Abstract
Macrophages, including alveolar macrophages, are primary phagocytic cells of the innate immune system. Many studies of macrophages and inflammation have been done in mouse models, where inducible nitric oxide synthase (NOS2) and nitric oxide (NO) are important components of the inflammatory response. Human macrophages, in contrast to mouse macrophages, express little detectable NOS2 and generate little NO in response to potent inflammatory stimuli. The human NOS2 gene is highly methylated around the NOS2 transcription start site. In contrast, mouse macrophages contain unmethylated cytosine-phosphate-guanine dinucleotides (CpGs) proximal to the NOS2 transcription start site. Further analysis of chromatin accessibility and histone modifications demonstrated a closed conformation at the human NOS2 locus and an open conformation at the murine NOS2 locus. In examining the potential for CpG demethylation at the NOS2 locus, we found that the human NOS2 gene was resistant to the effects of demethylation agents both in vitro and in vivo. Our data demonstrates that epigenetic modifications in human macrophages are associated with CpG methylation, chromatin compaction and histone modifications that effectively silence the NOS2 gene. Taken together, our findings suggest there are significant and under-appreciated differences in how murine and human macrophages respond to inflammatory stimuli.
Keywords: Gene regulation, macrophages, inflammation
Introduction4
Alveolar macrophages are primary phagocytic cells of the innate immune system (1, 2). They are responsible, among other functions, for clearing infectious, toxic and allergic particles from the respiratory tract. In the case of murine macrophages, the response to inflammatory stimuli is characterized by the production of large amounts of NO via NOS2 (3–5). NO produced by murine alveolar macrophages is a critical component in the removal of bacteria and foreign particles (6). It has been shown to be crucial to murine alveolar macrophage function and host survival (6–8).
The murine alveolar macrophage has been frequently used as a model for human alveolar macrophage physiology. NO production by murine macrophages has been inferred to explain human inflammatory pathophysiology. However, there is scant evidence describing NO production in human alveolar macrophages to support such an extrapolation (9, 10). What data there is consistently shows that, even when stimulated, healthy human macrophages make little to no NO (9, 11–13). In murine models, inhibiting NO production impairs clearance of intracellular pathogens. Similar treatment of human alveolar macrophages has no effect (14). Further complicating the current landscape is data showing that human alveolar macrophages recovered from patients with chronic inflammatory diseases are capable of making measurable, but small, amounts of NO (15–18). Thus, it remains unclear whether NO production is relevant to human macrophage physiology or macrophage-mediated pathophysiology. Given this apparent contrast with the central role NO plays in murine macrophage inflammation, we consider it essential to clarify whether these animal findings can be appropriately extrapolated to the human arena.
Epigenetic programming is one of the mechanisms that controls cell type specific transcription of genes. Epigenetic control involves multiple regulatory mechanisms that include untranslated RNA, histone acetylation, and methylation of histones and DNA CpG dinucleotide sequences (19–22). DNA methylation at the C-5 positions of cytosine (5mC) in the CpG dinucleotide is a well-characterized epigenetic modification controlling gene expression (23, 24). Methylation of CpG motifs in the promoter region has been repeatedly shown to influence DNA transcription (primarily decreasing transcription) and is an important step in both normal and malignant cell function (25–28). CpG methylation, coupled with subsequent histone deacetylation, condenses chromatin, and leads to gene silencing (29–31). This mechanism relies on methyl-CpG-binding domain proteins that recognize methylated cytosines in the CpG dinucleotides. Altering gene regulation through the reversal of DNA methylation and histone acetylation is a recent addition to clinical therapeutics. In some clinical trials, cancer chemotherapeutic agents are used specifically to promote gene transcription via CpG demethylation and histone deacetylation (32–34).
In this study, we examined the effect of classic NOS2 activating stimuli on NOS2 expression and NO production in human macrophages. We found that human macrophages failed to produce NO even after stimulation with potent inflammatory signals. Furthermore, we found that in contrast to mouse macrophages, human macrophages did not express detectable levels of NOS protein coded by the NOS2 gene. We found that CpG motifs proximal to the transcription start site (TSS) of the human NOS2 promoter region are fully methylated, consistent with an epigenetically repressed gene. In contrast, CpG motifs proximal to the TSS in mouse macrophage NOS2 were not methylated. Assays for chromatin accessibility and histone regulatory marks were consistent with the hypothesis that the human macrophage NOS2 gene is a silenced gene. These data describe previously unrecognized differences between human and mouse macrophages and suggests that murine data be used with caution when inferring links to human disease.
Materials and Methods
Cell lines
Human alveolar macrophages, human blood monocytes, a human monocytic leukemia cell line (THP-1), murine alveolar macrophage cell line (MHS) and murine peritoneal macrophage cell line (RAW 264.7) were used. THP-1 and MHS cells were cultured in RPMI supplemented with 5ml L-glutamine, 5ml 1M Hepes, 5ml 100mM sodium pyruvate, 5ml of 250mg/ml glucose, 5% fetal bovine serum (FBS), 5ml Pen/Strep per liter. Human monocytes were cultured in RPMI-1640 supplemented with gentamicin 40μg/ml and 5% FBS and human and murine alveolar macrophages were cultured in RPMI-1640 supplemented with gentamicin 40μg/ml.
Bronchoalveolar lavage
After informed consent was obtained under a Carver College of Medicine IRB-approved protocol, nonsmoking volunteer research subjects performed spirometry with exclusion criteria of an FEV1 or FVC < 80%. Next, they underwent standard flexible fiberoptic bronchoscopy. Premedication with intramuscular morphine 10mg injection was followed by local anesthesia with lidocaine instillation into the upper airway. Standard bronchoalveolar lavage was performed by serially instilling and suction retrieving 20 ml aliquots of normal saline five times in three different lung segments. The first collection was discarded to avoid possible contamination with upper airway secretions or lidocaine. The remaining lavage was filtered through sterile gauze and centrifuged at 200g for 5 minutes to pellet cellular material. The resulting pellet was suspended in phosphate buffered saline (PBS) and centrifuged at 200g for 5 minutes. A sample of the cells were labeled with Wright stain and microscopically examined to determine the proportion of the cells that were macrophages. Aliquots of 5×106 cells were stored at minus 80°C until RNA and DNA isolation procedure was performed. The procedure generated a relatively pure population of alveolar macrophages with fewer than 5% neutrophils or lymphocytes.
Human blood monocyte isolation
As part of the bronchoscopy protocol, some subjects underwent venipuncture with 180cc of blood drawn. Mononuclear cells were isolated from the gradient interface after centrifugation with Histopaque (Sigma-Aldrich, St. Louis, MO, USA). Monocyte isolation was performed with BigEasy Easy Sep system (StemCell, Vancouver, Canada) according to the manufacturer’s protocol and as described previously (35).
Mouse bronchoalveolar lavage
Mice were anesthetized, and the chest cavity was opened. The trachea was exposed, an incision made, and tubing inserted. The lungs were flushed with 3 × 1 ml PBS. Cells were pelleted. Slides were then made from the pelleted cells and determined to be greater than 95% macrophages by Wright stain.
Murine bone marrow derived macrophage isolation
To generate bone marrow derived macrophages, C57/Blk6 mice were euthanized and the tibias and femurs were harvested. Inside a sterile hood, bones were dipped in ethanol and submerged in DMEM supplemented with 10% fetal calf serum, L-glutamine and penicillin/streptomycin (DMEM). The ends of the bones were cut off with a sharp scissors and the bone marrow flushed out with DMEM using a syringe and 25G needle (approximately 3 mls per fibula and 2mls per tibia). Cells were centrifuged and resuspended in DMEM (6 ml/mouse) with an added 7 mls of DMEM and 2 mls of L cell conditioned medium (LCCM, obtained by culture of L929 cells in DMEM for 5 days). Cells were incubated for 6 days at 37°C and then removed from plates using .05% Trypsin/EDTA (Life Technologies). Cells were resuspended in DMEM at 1 million per ml and seeded onto the appropriate tissue culture dishes for the proposed experiments.
DNA and RNA isolation
DNA was isolated from macrophages using QIAgen DNAeasy Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. RNA was isolated using the mirVana miRNA Isolation kit (Applied Biosystems, Austin, TX, USA). The quantity and quality of the RNA samples was assessed using an Experion Automated Electrophoresis Station (Bio-Rad, Hercules, CA, USA). The RNA quality indicator was above 8 for all samples where values of greater than 8 indicate primarily intact RNA on a scale of 1–10. After preparation, RNA samples were stored at −80°C until use.
DNA methylation analysis
Determination of genome-wide methylation values was conducted under contract by the University of Minnesota BioMedical Genomics Center (Minneapolis, MN, USA) using the Illumina Infinium 450K Human Methylation array, which contains 485,000 probes that interrogate CpG residues in 99% of RefSeq annotated genes (NCBI, Bethesda, MD, USA). The resulting microarray data were inspected for complete bisulfite conversion of the DNA. Average β-values (i.e., average methylation) for each CpG residue were determined using the GenomeStudio V2009.2, methylation module version 1.5.5 version 3.2 (Illumina, San Diego, CA, USA). Comparison of β-values (i.e., methylation) between cases and controls was conducted using Student’s t-test, whereas comparisons of the relationship between overall values between individual arrays were conducted using Pearson’s correlation coefficient.
Primer design
We used Genome Browser (http://genome.ucsc.edu/index.html) to obtain the sequence of the NOS2 gene promoter region from −2000 base pairs upstream of TSS to the second exon. We used the publicly available online program Meth Primer (http://www.urogene.org/methprimer/index1.html) to obtain predicted bisulfite-converted sequence. Primers were designed based on this sequence (Supplemental Table 1).
Bisulfite sequencing
DNA was bisulfite modified then amplified using EZ DNA Methylation Kit (Zymo Research) according to the manufacturer’s instructions. The DNA samples were amplified using a touchdown nested PCR protocol (Supplemental Table 1) with primers designed for the human and murine NOS2 promoter regions (Supplemental Table 2). The PCR product was gel purified and extracted using QIAquick Gel Extraction Kit according to the manufacturer’s instructions. PCR products were then sequenced at the University of Iowa DNA Facility.
Western analysis
Whole cell protein was obtained by lysing the cells on ice for 20 minutes in 200 μl of lysis buffer (0.05 M Tris pH 7.4, 0.15 M NaCl, 1% NP-40) with added protease and phosphatase inhibitors: 1 protease minitab (Roche Biochemicals)/10 ml and 100 ul 100X phosphatase inhibitor cocktail (Calbiochem)/10 ml. The lysates were sonicated for 20 seconds, kept at 4°C for 30 minutes, spun at 15,000 g f.or 10 minutes and the supernatant saved. Protein determinations were made using the Bradford Protein assay from Bio-Rad. Cell lysates were stored at −70° until use.
Western analysis was performed on whole cell proteins. 30 μg of protein was mixed 1:1 with 2x sample buffer (20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.05% bromophenol blue and 1.25 M Tris pH 6.8, all chemicals from Sigma Chemical Co.) heated to 95o for 5 minutes and loaded onto a 10% SDSPAGE gel and run at 100 V for 90 minutes. Cell proteins were transferred to PVDF membranes (Bio-Rad Hercules, CA) by semi-dry transfer (BioRad). Equal loading of the protein groups on the blots was evaluated using Ponceaus S, a staining solution designed for protein quantification or by stripping and reprobing with antibodies to β-actin or GAPDH. The PVDF was dried and then incubated with the primary antibody overnight in 5% milk. The blots were washed four times with TTBS and incubated for 1 hour with horseradish-peroxidase conjugated anti-rabbit or mouse IgG antibody as a control. Immunoreactive bands were developed using a chemiluminescent substrate (ECL Plus, Amersham, Arlington Heights, IL). An autoradiograph was obtained, with exposure times of 10 seconds to 2 minutes.
Griess assay of nitric oxide production
Measurement of NO production was performed using the Griess reaction as described previously (13, 36) using Griess Reagent Kit for Nitrite Determination (Invitrogen, NY, USA) according to the manufacturer’s protocol.
Chromatin accessibility assay
Cells were cultured with or without stimulation with IFNγ and LPS, harvested and pelleted by centrifugation. Cells were washed with ice cold PBS and then lysed with lysis buffer (cold RSB + 0.10% NP-40). Nuclei were isolated by centrifugation. Nuclear DNA was digested with increasing concentrations of DNAse (0U, 2.5U, 5U, 10U) for 10 minutes at 37o. DNA was isolated with Qiagen DNAEasy Kit as described above. DNA was then purified and used for quantitative PCR. We designed primers that cover genomic sequences proximal to the TSS for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene; 60S ribosomal protein L30 (RPL30), a transcriptionally active gene; intercellular adhesion molecule 1 (ICAM1), an inflammatory response gene; β-globin gene (HBB) a known transcriptionally silent gene (silent in nonerythroid cells); tumor necrosis factor α (TNFα), an inflammatory response gene and NOS2 (Supplemental Table 2).
Chromatin immunoprecipitation (ChIP) assay
ChIP Assay was performed using a ChIP assay kit from Cell Signaling Technology (Beverly, MA, USA). Briefly, macrophage proteins and DNA were cross-linked with formaldehyde. Cells were lysed in SDS-lysis buffer and then sonicated. For sonication, the amplitude setting was 20%. The time setting was as follows: 1″ sonicate, 1″ off for a total of 10 each or 20″ total, rest 30″ on ice then repeat for a total of 3 times. Sonication was performed using a Sonics Vibra Cell Model CV18, Sonics and Materials, Inc. 50 ul of sheared chromatin was used to analyze chromatin digestion, with most of DNA sample found to be in the appropriate 500–600 bp size range. 10 ul of purified chromatin was stored as “input sample”. The remaining chromatin was diluted with provided ChIP buffer and then immunoprecipitation performed using anti-trimethyl-histone H3 (Lys27) (H3K27me3) polyclonal rabbit IgG (Cell Signaling, Beverly, MA), anti-trimethyl-histone H3 (Lys4) (H3K4me3) polyclonal rabbit IgG (Millipore, Temeluca, CA), or a negative control antibody (Normal Rabbit IgG #2729). ChIP-Grade Protein G Magnetic Beads were added to the incubation mix. After bead separation with a magnetic separation rack, chromatin was eluted from the antibody/Protein G beads with ChIP Elution Buffer. DNA was purified and used for quantitative PCR.
Demethylation and histone deacetylation treatment of therapy-resistant melanoma patients
DNA samples were obtained from conventional treatment resistant metastatic melanoma patients enrolled in an experimental protocol at the University of Iowa Hospitals and Clinics. The research treatment protocol included 5-azacytidine (a DNA methyltransferase inhibitor), a histone deacetylase inhibitor and an oral alkylating agent. Patients were consented and blood drawn at designated time points (day 0 and 2 to 3 weeks following the second treatment cycle) (Figure 7A). Demethylation of DNA was monitored by measuring serum hemoglobin F levels with high power liquid chromatography as described previously (38). For an analysis of DNA methylation at the NOS2 gene, DNA was isolated from subject blood mononuclear cells and bisulfite converted. CpG methylation was determined at the NOS2 locus by sequencing the region proximal to the TSS (primers are described in the Supplemental Files).
Results
Human macrophages do not increase NO production or NOS2 expression in response to inflammatory stimuli
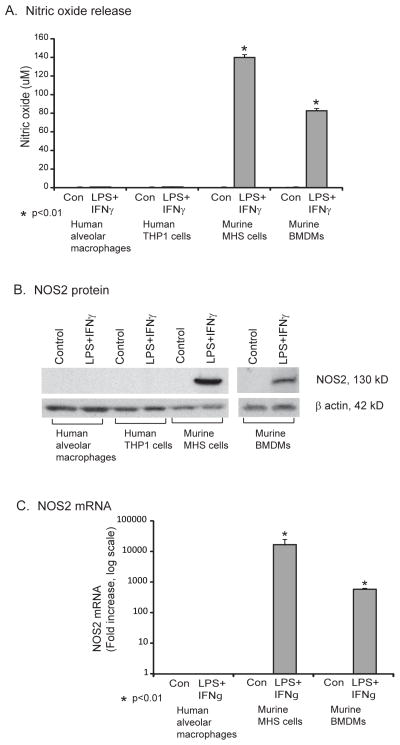
We studied primary human alveolar macrophages, the human macrophage cell line THP-1, bone marrow-derived murine macrophages, and the murine macrophage cell line MHS. Cells were stimulated with interferon gamma (IFNγ) and lipopolysaccharide (LPS) for twenty-four hours. Nitrite levels were measured in the cell culture supernatants. In human macrophages only minimal nitrite levels were released at baseline, with no sign of a response to the stimuli (Fig. 1A). In stark contrast, murine macrophages made large amounts of NO in response to stimulation (Fig. 1A). Next, we measured NOS2 protein levels in the cell lysates of human alveolar macrophages, human THP-1 cells, murine MHS macrophages and mouse bone marrow-derived macrophages after stimulation with IFNγ and LPS. NOS2 protein amounts were in line with our findings with NO showing a robust increase with stimulation in murine macrophages and no detectable NOS2 protein in human macrophages (Fig. 1B). NO measurements also correlated with NOS2 mRNA levels determined by qRT-PCR. There was an impressive increase in NOS2 mRNA with stimulation in bone marrow-derived mouse macrophages (> 500 fold) and in the murine MHS cell line (>10,000 fold). There were barely detectable levels of NOS2 mRNA present in human alveolar macrophages at baseline, with almost no increase following stimulation (Fig. 1C).
Figure 1. Murine, but not human, macrophages increase NO release and NOS2 expression after stimulation.
A. Human (alveolar macrophages obtained by bronchoscopy or THP-1 monocytic cell line) and murine cells (bone marrow-derived macrophages (BMDMs) or the mouse alveolar macrophage cell line, MHS) were put in culture at 1 million cells per ml and treated with and without LPS (100 ng/ml) and IFNγ (100 ng/ml) for 18 hours. Cells were cultured for 18 hours and supernatants collected for NO measurement (Greiss reaction). Data represents mean +/− standard error for three separate experiments. Significant differences from unstimulated control were evaluated using a Student’s t-test. B. The same human and mouse cells were cultured with and without LPS (100 ng/ml) and IFNγ (100 ng/ml) for 18 hours. Total cell protein was isolated and Western analysis for NOS2 performed. Shown is a representative blot from four replicate experiments. C. Human alveolar macrophages, MHS cells and murine bone marrow-derived macrophages were cultured with and without LPS (100 ng/ml) and IFNγ (100 ng/ml) for 4 hours. RNA was isolated and NOS2 and ICAM1 relative levels were analyzed using qRT-PCR). Data represent mean +/− standard error (SE) from three separate experiments. Significance was evaluated using Student’s t-test.
Human alveolar macrophages have an intact LPS/ IFNγ inflammatory pathway when stimulated
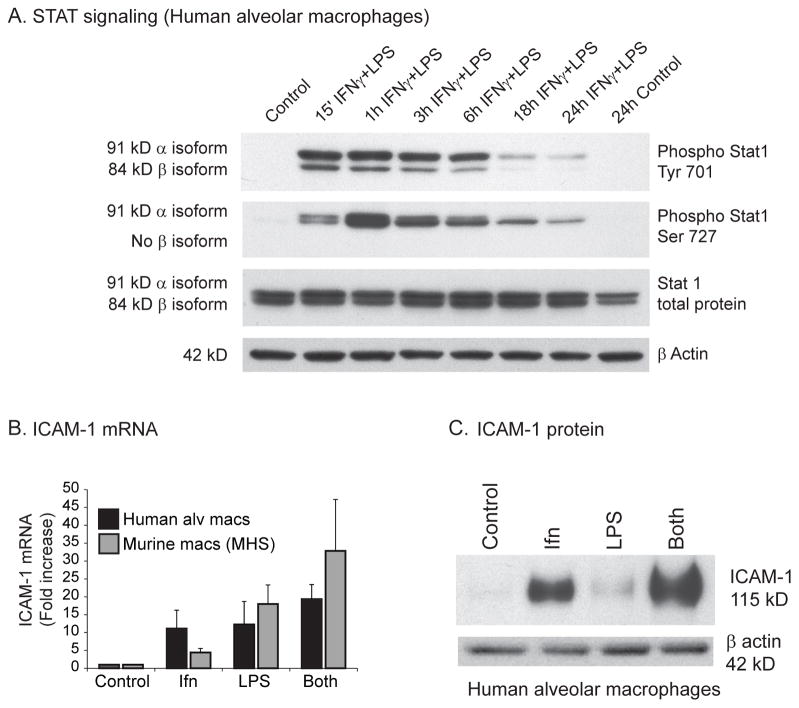
Having found that human alveolar macrophages lacked NOS2 expression, we next asked if the signaling cascade downstream of the LPS and IFNγ receptors was intact. To evaluate signaling events downstream of the LPS (TLR4) and IFNγ receptors, we evaluated STAT1 activation. STAT1 is a transcription factor that is activated via phosphorylation synergistically induced by the combination of LPS and IFNγ (37). Figure 2A demonstrates substantial activation of STAT1 by LPS/IFNγ reflected in the rapid increase in phosphorylated STAT1. To examine whether proteins other than NOS2 known to be induced by LPS/IFNγ were up-regulated in stimulated human alveolar macrophages, we examined intracellular adhesion molecule-1 (ICAM1). ICAM1 is a transmembrane protein that is expressed upon cytokine stimulation and facilitates macrophage migration past the endothelium (38). Stimulation of human alveolar macrophages with LPS/IFNγ led to significant upregulation of ICAM1 mRNA (Figure 2B); the murine macrophage cell line MHS showed a similar response. Stimulation of human alveolar macrophages also led to significant increase in ICAM1 protein (Figure 2C). These data demonstrate that there is not a global defect in LPS and IFNγ signaling in human macrophages and that the signaling cascade downstream of the LPS/IFNγ receptors in human alveolar macrophages is intact and upregulates inflammatory genes other than NOS2.
Figure 2. LPS and IFNγ signaling is intact in human alveolar macrophages.
A. Human alveolar macrophages were stimulated with and without LPS (100 ng/ml) and IFNγ (100 ng/ml) for varying time points. Whole cell protein was isolated and Western analysis performed using antibodies specific for phosphorylated STAT1 (tyrosine 701 and serine 727) and total STAT1. Shown is a representative Western from three separate experiments. B. Human alveolar macrophages and MHS cells were stimulated with and without LPS (100 ng/ml) and IFNγ (100 ng/ml) for 4 hours. RNA was isolated and ICAM1 levels determined with qRT-PCR. C. Human alveolar macrophages were stimulated with and without LPS (100 ng/ml) and IFNγ (100 ng/ml) for 18 hours. Whole cell protein was isolated and Western analysis performed for ICAM1. Shown is representative Western from three separate experiments.
CpG motifs in the NOS2 promoter region in human alveolar macrophages and monocytes are uniformly methylated
There is convincing evidence indicating that CpG methylation at the promoter region and first exon is an important epigenetic mechanism regulating the activation of gene transcription (39). Because we had found that intracellular signaling and downstream expression of other inflammatory genes was intact in stimulated human macrophages, we hypothesized that DNA methylation might be responsible for the lack of transcription of the NOS2 gene in human alveolar macrophages.
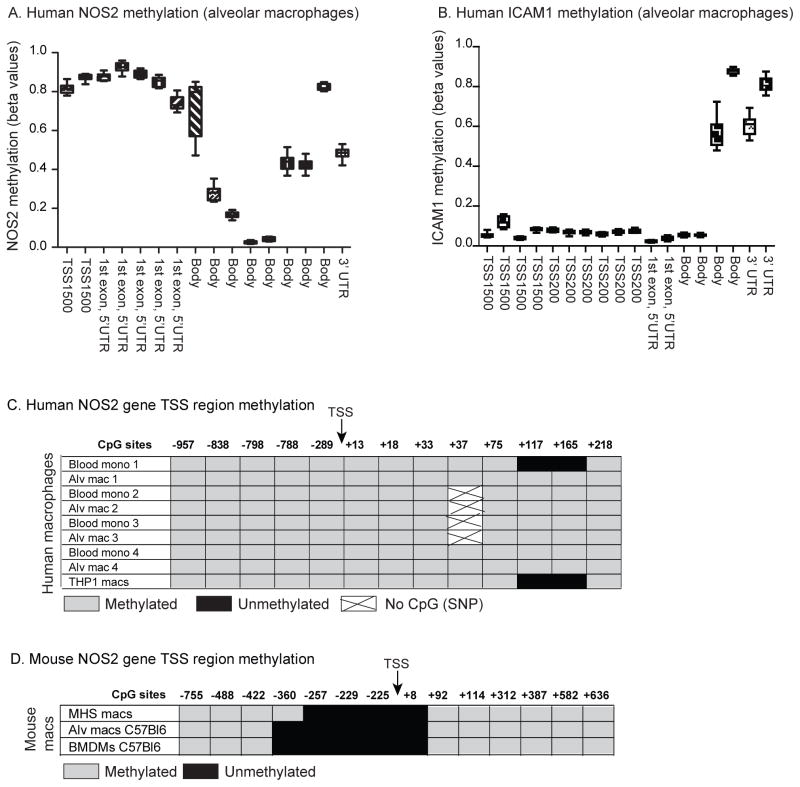
First, we assessed methylation levels at the NOS2 gene promoter region in data from an Illumina 450K array. The Illumina array allows for a broad analysis of methylation levels in more than 450,000 probes across the genome. The Illumina array includes a number of probes upstream of and leading into the NOS2 gene. In examining beta values (average methylation of 7 probes covering −1500bp upstream of TSS to the end of 1st exon) from an Illumina array (alveolar macrophages from 5 normal subjects), we found high levels of methylation at probes that clustered around the TSS of the NOS2 gene (Figure 3A). This was in stark contrast with the low methylation levels found at probes around the TSS of the LPS/ IFNγ inducible gene, ICAM1 (Fig 3 B).
Figure 3. The human NOS2 gene is methylated at the promoter region.
A+B. Human alveolar macrophages were obtained from normal volunteers. DNA was isolated and CpG methylation was determined using bisulfite conversion and the Illumina Infinium 450K Human Methylation array. Shown are representative beta values from 10 individuals for the NOS2 gene (A) and the ICAM1 gene (B). Probe sites from the Illumina array are listed in the Supplemental files. C. Individual bisulfite sequencing of CpG sites proximal to the NOS2 promoter was determined in paired samples of human blood monocytes and alveolar macrophages from 4 donors and THP-1 cells. DNA was isolated, bisulfite conversion performed and sequencing performed. Sites that sequenced as CG were considered methylated, while those that sequenced as TG were considered unmethylated. All CpGs from −957 (relative to TSS) to +218 of the human NOS2 gene were sequenced. D. Individual bisulfite sequencing of CpG sites proximal to the NOS2 promoter was determined in mouse macrophages (MHS cell line, C57Bl6 alveolar macrophages, and C5Bl6 bone marrow-derived macrophages). DNA was isolated, bisulfite conversion performed and the regions sequenced. All CpGs from −755 (relative to TSS) to +636 of the mouse NOS2 gene were sequenced.
Given the results from the Illumina methylation array, we next examined in greater detail the CpG motifs around the NOS2 TSS. To do this, we compared bisulfite sequencing of macrophage DNA from human (alveolar macrophages, blood monocytes, monocytic cell line THP-1) and murine sources (C5BL6 alveolar and bone marrow-derived macrophages and MHS macrophage cell line). The DNA was bisulfite converted and DNA sequencing of the NOS2 promoter region performed. Primers designed for the sodium bisulfite-treated DNA were used to sequence the region from approximately 1000 bp upstream of the TSS to the end first exon in the human and murine DNA. We found that across all the human macrophages, CpG motifs in that region were uniformly methylated (Fig 3C). For the murine cells, the data was distinctly different. By contrast, we found that CpGs in the region 400 bp upstream of the TSS and in exon 1 contained many unmethylated CpG motifs (Figure 3 D).
Based on previous data (40), we asked whether stimulation with cytokines could reverse CpG methylation. We stimulated THP-1 cells with a combination of cytokines and found unchanged methylation of CpG motifs in the promoter region after stimulation (data not shown).
Chromatin Immunoprecipitation Assay (ChIP) reveals silencing histone modification at the NOS2 gene in human alveolar macrophages
A ChIP assay generates data on whether a particular gene region is associated with repressive or transcriptionally active histone marks; this, in part, determines whether a given promoter is “open” or “closed” to transcription factors. Methylation of lysines on histones can result in either promotion or inhibition of transcription, based on the level of methylation and position of the lysine. It has been previously shown that, near gene transcription start sites, tri-methylation of lysine H3K4 is linked to active transcription while tri-methylation of lysine H3K27 is linked to transcriptional silencing. (41).
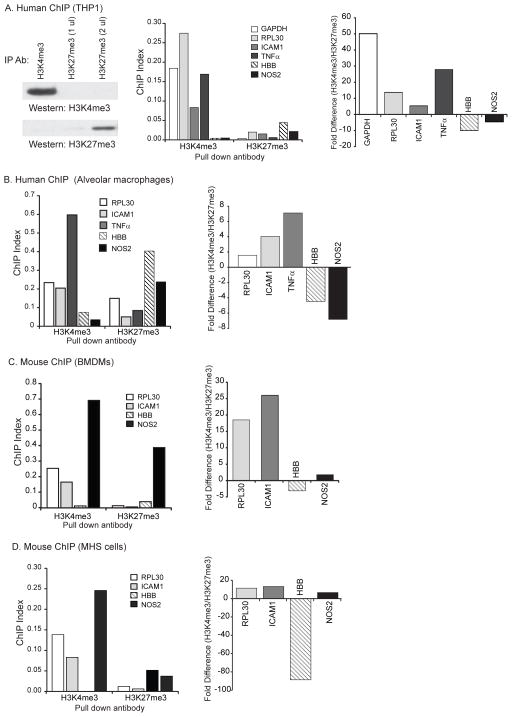
First, we established the efficacy of our immunoprecipitation using antibodies to H3K4me3 and H3K27me3 in lysates of THP-1cells (Fig 4A). Next, a ChIP assay was performed on formaldehyde fixed THP-1 cells and human alveolar macrophages.. We used antibodies to H3K4me3 and H3K27me3 to immunoprecipitate histone H3 protein. We then isolated DNA from the precipitated protein and performed qPCR using primers for NOS2, ICAM1, housekeeping gene GAPDH, RPL30, TNFα and a known silent gene, HBB. In both human alveolar macrophages and THP-1 cells, there was association of actively transcribed genes (GAPDH, ICAM1, RPL30, and TNFα) with the activating histone mark H3K4me3 and association of NOS2 and HBB with the silencing H3K27me3 modification (Fig. 4A & B). When these same studies were performed using mouse bone marrow-derived macrophages and the murine MHS cell line, NOS2 DNA was found associated with the activating histone modification H3K4me3 as would be expected of an accessible gene (Fig 4C & D). Thus, NOS2 has characteristics of a silenced gene in human macrophages and looks more like an open gene in mouse cells.
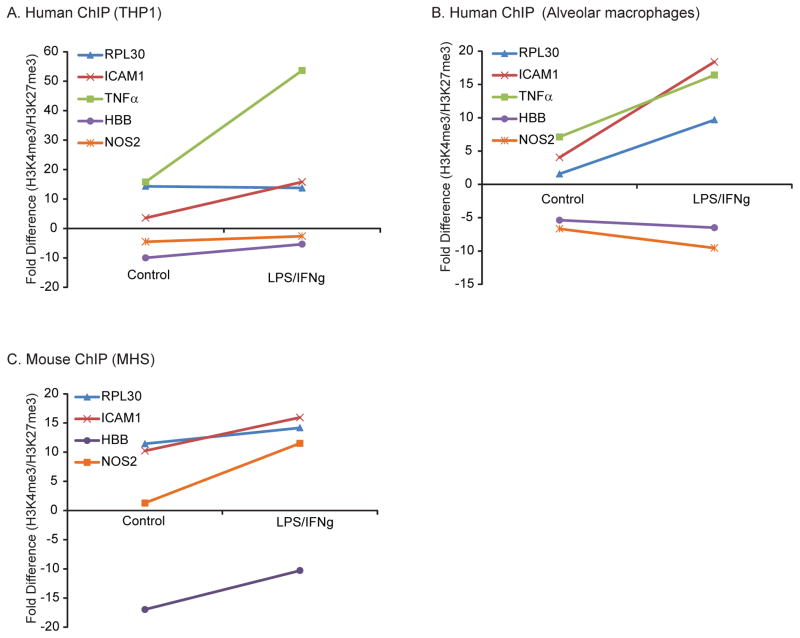
Figure 4. Human NOS2 gene is associated with silencing H3K27me3 marks; mouse NOS2 segregates with the activating histone mark H3K4me3.
A. THP-1 cells were processed for ChIP-Seq. Chromatin-associated DNA was pulled down with antibodies to either the activating histone modification, H3K4me3 or the silencing histone modification, H3K27me3. Shown is a Western analysis of proteins from immunoprecipitations with the two antibodies, demonstrating specific pull down of either H3K4me3 or H3K27me3. DNA was isolated and qRT-PCR performed using primers specific for the region proximal to the TSS of the GAPDH, RPL30, ICAM1, TNFa, HBB and NOS2. The graph on the left shows data from a representative experiment (of three) presented as a ChIP Index (2% × 2(C(T) 2% input sample − C(T) IP sample)). The graph on the right shows a comparison of the H3K4me3 pull down values and the H3K27me3 pull down values (H3K4me3 ChIP Index/H3K27me3 ChIP Index). If the ratio was less than 1, data was converted to a fold change ((1/value)( −1)). B, C, D. Identical experiments to those shown in Figure 5A were performed using human alveolar macrophages (B), Bone Marrow-Derived Macrophages from C57Bl6 mice (C) and the murine MHS macrophage cell line (D). The data is presented in the same format as A.
Association of the human NOS2 gene with H3K27me3 is resistant to inflammatory stimuli
In mice, macrophage NO expression increases dramatically with inflammatory stimuli. To determine if the association of the human NOS2 gene with the gene silencing histone modification, H3K27me3, was changed by inflammatory stimuli, human macrophages were stimulated with IFNγ and LPS. A ChIP assay was performed on control and stimulated cells. In both types of human macrophages, stimulation increased H3K4me3 association in the active genes (ICAM1, RPL30, and TNFα; Fig 5A). Stimulation did not increase activating histone marks associated with the known silenced gene HBB or NOS2 (Fig 5B); the activating/silencing mark ratio remained negative after stimulation. In comparison, mouse macrophages (MHS) stimulated with IFNγ and LPS demonstrated an increase in the association of the NOS2 gene with H3K4me3 (Fig 5C). Thus, NOS2 behaves like a “closed” gene that cannot be modulated by acute inflammatory signaling in human macrophages.
Figure 5. In human macrophages, LPS/IFNγ exposure does not increase NOS2 gene association with the activating histone mark H3K4me3.
In mouse macrophages, LPS/IFNγ exposure increased H3K4me3 association with the NOS2 locus. A. THP-1 cells were cultured with LPS (100 ng/ml) and IFNγ (100 ng/ml) for 18 hours. DNA and proteins were cross-linked and immunoprecipitation performed with antibodies to H3K4me3 and H3K27me3. DNA was isolated from the immunoprecipitated antibody/protein/DNA complex and qRT-PCR performed for the TSS proximal region of the RPL30, ICAM1, TNFα, HBB and NOS2 genes. The graph shows fold changes in the ChIP Index (2% × 2(C(T) 2% input sample − C(T) IP sample)). The data is representative of three separate experiments. B, C. Identical studies were performed in human alveolar macrophages and the murine macrophage cell line, MHS. The graph shows fold differences between the amount of gene specific DNA pulled down by the antibody to H3K4me3 and the antibody to H3K27me3. A positive fold difference implies an increase in association of gene promoter with activating histone marks. A negative fold difference implies an increase in association of gene promoter with silencing histone marks.
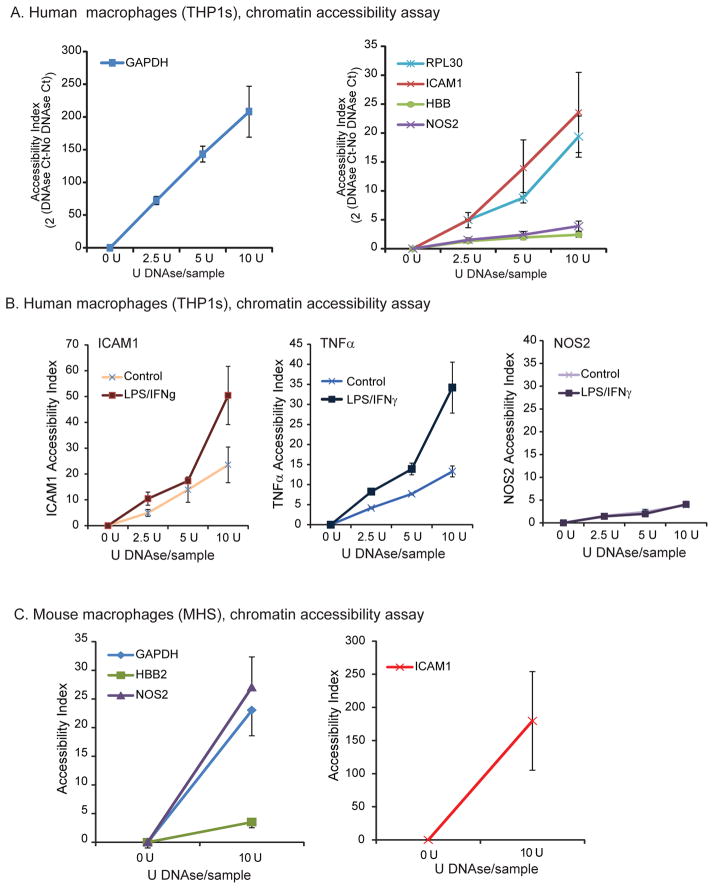
Chromatin accessibility assay indicates a closed chromatin conformation at the NOS2 locus in human alveolar macrophages
Because the ChIP assay suggested closed chromatin at the human NOS2 gene, we went on to directly measure chromatin accessibility. Chromatin accessibility assays provide data regarding the “closed” or “open” state of chromatin proximal to a particular gene. The assay tests the ability of DNAse to cleave and degrade the relevant DNA. Data is outlined as an Accessibility Index (accessibility index = 2^((Ct DNAse treated) - (Ct Uncut)). Thus, a higher number implies more degradation by DNase and, therefore, a more accessible gene. Previous research has shown strong inverse correlation between DNA methylation and chromatin accessibility (42). qPCR performed on DNAse treated human THP-1 cell nuclei revealed striking differences in chromatin accessibility between known actively transcribed genes (GAPDH, RPL30, ICAM1) and a known transcriptionally silent gene, HBB. In a series of 3 separate experiments, the NOS2 gene index was similar to the silent HBB gene (Fig. 6A). To ask if stimulation would change chromatin accessibility at the NOS2 locus, we stimulated human alveolar macrophages with IFNγ and LPS, followed by a chromatin accessibility assay. qPCR performed for ICAM1 and TNFα revealed even higher indices of accessibility after stimulation, while the NOS2 gene remained inaccessible regardless of stimulation (Fig. 6B). To ask if the murine NOS2 gene was more accessible than the human gene, we performed a chromatin accessibility assay on treated mouse MHS macrophages. In contrast with the human NOS2 gene, murine NOS2 gene segregated with known actively transcribed genes GAPDH and ICAM1 (Fig. 6C).
Figure 6. In human macrophages, NOS2 is a closed gene that remains inaccessible even after inflammatory stimulation.
A. Human THP-1 cells were treated with varying amounts of DNAse. DNA was isolated and qRT-PCR performed for GAPDH, RPL30, ICAM1 as representative actively transcribed “open” genes, HBB as a prototypical “closed” gene, and NOS2 to determine DNAse sensitivity at the loci. B. The effect of LPS/IFNγ exposure on chromatin accessibility was determined in THP-1 cells exposed to LPS (100 ng/ml) and IFNγ (100 ng/ml) for 18 hours. Pelleted cells were exposed to increasing levels of DNAse and qRT-PCR performed for ICAM1, TNFa and NOS2. Data is presented as Accessibility Index (2 (DNAse Ct-No DNAse Ct)). The data is mean +/− SE of three separate experiments. Significance was evaluated using Student’s t-test. C. Mouse MHS cells were treated with varying amounts of DNAse. DNA was isolated and qRT-PCR performed for GAPDH, ICAM1, HBB and NOS2 associated DNA to determine DNAse sensitivity at the loci. In human macrophages, NOS2 has a chromatin accessibility pattern similar to the closed gene HBB and does not become more accessible with stimulation.
These data collectively demonstrate that the difference between the mouse and human macrophage NO response to inflammatory signaling is due to epigenetic silencing of the gene in human cells.
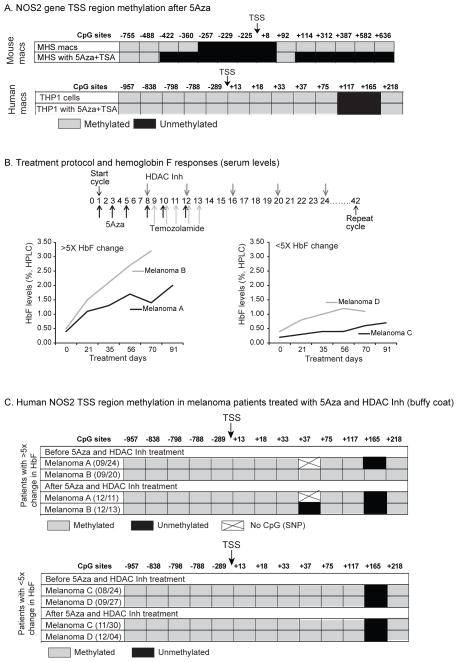
5-aza-2′-deoxycytidine (5-aza-dC) and Trichostatin A (TSA) induce changes in NOS2 promoter methylation profile in murine MHS cells
We tested the ability of the DNA methyltransferase inhibitor 5-aza-dC and the histone deacetylase inhibitor TSA to alter CpG methylation at the human NOS2 locus in vitro. The NOS2 TSS in THP-1 cell was fully methylated (except for two CpGs at +117 and +165) and we were unable to change the methylation profile with drug exposure (Fig 7A); this data was consistent with previous findings that showed an inability to demethylate the heavily methylated NOS2 promoter in human endothelial cells (43). This is consistent with our hypothesis that genes that are heavily methylated and associated with histone silencing marks are resistant to the effects of demethylating agents.
Figure 7.
Exposure to a demethylation agent and HDAC inhibitor increases the number of unmethylated CpGs in mouse macrophages, but has no effect on methylation of the NOS2 TSS region in human macrophages or circulating leukocytes from melanoma patients treated with 5Aza and an HDAC inhibitor. A. Mouse (MHS) and human (THP-1) macrophages were treated with 5-aza-dC (1 uM) on days 0 and 2. Trichostatin A was added on day 4 (5ng/ul). Cells were stimulated with IFNγ (100ng/ul), (LPS 100ng/ul) on day 5 and harvested on day 6. Control cells were stimulated on day 5 and harvested on day 6. DNA was isolated with bisulfite conversion and sequencing performed. Sites that sequenced as CG were considered methylated, while those that sequenced as TG were considered unmethylated. All CpGs from −755 (relative to TSS) to +636 of the mouse NOS2 gene were sequenced. B. Melanoma patients with resistant disease were treated at University of Iowa Hospitals and Clinics with 5-aza-dC, temozolamide and a histone deacetylase inhibitor throughout a 42-day cycle. Hemoglobin F (HbF) electrophoresis was performed from blood collected during the treatment. A 5-fold rise in HbF was used as a cut off for establishing expected drug effect. C. Whole blood was collected from melanoma patients undergoing experimental therapy outlined above before and after the cycle. DNA was isolated bisulfite conversion and sequencing performed. All CpGs from −957 (relative to TSS) to +218 of the human NOS2 gene were sequenced.
To ask if an already open locus (mouse NOS2) was sensitive to DNA methyltransferase inhibition, we treated the murine macrophage cell line, MHS, with 5-aza-dC and TSA. At baseline, sequencing of MHS DNA revealed a string of demethylated CpGs at the beginning of NOS2 promoter and upstream of it. Subjecting these cells to a protocol of 5-aza-dC and TSA treatment resulted in further demethylation of CpG targets upstream and downstream of the NOS2 promoter region (Figure 7A). This, again, supports our observations that the NOS2 gene is open for regulation in the mouse, while it is closed to manipulation in human macrophages.
5-aza-dC and TSA do not induce demethylation changes in NOS2 promoter methylation profile in human blood DNA during in vivo treatment
We next asked if in vivo exposure to a demethylating agent and inhibitor combination would alter CpG methylation at the NOS2 locus. 5-aza-dC and histone deacetylase inhibitors (HDAC) are being used in clinical trials for some malignancies, including melanoma. The hope is that the combined drug therapy will reverse methylation of tumor suppressor genes and slow the progression of the cancer. In a trial here at the University of Iowa, patients with advanced melanoma are treated with a protocol that includes 5-aza-dC and a histone deacetylase inhibitor (Figure 7A). We have collected blood before and after treatment and examined DNA methylation of the NOS2 gene. The clinical effect of the demethylating agents was confirmed by examining Hemoglobin F levels (HbF). During fetal development, HbF is the dominant hemoglobin isoform. From approximately 6 months post-partum through adulthood it is gradually replaced by Hemoglobin A. This process of “hemoglobin switching” is controlled epigenetically by methylation of the HbF (γ-globin) gene promoter in bone marrow erythroblasts (44). We followed four patients enrolled in the current trial. Two of these had a rise in HbF while the other two had unchanged levels of HbF during the course of treatment (Figure 7 B). We analyzed bisulfate-converted DNA for methylation of the NOS2 promoter in all 4 patients prior to treatment and then later post-treatment as outlined in the “Methods” section. We found two CpGs (+3 and +165) changed in one patient who showed a HbF response after treatment (Figure 7C). All other CpGs remained methylated. The second patient who responded to treatment with increase in HbF, along with 2 “non-responders” showed no changes in methylation pattern. This data suggests that the human NOS2 gene is resistant to the effects of a demethylating agent and HDAC inhibitor in vivo as well as in vitro.
Discussion
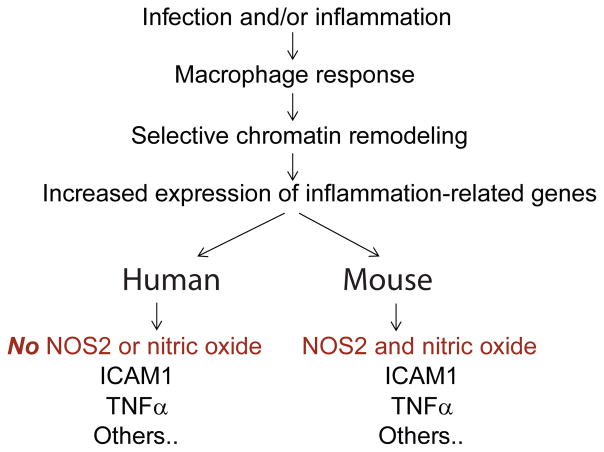
The major conclusion of our study is that human macrophages do not produce NO in response to acute inflammatory stimuli as a result of epigenetic regulation of the NOS2 gene (Figure 8). This conclusion is based on several complementary lines of experimental evidence. First, we demonstrate that despite an otherwise intact inflammatory response, human alveolar macrophages do not make NO or increase expression of NOS2 when stimulated. Next, we demonstrate uniform methylation of CpGs in the NOS2 promoter region in human macrophages. Consistent with the CpG methylation data, ChIP assays demonstrate that the human macrophage NOS2 gene is associated with a silencing histone mark and has tightly compacted chromatin rendering it inaccessible to transcription factors. Stimulation of human macrophages with LPS and IFNγ does not alter the NOS2 the ChIP assay results or chromatin accessibility. Finally, we show that this epigenetic silencing of the NOS2 region in human macrophages is resistant to the effects of demethylating agents both in circulating leukocytes in vivo and in a human macrophage cell line in vitro. Collectively, this experimental evidence strongly supports the overall concept that human macrophages, including primary alveolar macrophages, do not produce NO in response to inflammatory stimuli because of epigenetic silencing of the NOS2 gene.
Figure 8.
Diagram of study demonstrating effect of NOS2 silencing on inflammatory response in human macrophages compared to mouse macrophages.
Our human findings stand in stark contrast to the physiology of normal mouse macrophages. We find that in mouse macrophages the CpG motifs proximal to the NOS2 TSS are unmethylated. Furthermore, murine macrophage NOS2 segregates with “open” genes on chromatin accessibility assay and ChIP analysis reveals murine NOS2 is associated with H3K4me3, a histone mark that correlates with active transcription. Thus, it appears that the regulation of NO as an inducible inflammatory response in the mouse mononuclear phagocyte does not extend to human cells.
Our studies are consistent with a larger body of work on the overall differences in inflammation-related gene expression between mice and men (45). The literature is full of conflicting reports on the role of NO in human inflammation. Schneeman et al performed a comprehensive analysis of human monocytes in 1993 (11). They found that human mononuclear phagocytes exposed to an array of potent stimuli did not produce nitrite, consume L-arginine, produce L-citrulline or display NOS activity. Weinberg et al, found minimal immunoreactive NOS2 protein or NOS2 mRNA in human macrophages stimulated for 3 days in culture; secreted NO was not detected (46). This is similar to our studies demonstrating neither induction of NOS2 mRNA or protein with inflammatory stimuli. Any subtle discrepancies likely reflect differences in intensity of the stimulation used and/or purity of cell populations studied.
The actual role of NO in human lung disease remains incompletely defined. Exhaled NO is a marker of active inflammation in asthma, sarcoidosis and ARDS (18, 47–50). Whether NO is beneficial or harmful is unclear and the cells of origin not certain (51–55). There is evidence showing NO production in more chronic human lung diseases, such as granulomatous infections and pulmonary fibrosis (15–18). Immunoreactive NOS2 has been demonstrated in chronic granulomas, though absent in nodal or tissue mononuclear phagocytes (75). This suggests that in the appropriate chronic inflammatory context, epithelioid granulomas may uniquely acquire the ability to express functional NO. This does not, however, appear to be an important aspect of normal human macrophage physiology. Despite this defect in acute NO production, human alveolar macrophages have other mechanisms for effective pathogen killing (56, 57). This seems to stand in contrast with the purported central importance of high-level macrophage NO production in mouse lung defense (58, 59). Furthermore, there are conflicting data regarding the consequences of blocking NO production in murine models of acute lung injury and macrophage-dependent pathogen infection (5, 7). What these findings demonstrate is that we do not yet fully understand the role of NO in lung cell physiology across species. Identifying the molecular basis for this variant inflammatory response may yield important cross-species insights (9, 11, 60, 61).
This is the first study to explore epigenetic silencing of the NOS2 gene in human macrophages and to compare it with mouse macrophages. Our conclusion that differential CpG methylation contributes to decreased NO response in human macrophages is supported by a study in endothelial cells where methylation of the NOS2 core promoter areas contributed to decreased induction of the NOS2 gene in human endothelial cells (43). Extending this observation, we find that differential methylation upstream of the NOS2 TSS correlates with concurrent differences in chromatin accessibility and histone modifications that predict gene silencing. Human macrophages do not produce NO constitutively or in response to acute inflammatory signals because the NOS2 gene is epigenetically switched off.
Methylation of CpG motifs is one of several universal epigenetic mechanisms that control gene expression in both plants and animals (62–64). The preponderance of data linking promoter area CpG methylation and gene expression comes from studies on genes with CpG islands (an area longer than 500 bp with an observed CpG/expected CpG ratio ≥ 0.65) at the promoter region (65). While there is no CpG island in the NOS2 promoter, we have identified a number of CpGs proximal to the TSS that are differentially methylated between mouse and human macrophages. Recent literature supports the importance of these TSS-proximal, non-island, CpGs in regulation of gene expression (66–70). These particular studies identified the role of CpG methylation (proximal to the TSS without being in an island) in transcription of the IL-2, IFNγ, MMP13, IL1β and TNFα genes.
DNA methylation depends on an array of methyltransferases, enzymes capable of de novo methylation and demethylation of genomic DNA, along with maintenance of methylation in daughter cells (71, 72). Human and mouse macrophages express DNA methyltransferases DNMT1, 3A, and 3B that can be inhibited with 5-azacytidine with resultant reduced methylation in replicating daughter cells. Our efforts to demethylate the NOS2 TSS met with only modest success. In murine cells that already had many demethylated CpG motifs at baseline, in vitro inhibition of DNA methyltransferase was successful. Human cells, both in vivo and in vitro, proved more resistant to the demethylation agent. In some melanoma patients who received a DNA methylation inhibitor on an experimental treatment protocol a rise in blood HbF levels signaled demethylation at the bone marrow erythroblast level. The dose or duration of chemotherapeutic agents may not have been inadequate to demethylate the NOS locus in circulating leukocytes or the NOS2 gene may be more resistant to such manipulation. Toxicity of the DNA methyltransferase inhibitors can impair cell viability in vitro and demethylation agents work best on rapidly dividing cells. Our work is consistent with previous reports in which 5-aza-dC and TSA failed to alter DNA structure in tissue culture (73–75).
In this study we demonstrate silencing at the NOS2 gene at a number of levels: 1. CpG motifs proximal to the human TSS are methylated. 2. Histone modifications consistent with gene silencing are found at the NOS2 gene (H3K27me3). 3. Histone modifications consistent with active gene activity (H3K4me3) are in low abundance at the NOS2 promoter region. 4. The NOS2 locus is relatively protected from DNAse treatment suggesting closed chromatin. Our conclusion from these studies is that the NOS2 gene in human macrophages is silenced. Alternatively, production from this locus may be below the sensitivity of present measurement techniques or there may be transcription of a very unstable transcript. At this time, we can’t definitively rule out these possibilities, but consider them unlikely.
There are a number of potential weaknesses in this study. One of these is the lack of data showing that with demethylation of the NOS2 proximal CpGs, NOS2 is transcribed. Another weakness is the limited number of human cell types that are analyzed. It would be nice to know if the silencing is true for macrophages alone or is also true in other cell types such as airway epithelial cells. For the patient studies, this is only preliminary data and future trials with more patients and higher does of the DNMT and HDAC inhibitors may be more fruitful.
In future studies, we will be examining more cell types for silencing of the NOS2 locus. We will be asking whether this locus is dynamically regulated and further testing whether demethylation with a DNMT inhibitor is possible. This project was started because of our interest in human inflammation and its links to both chronic and acute diseases. There are some papers suggesting that mycobacterial infection generates nitric oxide from human macrophages (76, 77). There are also suggestions that nitric oxide plays an important role in COPD, ARDS, sarcoidosis, asthma and viral infections (78–82). In light of the present work, it will be important to determine the conditions under which macrophage NOS2 expression might change and how that might affect disease outcome.
Our findings demonstrate that the susceptibility to epigenetic modification is gene specific and may be predictable from methylation sequencing analysis. Future clinical trials utilizing treatments aimed at demethylation and/or histone modifications should consider the susceptibility of the target gene(s) of interest. In this study, we show that human macrophages, including primary alveolar macrophages, do not make NO at baseline or in response to acute inflammatory stimuli. The lack of NOS2 expression and subsequent NO secretion is explained by epigenetic changes that are consistent with silencing of the NOS2 transcription locus. We believe this study points out the pitfalls of cross species extrapolation in understanding inflammatory mechanisms. Future studies exploring how human macrophages kill pathogens and respond to inflammation should focus on alternative pathways to NOS2 and NO. To do otherwise is to risk having the best laid schemes of mice and men go awry (with apologies to Robert Burns).
Supplementary Material
Acknowledgments
We thank Richard Starr for technical assistance and Kathy Keck for aid in obtaining bone marrow derived macrophages from mice.
Footnotes
This project was supported by NIH R01 HL079901, NIH RO1 HL096625 and R21HL109589 to M. M. and by the National Institute for Environmental Health Sciences through the University of Iowa Environmental Health Sciences Research Center, NIEHS/NIH P30 ES005605 and Grant Number UL1RR024979 from the National Center for Research Resources (NCRR), a part of the National Institutes of Health (NIH).
The abbreviations used are: 5-aza-dC, 5-aza-2′deoxycytidine; BMDMs, bone marrow-derived macrophages; ChIP, Chromatin immunoprecipitation; CpG, cytosine-phosphate-guanine dinucleotides; FBS, fetal bovine serum; FEV1, Forced expiratory volume in 1 second; FVC, forced vital capacity; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; H3K27me3, trimethyl-histone H3 (Lys27); H3K4me3, trimethyl-histone H3 (Lys4); HBB, β-globin gene; HbF, hemoglobin F; HDAC, histone deacetylase; ICAM1, intercellular adhesion molecule 1; IFNγ, interferon gamma; LPS, lipopolysaccharide; MHS, murine alveolar macrophage cell line; NOS2, inducible nitric oxide synthase; NO, nitric oxide; PBS, phosphate buffered saline; RAW 264.7, murine peritoneal macrophage cell line; RPL30, 60S ribosomal protein L30; THP-1, human monocytic leukemia cell line; TNFα, tumor necrosis factor α; TSS, transcription start site.
References
- 1.Rubins JB. Alveolar Macrophages: Wielding the Double-Edged Sword of Inflammation. Am J Respir Crit Care Med. 2003;167:103–104. doi: 10.1164/rccm.2210007. [DOI] [PubMed] [Google Scholar]
- 2.Sibille Y, Reynolds HY. Macrophages and polymorphonuclear neutrophils in lung defense and injury. Am Rev Respir Dis. 1990;141:471–501. doi: 10.1164/ajrccm/141.2.471. [DOI] [PubMed] [Google Scholar]
- 3.Hanano R, Kaufmann SH. Nitric oxide production and mycobacterial growth inhibition by murine alveolar macrophages: the sequence of rIFN-gamma stimulation and Mycobacterium bovis BCG infection determines macrophage activation. Immunol Lett. 1995;45:23–27. doi: 10.1016/0165-2478(94)00193-u. [DOI] [PubMed] [Google Scholar]
- 4.Miles PR, Bowman L, Rengasamy A, Huffman L. Constitutive nitric oxide production by rat alveolar macrophages. American Journal of Physiology - Lung Cellular and Molecular Physiology. 1998;274:L360–L368. doi: 10.1152/ajplung.1998.274.3.L360. [DOI] [PubMed] [Google Scholar]
- 5.Farley KS, Wang LF, Razavi HM, Law C, Rohan M, McCormack DG, Mehta S. Effects of macrophage inducible nitric oxide synthase in murine septic lung injury. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2006;290:L1164–L1172. doi: 10.1152/ajplung.00248.2005. [DOI] [PubMed] [Google Scholar]
- 6.Skerrett SJ, Martin TR. Roles for tumor necrosis factor alpha and nitric oxide in resistance of rat alveolar macrophages to Legionella pneumophila. Infect Immun. 1996;64:3236–3243. doi: 10.1128/iai.64.8.3236-3243.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacMicking JD, Nathan C, Hom G, Chartrain N, Fletcher DS, Trumbauer M, Stevens K, Xie QW, Sokol K, Hutchinson N, et al. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell. 1995;81:641–650. doi: 10.1016/0092-8674(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 8.Anthony LS, Morrissey PJ, Nano FE. Growth inhibition of Francisella tularensis live vaccine strain by IFN-gamma-activated macrophages is mediated by reactive nitrogen intermediates derived from L-arginine metabolism. J Immunol. 1992;148:1829–1834. [PubMed] [Google Scholar]
- 9.Albina JE. On the expression of nitric oxide synthase by human macrophages. Why no NO? J Leukoc Biol. 1995;58:643–649. doi: 10.1002/jlb.58.6.643. [DOI] [PubMed] [Google Scholar]
- 10.Jungi TW, Adler H, Adler B, Thony M, Krampe M, Peterhans E. Inducible nitric oxide synthase of macrophages. Present knowledge and evidence for species-specific regulation. Vet Immunol Immunopathol. 1996;54:323–330. doi: 10.1016/s0165-2427(96)05690-5. [DOI] [PubMed] [Google Scholar]
- 11.Schneemann M, Schoedon G, Hofer S, Blau N, Guerrero L, Schaffner A. Nitric oxide synthase is not a constituent of the antimicrobial armature of human mononuclear phagocytes. J Infect Dis. 1993;167:1358–1363. doi: 10.1093/infdis/167.6.1358. [DOI] [PubMed] [Google Scholar]
- 12.Denis M. Human monocytes/macrophages: NO or no NO? J Leukoc Biol. 1994;55:682–684. doi: 10.1002/jlb.55.5.682. [DOI] [PubMed] [Google Scholar]
- 13.Amin AR, Attur M, Vyas P, Leszczynska-Piziak J, Levartovsky D, Rediske J, Clancy RM, Vora KA, Abramson SB. Expression of nitric oxide synthase in human peripheral blood mononuclear cells and neutrophils. J Inflamm. 1995;47:190–205. [PubMed] [Google Scholar]
- 14.Bermudez LE. Differential mechanisms of intracellular killing of Mycobacterium avium and Listeria monocytogenes by activated human and murine macrophages. The role of nitric oxide. Clin Exp Immunol. 1993;91:277–281. doi: 10.1111/j.1365-2249.1993.tb05895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson S, Bonecini-Almeida MdG, Lapa e Silva JR, Nathan C, Xie QW, Mumford R, Weidner JR, Calaycay J, Geng J, Boechat N, Linhares C, Rom W, Ho JL. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. The Journal of Experimental Medicine. 1996;183:2293–2302. doi: 10.1084/jem.183.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nozaki Y, Hasegawa Y, Ichiyama S, Nakashima I, Shimokata K. Mechanism of nitric oxide-dependent killing of Mycobacterium bovis BCG in human alveolar macrophages. Infect Immun. 1997;65:3644–3647. doi: 10.1128/iai.65.9.3644-3647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rich EA, Torres M, Sada E, Finegan CK, Hamilton BD, Toossi Z. Mycobacterium tuberculosis (MTB)- stimulated production of nitric oxide by human alveolar macrophages and relationship of nitric oxide production to growth inhibition of MTB. Tuber Lung Dis. 1997;78:247–255. doi: 10.1016/s0962-8479(97)90005-8. [DOI] [PubMed] [Google Scholar]
- 18.Saleh D, Barnes PJ, Giaid A. Increased production of the potent oxidant peroxynitrite in the lungs of patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 1997;155:1763–1769. doi: 10.1164/ajrccm.155.5.9154889. [DOI] [PubMed] [Google Scholar]
- 19.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 21.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic Silencing of MicroRNA-34b/c and B-Cell Translocation Gene 4 Is Associated with CpG Island Methylation in Colorectal Cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar_dominance. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razin A. CpG methylation, chromatin structure and gene silencing[mdash]a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 25.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 26.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 28.Belinsky SA. Silencing of genes by promoter hypermethylation: key event in rodent and human lung cancer. Carcinogenesis. 2005;26:1481–1487. doi: 10.1093/carcin/bgi020. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar S, Abujamra AL, Loew JE, Forman LW, Perrine SP, Faller DV. Histone deacetylase inhibitors reverse CpG methylation by regulating DNMT1 through ERK signaling. Anticancer Res. 2011;31:2723–2732. [PubMed] [Google Scholar]
- 30.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 31.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The Methyl-CpG-binding Protein MeCP2 Links DNA Methylation to Histone Methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 32.Ghoshal K, Datta J, Majumder S, Bai S, Dong X, Parthun M, Jacob ST. Inhibitors of Histone Deacetylase and DNA Methyltransferase Synergistically Activate the Methylated Metallothionein I Promoter by Activating the Transcription Factor MTF-1 and Forming an Open Chromatin Structure. Mol Cell Biol. 2002;22:8302–8319. doi: 10.1128/MCB.22.23.8302-8319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pryzbylkowski P, Obajimi O, Keen JC. Trichostatin A and 5 Aza-2′ deoxycytidine decrease estrogen receptor mRNA stability in ER positive MCF7 cells through modulation of HuR. Breast Cancer Res Treat. 2008;111:15–25. doi: 10.1007/s10549-007-9751-0. [DOI] [PubMed] [Google Scholar]
- 34.Chai G, Li L, Zhou W, Wu L, Zhao Y, Wang D, Lu S, Yu Y, Wang H, McNutt MA, Hu YG, Chen Y, Yang Y, Wu X, Otterson GA, Zhu WG. HDAC Inhibitors Act with 5-aza-2_-Deoxycytidine to Inhibit Cell Proliferation by Suppressing Removal of Incorporated Abases in Lung Cancer Cells. PLoS One. 2008;3:e2445. doi: 10.1371/journal.pone.0002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flaherty DM, Monick MM, Hinde SL. Human Alveolar Macrophages Are Deficient in PTEN: THE ROLE OF ENDOGENOUS OXIDANTS. J Biol Chem. 2006;281:5058–5064. doi: 10.1074/jbc.M508997200. [DOI] [PubMed] [Google Scholar]
- 36.Tsikas D. Analysis of nitrite and nitrate in biological fluids by assays based on the Griess reaction: appraisal of the Griess reaction in the L-arginine/nitric oxide area of research. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:51–70. doi: 10.1016/j.jchromb.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 37.Kovarik P, Stoiber D, Novy M, Decker T. Stat1 combines signals derived from IFN-[gamma] and LPS receptors during macrophage activation. EMBO J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dustin ML, Rothlein R, Bhan AK, Dinarello CA, Springer TA. Induction by IL 1 and interferon-gamma: tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1) The Journal of Immunology. 1986;137:245–254. [PubMed] [Google Scholar]
- 39.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 41.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan GC, Fish JE, Mawji IA, Leung DD, Rachlis AC, Marsden PA. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J Immunol. 2005;175:3846–3861. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 44.Saunthararajah Y, Hillery CA, Lavelle D, Molokie R, Dorn L, Bressler L, Gavazova S, Chen YH, Hoffman R, DeSimone J. Effects of 5-aza-2′-deoxycytidine on fetal hemoglobin levels, red cell adhesion, and hematopoietic differentiation in patients with sickle cell disease. Blood. 2003;102:3865–3870. doi: 10.1182/blood-2003-05-1738. [DOI] [PubMed] [Google Scholar]
- 45.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinberg JB, Misukonis MA, Shami PJ, Mason SN, Sauls DL, Dittman WA, Wood ER, Smith GK, McDonald B, Bachus KE, et al. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS): analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 1995;86:1184–1195. [PubMed] [Google Scholar]
- 47.BRETT SJ, EVANS TW. Measurement of Endogenous Nitric Oxide in the Lungs of Patients with the Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med. 1998;157:993–997. doi: 10.1164/ajrccm.157.3.9705060. [DOI] [PubMed] [Google Scholar]
- 48.Smith AD, Cowan JO, Brassett KP, Herbison GP, Taylor DR. Use of Exhaled Nitric Oxide Measurements to Guide Treatment in Chronic Asthma. N Engl J Med. 2005;352:2163–2173. doi: 10.1056/NEJMoa043596. [DOI] [PubMed] [Google Scholar]
- 49.Moodley Y, Chetty R, Lalloo U. Nitric oxide levels in exhaled air and inducible nitric oxide synthase immunolocalization in pulmonary sarcoidosis. Eur Respir J. 1999;14:822–827. doi: 10.1034/j.1399-3003.1999.14d17.x. [DOI] [PubMed] [Google Scholar]
- 50.Maziak W, Loukides S, Culpitt S, Sullivan P, Kharitonov SA, Barnes PJ. Exhaled nitric oxide in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;157:998–1002. doi: 10.1164/ajrccm.157.3.97-05009. [DOI] [PubMed] [Google Scholar]
- 51.Yang GG, Hsu YH. Nitric oxide production and immunoglobulin deposition in leptospiral hemorrhagic respiratory failure. J Formos Med Assoc. 2005;104:759–763. [PubMed] [Google Scholar]
- 52.KRISTOF AS, GOLDBERG P, AUBACH VL, HUSSAIN SNA. Role of Inducible Nitric Oxide Synthase in Endotoxin-induced Acute Lung Injury. Am J Respir Crit Care Med. 1998;158:1883–1889. doi: 10.1164/ajrccm.158.6.9802100. [DOI] [PubMed] [Google Scholar]
- 53.Lang JD, Chumley P, Eiserich JP, Estevez A, Bamberg T, Adhami A, Crow J, Freeman BA. Hypercapnia induces injury to alveolar epithelial cells via a nitric oxide-dependent pathway. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2000;279:L994–L1002. doi: 10.1152/ajplung.2000.279.5.L994. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi H, Hataishi R, Mitsufuji H, Tanaka M, Jacobson M, Tomita T, Zapol WM, Jones RC. Antiinflammatory properties of inducible nitric oxide synthase in acute hyperoxic lung injury. Am J Respir Cell Mol Biol. 2001;24:390–397. doi: 10.1165/ajrcmb.24.4.4218. [DOI] [PubMed] [Google Scholar]
- 55.Kubes P, Suzuki M, Granger DN. Nitric oxide: an endogenous modulator of leukocyte adhesion. Proceedings of the National Academy of Sciences. 1991;88:4651–4655. doi: 10.1073/pnas.88.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aston C, Rom WN, Talbot AT, Reibman J. Early inhibition of mycobacterial growth by human alveolar macrophages is not due to nitric oxide. Am J Respir Crit Care Med. 1998;157:1943–1950. doi: 10.1164/ajrccm.157.6.9705028. [DOI] [PubMed] [Google Scholar]
- 57.Hickman-Davis JM, O’Reilly P, Davis IC, Peti-Peterdi J, Davis G, Young KR, Devlin RB, Matalon S. Killing of Klebsiella pneumoniae by human alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2002;282:L944–956. doi: 10.1152/ajplung.00216.2001. [DOI] [PubMed] [Google Scholar]
- 58.Weinberg JB. Nitric oxide production and nitric oxide synthase type 2 expression by human mononuclear phagocytes: a review. Mol Med. 1998;4:557–591. doi: 10.1007/BF03401758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bukrinsky MI, Nottet HS, Schmidtmayerova H, Dubrovsky L, Flanagan CR, Mullins ME, Lipton SA, Gendelman HE. Regulation of nitric oxide synthase activity in human immunodeficiency virus type 1 (HIV-1)-infected monocytes: implications for HIV-associated neurological disease. J Exp Med. 1995;181:735–745. doi: 10.1084/jem.181.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneemann M, Schoeden G. Macrophage biology and immunology: man is not a mouse. J Leukoc Biol. 2007;81:579. doi: 10.1189/jlb.1106702. [DOI] [PubMed] [Google Scholar]
- 61.Schneemann M, Schoedon G. Species differences in macrophage NO production are important. Nat Immunol. 2002;3:102. doi: 10.1038/ni0202-102a. [DOI] [PubMed] [Google Scholar]
- 62.Saze H, Scheid OM, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 63.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 64.Jones PA, Takai D. The Role of DNA Methylation in Mammalian Epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 65.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hashimoto K, Otero M, Imagawa K, de Andres MC, Coico JM, Roach HI, Oreffo RO, Marcu KB, Goldring MB. Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1beta (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J Biol Chem. 2013;288:10061–10072. doi: 10.1074/jbc.M112.421156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE. G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. J Biol Chem. 2008;283:32198–32208. doi: 10.1074/jbc.M803446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berkley AM, Hendricks DW, Simmons KB, Fink PJ. Recent thymic emigrants and mature naive T cells exhibit differential DNA methylation at key cytokine loci. J Immunol. 2013;190:6180–6186. doi: 10.4049/jimmunol.1300181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177:1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 70.Williams CL, Schilling MM, Cho SH, Lee K, Wei Aditi M, Boothby M. STAT4 and T-bet are required for the plasticity of IFN-gamma expression across Th2 ontogeny and influence changes in Ifng promoter DNA methylation. J Immunol. 2013;191:678–687. doi: 10.4049/jimmunol.1203360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 72.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Narayan G, Goparaju C, Arias-Pulido H, Kaufmann A, Schneider A, Durst M, Mansukhani M, Pothuri B, Murty V. Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. Mol Cancer. 2006;5:16. doi: 10.1186/1476-4598-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo DL, Zhang J, Yuen ST, Tsui WY, Chan ASY, Ho C, Ji J, Leung SY, Chen X. Reduced expression of EphB2 that parallels invasion and metastasis in colorectal tumours. Carcinogenesis. 2006;27:454–464. doi: 10.1093/carcin/bgi259. [DOI] [PubMed] [Google Scholar]
- 75.Chan GC, Fish JE, Mawji IA, Leung DD, Rachlis AC, Marsden PA. Epigenetic Basis for the Transcriptional Hyporesponsiveness of the Human Inducible Nitric Oxide Synthase Gene in Vascular Endothelial Cells. The Journal of Immunology. 2005;175:3846–3861. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 76.Jung JY, Madan-Lala R, Georgieva M, Rengarajan J, Sohaskey CD, Bange FC, Robinson CM. The intracellular environment of human macrophages that produce nitric oxide promotes growth of mycobacteria. Infect Immun. 2013;81:3198–3209. doi: 10.1128/IAI.00611-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mattila JT, Ojo OO, Kepka-Lenhart D, Marino S, Kim JH, Eum SY, Via LE, Barry CE, 3rd, Klein E, Kirschner DE, Morris SM, Jr, Lin PL, Flynn JL. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J Immunol. 2013;191:773–784. doi: 10.4049/jimmunol.1300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brett SJ, Evans TW. Measurement of endogenous nitric oxide in the lungs of patients with the acute respiratory distress syndrome. Am J Respir Crit Care Med. 1998;157:993–997. doi: 10.1164/ajrccm.157.3.9705060. [DOI] [PubMed] [Google Scholar]
- 79.Brindicci C, Kharitonov SA, Ito M, Elliott MW, Hogg JC, Barnes PJ, Ito K. Nitric oxide synthase isoenzyme expression and activity in peripheral lung tissue of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;181:21–30. doi: 10.1164/rccm.200904-0493OC. [DOI] [PubMed] [Google Scholar]
- 80.Moodley YP, Chetty R, Lalloo UG. Nitric oxide levels in exhaled air and inducible nitric oxide synthase immunolocalization in pulmonary sarcoidosis. Eur Respir J. 1999;14:822–827. doi: 10.1034/j.1399-3003.1999.14d17.x. [DOI] [PubMed] [Google Scholar]
- 81.Raza MW, Essery SD, Weir DM, Ogilvie MM, Elton RA, Blackwell CC. Infection with respiratory syncytial virus and water-soluble components of cigarette smoke alter production of tumour necrosis factor alpha and nitric oxide by human blood monocytes. FEMS Immunol Med Microbiol. 1999;24:387–394. doi: 10.1111/j.1574-695X.1999.tb01310.x. [DOI] [PubMed] [Google Scholar]
- 82.Smith AD, Cowan JO, Brassett KP, Herbison GP, Taylor DR. Use of exhaled nitric oxide measurements to guide treatment in chronic asthma. N Engl J Med. 2005;352:2163–2173. doi: 10.1056/NEJMoa043596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.