Abstract
GE2270 is a thiopeptide antibiotic generated by extensive posttranslational modifications of a ribosomally generated precursor peptide. Thiopeptides are especially active against Gram-positive bacteria, including methicillin resistant Staphylococcus aureus (MRSA). In this study the GE2270 biosynthetic gene cluster (pbt) from Planobispora rosea ATCC 53733 was successfully expressed in the heterologous host strain Streptomyces coelicolor M1146. Notably, exconjugants containing the pbt gene cluster could only be obtained after deletion of the major part of the ribosomal genes flanking the gene cluster. This is a striking example that genes belonging to primary metabolism can prevent the successful conjugative transfer of DNA from phylogenetic distant species and thus complicate heterologous expression of secondary metabolite gene clusters. GE2270 production in the heterologous producer strain increased after introduction of the constitutive ermE* promoter upstream of the GE2270 resistance gene tuf from P. rosea. Insertion of the inducible tcp830 promoter resulted in inducible GE2270 production. When the regulatory gene pbtR was deleted, the resulting strain ceased to produce GE2270, suggesting an essential role of PbtR as a putative transcriptional activator of GE2270 expression.
Introduction
Thiopeptide antibiotics comprise about one hundred natural compounds, including the first representative of the class, micrococcin, discovered in 1948 [1], as well as thiostrepton, thiomuracin or berninamycin. They derive from ribosomally generated precursor peptides with a length of approximately 50–60 amino acids, which are shortened to the 14–18 amino acids C-terminal region forming the final antibiotic after extensive posttranslational modifications [2]–[4].
Thiopeptides are especially active against Gram-positive bacteria, including MRSA, but have also been shown to be active against malaria parasites [5], [6] and to have antiproliferative activity against human cancer cells [7], [8]. They exhibit their antibiotic activity by one of two mechanisms, both inhibiting bacterial protein biosynthesis. One subclass of thiopeptides targets the bacterial elongation factor Tu (EF-Tu), preventing its delivery of aminoacyl-tRNAs to the ribosome. This subclass includes, among others, GE2270, thiomuracin and GE37468 [9]. The other subclass targets the 50S subunit of the ribosome and affects the loops defined by 23S rRNA and the L11 protein. Prominent members of this class are for instance thiostrepton and the thiocillins [10].
Their interesting biological activities make thiopeptides promising drug candidates. The low water solubility and the poor pharmacokinetics of thiopeptides have so far prevented their clinical use, but derivatives of the thiopeptide antibiotic GE2270, produced by the rare actinomycete Planobispora rosea ATCC 53733, have now entered clinical testing for the topical treatment of acne [11] or Clostridium difficile infections [12]. Structural modification of the GE2270 molecule by genetic engineering may provide a strategy to generate additional compounds of this class for medical use. This has prompted recent efforts directed at the heterologous expression and genetic modification of thiazolylpeptides [13]–[15].
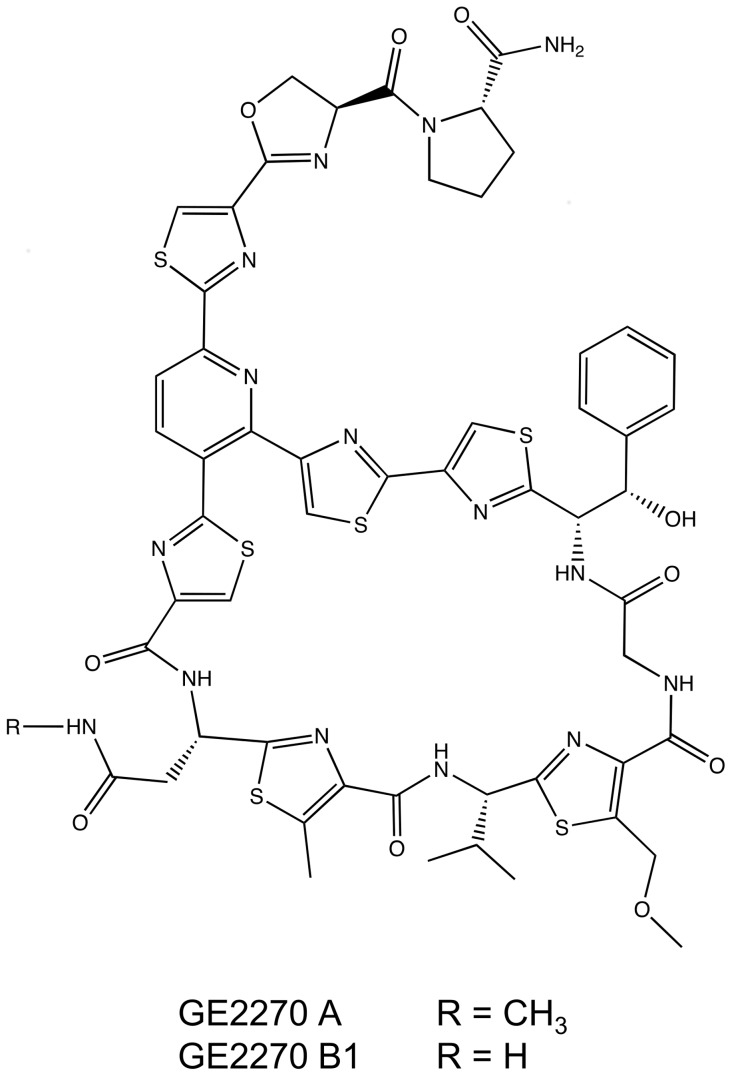
GE2270 is naturally produced by Planobispora rosea in the form of 10 closely related congeners [16], which differ mainly in their methylation state. The two of them discussed in this study are shown in Figure 1. The main compound produced is GE2270A, which has the highest methylation state and antibacterial activity. All other congeners also exhibit activity against Gram-positive bacteria and their minimum inhibitory concentrations (MIC) are only slightly lower than that of GE2270A [16], [17].
Figure 1. Structures of the two GE2270 congeners described in this study.
Recently, the biosynthetic gene cluster of GE2270 (pbt) in P. rosea was discovered and heterologously expressed in Nonomuraea sp. ATCC 39727, but attempts on heterologous expression in Streptomycetes, namely Streptomyces lividans and Streptomyces albus, remained unsuccessful [15]. This cluster shows high sequence identity (between 64–92% on amino acid level) to the GE2270 biosynthetic gene cluster (tpd) previously described from Nonomuraea sp. WU8817 [18]. It comprises 17 genes, pbtR, pbtG1, pbtB1, pbtO, pbtX, pbtM1, pbtM2, pbtA-H, pbtM3 and pbtM4, all transcribed in the same direction. The putative resistance gene tuf (hereafter referred to as tufR), coding for a resistant form of EF-Tu, is found in short distance downstream of the cluster.
The gene cluster for another thiopeptide targeting EF-Tu, GE37468 from Streptomyces sp. ATCC 55365, has been expressed in Streptomyces lividans, as reported by the group of C. T. Walsh in 2011 [13]. However the yield (2–3 mg l−1) was lower than in the wild type strain and much lower than GE2270 production in Nonomuraea sp. ATCC 39727 (250 mg l−1) [15].
Our laboratory successfully employed Streptomyces coelicolor host strains (M512, M1146, M1152, M1154) [19], for the heterologous expression of several antibiotic biosynthetic gene clusters, e.g. of aminocoumarins and caprazamycin [21], [22]. Particularly S. coelicolor M1146, derived from S. coelicolor M145 but lacking the gene clusters for actinorhodin (act), undecylprodigiosin (red), calcium dependent antibiotic (cda) and coelimycin P1 (cpk) [19] proved to be a valuable expression host due to its good growth and sporulation and the lower background production of other secondary metabolites. In some cases production of the heterologously expressed antibiotic was achieved or increased by additional use of constitutive [23] or inducible promoters [24].
Especially the successful heterologous production of GE2270 in Nonomuraea sp. ATCC 39727 provokes the question why heterologous expression of the pbt gene cluster failed in Streptomyces strains. Thus, we addressed this question employing the engineered expression host S. coelicolor M1146 and the inducible tcp830 promoter [25]. Heterologous expression of GE2270 in this strain would open a convenient route to the generation of new GE2270 derivatives by genetic methods, since genetic modification and handling is easier for this strain than for Nonomuraea sp. ATCC 39727.
Materials and Methods
Bacterial strains, cosmids, plasmids and general methods
Bacterial strains, cosmids and plasmids used in this study are listed in Table S1.
Escherichia coli strains were cultivated in LB medium [26], Streptomyces coelicolor strains were maintained and grown on MS medium [27]. Standard procedures for DNA isolation and manipulation were carried out as described by Sambrook and Russell for E. coli [26] and Kieser et al. for Actinomycetes [27].
Production, extraction and detection of GE2270 congeners
Homogenized frozen inoculum was prepared as described before [22], [28]. For standard cultivation each well of 24-square deepwell plates was filled with 3 ml of production medium, as described by Siebenberg et al. [28] and inoculated with 7.5 µl of homogenized, frozen inoculum. Cultivation was carried out at 30°C and 300 rpm. Cultivations in 300 ml baffled Erlenmeyer flasks were carried out in 50 ml medium at 30°C and 200 rpm.
Six typical thiopeptide production media (namely “main production medium” described by Selva et al. [29], RARE3 [30], Fermentation medium C [31], M8 [32], GE37468 production medium [13] and AF medium [33]), four production media proven for production of secondary metabolites (namely chemically defined medium (CDM) described by Kominek [34], liquid MS medium [27], GYM medium [35] and phenazine production medium [36]) and further 31 production media according to a systematically selected screen media list kindly provided by EntreChem S. L. (Oviedo, Spain) were prepared and tested for GE2270 production. For GE2270 detection 2 ml of culture were pelleted and extracted with 650 µl of methanol by shaking at 1400 rpm and 40°C for 1 hour. 80 µl of the centrifuged supernatant were analyzed by HPLC on an Agilent 1100 system equipped with a reverse phase column (Reprosil-Pur C18 AQ (5 µm), 250 mm×4.6 mm) at a flow rate of 1 ml min−1 with a linear gradient from 10% to 90% acetonitrile and 3.16 g l−1 ammonium formate in H2O over 30 min and UV detection at 310 nm. HPLC-MS analysis was carried out as described before [15].
Construction of Streptomyces coelicolor M1146 mutants containing the GE2270 biosynthetic gene cluster
The original cosmid 2F7 was identified from a SuperCos3 based cosmid library; the vector backbone contains an apramycin and an ampicillin resistance for antibiotic selection [15]. The insert covers the GE2270 biosynthetic gene cluster (pbt) from the source strain Planobispora rosea ATCC 53733. Additionally it contains the resistance gene EF-Tu and 25 additional ribosomal genes downstream of the cluster, as well as a gene coding for the ß′-subunit of DNA dependent RNA polymerase (rpoC) upstream of the cluster (GenBank accession number KF366381.2).
In a first step the apramycin resistance gene (aac(3)IV) on the SuperCos3 backbone was exchanged with a chloramphenicol resistance gene (cat) from pACYC184 [37]. This was necessary as most planned modifications of cosmid 2F7 were based on cassettes containing an apramycin resistance gene. The cat gene was amplified with primer pair targcat_fwd (TGGGCTACGTCTTGCTGGCGTTCGTTTTCCGGATCGCG A CTAGTCACTAAATCAGTAAGTTGGC) and targcat_rev (CGCGACCTTGCCCCTCCAACGTCATCTCGTTCTCCGCTC ACTAGTATTATCACTTATTCAGGCG), both with SpeI restriction sites marked in bold. Italic letters indicate 39 nucleotides homologous to cosmid 2F7 sequences (as for the rest of the described primers) according to the following Red/ET-mediated recombination [38] resulting in cosmid 2F7cat, which was confirmed by restriction analysis. To construct cosmid pbtKA01 the aac(3)IV-tcp830 cassette derived from pMS80 [25] was amplified with primer pair targC/D_fwd (GAAGAGATCACCCGCGACATCCCCAACGTCTCCGAGGAG GTGTAGGCTGGAGCTGCTTC) and targC_rev (TCGGACCACTCGTTCGGCATTACTGCGCGAGCTCGTCAC TCTAGACCTCCGACGTACGC). The resulting PCR product was utilized in Red/ET mediated recombination to replace the first 5418 bp of the insert of cosmid 2F7cat, consisting mainly of the coding sequence for RNA polymerase subunit β′, and thus placing the tcp830 promoter in front of pbtR. For the construction of pbtKA02 primer targC/D_fwd was reused in combination with primer targD_rev (CCTCGAAACCGGAGAAAGATATGCCGGCGCCACGGTCAT TCTAGACCTCCGACGTACGC) to replace the first 6128 bp of the insert of cosmid 2F7cat by Red/ET mediated recombination. Thus, the tcp830 promoter was placed upstream of the first gene involved in the biosynthesis of GE2270, pbtG1, in the same step deleting the transcriptional regulator pbtR. Both cosmids were confirmed via restriction analysis and partial sequencing.
For the construction of cosmid pbtCK01 the apramycin resistance cassette [aac(3)IV] on plasmid pIJ774 was amplified [39] using the primer pair targA-rib_fwd (ACCGTCGGCGCCGGCCGCGTCACCAAGATCCTCAAGTAG GTATACATTCCGGGGATCCGTCGACC) including a restriction site for BstZ171 (bold) and targA-rib_rev (GACCAGGATCTCCTCGTCCGCGTAGAAGGTGATGAGCGG TTATAATGTAGGCTGGAGCTGCTTCG) with a restriction site for PsiI (bold). The resulting PCR product replaced 12,354 bp in cosmid 2F7cat via Red/ET-mediated recombination [38], comprising 22 ribosomal genes between the tufR gene encoding EF-Tu and the SuperCos3 vector. The resulting cosmid pbtCK01 was confirmed by restriction analysis and partial sequencing.
To obtain cosmid pbtCK02, first the apramycin resistance cassette was removed from pbtCK01 in vitro by application of Cre recombinase (New England Biolabs, Frankfurt am Main, Germany) according to the manufacturer's manual utilizing the loxP recognition sites flanking the apramycin resistance cassette [40]. The correct excision of the cassette was proven by restriction analysis and PCR. In order to insert the constitutive ermE* promoter in front of the tufR gene, a 1780-bp fragment was amplified from a previously published pUWL201 derivative [23], [41], using primer pair targB-rib_ermE_f (GGCAACTCGCTCATCGAGTGCGTGGTCACCGGGGGCTGA GTATACTCAGGCGCCGGGGGCGGTG) including a restriction site for BstZ171 (bold) and targB-rib_ermE_r (TGGTCCGCTCGAACTTGGCCTTGGCCACTGTCTGTCTCC TTATAAATCCTACCAACCGGCACGATTG) including a restriction site for PsiI (bold). This PCR- product contained the ermE* promoter and the hygromycin resistance gene (hyg) and was inserted into the derivative of cosmid pbtCK01 lacking the [aac(3)IV] cassette by Red/ET-mediated recombination. This resulted in the loss of 3560 bp, containing the ribosomal genes rpsL, rpsG and fusA, and in placing the tuf gene under control of the ermE* promoter. Restriction analysis and partial sequencing verified the accuracy of pbtCK02.
Cosmids pbtCK03, pbtCK04 and pbtCK05 are derivatives of pbtCK02, containing the synthetic tetracycline-inducible promoter tcp830 from plasmid pMS80 [25]. To construct cosmid pbtCK03(apra) the [aac(3)IV-tcp830] cassette was amplified and utilized in Red/ET mediated recombination as described for pbtKA01, but targeting the insert of cosmid pbtCK02, thus placing the tcp830 promoter again in front of pbtR. In the same way the construction of pbtCK04(apra) was done as described for pbtKA02, but targeting cosmid pbtCK02 instead of 2F7cat. A third primer pair, targE_fwd (GTCGACCTGTACGCCGCGACCGAGCAGATCGGCGGATGA GTGTAGGCTGGAGCTGCTTC) and targE_rev (CATGGGCAGGTCGTTAAGGTTCAACTCCATCTCGCTCAT TCTAGACCTCCGACGTACGC), was used to construct pbtCK05(apra) by amplifying the [aac(3)IV-tcp830] cassette for its insertion in front of pbtA, coding for the precursor peptide of GE2270, under loss of the 125-bp intergenic region between pbtM2 and pbtA. After successful construction of cosmids pbtCK03(apra), pbtCK04(apra) and pbtCK05(apra) the apramycin resistance cassette was removed from each of them via FLP-mediated excision [42], resulting in cosmids pbtCK03, pbtCK04 and pbtCK05. All three cosmids were confirmed via restriction analysis and partial sequencing.
In addition a negative control cosmid was constructed, which is lacking the entire pbt gene cluster but retains the ribosomal genes. For that purpose the apramycin resistance cassette [aac(3)IV] from plasmid pIJ774 [39] was amplified with primer pair targZ_fwd (GAAGAGATCACCCGCGACATCCCCAACGTCTCCGAGGAG ATTCCGGGGATCCGTCGACC) and targZ_rev (CCCCCGTCGCGAACCCCGGAAGGAGCCGCCGGGCAGGCC TGTAGGCTGGAGCTGCTTCG). The 1,441-bp PCR fragment was inserted into 2F7cat by Red/ET-mediated recombination replacing the 26,831 bp comprising the entire GE2270 gene cluster.
Cosmids pbtCK01-pbtCK05 and pbtCK08 were transformed into non-methylating E. coli ET12567/pUB307 [43] and introduced into S. coelicolor M1146 by conjugation [27].
Agar diffusion test
Approximately 105 spores of S. coelicolor M1146, S. coelicolor M1146(pbtCK01) and S. coelicolor M1146(pbtCK02) were streaked on 20 ml DNA agar plates (0.46 g Difco Nutrient Agar + 20 ml distilled water). GE2270 standard solved in acetonitrile: water (70∶30) was spotted on the plates in final amounts of 0.4 µg, 4 µg, 8 µg and 12 µg. 20 µg Kanamycin were used as positive control, GE2270 solvent was used as negative control, respectively. Inhibition zones were analyzed after 48 hours of cultivation at 30°C.
Construction of the phylogenetic tree
A neighbor-joining cladogram was constructed with MEGA5 [44]. Alignment of the sequences was done with ClustalW, bootstrap values (in percent) are calculated from 1000 resamplings. The tree was rooted to the 16S rRNA sequence from Bacillus cereus ATCC 14579 (NR_074540.1).
16S rRNA sequences of selected actinomycetes were obtained from GenBank, despite Streptomyces coelicolor A3(2) (http://strepdb.streptomyces.org.uk) and Planobispora rosea ATCC 53733 (Naicons, see Supporting Information S1), Catenulispora acidiphila DSM 44928 (accession number (acc. No.): NR_074457.1), Micromonospora pallida DSM 43817 (acc. No.: NR_044884.1), Nocardia aobensis IFM 0372 (acc. No.: AB126876.1), Nonomuraea sp. ATCC 39727 (acc. No.: AJ582011.2), Propionibacterium acnes DSM 1897 (acc. No.: X53218.1), Saccharopolyspora erythraea NRRL 2338 (acc. No.: NR_074095), Salinispora tropica CNB-440 (acc. No.: NR_074502.1), Streptomonospora salina YIM90002 (acc. No.: NR_025042.1) and Streptomyces lividans NBRC 15678 (acc. No.: AB184694.1).
Results
Failure of conjugative transfer of the GE2270 biosynthetic gene cluster with flanking ribosomal genes into S. coelicolor M1146
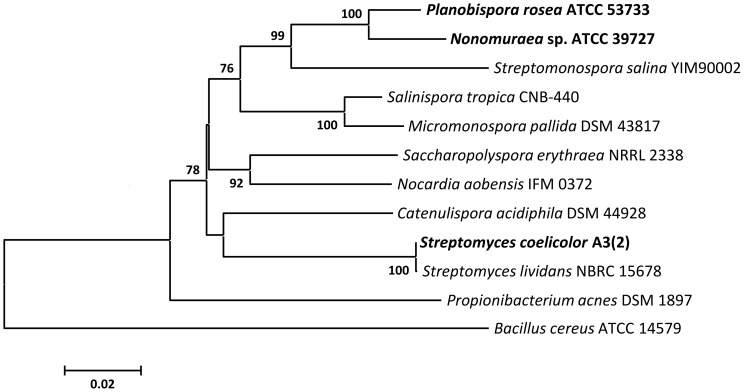
We focused our interest on the obvious difference between the successful heterologous expression of GE2270 in Nonomuraea sp. ATCC 39727 and the unsuccessful attempts in S. lividans and S. albus observed by Tocchetti et al. [15]. As all experiments in this study were based on the same cosmid (2F7), we concluded that the cluster was intact and that the unsuccessful heterologous expression might be due to a lack of transcription of the GE2270 biosynthetic gene cluster (pbt) in Streptomyces. This lack of transcription could be due to the phylogenetic relationship of the species; Nonomuraea sp. ATCC 39727 and Planobispora rosea are two closely related species, whereas Streptomycetes are a more distantly related genus of Actinomycetes (Figure 2).
Figure 2. Phylogenetic tree based on 16S rRNA gene sequences of selected actinomycetes.
Neighbor-joining cladogram constructed in MEGA5 [44]; alignment of the sequences was done with CLUSTALW, bootstrap values (in percent) are calculated from 1000 resamplings and shown at the respective nodes for values >50%, the scale bar represents 0.02% sequence divergence. The tree is rooted to 16S rRNA from Bacillus cereus ATCC 14579.
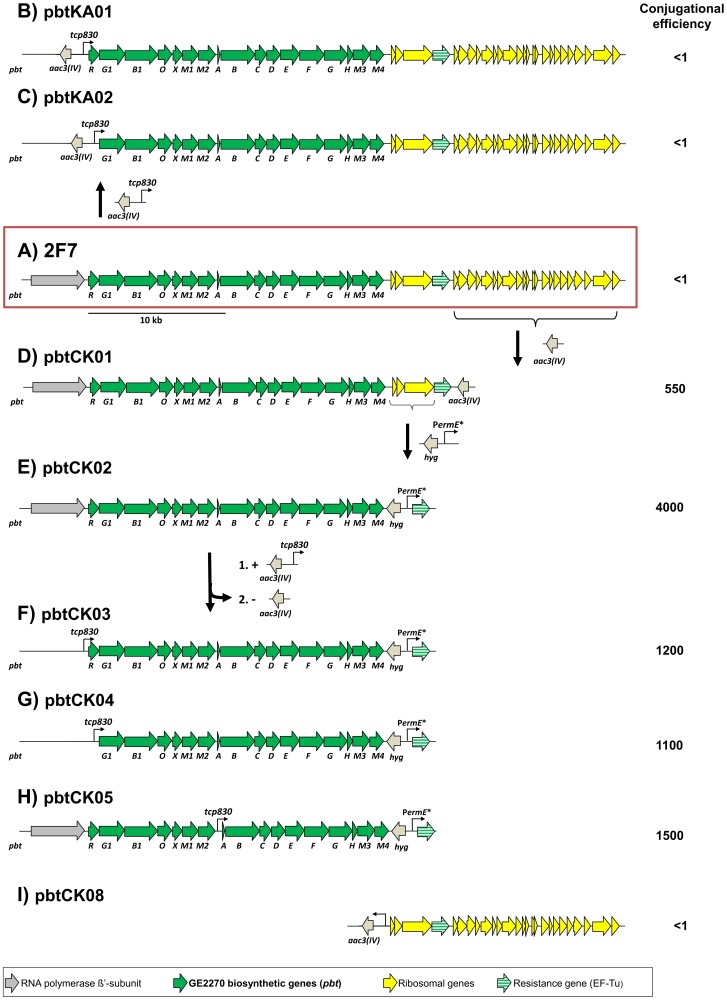
All genes of the pbt gene cluster are orientated in the same direction (Figure 3), as it is the case for the novobiocin gene cluster, for which the suitability of the inducible tcp830 promoter for induction of transcription and thus expression of novobiocin was successfully demonstrated [24]. The tcp830 promoter cassette derived from pMS80 contains an apramycin resistance gene as selection marker. Therefore, we first exchanged the apramycin resistance gene in the SuperCos3 backbone of cosmid 2F7 [15] for a chloramphenicol resistance gene (cat), generating cosmid 2F7cat. Then we constructed two derivatives of this cosmid (Figure 3B and C), one placing the tcp830 promoter in front of the first gene of the pbt cluster, encoding the TetR family regulator PbtR (generating pbtKA01), and a second one placing the tcp830 promoter in front of the first biosynthetic gene of the pbt cluster, pbtG1, while in the same step deleting the regulatory gene pbtR (generating pbtKA02). In order to minimize insert size, we deleted the sequence upstream of the pbt cluster, containing the rpoC gene encoding the β′-subunit of DNA-dependent RNA polymerase, in both constructs in the same step. Both cosmids were then conjugated into S. coelicolor M1146 (see introduction). Unexpectedly, the conjugation of both constructs as well as of the original cosmid 2F7 into S. coelicolor M1146 failed and no exconjugants were detectable in repeated experiments (Figure 3B and C), suggesting a principal problem during conjugation rather than of transcription.
Figure 3. Insert of cosmid 2F7 and construction of cosmids derived from 2F7.
A) Insert of 2F7 and 2F7cat comprising the GE2270 biosynthetic gene cluster (pbt), the tufR gene coding for the GE2270-resistant EF-Tu, 25 adjacent ribosomal genes and rpoC coding for RNA polymerase β′-subunit (see Table S2). B) Introduction of the inducible tcp830 promoter in front of the regulatory gene pbtR resulting in cosmid pbtKA01. C) For the construction of pbtKA02 the tcp830 promoter was introduced in front of pbtG1 under loss of pbtR. D) Replacement of 22 genes encoding ribosomal proteins with an apramycin resistance cassette (aac(3)IV) resulted in cosmid pbtCK01. E) pbtCK02 was constructed by introduction of the constitutive promoter ermE* and associated replacement of the ribosomal genes rpsL, rpsG and fusA with a hygromycin resistance cassette (hyg). F–H) Introduction of the inducible tcp830 promoter at three distinct positions in each case followed by subsequent removal of the employed aac(3)IV cassette. F) tcp830 introduced in front of the regulatory gene pbtR resulting in cosmids pbtCK03. G) tcp830 introduced in front of pbtG1 under loss of pbtR resulting in cosmid pbtCK04 and H) tcp830 introduced in front of the structural gene pbtA resulting in cosmid pbtCK05. I) negative control cosmid pbtCK08 was constructed by replacement of the pbt biosynthetic genes of cosmid 2F7cat by an apramycin resistance cassette (aac(3)IV). For each construct, the efficiency of conjugal transfer into Streptomyces coelicolor M1146 is expressed as number of exconjugants per 1 million spores.
Manipulation of cosmid 2F7cat to facilitate its conjugation and heterologous production of GE2270 in Streptomyces coelicolor M1146
The insert sequence of cosmid 2F7cat (and 2F7) covers not only the GE2270 biosynthetic gene cluster (pbt) from the source strain Planobispora rosea ATCC 53733, but also the resistance gene tufR, encoding a resistant EF-Tu, 25 additional ribosomal genes downstream of the cluster, as well as the already described gene rpoC coding for the ß′-subunit of DNA dependent RNA polymerase upstream of the cluster.
We speculated that either the toxicity of the heterologously formed GE2270 or a detrimental effect of the 25 ribosomal genes from P. rosea (Table S2) might prevent the successful conjugative transfer of this cosmid into S. coelicolor, and thus we further modified cosmid 2F7cat.
In order to delete most of the ribosomal genes which might interfere with the translational machinery of the expression host, the 22 ribosomal genes downstream of the tufR gene were replaced by an apramycin resistance cassette, resulting in cosmid pbtCK01 (Figure 3D). The applied apramycin resistance cassette obtained from pIJ774 [39] is flanked by loxP sites enabling subsequent in vitro removal of the cassette with the help of Cre recombinase.
To ensure expression of the GE2270 resistance gene tufR in the heterologous expression strain S. coelicolor M1146, the strong constitutive ermE* promoter [23], [45] was introduced in front of the tufR gene (Figure 3E) resulting in cosmid pbtCK02. The last three remaining ribosomal genes rpsL, rpsG and fusA were eliminated at the same time.
To provide unequivocal evidence whether or not the ribosomal genes of P. rosea are responsible for the failure of the conjugative transfer of cosmid 2F7 and its derivatives shown in Figure 3B and C into S. coelicolor, a further cosmid termed pbtCK08 was constructed, which contained only the 26 ribosomal genes, but not the pbt genes encoding GE2270 biosynthesis (Figure 3I).
To ensure transcription of the pbt gene cluster in S. coelicolor M1146 three additional constructs (pbtCK03, pbtCK04 and pbtCK05) were generated by introducing the tetracycline inducible promoter tcp830 into distinct positions of the GE2270 gene cluster in cosmid pbtCK02, namely: (i) upstream of the first gene of the cluster, which is the putative regulatory gene pbtR, belonging to the tetR family of transcriptional regulators, which mostly act as transcriptional repressors [46] (pbtCK03, see Figure 3F); (ii) in front of the second gene of the cluster, pbtG1, under simultaneous deletion of the putative transcriptional repressor pbtR (pbtCK04, see Figure 3G); (iii) in front of pbtA, the structural gene encoding the precursor peptide of GE2270 (see Figure 3H).
Influence of ribosomal genes from P. rosea on conjugation efficiency in S. coelicolor M1146
We introduced the seven cosmids depicted in Figure 3A, D-I into S. coelicolor M1146 via conjugation as described by Gust et al. [38]. For each construct the number of exconjugants was determined from three replicate experiments. Conjugation efficiency was calculated as the number of exconjugants per 106 recipient spores and is given for each cosmid in Figure 3.
As observed previously, the conjugal transfer of cosmid 2F7 was unsuccessful. However, after deletion of the 22 ribosomal genes downstream of the tufR gene in cosmid pbtCK01 (Figure 3D) exconjugants were readily obtained (550 per 106 spores). Thus the unsuccessful conjugation of cosmid 2F7 was not caused by a lack of resistance to GE2270 as assumed initially, but indeed seemed to be caused by a conflict of S. coelicolor with the ribosomal genes of P. rosea. After additional deletion of the remaining three ribosomal genes rpsL, rpsG and fusA and introduction of the ermE* promoter (cosmid pbtCK02, Figure 3E), conjugation efficiency increased again 7 to 8-fold; reaching 4000 exconjugants per 106 spores. As expected, similar results were obtained with cosmids pbtCK03-05, depicted in Figure 3F, G and H.
Cosmid pbtCK08, containing all ribosomal genes, but lacking the GE2270 biosynthetic gene cluster proved to be as unsuitable for conjugal transfer as the original unmodified cosmid 2F7. This confirms that the ribosomal genes flanking the pbt gene cluster are indeed the reason for the unsuccessful attempts at conjugation of cosmid 2F7 into S. coelicolor.
Media screen and heterologous production of GE2270A in S. coelicolor M1146(pbtCK01)
For each of the five cosmids transferred efficiently into S. coelicolor M1146, three exconjugants were tested for production of GE2270 using published production media for thiazolylpeptides, namely the “main production medium” described by Selva et al. [29], RARE3 [30], Fermentation medium C [31], M8 [32], GE37468 production medium [13] and AF medium [33]. Anhydrotetracycline (aTc) was added to induce the tcp830 promoter, where required. However, no production of GE2270 congeners could be detected via HPLC and LC-MS in any of these exconjugants, whereas GE2270 production was detected readily in each of these media in the native producer P. rosea. To proof sample treatment and analytical methods, the HPLC detection limit was ascertained to be at 0.02 µg corresponding to a production rate of 0.08 mg l−1. Extraction of prior added GE2270A standard to the culture resulted in a recovery rate of 73%.
To investigate whether the pbt gene cluster on cosmid pbtCK01 was still intact, we conjugated this cosmid into Nonomuraea sp. ATCC 39727 and compared its production to Nonomuraea sp. ATCC 39727 containing cosmid 2F7. Both strains produced comparable amounts of GE2270A over time, demonstrating the GE2270 cluster on cosmid pbtCK01 to be fully intact (Figure S1).
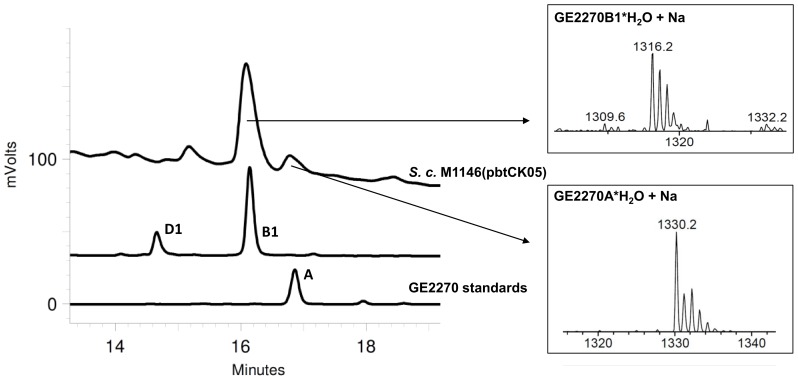
Since no GE2270 congeners were produced by the heterologous expression strains in previously published thiopeptide production media, we decided to investigate GE2270 production in 35 additional production media, 31 derived from a screen media list kindly provided by EntreChem, S. L. (Oviedo, Spain), as well as four media previously employed in our laboratory for antibiotic production in S. coelicolor M1146, i.e. chemically defined medium (CDM) described by Kominek [34], MS medium, [27] GYM medium [35] and phenazine production medium [36]. S. coelicolor M1146(pbtCK01) was cultivated in each of these media. Three replicate cultures were prepared for each medium using 24-square deepwell plates. After 3 and 6 days of cultivation culture extracts were analyzed via HPLC. GE2270 production was detected only in a single medium out of the investigated 35 media, namely in screening medium 13 (SM13), originally described by Imada et al. [47] and named “main fermentation medium”. The presence and identity of at least two GE2270 congeners, GE2270A and GE2270B1, was confirmed by HPLC-MS (see Figure 4) with a mean production of GE2270A of about 1 mg l−1. The ingredients of this medium are glucose, glycerol, Pharmamedia®, soluble starch, corn steep liquor, CaCO3, peptone and NaCl. All of these ingredients are also contained in some of the other screened media, thus it was not possible to conclude why only this medium and none of the others gave rise to GE2270 production in S. coelicolor M1146.
Figure 4. HPLC-MS analysis of a methanolic extract of S. coelicolor M1146(pbtCK05).
Data confirms the presence of GE2270A; GE2270B1 is identified as the main product.
Addition of 0.6% of the water-soluble siloxylated ethylene oxide/propylene oxide copolymer Q2-5247 (Dow Corning, USA), suggested to act as oxygen carrier, has been shown to increase secondary metabolite production in some heterologous expression strains [22], [48]. However, it had no influence on GE2270 production (Data not shown).
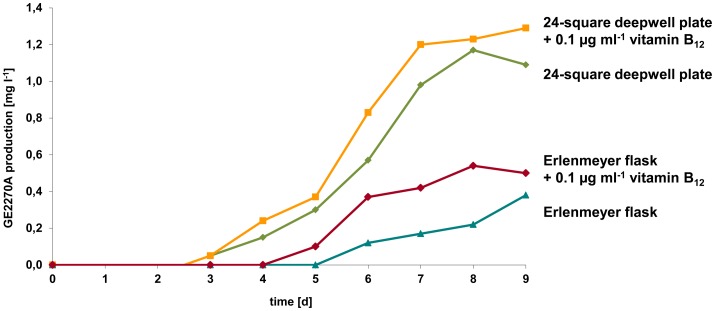
Addition of vitamin B12 (cobalamin) during cultivation of the natural producer strain P. rosea [31] has been reported to lead to a doubling of total GE2270 complex production and to shift the relative composition towards the most methylated compound, GE2270A. Therefore we added vitamin B12 (0.1 µg ml−1) to SM13 medium and analyzed GE2270 production in 24-square deepwell plates and Erlenmeyer flasks (Figure 5). GE2270A production is nearly 3-fold higher in 24-square deepwell plates than in Erlenmeyer flasks. However, supplementation of vitamin B12 resulted only in a slightly higher formation of the completely methylated congener, GE2270A.
Figure 5. GE2270A production over time in S. coelicolor M1146(pbtCK01).
Cultivation was performed in SM13 medium in 300-square deepwell plates [28].
Influence of constitutive expression of the resistance gene and of introduction of the tcp830 promoter at different positions of the cluster on heterologous production of GE2270
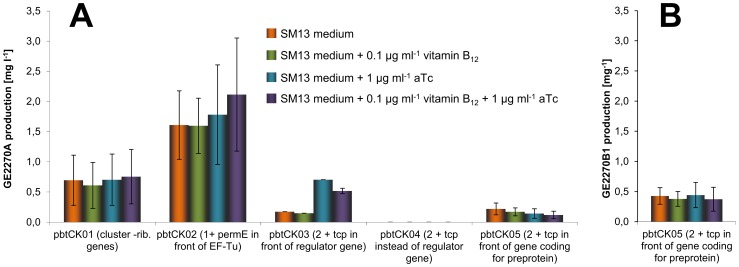
After establishing heterologous expression of GE2270 in S. coelicolor M1146(pbtCK01), we examined GE2270 production resulting from expression of the modified pbt cosmids shown in Figure 3E, F, G and H. All strains were grown in SM13 medium using 24-square deepwell plates and cultivating three independent exconjugants for each construct. Cultivation was carried out with and without addition of anhydrotetracycline (aTc), which induces transcription from the tcp830 promoter [24], [25], and with and without addition of vitamin B12. The results are depicted in Figure 6.
Figure 6. GE2270 production of S. coelicolor M1146 strains containing different constructs.
A) GE2270A production of S. coelicolor M1146 strains containing different constructs of the GE2270 biosynthetic gene cluster (see Figure 3). The amount of GE2270A was determined after 8 days of cultivation in 24-square deepwell plates [28]. B) GE2270B1 production in S. coelicolor M1146(pbtCK05). Values are mean ± SEM from triplicated cultivation of three individual exconjugants each.
Cultivating S. coelicolor M1146(pbtCK01), GE2270A production was approximately 0.7 mg l−1 under all tested conditions (Figure 6A). As expected, addition of aTc did not have any effect on GE2270 production, as there is no inducible promoter present in this construct.
In S. coelicolor M1146(pbtCK02), in which the constitutive ermE* promoter has been inserted in front of the resistance gene tufR, coding for the GE2270 insensitive EF-Tu, production was 2.5-fold higher compared to S. coelicolorM1146(pbtCK01) under all measured conditions (Figure 6A).
Insertion of the inducible tcp830 promoter upstream of the gene cluster (pbtCK03, Figure 3F) resulted in very low GE2270A production (0.17 mg l−1) when the heterologous expression strain was cultivated without aTc, which was to be expected, as the tcp830 promoter is not fully repressed in S. coelicolor [25]. However, when the inducer aTc was added, production increased about 4-fold to 0.7 mg l−1, but remained lower than in S. coelicolor M1146(pbtCK02) (Figure 6A).
Deletion of pbtR, assumed to encode a transcriptional repressor belonging to the TetR-family, together with a placement of the first biosynthetic gene pbtG1 under control of the tcp830 promoter (S. coelicolor M1146(pbtCK04)), unexpectedly resulted in a complete abolishment of GE2270 biosynthesis (Figure 6A). This suggests that PbtR, though belonging to a family known as transcriptional repressors, is strictly needed for GE2270 production.
In the last construct the tcp830 promoter was introduced in front of the structural gene pbtA, encoding the precursor peptide of GE2270 (S. coelicolor M1146(pbtCK05)). Surprisingly, production of GE2270A was not inducible with aTc and, with an average of 0.14 mg l−1, corresponding to the amount detected in S. coelicolor M1146(pbtCK03) without induction. In contrast to all other strains, GE2270A was not the main congener produced by this strain. Another GE2270 derivative was detected to accumulate in the extracts in amounts of 0.5 mg l−1 (Figure 6B). HPLC-MS confirmed this main product to be GE2270B1, a congener lacking a N-methyl group (Figures 1 and 4). Gene pbtM1 encodes the N-methyltransferase likely responsible for the conversion of GE2270B1 into GE2270A and is situated upstream of the tcp830 promoter in this construct. Production of GE2270B1 was not induced by addition of aTc.
GE2270A resistance of S. coelicolor M1146 and derivatives
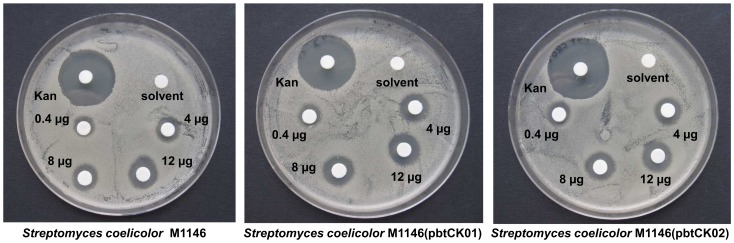
To clarify whether the resistant EF-Tu from P. rosea also conveys resistance against GE2270A to the heterologous producer strains, an agar diffusion test was carried out. Resistance of S. coelicolor M1146, S. coelicolor M1146(pbtCK01) and S. coelicolor M1146(pbtCK02) against GE2270A was investigated. After two days of cultivation, growth inhibition was clearly visible for all three strains, even at the lowest amount of 0.4 µg (Figure 7), proving that they are equally sensitive against GE2270A. No relevant difference was observable between S. coelicolor M1146, not carrying the resistance gene tufR of P. rosea, S. coelicolor M1146(pbtCK01) carrying the tufR gene coding for the resistant EF-Tu under control of its own promoter and S. coelicolor M1146(pbtCK02) which carries the tufR gene under control of the constitutive ermE* promoter. Therefore, tufR which has been proven to convey GE2270 resistance to P. rosea [49], [50] does not convey significant resistance to S. coelicolor, and obviously this may limit GE2270 production rates in the heterologous expression strain.
Figure 7. Agar diffusion test to determine the resistance of Streptomyces coelicolor strains against GE2270A.
GE2270A was applied in amounts of 0.4 µg to 12 µg; 20 µg kanamycin (Kan) were applied as positive control.
Discussion
In this study we successfully solved the problem of conjugative transfer of the GE2270 biosynthetic gene cluster into Streptomycetes, allowing for the first time heterologous production of GE2270 in a Streptomyces strain. We thereby demonstrated that genes belonging to primary metabolism of phylogenetic distant donor species may prevent conjugal transfer of the desired biosynthetic gene cluster.
Our first attempt to achieve heterologous production of GE2270 in S. coelicolor M1146 targeted the transcription of the pbt cluster by introduction of the inducible tcp830 promoter, which has been used very successfully to induce heterologous production of novobiocin in S. coelicolor [24]. Two new cosmids were constructed, placing either the regulatory gene pbtR (pbtKA01, Figure 3B) or the first of the biosynthetic genes (pbtKA02, Figure 3C) under control of the tcp830 promoter. Neither the two constructs nor the original cosmid 2F7 could be conjugated into S. coelicolor M1146. These results indicated a possible toxicity issue caused either by a toxic effect of GE2270 on S. coelicolor M1146, or by a detrimental effect of the ribosomal genes of P. rosea, which are contained in these cosmids.
To solve this problem, two new cosmids were constructed, one by deleting most of the ribosomal genes (pbtCK01, Figure 3D) and the other by placing the resistance gene tufR under control of the constitutive ermE* promoter, at the same time deleting the remaining three ribosomal genes (pbtCK02, Figure 3E). Interestingly, we achieved normal amounts of exconjugants in S. coelicolor M1146 already with cosmid pbtCK01, after deletion of most of the ribosomal genes. The number of exconjugants was further increased with cosmid pbtCK02, probably due to the deletion of the three remaining ribosomal genes rather than the constitutive expression of tufR, as production of GE2270 was only detected in screening medium 13. As expected, similar conjugation efficiencies could be obtained for the cosmids pbtCK03, pbtCK04 and pbtCK05 (Figure 3F-H), but not with cosmid pbtCK08 (Figure 3I), which lacks the entire biosynthetic gene cluster, but still contains all ribosomal genes. In contrast, conjugation of 2F7 and pbtCK01 into Nonomuraea sp. ATCC 39727, a strain closely related to Planobispora rosea (Figure 2), was successful. This demonstrates that indeed the ribosomal genes on cosmid 2F7 are responsible for its detrimental effect and prevent conjugation into Streptomyces, but not into Nonomuraea sp. ATCC 39727. In this heterologous producer no differences in GE2270A production were visible, whether the original cosmid 2F7 or pbtCK01 was integrated into its genome (Figure S1). The greater tolerance towards the foreign ribosomal genes observed by Nonomuraea sp. ATCC 39727 seems to be based on the closer phylogenetic relationship of the two strains. Possibly, these foreign ribosomal genes impair formation of functional ribosomes in exconjugants of S. coelicolor and render them non-viable.
In contrast, exconjugants obtained from S. coelicolor M1146 with the modified cosmids pbtCK01 and pbtCK02 were clearly viable, but nonetheless GE2270 production could not be detected in previously described thiopeptide production media. Eventually, we detected GE2270 production in only a single medium out of 41. This emphasizes the importance of testing a variety of media in heterologous expression experiments. Production in the Streptomyces host was lower (max. 3 mg l−1 GE2270A under optimal conditions using pbtCK02) than in the native producer P. rosea or in the heterologous producer Nonomuraea sp. ATCC 39727. A similar observation has been reported for the heterologous expression of the related thiopeptide GE37468 from Streptomyces ATCC 55365 in Streptomyces lividans, where production only reached 2–3 mg l−1 [13].
Notably, a 2.5-fold higher GE2270 production was observed using pbtCK02 as compared to pbtCK01. This increase might be due to the deletion of the remaining three ribosomal genes in pbtCK02 rather than to the constitutive expression of the resistance gene tufR.
In agar diffusion tests S. coelicolor M1146, as well as the strains S. coelicolor M1146(pbtCK01) and S. coelicolor M1146(pbtCK02) were sensitive to GE2270A; despite of the fact that the latter two strains contain the resistance gene tufR [49] under control of either its native promoter or the constitutive ermE* promoter. Thus, the resistance gene tufR is apparently not able to confer full resistance against GE2270 to the heterologous host strain S. coelicolor M1146. Elongation factor Tu is a very abundant protein, playing a central role in ribosomal protein synthesis by delivering aminoacyl-tRNA to the growing protein chain. Possibly, the efficiency of the heterologous, GE2270-resistant EF-Tu for protein biosynthesis in S. coelicolor is quite low, or its level of expression is too low to confer full resistance.
In this study, we also tried to achieve inducible GE2270 production by introducing the tetracycline-inducible tcp830 promoter [25]. Application of this promoter/inducer system has been shown to be free of unwanted pleiotropic effects [25]. The insertion of tcp830 upstream of the entire gene cluster indeed results in an aTc inducible 4-fold increase in GE2270 production. Placement of the tcp830 promoter in front of the structural gene pbtA in construct pbtCK05 leads to the formation of GE2270B1 as main product but also to smaller amounts of GE2270A. This can be explained by the organization of the pbt gene cluster, where some of the responsible genes for the biosynthesis of the final product, GE2270A, are located upstream of the introduced promoter and thus GE2270B1 is accumulated as an intermediate in GE2270A biosynthesis. Surprisingly, GE2270 production was not inducible by aTc in this construct. In S. coelicolor M1146(pbtCK04) in addition to the insertion of the tcp830 promoter, pbtR, the regulator gene of the cluster, was deleted. This resulted in a total abolishment of GE2270 production. PbtR is a TetR-like protein with 64% sequence identity to TpdR, the transcriptional regulator of the tpd cluster from the thiomuracin- and GE2270-producer strain Nonomuraea sp. WU8817 [18]. In that study quantitative RT-PCR showed a strong induction of expression of tpdR and tpdA (the homolog of pbtA) in correlation with the onset of antibiotic production. This observation, in combination with the abolishment of GE2270 production in construct pbtCK04 shown in the present study, suggests a role as positive regulator for PbtR. While most TetR-family regulators act as transcriptional repressors, there are some which activate transcription [46].
Neither of our three constructs improved GE2270 production to levels similar or even higher to those observed for S. coelicolor M1146(pbtCK02). In conjunction with the missing inducibility of pbtCK05, this suggests a more complex regulation of the GE2270 biosynthetic gene cluster by more than one promoter region.
In conclusion, in this study we identified and solved a principal problem in the heterologous expression of the GE2270 biosynthetic gene cluster in Streptomyces. It was shown that genes belonging to the primary metabolism, in our case the ribosomal genes flanking the pbt gene cluster, can completely prevent the success of heterologous expression experiments. This may be of considerable importance for future studies, especially studies using recently developed methods for heterologous expression of large secondary metabolite gene clusters, e.g. by use of P1-derived artificial chromosome (PAC) vectors, which can tolerate inserts up to a size of approximately 200 kb [51].
Supporting Information
Comparison of the original cosmid 2F7 and pbtCK01 lacking 22 ribosomal genes concerning GE2270A production over time in Nonomuraea sp. ATCC 39727.
(PDF)
Sequence of 16S rRNA of Planobispora rosea ATCC 53733.
(DOCX)
List of strains, plasmids and cosmids employed in this study.
(PDF)
List of ribosomal proteins contained in cosmid 2F7 (GenBank accession number KF366381.2) compared to their orthologous in S. coelicolor A3(2).
(DOCX)
Acknowledgments
We thank EntreChem S. L. (Oviedo) for providing the media screen list. We are grateful to Lutz Heide for helpful discussions, critical reading of the manuscript and valuable comments; to Andreas Kulik for HPLC-MS measurements and Franziska Leipoldt for helpful advices concerning phylogenetic trees and the MEGA5 software.
Funding Statement
Financial support was provided from the German Federal Ministry of Education and Research (ERA-IB GenoDrug, FK20315930). AT and MS were partially supported by grants from Italian MIUR and Regione Lombardia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Su TL (1948) Micrococcin, an antibacterial substance formed by a strain of Micrococcus . Br J Exp Pathol 29: 473–481. [PMC free article] [PubMed] [Google Scholar]
- 2. Liao R, Duan L, Lei C, Pan H, Ding Y, et al. (2009) Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol 16: 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, et al. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30: 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Q, Liu W (2013) Biosynthesis of thiopeptide antibiotics and their pathway engineering. Nat Prod Rep 30: 218–226. [DOI] [PubMed] [Google Scholar]
- 5. McConkey GA, Rogers MJ, McCutchan TF (1997) Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J Biol Chem 272: 2046–2049. [DOI] [PubMed] [Google Scholar]
- 6. Rogers MJ, Cundliffe E, McCutchan TF (1998) The antibiotic micrococcin is a potent inhibitor of growth and protein synthesis in the malaria parasite. Antimicrob Agents Chemother 42: 715–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhat UG, Halasi M, Gartel AL (2009) Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One 4: e5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gartel AL (2013) Thiazole antibiotics siomycin A and thiostrepton inhibit the transcriptional activity of FOXM1. Front Oncol 3: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anborgh PH, Parmeggiani A (1991) New antibiotic that acts specifically on the GTP-bound form of elongation factor Tu. EMBO J 10: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bagley MC, Dale JW, Merritt EA, Xiong X (2005) Thiopeptide antibiotics. Chem Rev 105: 685–714. [DOI] [PubMed] [Google Scholar]
- 11. Butler MS (2005) Natural products to drugs: natural product derived compounds in clinical trials. Nat Prod Rep 22: 162–195. [DOI] [PubMed] [Google Scholar]
- 12. LaMarche MJ, Leeds JA, Amaral A, Brewer JT, Bushell SM, et al. (2012) Discovery of LFF571: an investigational agent for Clostridium difficile infection. J Med Chem 55: 2376–2387. [DOI] [PubMed] [Google Scholar]
- 13. Young TS, Walsh CT (2011) Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans . Proc Natl Acad Sci U S A 108: 13053–13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malcolmson SJ, Young TS, Ruby JG, Skewes-Cox P, Walsh CT (2013) The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proc Natl Acad Sci U S A 110: 8483–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tocchetti A, Maffioli S, Iorio M, Alt S, Mazzei E, et al. (2013) Capturing linear intermediates and C-terminal variants during maturation of the thiopeptide GE2270. Chem Biol 20: 1067–1077. [DOI] [PubMed] [Google Scholar]
- 16. Selva E, Ferrari P, Kurz M, Tavecchia P, Colombo L, et al. (1995) Components of the GE2270 complex produced by Planobispora rosea ATCC 53773. J Antibiot (Tokyo) 48: 1039–1042. [DOI] [PubMed] [Google Scholar]
- 17. King A, Bethune L, Phillips I (1993) In vitro activity of MDL 62,879 (GE2270 A) against aerobic gram-positive and anaerobic bacteria. Antimicrob Agents Chemother 37: 746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, et al. (2009) Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc 131: 5946–5955. [DOI] [PubMed] [Google Scholar]
- 19. Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Floriano B, Bibb M (1996) afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 21: 385–396. [DOI] [PubMed] [Google Scholar]
- 21. Li SM, Heide L (2006) The biosynthetic gene clusters of aminocoumarin antibiotics. Planta Medica 72: 1093–1099. [DOI] [PubMed] [Google Scholar]
- 22. Flinspach K, Westrich L, Kaysser L, Siebenberg S, Gomez-Escribano JP, et al. (2010) Heterologous expression of the biosynthetic gene clusters of coumermycin A(1), clorobiocin and caprazamycins in genetically modified Streptomyces coelicolor strains. Biopolymers 93: 823–832. [DOI] [PubMed] [Google Scholar]
- 23. Saleh O, Bonitz T, Flinspach K, Kulik A, Burkard N, et al. (2012) Activation of a silent phenazine biosynthetic gene cluster reveals a novel natural product and a new resistance mechanism against phenazines. Med Chem Commun 3: 1009–1019. [Google Scholar]
- 24. Dangel V, Westrich L, Smith MC, Heide L, Gust B (2010) Use of an inducible promoter for antibiotic production in a heterologous host. Appl Microbiol Biotechnol 87: 261–269. [DOI] [PubMed] [Google Scholar]
- 25. Rodriguez-Garcia A, Combes P, Perez-Redondo R, Smith MCA, Smith MCM (2005) Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces . Nucleic Acids Res 33: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW (2001) Molecular Cloning. A Laboratory Manual. New York Cold Spring Harbor Laboratory Press.
- 27.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. Norwich, UK: John Innes Foundation.
- 28. Siebenberg S, Bapat PM, Lantz AE, Gust B, Heide L (2010) Reducing the variability of antibiotic production in Streptomyces by cultivation in 24-square deepwell plates. J Biosci Bioeng 109: 230–234. [DOI] [PubMed] [Google Scholar]
- 29. Selva E, Beretta G, Montanini N, Saddler GS, Gastaldo L, et al. (1991) Antibiotic GE2270A: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J Antibiot (Tokyo) 44: 693–701. [DOI] [PubMed] [Google Scholar]
- 30. Sosio M, Stinchi S, Beltrametti F, Lazzarini A, Donadio S (2003) The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem Biol 10: 541–549. [DOI] [PubMed] [Google Scholar]
- 31. Gastaldo L, Marinelli F (2003) Changes in GE2270 antibiotic production in Planobispora rosea through modulation of methylation metabolism. Microbiology 149: 1523–1532. [DOI] [PubMed] [Google Scholar]
- 32. Pozzi R, Simone M, Mazzetti C, Maffioli S, Monciardini P, et al. (2011) The genus Actinoallomurus and some of its metabolites. J Antibiot (Tokyo) 64: 133–139. [DOI] [PubMed] [Google Scholar]
- 33. Donadio S, Monciardini P, Sosio M (2009) Chapter 1. Approaches to discovering novel antibacterial and antifungal agents. Methods Enzymol 458: 3–28. [DOI] [PubMed] [Google Scholar]
- 34. Kominek LA (1972) Biosynthesis of novobiocin by Streptomyces niveus . Antimicrob Agents Chemother 1: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shima J, Penyige A, Ochi K (1996) Changes in patterns of ADP-ribosylated proteins during differentiation of Streptomyces coelicolor A3(2) and its development mutants. J Bacteriol 178: 3785–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sedmera P, Pospisil S, Novàk J (1991) New furanonaphthoquinone from Streptomyces cinnamonensis . Journal of Natural Products 54: 870–872. [Google Scholar]
- 37. Chang AC, Cohen SN (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134: 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100: 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khodakaramian G, Lissenden S, Gust B, Moir L, Hoskisson PA, et al. (2006) Expression of Cre recombinase during transient phage infection permits efficient marker removal in Streptomyces . Nucleic Acids Res 34: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fedoryshyn M, Welle E, Bechthold A, Luzhetskyy A (2008) Functional expression of the Cre recombinase in Actinomycetes. Appl Microbiol Biotechnol 78: 1065–1070. [DOI] [PubMed] [Google Scholar]
- 41. Doumith M, Weingarten P, Wehmeier UF, Salah-Bey K, Benhamou B, et al. (2000) Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea . Mol Gen Genet 264: 477–485. [DOI] [PubMed] [Google Scholar]
- 42. Fedoryshyn M, Petzke L, Welle E, Bechthold A, Luzhetskyy A (2008) Marker removal from actinomycetes genome using Flp recombinase. Gene 419: 43–47. [DOI] [PubMed] [Google Scholar]
- 43. Flett F, Mersinias V, Smith CP (1997) High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155: 223–229. [DOI] [PubMed] [Google Scholar]
- 44. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bibb MJ, Janssen GR, Ward JM (1985) Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus . Gene 38: 215–226. [DOI] [PubMed] [Google Scholar]
- 46. Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, et al. (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69: 326–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Imada A, Nozaki Y, Hasegawa T, Mizuta E, Igarasi S, et al. (1978) Carriomycin, a new polyether antibiotic produced by Streptomyces hygroscopicus . J Antibiot (Tokyo) 31: 7–14. [DOI] [PubMed] [Google Scholar]
- 48. Dey ES, Norrlow O, Liu Y (2004) Artificial carrier for oxygen supply in biological systems. Appl Microbiol Biotechnol 64: 187–191. [DOI] [PubMed] [Google Scholar]
- 49. Sosio M, Amati G, Cappellano C, Sarubbi E, Monti F, et al. (1996) An elongation factor Tu (EF-Tu) resistant to the EF-Tu inhibitor GE2270 in the producing organism Planobispora rosea . Mol Microbiol 22: 43–51. [DOI] [PubMed] [Google Scholar]
- 50. Mohrle VG, Tieleman LN, Kraal B (1997) Elongation factor Tu1 of the antibiotic GE2270A producer Planobispora rosea has an unexpected resistance profile against EF-Tu targeted antibiotics. Biochem Biophys Res Commun 230: 320–326. [DOI] [PubMed] [Google Scholar]
- 51. Jones AC, Gust B, Kulik A, Heide L, Buttner MJ, et al. (2013) Phage p1-derived artificial chromosomes facilitate heterologous expression of the FK506 gene cluster. PLoS One 8: e69319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the original cosmid 2F7 and pbtCK01 lacking 22 ribosomal genes concerning GE2270A production over time in Nonomuraea sp. ATCC 39727.
(PDF)
Sequence of 16S rRNA of Planobispora rosea ATCC 53733.
(DOCX)
List of strains, plasmids and cosmids employed in this study.
(PDF)
List of ribosomal proteins contained in cosmid 2F7 (GenBank accession number KF366381.2) compared to their orthologous in S. coelicolor A3(2).
(DOCX)