Abstract
Disease outcome is known to be influenced by defined subsets of invariant Natural Killer T (iNKT) cells residing in distinct locations within peripheral tissue. However, the factors governing the development of these unique iNKT sublineages during thymic development are unknown. Here we explored the mechanism by which E protein transcription factors and their negative regulators, the Id proteins control the development of iNKT sublineages after positive selection. We found that E proteins directly bound the PLZF promoter and were required for expression of this lineage-defining transcription factor and for the maturation and expansion of thymic iNKT cells. Moreover, expression of the negative regulators of E proteins, Id2 and Id3, defined distinct iNKT cell sublineages. Id3 was expressed in PLZFhigh NKT2 cells and loss of Id3 allowed for increased thymic iNKT cell expansion and abundance of the PLZF+ NKT2 sublineage. Id2 was expressed in TBET+ NKT1 cells and both Id proteins were required for the formation of this sublineage. Thus, we provide insight into E and Id protein regulation of iNKT cell proliferation and differentiation to specific sublineages during development in the thymus.
Introduction
Natural Killer T (NKT) cells are a unique subset of T cells able to recognize glycolipid antigens presented by the MHC class I-like molecule CD1d. The best-studied NKT cell population utilizes an invariant T cell receptor (TCR) α-chain comprised of the variable region 14 and the joining region 18 (Vα14-Jα18) gene segments, and these cells are therefore termed invariant NKT (iNKT) cells. Within hours of activation, iNKT cells produce large amounts of numerous cytokines and thus play an important role in the early immune response to microbial pathogens. In addition, iNKT cells are involved in protection from cancer and have been implicated in autoimmune diseases such as ulcerative colitis and type 1 diabetes (1-3). As iNKT cell number and function are associated with these diseases and vary broadly in humans and different mouse strains (4, 5), it is essential to understand the mechanisms driving iNKT cell maturation and differentiation.
iNKT cells undergo positive selection, expansion and early maturation in the thymus where four developmental stages have been defined based on the expression of CD24, CD44 and NK1.1; this understanding of iNKT cell development is used by many studies (2, 6, 7). Upon rearrangement of the canonical Vα14-Jα18 TCR and positive selection by CD1d-expressing cortical thymocytes, commitment to the iNKT cell lineage is observed by cells expressing CD24 (stage 0) (2, 6, 7). Subsequently, iNKT cells downregulate CD24 expression transitioning to the highly proliferative CD24-CD44-NK1.1- stage 1, a process dependent on both EGR2 and NF-κB transcription factors (6, 8, 9). EGR2 is involved in direct activation of PLZF expression, the lineage-defining transcription factor of the NKT cell program, and the presence of PLZF allows iNKT cell progression from stage 1 to CD44+NK1.1- stage 2 (9-11). At stages 1 and 2, iNKT cells undergo extensive proliferation, which is abrogated in the absence of the transcription factor c-MYC (12, 13). Subsequently, many stage 2 iNKT cells exit the thymus to complete maturation from stage 2 to stage 3 in peripheral tissue, although a subfraction will mature and remain in the thymus (14). IL-15 and expression of the transcription factor TBET are essential for this transition from stage 2 to stage 3, which is characterized by upregulation of NK1.1 (15, 16).
This concept of sequential, well-defined developmental stages of iNKT cells has recently been modified in the context of new findings. It is now appreciated that within the CD44+NK1.1- stage 2 population, there exists three subsets of iNKT cells: (1) Cells that continue to differentiate, upregulating TBET while downregulating PLZF, and produce IFNγ upon stimulation (NKT1 cells), (2) Cells that retain PLZF expression, and produce IL-4 and IL-13 (NKT2 cells), and (3) Cells that upregulate expression of RORγt, while remaining low for PLZF and TBET, and produce IL-17 (NKT17 cells) (1, 17, 18). Thus, it is likely that alterations in iNKT cell maturation that affect the transition from stage 2 to stage 3, will also affect differentiation of all three sublineages of iNKT cells. Currently, many of the factors that regulate the development of these individual subpopulations remain unknown.
E proteins are basic helix-loop-helix transcription factors. In lymphocytes, E47 and E12 (Tcf3), HEB (Tcf12) and E2-2 (Tcf4) bind to canonical E-box binding sites as homo- or heterodimers. In contrast, Inhibitor of DNA binding (Id) proteins are able to heterodimerize with E proteins but, as they lack a DNA binding domain, act as dominant-negative regulators of E protein transcriptional activity (19). Of the four Id proteins, Id2 and Id3 are most highly expressed in lymphocytes. E and Id proteins influence T cell development and formation of immunological memory (20-23). Furthermore, Id2 is required for survival of iNKT cells in the liver and bone marrow, whereas absence of Id3 leads to abundance of innate-like γδ T cells (24-27). In addition, we previously found that the E protein transcription factor HEB allows for survival of CD4+CD8+ double positive (DP) thymocytes. HEB promoted RORγt and BCL-XL expression, allowing secondary TCRα rearrangements including the canonical iNKT cell Vα14-Jα18 TCRα gene rearrangement to occur (28). One finding of note in our previous study was that rescue of HEB-deficient mice with a Vα14-Jα18 transgene did not lead to complete recovery of iNKT cells, particularly in terms of their maturation state. Moreover, we observed high mRNA expression levels of both E2A and HEB at stage 0 during iNKT cell development. In the current study, we explore the mechanisms by which E and Id proteins control iNKT cell development and differentiation beyond TCRα rearrangement.
Materials and Methods
Mice
Mice were bred and housed in specific pathogen-free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of California San Diego. Tcf3f/fTcf12f/fCd4cre- and Tcf3f/fTcf12f/fCd4cre+ mice were a gift from Y. Zhuang (Duke University), generated as previously described (29). Vα14-Jα18 TCR transgenic mice were a gift from A. Bendelac (University of Chicago) and M. Kronenberg (La Jolla Institute for Allergy and Immunology). Id3-GFP mice were a gift from C. Murre and were generated as described (22). Id2-YFP mice were generated as previously described (21).
Fetal liver chimeras were generated by transferring 5-10 × 106 E14.5 fetal liver cells into lethally irradiated (1000 rad) CD45.1.2 recipient mice. Id2Y/Y donor cells were CD45.2, WT donor cells were CD45.1, Id3G/G donor cells were CD45.2, and recipient mice were CD45.1.2.
Mixed bone marrow chimeras were generated by transferring a mixture of 6 × 106 B220−CD4−CD8− bone marrow cells of a CD45.1 wild-type donor and 3 × 106 B220−CD4−CD8− bone marrow cells of CD45.2 Id3G/G donor into lethally irradiated (1000 rad) CD45.1.2 recipient mice. All chimeras were rested for at least eight weeks to allow reconstitution of the host.
Flow Cytometry
Single-cell suspensions were prepared from indicated tissues. Hepatic NKT cells were isolated from liver as previously described (28) using 34% Percoll solution (Sigma). The following antibodies were used: TCRβ FITC (clone H57-597), CD4 FITC (L3T4), CD8 FITC (53-6.7) (Biolegend), CD24 FITC (M1/69) (BD Pharmingen), CD45.1 FITC (A20), CD45.2 FITC (104), CD1d Tetramer Alexa 488 (NIH Tetramer Core Facility (mCD1d/PBS57)), CD1d Tetramer PE (NIH Tetramer Core Facility (mCD1d/PBS57)), TCRβ PE (H57-597), PLZF PE (Mags.21F7), NK1.1 PerCP Cy5.5 (PK136), CD45.2 PerCP Cy5.5 (104), NK1.1 APC (PK136), TCRβ APC (H57-597), CD45.2 APC (104), CD44 APC (IM7), CD4 APC (L3T4) (BD Pharmingen), CD8 APC (53-6.7) (Biolegend), TBET APC (4B10), CD1d Tetramer Alexa 647 (NIH Tetramer Core Facility (mCD1d/PBS57)), CD19 PeCy7 (ID3), CD45.1 PeCy7 (A20), CD45.2 Alexa 750 (104), Ki-67 Pacific Blue (SolA15). All antibodies were purchased from eBioscience unless otherwise specified. Samples were collected on a FACS LSRII, FACS Fortessa or FACS Aria (BD Biosciences) fitted with custom mirrors from Omega Filters (510/21 with 502LP or 505LP for GFP, 530/30 with 525LP for YFP) and were analyzed with FlowJo software (TreeStar).
Quantitative PCR and Chromatin Immunoprecipitation
Cells were sorted as indicated. RNA was extracted with TRIzol reagent (Invitrogen), treated with genomic DNA wipeout reagent (Qiagen) and cDNA was generated using cDNA kit (Qiagen). The abundance of mRNA was assessed by quantitative PCR with nonspecific product detection (SYBR Green; Stratagene) using primers that amplify in a linear relationship with primers for ‘housekeeping’ genes. Results were normalized to expression of HPRT transcripts. Chromatin-immunoprecipitation assays were performed as described (28). A polyclonal antibody specific for E2A (V-18X) or HEB (A-20X) (Santa Cruz Biotechnology) was used to precipitate E2A-DNA or HEB-DNA complexes, and rabbit immunoglobulin G antibody (2729; Cell Signaling Technology) was used as a negative control. Immunocomplexes were bound to protein G–Sepharose beads (Cell Signaling Technology) and were washed four times. DNA was eluted and purified and was analyzed by quantitative PCR to detect putative E protein binding sites in the zbtb16 gene. Zbtb16 ChIP primer sequences E box site 1: 5′ gggttctctggttgctgct and 3′agcccttgcctgtacaaaga. Zbtb16 ChIP primer sequences E box site 2: 5′ caccggaatgcacaggag and 3′ gggagaaaaggatgcacaaa.
Statistical Analysis
Differences between data sets were analyzed by an unpaired two-tailed student's t-test, Mann Whitney U test, one-way ANOVA or Bonferroni post-hoc test where applicable.
Results
E proteins are required for iNKT cell development
While we previously detected high expression levels of E2A and HEB mRNA at stage 0 of iNKT cell development, indicating a possible requirement for E proteins during iNKT cell thymic development, we showed loss of E protein expression led to impaired rearrangement of the canonical Vα14-Jα18 iNKT cell TCR (28). Here we crossed Vα14-Jα18 transgenic (Vα14tg) mice to mice conditionally deficient for Tcf3 (E2A) and Tcf12 (HEB) at the DP stage of thymocyte development (Tcf3f/fTcf12f/fCd4cre+), thus bypassing the requirement for E proteins in TCR rearrangement. Both the frequency (Fig. 1A and 1B) and absolute number (Fig. 1C) of iNKT cells in the thymus, spleen and liver were reduced in the E protein-deficient mice relative to wild type mice, showing impaired iNKT cell development in the absence of E proteins.
Figure 1. Loss of E protein expression impairs iNKT cell maturation.
Tcf3+/+Tcf12+/+Cd4cre- or Tcf3f/f Tcf12f/f Cd4cre+ mice expressing a Vα14-Jα18 TCR transgene were characterized. (A) Flow cytometric analysis of iNKT cell frequency in Vα14-Jα18tg+Tcf3+/+Tcf12+/+Cd4cre- and Vα14-Jα18tg+Tcf3f/fTcf12f/fCd4cre+ lymphocytes from indicated tissues. Numbers indicate percentage of cells within the drawn gate. (B) Average frequency or (C) absolute number (±SEM) of CD1d-tet+TCRβ+ gated iNKT cells from the indicated genotype and tissue. (D) Flow cytometric analysis of CD1d-tet+CD24+ expression in the thymus and CD44+NK1.1+ expression by gated iNKT lymphocytes from the indicated genotype in the indicated tissue. (E) Average frequency and absolute number (±SEM) of maturation stages as defined by CD1d-tet and CD24 or CD44 and NK1.1 expression on CD1d-tet+TCRβ+ gated iNKT cells from the indicated genotype in the thymus. Stage 0 cells are defined by CD1d-tet+CD24+ expression. Gated CD1d-tet+CD24- cells define stages 1, 2 and 3. Graphs are the average of 11 mice from 8 independent experiments. Statistical significance was determined using unpaired two-tailed t test, ns = not significant, *, P < 0.05, **, P < 0.005, ***, P < 0.0005, ****, P < 0.0001.
Closer analysis revealed E protein-deficient iNKT cell development was blocked at stage 0 (CD1d-tet+CD24+) and stage 1 (CD1d-tet+CD24-CD44-NK1.1-) (Fig. 1D). This defect at stage 1 was also observed in iNKT cells in peripheral tissue (Fig. 1D). Interestingly, examination of absolute cell numbers in the thymus showed equal numbers of stage 0 and stage 1 iNKT cells in the absence of E proteins while there was a significant loss of cells at stages 2 and 3 (Fig. 1E). These data indicated a failure of thymic iNKT cells to progress through development in the absence of E2A and HEB and suggested a role for these proteins during iNKT cell differentiation, after positive selection to the lineage.
E proteins regulate proliferation and PLZF expression during development of iNKT cells
Given that iNKT cells failed to progress to stages 2 and 3 during thymic development, we next asked how E proteins might influence this process. Staining for Ki-67, a marker of actively dividing cells, we observed that the number of proliferating E protein-deficient cells at stage 1 was reduced by 50% compared wild type cells (Fig. 2A). E protein-deficient cells at stage 0 also showed a slight increase in Ki-67 expression (Fig. 2A). Thus, failure of iNKT cells to accumulate at later stages of development in the absence of E proteins was attributable in part to the inability of iNKT cells to undergo the ‘proliferative burst’ typically observed in these cells at stage 1. Tcf3f/fTcf12f/fCd4cre+ iNKT cells did not show an increase in active caspase 3 and 7 positive iNKT cells at stage 0 and 1 relative to wild type cells (data not shown), indicating E proteins influence iNKT cell proliferation rather than survival at stage 1.
Figure 2. E proteins regulate proliferation and PLZF expression in thymic iNKT cells.
(A) Ki-67 expression by stage 0 and stage 1 CD1d-tet+TCRβ+ gated iNKT thymocytes from the indicated genotypes. Numbers indicate percentage Ki-67+ cells within the gate. Bar graphs indicate average frequency (±SEM) of CD1d-tet+TCRβ+ gated iNKT Ki-67+ cells at stage 0 and 1. Graphs are an average of 6 mice from 3 individual experiments. Statistical significance was determined using unpaired two-tailed t test, *, P < 0.05. (B) Bar graphs indicate relative expression of zbtb16 mRNA by stage 0 and 1 CD1d-tet+TCRβ+ gated iNKT cells determined by qPCR. Data are normalized to Vα14-Jα18tg+Tcf3+/+Tcf12+/+Cd4cre- cells and are the average of 3 individual samples from 3 independent experiments. Histogram plots indicate PLZF expression by stage 0 and 1 CD1d-tet+TCRβ+ CD1d-tet+TCRβ+ gated iNKT cells from the indicated genotypes. Numbers indicate percentage of PLZFhigh cells within the gate. Data are representative of 3 independent experiments with n = 1 – 3 mice per group. Statistical significance was determined using unpaired two-tailed t test, **, P < 0.005, ****, P < 0.0001. (C) The zbtb16 promoter is a direct target of E2A and HEB. Schematic indicating position of E box sites in the zbtb16 promoter relative to the ATG start site. Thymic iNKT cells from the indicated genotypes were sorted at stage 0 (CD1d-tet+TCRβ+CD24+) and chromatin was precipitated with anti-E2A, anti-HEB or control IgG antibodies. The relative amounts of input or precipitated DNA containing the indicated E box sites were determined by qPCR. Fold change of Vα14-Jα18tg+Tcf3+/+Tcf12+/+Cd4cre- pull-down over Vα14-Jα18tg+Tcf3f/f Tcf12f/f Cd4cre+ control are graphed and are an average of sorted cells from 3 mice for Vα14-Jα18tg+Tcf3+/+Tcf12+/+Cd4cre- and 2 mice for Vα14-Jα18tg+Tcf3f/fTcf12f/f Cd4cre+.
We were next interested in identifying the targets of E protein transcription factors during iNKT cell differentiation. One of the most prominent transcription factors, often referred to as the ‘master regulator’ of iNKT cell development, is the protein PLZF encoded by the gene Zbtb16. Examining mRNA expression, we observed a 90% reduction of Zbtb16 expression in sorted iNKT cells from E protein-deficient animals relative to their wild type counterparts at stages 0 and 1 when Zbtb16 is most highly expressed (Fig. 2B). Moreover at the protein level, PLZF expression was also diminished by 84% in Tcf3f/fTcf12f/fCd4cre+ iNKT cells at stage 1 (Fig. 2B). Examination of the Zbtb16 promoter revealed two conserved E box sites, one in the 5′ upstream region (E box site 1) and one in the first intron upstream of the ATG start site (E box site 2) (Fig. 2C). Using chromatin immunoprecipitation (ChIP), we showed binding of E2A and HEB at both of these E box sites in the Zbtb16 promoter. With sorted Tcf3f/fTcf12f/fCd4cre+ iNKT cells at stage 0 as a negative control, we showed E protein binding was specific at these two E box sites (Fig. 2C). Although PLZF is required for iNKT cell development, notably it does not influence the proliferative burst observed as iNKT cells proceed through development (Kovalovsky et al., 2008). Thus, we conclude E protein transcription factors dually influence iNKT cell development by regulating expression of the lineage-defining transcription factor PLZF and by regulating proliferation.
Id2 and Id3 expression define developmental stages and sublineages in thymic iNKT cells
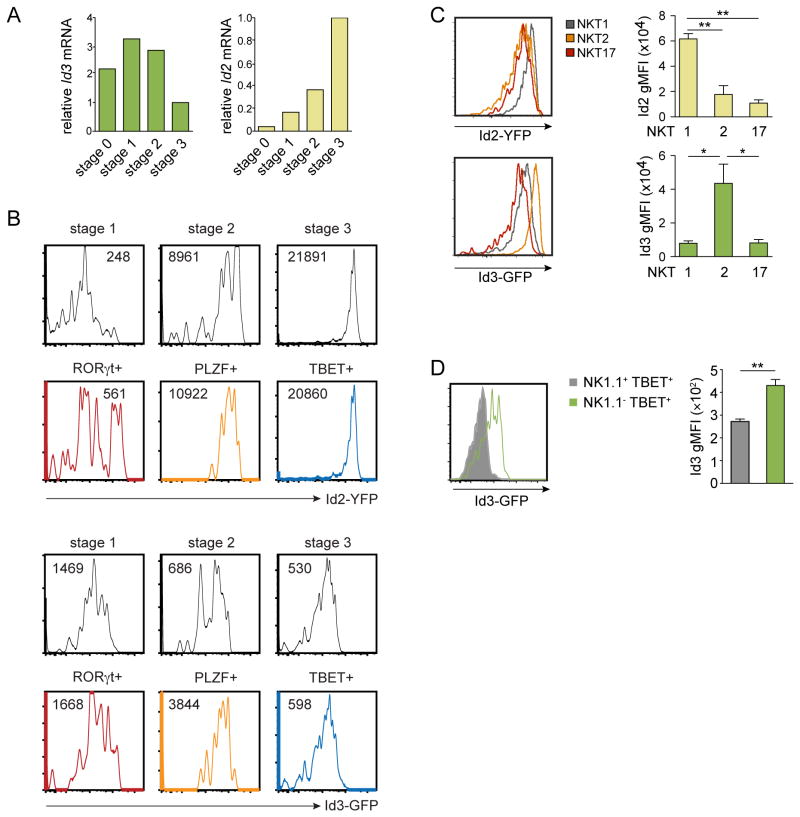
E proteins are negatively regulated by the Id proteins, of which Id2 and Id3 are the Id family members most highly expressed in thymocytes. We previously showed the transcriptional regulator Id2 is essential for peripheral iNKT cell survival in hepatic tissue (25). Here we wanted to explore the contribution of both Id2- and Id3-mediated control of E proteins to iNKT cell thymic development. Examining mRNA expression in sorted iNKT cells from different stages of thymic development, we noted Id3 was most highly expressed at the early stages (stages 1 and 2) while Id2 was expressed more prominently in stage 3 iNKT cells (Fig. 3A). Using reporter mice in which YFP or GFP was knocked into the first exon of the Id2 or Id3 gene respectively, we similarly observed Id3-GFP expression is highest at the earliest stages of iNKT cell development, while Id2-YFP expression was most readily detected as the iNKT cells progressed through maturation stages in the thymus (Fig. 3B).
Figure 3. Id2 and Id3 expression define iNKT cell developmental stages and sublineages in the thymus.
(A) Relative Id3 and Id2 mRNA expression levels by sorted wild-type thymic CD1d-tet+TCRβ+ iNKT cells. Relative expression indicated is normalized to stage 3 mRNA levels. Data are representative of three independent experiments. (B) Histograms showing Id3-GFP and Id2-YFP reporter expression by CD1d-tet+TCRβ+ gated iNKT cells at stage 1 (CD24-CD44-NK1.1-), stage 2 (CD24-CD44+NK1.1-) and stage 3 (CD24-CD44+NK1.1+) or by iNKT cells expressing TBET, PLZF or RORγt in the thymus. Numbers in histograms indicate mean fluorescence intensity (MFI). Data are representative of 3 mice per group from 3 independent experiments. (C) Histograms showing Id2-YFP and Id3-GFP expression in gated CD1d-tet+TCRβ+ TBET+ NKT1, PLZFhigh NKT2 and RORγt+ NKT17 cells respectively. Bar graphs indicate average Id2-YFP and Id3-GFP MFI of indicated sublineages. (E) Histogram showing Id3-GFP expression of NK1.1- TBET+ (green line) and NK1.1+ TBET+ (grey line) cells. Bar graph indicates average Id3-GFP MFI of indicated cell type. Data are average (±SEM) of 3 mice from 3 independent experiments. Statistical significance was determined using one-way ANOVA and Bonferroni post-hoc test, *, P < 0.05, **, P < 0.005.
Gating on the recently described iNKT cell sublineages, defined by expression of unique transcription factors in the thymus (1, 17, 18), we observed that each sublineage had a distinct Id protein expression profile; Id2-YFP expression was highest in TBET+ NKT1 cells, while Id3-GFP remained high in PLZFhigh NKT2 cells. RORγt+ NKT17 cells were intermediate for both Id2 and Id3 (Fig. 3B and 3C). Finally, gating on NK1.1- cells, we could identify a population of TBET+ cells, potentially representing a population of immature NKT1 cells. Comparing NK1.1-TBET+ to NK1.1+TBET+ cells in the thymus, we noted Id3 expression was significantly higher in the NK1.1-TBET+ immature iNKT cells (Fig. 3D). We speculate that as NKT1 cells mature and upregulate NK1.1 expression, Id3 expression is downregulated while Id2 expression continues to be upregulated. We found that Id2 and Id3 mark these different NKT sublineages, implying distinct regulation and/or function of these Id protein family members.
Loss of Id2 and Id3 affect iNKT cell abundance and development
To examine the roles of Id2 and Id3 in thymic iNKT cell development, we used mice homozygous for the knock-in sequence encoding YFP into Id2 (Id2Y/Y) or GFP into Id3 (Id3G/G), thus generating Id2- or Id3-deficient mice. Mice deficient for Id2 die at approximately two weeks of age on the B6 background; therefore we generated fetal liver chimeras (FLC), transferring congenically marked E14.5 fetal liver cells into irradiated hosts. As we have previously shown, using Id2-germline knock-out FLC (Id2Y/Y), thymic development of iNKT cells in the absence of Id2 is relatively normal and Id2 expression is essential for the survival of peripheral iNKT cells (25). Interestingly, loss of Id2 minimally affected thymic development of iNKT cells, most likely due to a role of Id3 during the early stages of iNKT cell development (Fig. 4A). In Id3-deficient FLC (Id3G/G), we similarly observed no significant difference in the frequency or cell number of iNKT cells in the thymus, however Id3-deficiency led to a decrease in the absolute number of iNKT cells in peripheral tissues (data not shown).
Figure 4. Compensation by Id2 and Id3 as iNKT cells differentiate in the thymus.
Flow cytometric analysis showing CD44+NK1.1+ expression by CD1d-tet+TCRβ+ gated iNKT cells from (A) wild type (WT), Id2-deficient (Id2Y/Y) and Id2-deficient Id3-heterozygote (Id2Y/YId3G/+) fetal liver chimeras and (B) wild type (WT), Id3-deficient (Id3G/G) and Id3-deficient Id2-heterozygote (Id3G/GId2Y/+) fetal liver chimeras. Bar graphs indicate iNKT cell frequency and absolute number at stages 1, 2 and 3 of the indicated genotype. (C) Histograms and bar graphs indicate Id3-GFP expression by Id2-deficient cells and Id2-YFP expression by Id3-deficient cells for CD1d-tet+TCRβ+ gated iNKT cells in the thymus, spleen and liver. Data shown are an average (±SEM) of 4 fetal liver chimeras per group, representative of 2 independent experiments. Statistical significance was determined using one-way ANOVA or unpaired two-tailed t test, *, P < 0.05, **, P < 0.005, ***, P < 0.0005, ****, P < 0.0001.
We next asked how iNKT cell development would be affected by loss of both Id2 and Id3. However, our efforts to generate Id2-Id3-double deficient FLC were unsuccessful, most likely due to requirement for these proteins during embryonic development. Instead, we continued our analysis of compound homo- and heterozygote Id2- and Id3-deficient mice and examined iNKT cell thymic development in Id2-deficient Id3-heterozygote (Id2Y/YId3G/+) FLC relative to wild-type (WT) and Id2-deficient (Id2Y/Y) FLC (Fig. 4A). Here we observed a 5-fold increase in frequency and 2-fold increase in absolute number of iNKT cells at stage 1 relative to wild-type cells and a significant decrease (P < 0.0005) in iNKT cells at stage 3 (Fig. 4A). This phenotype was more pronounced in Id3-deficient Id2-heterozygote (Id3G/GId2Y/+) iNKT cells (Fig. 4B). Id3G/GId2Y/+ iNKT cells were significantly increased in frequency and absolute number (P < 0.005) at stage 1 and were decreased more than 60% at stage 3 relative to wild-type cells (Fig. 4B). We postulate that in thymic iNKT cells, Id proteins are required to regulate iNKT cell abundance and allow for progression to stage 3 while loss of three Id protein alleles led to a decrease in CD44+ NK1.1+ mature iNKT cells (Fig. 4A and 4B).
We have previously shown that Id3 expression increased in CD8+ effector T cells in the absence of Id2 (21). Thus, we were interested in addressing the possible compensatory roles for Id2 and Id3 during iNKT cell development. Examining Id3-GFP expression in Id2-deficient iNKT cells (Id2Y/Y), we noted that Id3 expression was dramatically increased in these cells in the thymus, spleen and liver (Fig. 4C). Moreover, Id2-YFP expression decreased slightly with loss of Id3 (Id3G/G) (Fig. 4C). Thus, as in the case of CD8+ effector T cells, it appears Id2 can negatively regulate Id3 expression while Id3 may positively regulate Id2 expression (21).
Id3 limits abundance of iNKT cells
Id3 germline deficiency results in a complex T and B cell phenotype with an increase in γδ T cells as well as innate-like and effector-memory-like T cells (22, 24, 26, 27). In order to assess the cell-intrinsic role of Id3 in iNKT cell development without possible influence of these developmental defects, we generated mixed bone marrow chimeras, using equal numbers of Id3-sufficient (Id3+/+) and Id3-deficient (Id3G/G) bone marrow cells to reconstitute irradiated host mice. Loss of Id3 in these mixed bone marrow chimeras led to a sharp increase in both the frequency and number of iNKT cells in the thymus, spleen and liver while limited numbers of WT iNKT cells were recovered, making comparative analysis of iNKT cell subsets difficult (data not shown). It is also possible that in this system increased numbers of γδ T cells might influence the development of both the WT and Id3-deficient iNKT cells. Thus, in order to obtain sufficient numbers of WT iNKT cells, we next mixed Id3 WT and Id3-deficient bone marrow cells at a 2:1 ratio. We, again, observed increased frequency and absolute number of the Id3-deficient iNKT cells (Fig. 5A and 5B), whereas conventional T cell numbers reflected the 2:1 ratio of the engrafted bone marrow cells in these mice (Fig. 5B). The difference in iNKT cell numbers between the germline-knock-out FLC and the Id3+/+/Id3G/G mixed bone marrow chimeras may be accounted for by the presence of increased innate-like γδ T cell numbers in the germline knock out FLC, as these cells compete with iNKT cells for space within the thymic niche (30, 31). Further analysis of iNKT cell differentiation indicated stage 0 cells were unaffected by Id3 deficiency in the mixed chimeras (Fig. 5C and 5D). However, the Id3-deficient iNKT cells recovered from the mixed bone marrow chimeras were present in increased absolute numbers at stages 2 and 3, as they developed in the thymus (Fig. 5D). Thus, we conclude that negative regulation of E proteins by Id3 is an essential process that must occur as iNKT cells progress through thymic differentiation and that loss of Id3 and continued E protein activity leads to abundance of iNKT cells.
Figure 5. Id3 controls iNKT cell accumulation and maturation.
Mixed bone marrow chimeras were generated by transfer of a 2:1 mix of CD45.1 Id3+/+ with CD45.2 Id3G/G bone marrow into irradiated recipients. (A) Flow cytometry plots showing iNKT cell frequency in wild-type Id3+/+and Id3-deficient Id3G/G lymphocytes from indicated tissues. Numbers indicate percentage of cells within the gate. (B) Average frequency and absolute number (±SEM) of Id3+/+ and Id3G/G CD1d-tet+TCRβ+ iNKT cells or conventional αβ T cells from indicated tissues. (C) CD1d-tet+CD24+ and CD44+NK1.1+ expression by CD1d-tet+TCRβ+ gated Id3+/+ and Id3G/G iNKT thymocytes assessed by flow cytometry. Numbers indicate percentage of cells within the gate. (D) Average frequency and absolute number (±SEM) of maturation stages for CD1d-tet+TCRβ+ gated Id3+/+ and Id3G/G iNKT thymocytes. Data are average (±SEM) of 3 mice, representative of 3 independent experiments with n = 3 – 4 mice. Statistical significance was determined using unpaired two-tailed t test, ns = not significant, *, P < 0.05, **, P < 0.005, ***, P < 0.0005.
Id2 and Id3 regulate iNKT cell proliferation and sublineage commitment
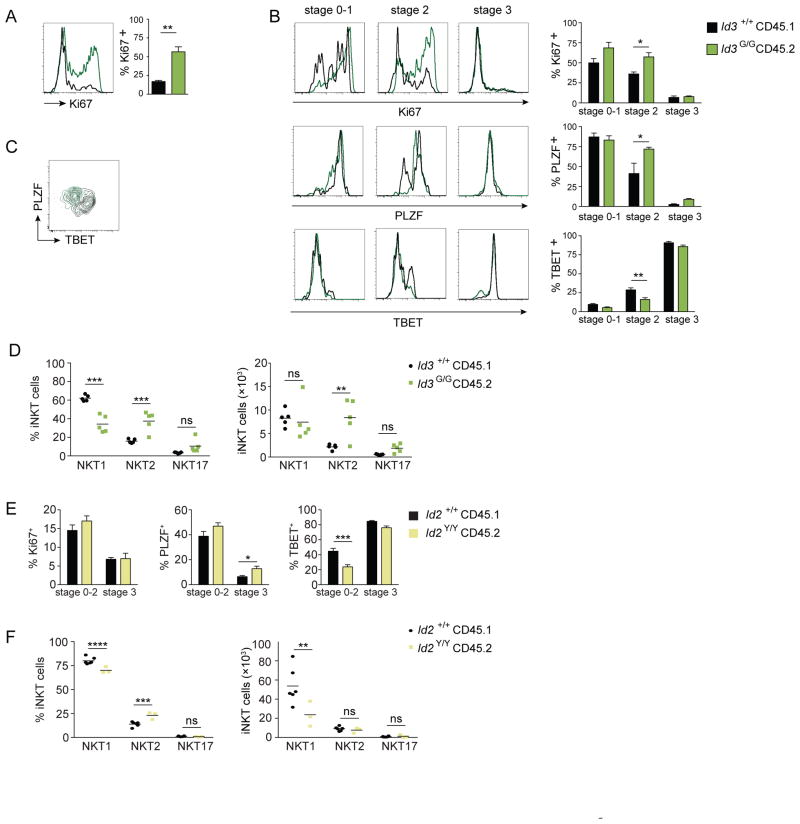
Since we observed higher iNKT cell numbers during thymic development in the absence of Id3 (Fig. 5D), we examined proliferation and cell death of Id3-deficient iNKT cells in the mixed bone marrow chimeras. An abundance of Ki-67+ iNKT cells could be readily detected among Id3-deficient iNKT cells relative to wild type cells in the thymi of mixed bone marrow chimeras, while proliferation remained unchanged in peripheral organs (Fig. 6A and data not shown). Observing different stages of iNKT cell development in the thymus, we found that more Id3G/G cells were actively dividing than their wild type counterparts at stages 0-1 and stage 2, in particular (Fig. 6B). Interestingly, in Id2 and Id3 reporter mice, proliferating iNKT cells in the thymus expressed more Id3 but less Id2 than non-proliferating NKT cells (data not shown). Thus loss of Id3, which should result in enhanced E protein activity, led to increased and prolonged proliferation at the early stages of iNKT cell development, conversely loss of E protein activity abrogated proliferation at this crucial stage of development (Fig. 6B and 2A).
Figure 6. Id3 regulates proliferation and sublineage commitment in iNKT cells.
Mixed bone marrow chimeras were generated by transfer of a 2:1 mix of Id3+/+ to Id3G/G bone marrow to irradiated recipients. (A) Histograms showing Ki-67 staining by gated wild-type (Id3+/+) (black line) and Id3-deficient (Id3G/G) (green line) iNKT cells from the thymus. Bar graphs indicate average frequency (±SEM) of Ki-67+ iNKT cells. (B) Histograms showing Ki-67, PLZF and TBET expression by CD1d-tet+TCRβ+ gated wild-type (Id3+/+) (black line) and Id3-deficient (Id3G/G) (green line) iNKT cells at stage 0–1, stage 2 and stage 3 in the thymus. Bar graphs indicate frequency of Ki-67+, PLZFhigh and TBET+ cells by CD1d-tet+TCRβ+ gated Id3+/+ and Id3G/G iNKT thymocytes at indicated developmental stages. Data are average (±SEM) of 3 mice, representative of 3 independent experiments. (C) Flow cytometry plot indicating PLZF and TBET expression by wild-type (Id3+/+) (black) and Id3-deficient (Id3G/G) (green) iNKT thymocytes. (D) Graphs of relative frequency and absolute number of TBET+ NKT1, PLZFhigh NKT2 and RORγt+ NKT17 cells of total gated Id3+/+ and Id3G/G iNKT thymocytes. Data are average of 5 mice, representative of 3 independent experiments. (E) Bar graphs indicate frequency of Ki-67+, PLZF+ and TBET+ cells by CD1d-tet+TCRβ+ gated Id2+/+ and Id2Y/Y iNKT thymocytes at indicated developmental stages. Data are average (±SEM) of 6 mice, representative of 2 independent experiments. (F) Graphs of relative frequency and absolute number of TBET+ NKT1, PLZFhigh NKT2 and RORγt+ NKT17 cells of total gated Id2+/+ and Id2Y/Y iNKT thymocytes. Data are average of 6 mice (Id2+/+) and 3 mice (Id2Y/Y), representative of 3 independent experiments. Statistical significance was determined using Mann Whitney U test and unpaired two-tailed t test where applicable, ns = not significant, *, P < 0.05, **, P < 0.005, ***, P < 0.0005.
We next examined the iNKT cell sublineages in the Id3+/+/Id3G/G mixed bone marrow chimeras. More iNKT cells expressed PLZF in the absence of Id3, in particular iNKT cells at stage 2, reinforcing our data indicating PLZF expression is positively regulated by E proteins (Fig. 6B, 2B and 2C). In contrast, fewer Id3-deficient iNKT cells at stage 2 expressed TBET, consistent with the reduced frequency of Id3G/G iNKT cells reaching stage 3 (Fig. 5D and 6B). Examining PLZF and TBET expression together, we clearly observed more Id3-deficient iNKT cells expressing PLZF alone (Fig. 6C). These data suggest E proteins or E protein-driven PLZF expression may negatively impact TBET expression during iNKT cell development. RORγt+ cells were infrequent in both WT and Id3-deficient iNKT cells but trended towards a slightly higher frequency in the Id3-deficient population. Consequently, we examined the frequency of TBET+ NKT1, PLZFhigh NKT2, and RORγt+ NKT17 cells in the presence or absence of Id3. The relative frequency of the TBET+ NKT1 population was decreased in the absence of Id3, while the PLZFhigh NKT2 population was increased in the thymus (Fig. 6D). Moreover, the absolute number of PLZFhigh NKT2 cells was increased in the absence of Id3 in the thymus (Fig. 6D). Thus, our analyses indicate Id3-deficiency results in a significant increase in PLZFhigh NKT2 cells, most likely accounted for by uncontrolled E protein activity within this sublineage.
Finally, we examined the iNKT cell sublineages in either Id2+/+ or Id2Y/Y bone marrow chimeras. As expected, we observed no significant difference in proliferation at any stage during iNKT cell development in the thymus (Fig. 6E). However, Id2-deficiency resulted in an increase in PLZF+ cells at stage 3 and a decrease in the frequency of TBET+ expressing cells prior to stage 3, suggesting that uncontrolled E protein activity positively regulates PLZF expression and might negatively regulate TBET expression (Fig. 6E). These data are supported by previously published data showing Id2-deficient CD8+ effector T cells fail to upregulate TBET expression during infection (32). Examining the overall frequency and absolute number of TBET+ NKT1, PLZFhigh NKT2, and RORγt+ NKT17 iNKT cells in the presence or absence of Id2, we consistently observed a decrease in the TBET+ NKT1 population in the absence of Id2 (Fig. 6F). Thus, we conclude Id2 marks and is necessary for the differentiation and maturation of the TBET+ NKT1 cells in the thymus.
Discussion
Here we have shown E protein transcription factors and Id proteins together act to orchestrate the differentiation, proliferation and survival of iNKT cells during thymic development. E2A and HEB both bound the promoter of Zbtb16, initiating expression of the transcription factor PLZF during the earliest stages of iNKT cell development. Furthermore, E proteins promoted iNKT cell proliferation at this crucial developmental stage and loss of E2A and HEB led to a paucity of mature iNKT cells in peripheral tissues. Id proteins, in turn, influenced subsequent stages of iNKT cell development. Id3 was required for regulating iNKT cell expansion and its loss led to a dramatic increase in iNKT cell numbers. Moreover, a sublineage of iNKT cells identified by constitutively high PLZF expression, PLZFhigh NKT2 cells, could also be identified by high Id3 expression; Id3 deficiency increased the abundance of PLZFhigh NKT2 cells indicating that sustained E protein DNA-binding activity positively regulates this sublineage. Finally, high Id2 expression denoted NK1.1+TBET+ NKT1 cells and was essential for the survival of these cells in the periphery.
Both E and Id proteins influence the proliferative burst that occurs by newly committed iNKT cells after positive selection. We observed lower proliferative capacity in the absence of E proteins while conversely an increase in proliferation was observed in the absence of Id3, particularly at stages 1 and 2. Our previous work indicated that the E protein HEB was necessary for expression of BCL-XL at the DP stage of T cell development, and indeed cells were susceptible to death in the absence of HEB (28). Here we find E proteins also influence proliferation at subsequent stages of iNKT cell development, resulting in a similar phenotype to that observed due to deficiency in the transcription factor c-MYC (12, 13). It is possible that E proteins regulate c-MYC expression directly or that both proteins collaborate to promote the proliferative burst. Interestingly, such a synergy has recently been suggested to occur during the development of Burkitt lymphoma (33). Furthermore, c-MYC and E protein transcription factors are able to directly bind to the promoter of Id3, consistent with our observation that proliferating thymic iNKT cells express elevated levels of Id3 (33, 34). Thus, we hypothesize that E protein-controlled expression of Id3 establishes a negative feedback loop to limit excessive iNKT cell proliferation during maturation.
Recent studies have begun uncovering how expression of the iNKT cell lineage-defining transcription factor PLZF is regulated. The transcription factor EGR2 is capable of binding the Zbtb16 promoter and indeed regulates expression of PLZF (9). We did not find evidence for regulation of EGR2 expression by E proteins, and thus favor the idea that EGR2 and E proteins independently regulate PLZF expression during iNKT cell development. Unlike E protein-deficient iNKT cells that do not express PLZF, iNKT cells expressing Id3 are positive for PLZF. This suggests that Id3 reduces but does not abolish E protein transcriptional activity completely.
The regulation of Id2 expression in iNKT cells during development is still being elucidated. It has been reported that in myeloid cells PLZF can directly bind the Id2 promoter, while in iNKT cells EGR2 expression was essential for the upregulation of Id2 (9, 35) Furthermore, we have previously shown that active STAT4 and STAT5 directly bind the Id2 promoter to increase its expression in T cells and dendritic cells (21, 36). Interestingly, EGR2 directly induces transcription of Il2rb in iNKT cells and both IL-2RB (CD122) and its ligand IL-15 are essential for the formation of stage 3 and presumably NKT1 cells (9, 37). We therefore hypothesize that EGR2 enables IL-15 signaling, which in turn activates STAT5 to induce Id2 expression. Thus, Id2 and Id3 differentially regulate E protein binding to DNA, potentially forming a gradient of transcriptional activity that influences iNKT cell development and proliferation.
A paradigm shift has recently been appreciated in the differentiation program that occurs as iNKT cells undergo development in the thymus. Terminally differentiated PLZFhigh NKT2 and RORγt+ NKT17 cells were found within the stage 2 CD44+NK1.1- population (1, 17, 18, 38). We found that whereas Id2 is highly expressed by TBET+ NKT1 cells, Id3 is expressed by PLZFhigh NKT2 cells. We propose, as recently suggested by Hu and colleagues, that Id3 may be a unique transcriptional regulator associated with the NKT2 sublineage (39). Although the cytokine milieu required for the maintenance of Id3+ NKT2 cells has not been clearly identified, our previous analysis in CD8+ effector cells showed Id3 expression was downregulated in the presence of IL-2, IL-12 and IL-21 (21). It is possible that Id3 expression is maintained by IL-25, as NKT2 cells reportedly express high levels of IL-17RB (5, 39, 40). Indeed, preliminary data indicate Id3-GFP+ iNKT cells have higher expression levels of IL-17RB than Id3-GFP- iNKT cells (unpublished observation and (39)). Our previous data further support the notion that Id2+ NKT1 cells may be sensitive to IL-12 signaling, as these cells constitutively express IL-12R (1, 17), CD8+ effector T cells upregulate Id2 expression upon stimulation with IL-12, and STAT4 can bind and promote Id2 expression (21). Indeed, recent data by Watarai and colleagues showed Stat4 mRNA levels were upregulated in NKT1 cells relative to NKT2 cells (5). Thus, we postulate that in the periphery the cytokine milieu within a particular organ in which the iNKT cell sublineage resides, will influence the homeostasis and survival of both Id2+ NKT1 and Id3+ NKT2 cells.
Interestingly, RORγt+ NKT17 cells displayed comparatively low expression of both Id2 and Id3, implying increased E protein activity in this sublineage. This observation is consistent with our previous finding that HEB can directly bind to the Rorc promoter to induce RORγt expression (28). However, with loss of Id3 and increased E protein transcriptional activity, we found only a slight increase in the frequency of the RORγt+ NKT subset, suggesting that E protein activity alone is not sufficient for commitment to this sublineage. Other transcription factors are likely to block RORγt+ NKT commitment in a dominant fashion. Indeed, the transcription factor Th-POK has recently been described to inhibit RORγt expression and NKT17 sublineage commitment (41).
We consistently found that loss of three Id protein alleles led to a decrease in both the frequency and absolute number of mature CD44+ NK1.1+ iNKT cells, while loss of Id3 alone affected immature iNKT cell populations. Loss of both Id2 alleles affected mature CD44+ NK1.1+ iNKT cells, a defect exacerbated by loss of one allele of Id3. Thus, although Id2 and Id3 can compensate for one another to some degree during early iNKT cell development, Id2 is required for formation of mature CD44+ NK1.1+ iNKT cells while Id3 is necessary for formation of NKT2 cells, roles that cannot be compensated for by unhindered expression of Id3 or Id2, respectively. These data are consistent with two recent publications. Continuous E protein activity, using a conditional mouse model, led to an increase in NKT2 and NKT17 cells with a corresponding loss of NKT1 cells (39). Similarly, loss of both Id2 and Id3, using a conditional knock-out approach, led to increased iNKT cell number (31). Thus, it appears Id2 and Id3 play unique non-compensatory roles in the formation of distinct sublineages of iNKT cells.
With loss of Id3 and increased E protein activity, we found a considerable expansion of the PLZFhigh NKT2 subset at the expense of the TBET+ NKT1 subset. Such changes in iNKT cell polarization imply higher susceptibility to development of colitis or asthma while maintaining increased protection from Th2-driven infections. NKT2 cells have been reported to play a role in the induction of airway hyperreactivity (5, 40, 42). With abundant PLZFhigh NKT2 cells, we would predict an increased susceptibility to AHR. Similarly, a greater frequency of PLZFhigh NKT2 cells was identified in the mesenteric lymph nodes, particularly in BALB/c mice (5). NKT cells have been associated with increased pathology in mouse models of ulcerative colitis and these cells reportedly produce high levels of IL-4 and IL-13, suggesting that the iNKT cells residing in the large intestine may be part of the PLZFhigh NKT2 sublineage (43, 44).
E protein transcription factors and their negative regulators the Id proteins have many important functions during lymphocyte development. We show here that E proteins control proliferation after positive selection and regulation of PLZF expression. Id2 and Id3 define and influence commitment to the TBET+ NKT1 and PLZFhigh NKT2 populations, respectively. Our findings indicate that E proteins and Id proteins dictate the development, differentiation and eventual iNKT cell polarization and abundance observed in health and disease.
Acknowledgments
We would like to thank Drs. Engel, Best and Omilusik for critical reading of the manuscript.
Footnotes
FOOTNOTES: This work was supported by PEW Scholars (A.W.G.), Leukemia and Lymphoma Society Scholar (A.W.G) and Fellow (L.M.D) awards and grants from the NIH (AI067545 and AI072117) to A.W.G.; KAI097286A to L.M.D. and the Austrian Science Fund (FWF): [J 3189-B11] to M.H.S.
References
- 1.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 3.Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: natural killer T cell defects and human disease. Nat Rev Immunol. 2011;11:131–142. doi: 10.1038/nri2904. [DOI] [PubMed] [Google Scholar]
- 4.Montoya CJ, Pollard D, Martinson J, Kumari K, Wasserfall C, Mulder CB, Rugeles MT, Atkinson MA, Landay AL, Wilson SB. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology. 2007;122:1–14. doi: 10.1111/j.1365-2567.2007.02647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H, Taniguchi M. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nature immunology. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 7.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 8.Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, Glimcher LH. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nature immunology. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, Singh H, Bendelac A. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nature immunology. 2012;13:264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant'Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dose M, Sleckman BP, Han J, Bredemeyer AL, Bendelac A, Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci U S A. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mycko MP, Ferrero I, Wilson A, Jiang W, Bianchi T, Trumpp A, MacDonald HR. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J Immunol. 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 14.Allende ML, Zhou D, Kalkofen DN, Benhamed S, Tuymetova G, Borowski C, Bendelac A, Proia RL. S1P1 receptor expression regulates emergence of NKT cells in peripheral tissues. FASEB J. 2008;22:307–315. doi: 10.1096/fj.07-9087com. [DOI] [PubMed] [Google Scholar]
- 15.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 16.Ranson T, Vosshenrich CA, Corcuff E, Richard O, Laloux V, Lehuen A, Di Santo JP. IL-15 availability conditions homeostasis of peripheral natural killer T cells. Proc Natl Acad Sci U S A. 2003;100:2663–2668. doi: 10.1073/pnas.0535482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Current opinion in immunology. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nature immunology. 2013 doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murre C. Helix-loop-helix proteins and lymphocyte development. Nature immunology. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 20.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nature immunology. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 21.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D'Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8(+) T cell subsets. Nature immunology. 2011 doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nature immunology. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Cruz LM, Lind KC, Wu BB, Fujimoto JK, Goldrath AW. Loss of E protein transcription factors E2A and HEB delays memoryprecursor formation during the CD8+ T-cell immune response. Eur J Immunol. 2012;42:2031–2041. doi: 10.1002/eji.201242497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent “innate” gammadelta T cells. PLoS One. 2010;5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monticelli LA, Yang Y, Knell J, D'Cruz LM, Cannarile MA, Engel I, Kronenberg M, Goldrath AW. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc Natl Acad Sci U S A. 2009;106:19461–19466. doi: 10.1073/pnas.0908249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, Sant'Angelo DB. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nature immunology. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira P, Boucontet L. Innate NKTγδ and NKTαβ cells exert similar functions and compete for a thymic niche. Eur J Immunol. 2012;42:1272–1281. doi: 10.1002/eji.201142109. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Wu D, Jiang N, Zhuang Y. Combined Deletion of Id2 and Id3 Genes Reveals Multiple Roles for E Proteins in Invariant NKT Cell Development and Expansion. J Immunol. 2013 doi: 10.4049/jimmunol.1301252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, Speed TP, Busslinger M, Nutt SL, Belz GT. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol. 2013;190:4585–4594. doi: 10.4049/jimmunol.1300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, Liu X, Powell J, Yang Y, Xu W, Zhao H, Kohlhammer H, Rosenwald A, Kluin P, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Ogwang MD, Reynolds SJ, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pittaluga S, Wilson W, Waldmann TA, Rowe M, Mbulaiteye SM, Rickinson AB, Staudt LM. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seitz V, Butzhammer P, Hirsch B, Hecht J, Gutgemann I, Ehlers A, Lenze D, Oker E, Sommerfeld A, von der Wall E, Konig C, Zinser C, Spang R, Hummel M. Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS One. 2011;6:e26837. doi: 10.1371/journal.pone.0026837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doulatov S, Notta F, Rice KL, Howell L, Zelent A, Licht JD, Dick JE. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 2009;23:2076–2087. doi: 10.1101/gad.1788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li HS, Yang CY, Nallaparaju KC, Zhang H, Liu YJ, Goldrath AW, Watowich SS. The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood. 2012;120:4363–4373. doi: 10.1182/blood-2012-07-441311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, Kronenberg M. Homeostasis of V alpha 14i NKT cells. Nature immunology. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 38.Michel ML, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, Eberl G, Leite-de-Moraes MC. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19845–19850. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu T, Wang H, Simmons A, Bajana S, Zhao Y, Kovats S, Sun XH, Alberola-Ila J. Increased Level of E Protein Activity during Invariant NKT Development Promotes Differentiation of Invariant NKT2 and Invariant NKT17 Subsets. J Immunol. 2013 doi: 10.4049/jimmunol.1301546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, Iwamura C, Nakajima H, Nakayama T, Taniguchi M. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J Exp Med. 2008;205:2727–2733. doi: 10.1084/jem.20080698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engel I, Zhao M, Kappes D, Taniuchi I, Kronenberg M. The transcription factor Th-POK negatively regulates Th17 differentiation in Valpha14i NKT cells. Blood. 2012;120:4524–4532. doi: 10.1182/blood-2012-01-406280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock P, Lombardi V, Kohlrautz V, Akbari O. Induction of airway hyperreactivity by IL-25 is dependent on a subset of invariant NKT cells expressing IL-17RB. J Immunol. 2009;182:5116–5122. doi: 10.4049/jimmunol.0804213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756–1767. doi: 10.1053/j.gastro.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. 2002;17:629–638. doi: 10.1016/s1074-7613(02)00453-3. [DOI] [PubMed] [Google Scholar]