, where a fraction of FGFR1 was detected in the mitochondria (Figure 6K; left). Consistently, stimulation with EGF treatment resulted in increased Y381 phosphorylation of mitochondrial PDP1 in 3T3 cells, where mitochondria-localized EGFR was detected (Figure 6K; right). Thus, these results suggest that tyrosine kinase signaling commonly regulates Y381 phosphorylation level of PDP1 to control mutually exclusive recruitment of ACAT1 and SIRT3 to PDC.
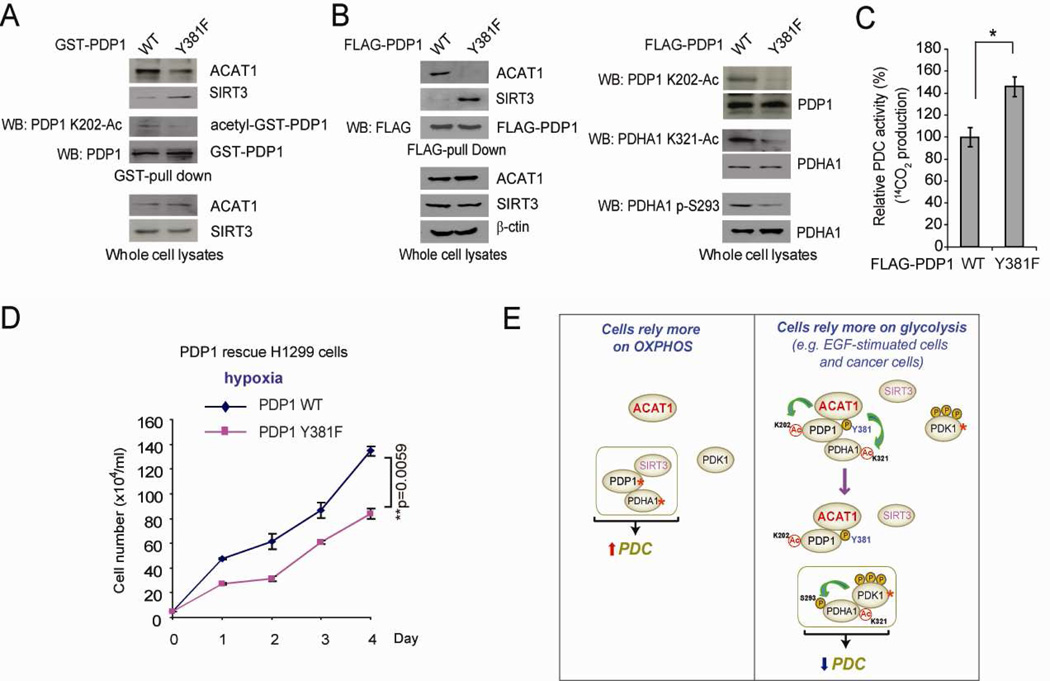
In addition, as shown in Figures 7A and 7B, respectively, both GST-PDP1 Y381F mutant expressed by transient transfection in FGFR1-expressing H1299 cells and FLAG-PDP1 Y381F stably expressed in “rescue” H1299 cells (Figure S7A) have decreased K202 acetylation with reduced binding ability to ACAT1 but increased binding ability to SIRT3, compared to control PDP1 WT. Moreover, rescue expression of PDP1 Y381F also results in reduced K321 acetylation and S293 phosphorylation of PDHA1 (Figure 7B, right), along with increased PDC flux rate and decreased cell proliferation rate under hypoxia (Figures 7C and 7D, respectively) as well as decreased lactate production and increased oxygen consumption rate (Figures S7B and S7C, respectively), similar to the phenotypes of K→R rescue cells of PDHA1 and PDP1 (Figure 2). These data together suggest that Y381 phosphorylation of PDP1 is sufficient and important to regulate lysine acetylation of PDP1 and PDHA1 as well as subsequent cancer cell metabolism and cell proliferation, by altering recruitment of ACAT1 and SIRT3 to PDC.
Figure 7. Y381 phosphorylation of PDP1 is important for lysine acetylation of PDP1 and PDHA1 and contributes to the enhanced glycolysis in cancer cells.
(A) GST-PDP1 WT and Y381F were expressed in H1299 cells by transient transfection. GST-pull-down was performed, followed by Western blot to detect endogenous ACAT1 and SIRT3 bound to GST-PDP1 as well as K202 acetylation level of GST-PDP1 protein.
(B) Left: FLAG-PDP1 WT and Y381F were pulled down from rescue cells and applied to Western blot for detection of endogenous ACAT1 and SIRT3 bound to FLAG-PDP1 variants. Right: Western blot was performed to detect K202 acetylation level of FLAG-PDP1 and K321 acetylation and S293 phosphorylation levels of endogenous PDHA1.
(C–D) PDP1 Y381 rescue cells and control WT cells were tested for PDC flux rate (C) and cell proliferation rate under hypoxia (D).
(E) Proposed model shows that Y381 phosphorylation of PDP1 toggles recruitment of ACAT1 and PDP1 to regulate lysine acetylation status of PDHA1 and PDP1, which contributes to phosphorylation-dependent regulation of PDHA1 and subsequent PDC activity (active enzymes are marked by asterisks). The error bars represent mean values +/− SD.
Also see Figure S7.
DISCUSSION
Our findings provide new insight into the molecular mechanisms underlying PDC regulation, which complements the currently accepted model in which PDHA1 and subsequent PDC activity is regulated through cyclic serine phosphorylation/dephosphorylation. As proposed in Figure 7E, our results suggest that, in normal, differentiated cells that predominantly rely on OXPHOS, PDC is physiologically activated with the core of PDC consisting of active PDP1 and PDHA1, which have only basal lysine acetylation levels due to association of SIRT3. ACAT1 and PDK1 in such cells are either not part of PDC or their presence in PDC is relatively low. In contrast, in glycolytic, EGF-stimulated cells where EGFR is activated and cancer cells where tyrosine kinase signaling is commonly upregulated, Y381 phosphorylation of PDP1 occurs, leading to SIRT3 dissociation and ACAT1 recruitment. ACAT1 acetylates PDP1 at K202 and PDHA1 at K321, leading to dissociation of PDP1 from PDHA1 and recruitment of active PDK1 to PDHA1, respectively. The core of PDC thus consists of inactive PDHA1 with high levels of inhibitory S293 phosphorylation due to PDK1 recruited by K321 acetylation, which results in less active PDC. Inhibition of PDC facilitates a metabolic alteration to allow cells rely more on glycolysis in these cells, where presence of PDK1 is increased in the PDC, while ACAT1, SIRT3 and PDP1 are either not part of PDC or the presence of these proteins in the PDC is at a basal level. Our results suggest that disruption of lysine acetylation of PDHA1 and PDP1 or knockdown of ACAT1 renders cells more reliant on OXPHOS for ATP production. Thus, under normoxia when oxygen is sufficient, cells expressing K→R mutants of PDP1 or PDHA1 and ACAT1 KD cells can maintain ATP production and proliferation rates that are comparable to the control cells, whereas under hypoxia when oxygen is insufficient to fulfill the request of OXPHOS, these cells show decreased ATP levels and consequently reduced proliferation compared to controls cells.
Moreover, our studies showcase the beauty of complex signal transduction-based regulation of cellular processes. We provide evidence to support a concept that hierarchical, distinct post-translational modifications act in concert to provide precise regulation of a series of sequential events, wherein tyrosine phosphorylation status of PDP1 regulates lysine acetylation levels of PDP1 and PDHA1, which acts in concert with tyrosine phosphorylation-activated PDK1 (Hitosugi et al., 2011) to regulate inhibitory serine phosphorylation of PDHA1. In addition, our findings that diverse tyrosine kinases (TKs) inhibit PDP1 (current manuscript) but activate PDK1 (Hitosugi et al., 2011) via tyrosine phosphorylation suggest that upstream tyrosine kinase signaling pathways may regulate a functional protein complex (PDC) not only by coordinating different components with opposite functions (PDP1 and PDK1) but also providing multiple layers of regulation that ensure appropriate control of the complex in response to cellular events. The latter notion is supported by our finding that Y381 phosphorylation of PDP1 regulates PDK1 binding to PDHA1 by recruiting ACAT1. Furthermore, we and others previously reported that a fraction of different TKs including EGFR, FGFR1, FLT3 and JAK2 also localize in mitochondria (Boerner et al., 2004; Hitosugi et al., 2011), where they may phosphorylate PDP1. Given the fact that these TKs are also frequently dysregulated in diverse human cancers (Krause and Van Etten, 2005), these findings together suggest that phosphorylation of Y381 of PDP1 represents a common mechanism to mediate upstream tyrosine kinase signaling-dependent regulation to mitochondria PDC in EGF-stimulated cells and cancer cells.
We also identified mitochondrial ACAT1 and SIRT3 as the upstream acetyltransferase and deacetylase of PDHA1 and PDP1. Part of this finding is consistent with a recent report in which loss of SIRT3 in muscle cells results in highly lysine-acetylated PDHA1 with decreased enzyme activity (Jing et al., 2013). However, SIRT3 was suggested to deacetylate the K336 site of PDHA1. Interestingly, acetylated K321 of PDHA1 was detected in cells of many different cancer types, while PDHA1 K366 was detected as acetylated in human myeloid leukemia MV4–11 cells (http://www.phosphosite.org/proteinAction.do?id=1271&showAllSites=true). These findings may suggest that, although lysine acetylation of PDHA1 represents a common regulatory mechanism in human cells, PDHA1 might be differentially acetylated in different cell types. In addition, Park et al recently reported that SIRT5 regulates PDH activity through lysine desuccinylation (Park et al., 2013). These findings, together with our own, suggest that different post-translational modifications may coordinate to regulate PDH and consequent PDC activity.
Our findings also add to emerging evidence which has shown that lysine acetylation of metabolic enzymes is common and important to link cell signaling pathways to metabolic pathways in cancer cells (Choudhary et al., 2009; Kim et al., 2006b; Wang et al., 2010; Zhao et al., 2010). For example, Lv et al. reported that pyruvate kinase M2 isoform (PKM2) is acetylated at K305 and K433, leading to inhibition of PKM2 enzyme activity but enhanced nuclear accumulation and protein kinase activity of PKM2 in cancer cells (Lv et al., 2011; Lv et al., 2013). In addition, K5 acetylation promotes degradation of lactate dehydrogenase A (LDH-A), which is regulated by its upstream deacetylase SIRT2, and K5 acetylation status of LDH-A reversely correlates with protein expression level of SIRT2 in tumor tissue samples from pancreatic cancer patients (Zhao et al., 2013). Increased lysine acetylation of ATP-citrate lyase (ACLY), a key enzyme in lipid biosynthesis, was found in human lung cancers, which stabilize ACLY to promote fatty acid biosynthesis and consequent tumor growth (Lin et al., 2013).
ACAT1 is a key enzyme in ketogenesis by converting two acetyl-CoA to acetoacetyl-CoA and CoA (Balasse and Fery, 1989). To our knowledge, ACAT1 is the first mitochondrial protein acetyltransferase that is linked to cancer metabolism and tumor growth. In contrast, SIRT3 has been previously suggested to function as a tumor suppressor (Bell et al., 2011; Kim et al., 2010; Xiao et al., 2013) as well as a repressor of the Warburg effect through HIF1α destabilization (Finley et al., 2011), which is consistent with our findings that deacetylation of PDHA1 and PDP1 by SIRT3 provides a disadvantage to cancer cell metabolism and cell proliferation. Future studies are warranted to explore how oncogenic signals coordinate the enzyme activity of ACAT1 and SIRT3 to regulate lysine acetylation levels of PDHA1 and PDP1 to promote glycolysis, which provides a metabolic advantage to cancer cell proliferation and tumor growth. Moreover, our findings that ACAT1 predominantly signals through PDP1 and PDHA to promote the Warburg effect and tumor growth suggest that the ACAT1-PDP1-PDHA axis represents a promising anti-cancer target. Functional attenuation of ACAT1 or activation of its downstream effectors PDP1 and PDHA1 could lead to attenuated cancer cell proliferation and tumor growth due to metabolic defects.
EXPERIMENTAL PROCEDURES
Screening for upstream acetyltransferase(s) of PDP1
The identities of known acetyltransferases in the human genome were provided by http://www.phosphosite.org/psrSearchAction.do. We constructed a “targeted shRNA library” that target 50 out of 71 acetyltransferases genes, which are available in the shRNA library targeting the whole human genome (OpenBioSystems). Each acetyltransferase gene was targeted by a shRNA pool that contains 2–5 different lentiviral-based shRNA constructs that target different regions of the target gene. A PDP1 phosphatase assay-based screening strategy was designed to identify the upstream acetyltransferase(s) of PDP1 using this shRNA library. In brief, H1299 lung cancer cells that were infected with lentiviruses targeting each acetyltransferase. Four days after lentiviral infection, H1299 cells were treated with NAM+TSA for 16 hours. PDP1 assay was performed as described below using recombinant PDP1 incubated with 20 µg cell lysates.
Xenograft studies and primary tissue samples from leukemia patients and healthy donors
Approval of use of mice and designed experiments was given by the Institutional Animal Care and Use Committee of Emory University. Approval of use of human specimens was given by the Institutional Review Board of Emory University School of Medicine. All clinical samples were obtained with informed consent with approval by the Emory University Institutional Review Board. Clinical information for the patients was obtained from the pathologic files at Emory University Hospital under the guidelines and with approval from the Institutional Review Board of Emory University School of Medicine and according to the Health Insurance Portability and Accountability Act. Detailed experimental procedures using mice and human primary tissue samples are described in Supplemental Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Lysine acetylation promotes PDHA/PDK binding but disrupts PDHA/PDP association
Lysine acetylation of PDHA1 and PDP1 promotes the Warburg effect
ACAT1 acetylates and SIRT3 deacetylates PDHA1 and PDP1
Tyrosine phosphorylation of PDP1 dissociates SIRT3 and recruits ACAT1 to PDC
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants CA140515 (J.C.) and the Pharmacological Sciences Training Grant T32 GM008602 (S.E.), DoD grant W81XWH-12-1-0217 (J.C.), and the Hematology Tissue Bank of the Emory University School of Medicine and the Georgia Cancer Coalition (H.J.K.). J.X., M.T., T.-L.G., M.A., and S.L. are employees of Cell Signaling Technology, Inc. S.E. is an NIH pre-doctoral fellow and an ARCS Foundation Scholar. H.J.K., F.R.K., S.K. and J.C. are Georgia Cancer Coalition Distinguished Cancer Scholars. S. K. is a Robbins Scholar. S.K. and J.C. are American Cancer Society Basic Research Scholars. J.C. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes detailed experimental procedures and seven figures.
REFERENCES
- Balasse EO, Fery F. Ketone body production and disposal: effects of fasting, diabetes, and exercise. Diabetes Metab Rev. 1989;5:247–270. doi: 10.1002/dmr.5610050304. [DOI] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Molecular and cellular biology. 2004;24:7059–7071. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Bowker-Kinley MM, Huang B, Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Advances in enzyme regulation. 2002;42:249–259. doi: 10.1016/s0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- Hiromasa Y, Fujisawa T, Aso Y, Roche TE. Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J Biol Chem. 2004;279:6921–6933. doi: 10.1074/jbc.M308172200. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Molecular cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochemical Society transactions. 2003;31:1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- Jing E, O’Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, et al. Sirt3 Regulates Metabolic Flexibility of Skeletal Muscle through Reversible Enzymatic Deacetylation. Diabetes. 2013 doi: 10.2337/db12-1650. 2013 Jul 8.[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor I and dysregulated c-myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Molecular and Cellular Biology. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006a;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006b;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. The New England journal of medicine. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Lin R, Tao R, Gao X, Li T, Zhou X, Guan KL, Xiong Y, Lei QY. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Molecular cell. 2013;51:506–518. doi: 10.1016/j.molcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and cellular biology. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Molecular cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, Jiang Y, Zhou X, Li TT, Guan KL, et al. Mitogenic and Oncogenic Stimulation of K433 Acetylation Promotes PKM2 Protein Kinase Activity and Nuclear Localization. Molecular cell. 2013;52:340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick DT, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Molecular pharmacology. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Molecular cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Roche TE, Baker JC, Yan YH, Hiromasa Y, Gong XM, Peng T, Dong JC, Turkan A, Kasten SA. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Re. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- Sumi S, Ichihara K, Kono N, Nonaka K, Tarui S. Insulin and epidermal growth factor stimulate glycolysis in quiescent 3T3 fibroblasts with no changes in key glycolytic enzyme activities. Endocrinol Jpn. 1984;31:117–125. doi: 10.1507/endocrj1954.31.117. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Xiao K, Jiang J, Wang W, Cao S, Zhu L, Zeng H, Ouyang R, Zhou R, Chen P. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncology reports. 2013 doi: 10.3892/or.2013.2604. [DOI] [PubMed] [Google Scholar]
- Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, Xu YH, Dong B, Xiong Y, Lei QY, Guan KL. Lysine-5 acetylation negatively regulates lactate dehydrogenase a and is decreased in pancreatic cancer. Cancer cell. 2013;23:464–476. doi: 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.