SUMMARY
Nature combines existing biochemical building blocks, at times with subtlety of purpose. RNA modifications are a prime example of this, where standard RNA nucleosides are decorated with chemical groups and building blocks that we recall from our basic biochemistry lectures. The result: a wealth of chemical diversity whose full biological relevance has remained elusive despite being public knowledge for some time. Here, we will highlight a number of modifications that, because of their chemical intricacy, rely on seemingly unrelated pathways to provide co-factors for their synthesis. Besides their immediate role in affecting RNA function, modifications may act as sensors and transducers of information that connect a cell's metabolic state to its translational output, carefully orchestrating a delicate balance between metabolic rate and protein synthesis at a system's level.
INTRODUCTION
Basic metabolic pathways use ubiquitous metabolites and coenzymes to transfer methyl groups, acetyl groups, aminoacids, isoprenoids, sugars, phosphate and the like. Many metabolite–RNA conjugates are known, or likely, to enhance the chemical functionality of their targets (Figure 1). Despite significant advances, technical bottlenecks have throttled progress in the modification field, including: (i) the lack of sensitive analytic tools for the chemical or physicochemical identification and detection of modified nucleotides in limiting sample amount and (ii) the absence of focused interest, by bioorganic chemists, to synthesize and generate authentic standards for the selective detection of existing as well as newly discovered modifications. However, recent developments in mass spectrometry and nucleoside chemistry have allowed more sensitive detection and quantification (Li and Limbach, 2012). Concomitantly, interest has resurfaced and connections are being made between tRNA modifications, a cell's overall stress response, cell development and its protein synthesis capacity (Chan et al., 2010; Chan et al., 2012).
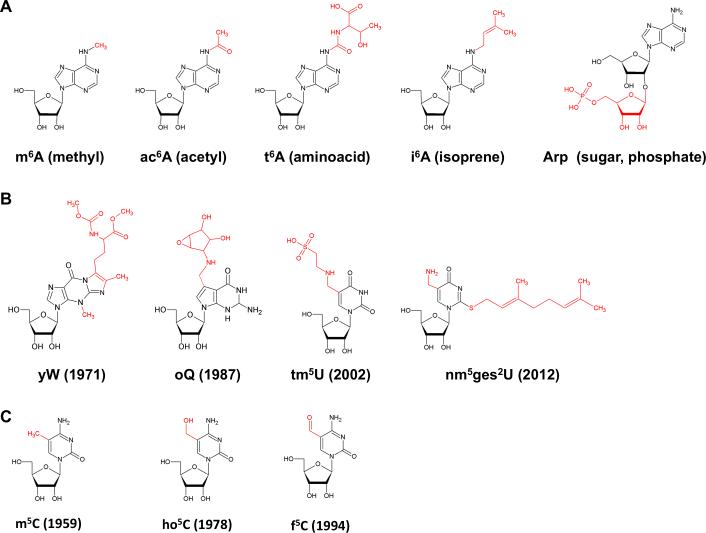
Figure 1. Selected Modified Ribonucleosides.
(A) Adenosine modifications resulting from conjugation to ubiquitous electrophilic metabolites. (B) Chemically sophisticated hypermodifications discovered throughout the past five decades. The year of publication is given in brackets. (C) C5-modified cytidines related to so call epigenetic DNA modifications recently discovered. m6A, 6-methyladenosine; ac6A, 6-acetyladenosine; t6A, 6-threonyladenosine; i6A, 6-isopentenyladenosine; Arp, 2’-O-ribosyladenosine phosphate; yW, wybutosine; oQ, epoxyqueuosine; tm5U, 5-taurinomethyluridine; nm5ges2U, 5-aminomethyl-2-geranyluridine; 5-methyluridine; ho5C, 5-hydroxycytidine; f5C, 5-formyluridine.
Renewed interest has also been driven by the advent of methods for transcriptome-wide detection of simple RNA modifications, for example the case of 6-methyladenosine (m6A) (Dominissini et al., 2012; Meyer et al., 2012), 5-methylcytosine (m5C) (Squires et al., 2012) and inosine (I) (Sakurai et al., 2010). Significant crosstalk from the DNA field has also promoted interest in RNA modifications, mainly arising from “newly discovered” DNA modifications that deservedly received enormous attention in the field of epigenetics (Calo and Wysocka, 2013).
In this review, we will highlight modifications that depend on building blocks from a number of interconnected metabolic pathways and as such may help coordinate protein synthesis and metabolism. Possible relationships have been established between tRNA modifications and environmental changes. Here we emphasize that, although such connections have been made under conditions of environmental stress (i.e. oxidative stress), these may not be limited to environmental extremes and could be part of a cells normal program of growth.
Combinatorial modifications and modification cascades
With increasingly complex modifications also comes sophisticated chemistry, presenting a synthetic challenge for the organic chemist. Nature, as usual, has elegantly solved chemical complications by breaking down the task into a series of simple steps, each catalyzed by specific enzymes. It is now clear that the overwhelming chemical diversity, found in nucleoside modifications, is actually created by versatile combinations of a limited sub-set of chemical reactions, e.g. group transfer reactions.
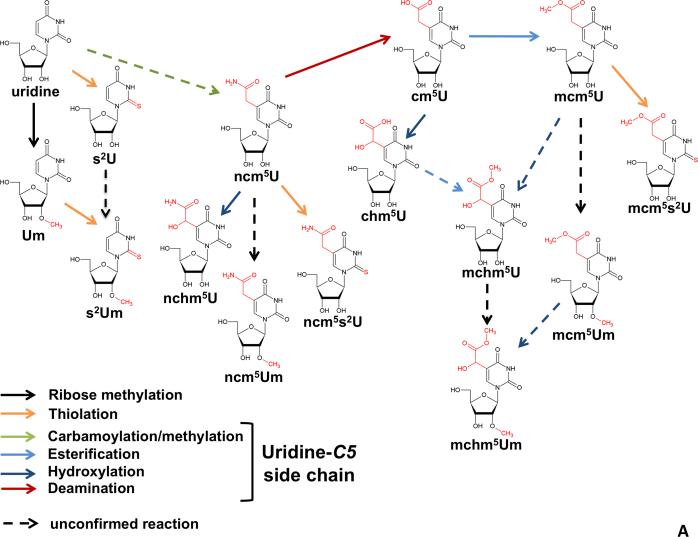
In most group-transfer reactions, various nucleophilic sites in RNA are modified by activated variegated electrophiles that are part of the cell's typical metabolic repertoire, and as such have co-evolved with specific enzymes and catalytic motifs. The range of chemical reactions is not restricted to simple nucleophilic substitution, but extends to oxidative reactions and CH-activation by transition metals, thiolation, selenation, amidation, esterification, as well as Michael, Schiff and Mannich chemistry. This has led to modification diversity, where some modifications take place at several sites of a given standard nucleoside in a combinatorial fashion, with little cross talk between the progress at two given sites and each pathway proceeding independently of each other. For example, two pathways operate rather independently at positions 2 and 5 of the uracil ring (Figure 2A). Thiolation at position 2 may precede or follow the synthesis of the side chain at position 5. As a result, these “orthogonal” transformations are interchangeable in their order of occurrence. Thiolation may also be followed by further modifications at the 2 position, e.g. geranylation. Methylation at the ribose 2’OH position also occurs at several uridine derivatives, further compounding the variety of generated permutations. Among these, and regardless of the actual reaction mechanism, which we will discuss below, C5-substituted uridines are the largest contributors to the hitherto described chemical diversity (Machnicka et al., 2013).
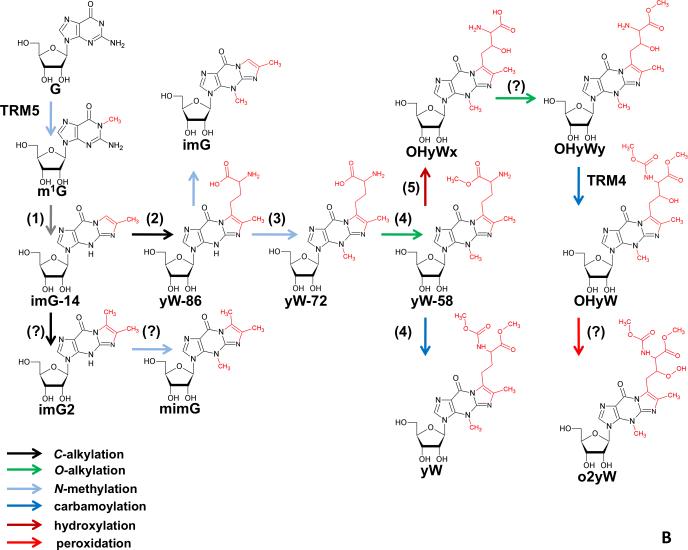
Figure 2. Hypermodification Pathways.
(A) Eukaryotic modification network of uridines at the wobble position 34. The weave results from the overlay of independently operating modification pathways, which modify at positions 2’-O of the ribose, C5 of the uridines, or O2 of the uridines. The lack of a fixed order of action generates a multitude of permutations. (B) Biosynthesis pathway of wybutosine derivatives. Note that transformations occur in a defined serial manner. Since branching pathways do not reunite, the pathway by which a given modification is generated is unambiguous. Numbers in parentheses refer to the TYW1-5 enzymes involved in each step and referred to in the main text. Question marks (?) indicate reactions for which the given enzyme is not known.
In contrast to the partially “random” order of uridine modification (Figure 2A), other hypermodifications are executed in precise order, since the reaction steps, rather than being orthogonal, directly build on one another. For example, the biosynthesis of wybutosine (Figure 2B) which, in clear contrast to the wobble uridine pathways, splits up into several branches, but there is no reunification of any two branches, meaning that a substrate is committed to a defined sequence of modification steps after entering a branch. Thus, modifications can occur independently of each other between sites or be part of cascades at individual sites or even at different sites.
Fancy chemistry at the business end: the tRNA anticodon
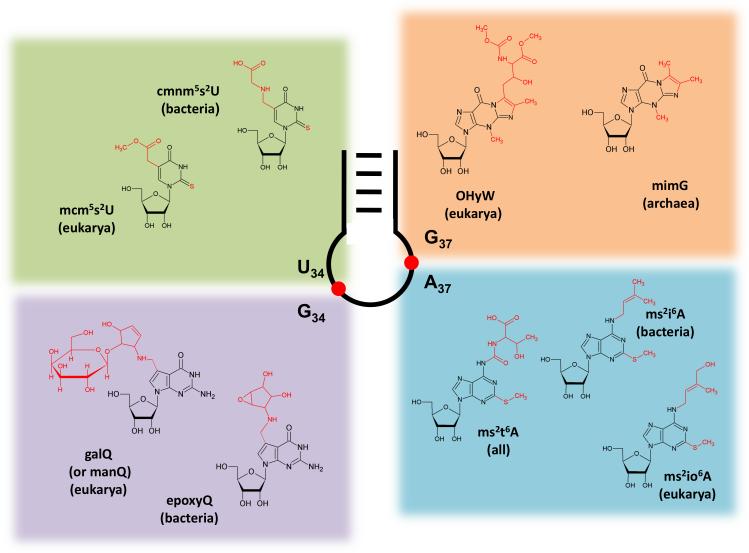
By far the greatest diversity of hyper-modified nucleotides occurs at positions 34 and 37 of the anticodon of tRNAs. Modifications at these positions presumably evolved both to enhance base-pairing flexibility during wobble decoding as needed (via position 34) and to ensure reading frame maintenance (by modifications at position 37) (El Yacoubi et al., 2012; Gustilo et al., 2008). The key roles played by anticodon modifications can be highlighted here by examples of enzymatic machineries that generate four well-studied anticodon modifications: 1) C5 methyluridine derivatives and queuosine (Q) at the wobble nucleotide (first position of the anticodon); 2) wybutosine derivatives and hyper-modified adenosines neighboring the anticodon (position 37), (Figure 4).
Figure 4. Hypermodifications at Positions 34 and 37 in the Anticodon Loop.
Positions 34 and 37 of the anticodon loop undergo by far the largest diversity of post-transcriptional modifications. Highlighted are modified uridines (upper left panel) and guanosines (lower left panel); ubiquitous hypermodifications ensuring correct decoding at the wobble position. Sophisticated purine modifications found at position 37 (upper and lower right panels) play roles in reading frame maintenance.
Key modifications at a tRNA's corporate headquarter: methyluridine derivatives and queuosine at position 34
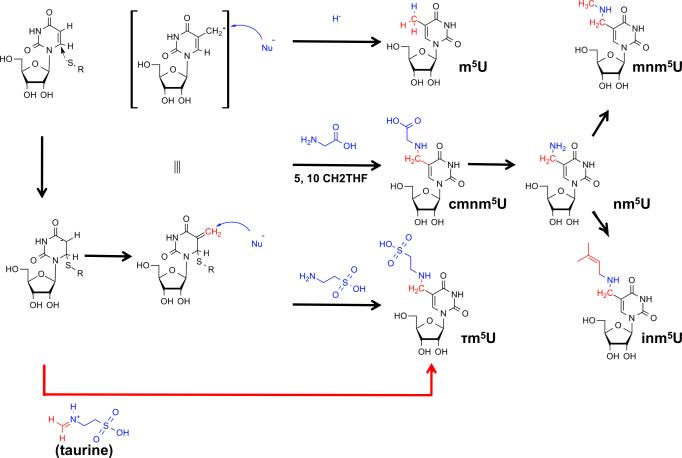
Together, the U34 modifications tend to increase the stability of the tRNA-mRNA interaction during decoding by increasing anticodon rigidity and serve as determinants of amino acylation, translation efficiency, and fidelity (Gustilo et al., 2008). As mentioned, modifications of uridine 34 (first anticodon position), may start with thiolation of O2, or with alkylation at C5 (Figure 2A). In Bacteria, the end product is either 5-carboxymethylaminouridine (cmnm5U) or 5-methylaminomethyluridine (mnm5U). In mitochondria, owing to their bacterial ancestry, similar modifications occur, mammals representing a notable exception where the final product involves the incorporation of the amino acid taurine to form 5-taurinomethyluridine (τm5U) (Suzuki et al., 2001) (Figure 1B). In eukaryotes, different versions of these modifications are then obtained by further 2’-O-methylation and/or 2-sulfurylation to xm5s2Um (where x indicates any hypermodification of m5U, for example mnm5U, Figure 2A) (Armengod et al., 2012). Early studies in Bacteria implicated the involvement of a heterotetrameric enzyme complex formed by the proteins TRME (mnmE) and GidA in C5 modifications (Bregeon et al., 2001). Mutation of any of these genes leads to the complete disappearance of the C-5 modifications. The crystal structure of TRME, a GTP-binding protein, suggested that it could utilize 5-formyltetrathydrofolate, thus a C1-moiety at the oxidation state +II, (5-CHO-THF, corresponding to an equivalent of formic acid) as the source of the C-1 moiety necessary for the first step of the reaction (Scrima et al., 2005). However, several studies, including a biochemical reconstitution of the first step of the reaction, demonstrated a requirement for TRME/GidA and 5,10-methylene THF, thus a C-1 donor at the oxidation state 0 (corresponding to an equivalent of formaldehyde) (Moukadiri et al., 2009) (Figure 5). This step is then followed by glycine addition by the second enzyme in the pathway, TRMC (MnmC1, MnmC2). GTP was proposed to effect a conformational change in the 5,10-methylene THF-binding site (Prado et al., 2013). This change is needed to bring together the C-1 moiety of 5,10-methylene-THF and the C5 position of uridine, which are otherwise separated by a distance of 11Å in the TRME crystal structure. Although the actual reaction mechanism has not been conclusively proven, structural and biochemical data, as well as the analogy to related enzymes, imply that TRME forms a covalent bond by Michael addition of a cystein thiolate to C6 of U34. This leads to activation of the C5 position and sets it on path for a nucleophilic attack onto the C1 electrophile (Meyer et al., 2009). The overall mechanism involves redox cofactors FAD and possibly NAD+, but their precise oxidation state and role is yet to be determined. GidA is also involved in the glycine addition step to form cmnm5U (Figure 5). In other cases where mnm5U is found instead, TRMC removes the 2-carbon skeleton of the glycine residue from cmnm5U in an FAD-dependent manner, and further methylates the resulting nm5U to mnm5U (Figure 3)(Armengod et al., 2012; Prado et al., 2013).
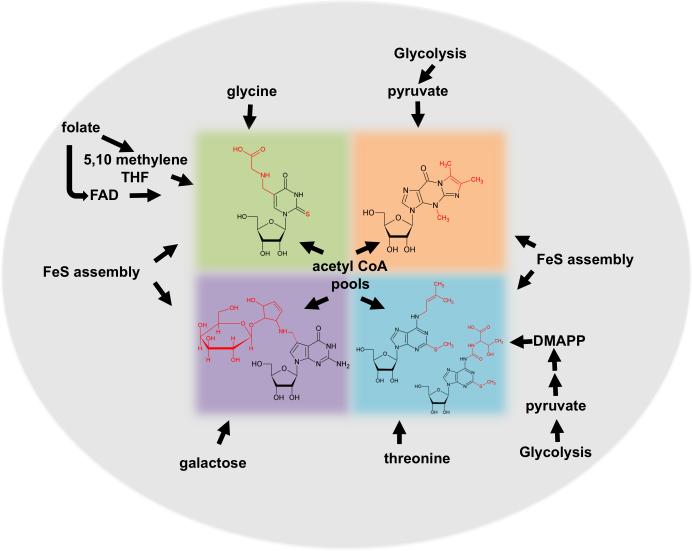
Figure 5. Various Metabolic Pathways Contribute Co-factors for Modification.
The figure shows hyper-modifications highlighted in this review that require building blocks from common metabolic pathways. For example, pyruvate derived from glycolysis. These interconnections may suggest coupling between metabolic and translational rates. The individual color boxes correspond to those modifications shown in figure 4.
Figure 3. Chemical Strategies for C5 Modification of Uridines in Bacteria.
The C5 position of uridines is rendered nucleophilic by Michael addition of a cysteine thiolate to the 5,6 double bond. 5,10-methylene tetrahydrofolate (5,10-CH2THF) provides a C1-body at the oxidation state of formaldehyde which acts as electrophile (red), leading to an electrophilic 5-methyleneuridine intermediate. This intermediate can be attacked by a variety of nucleophiles (blue), which may include hydride (from THF), or different aminoacids such as glycine (bacteria) or taurine (metazoan mitochondria). After dealkylation of cmnm5U, the resulting 5-aminomethyluridine (nm5U) can be methylated to mnm5U or prenylated to inm5U. The latter reaction is unconfirmed.
A different situation occurs in the eukaryotic cytoplasm, where C5 of U34 is usually hypermodified to methoxycarbonylmethyluridine (mcm5U). Although lack of an in vitro system has hampered the biochemical elucidation of the reaction mechanism, ample genetic data links the enzymes involved in this pathway to the so-called elongator complex (Elp) (Huang et al., 2005; Mehlgarten et al., 2010). Elp is comprised of 6 proteins in yeast (Elp 1-6), originally identified as sub-complexes that serve as accessory factors to RNA polymerase II. Orthologs of Elp 2-4 exist in humans with two additional proteins that are presumably analogous to Elp5 and 6 from yeast (Hawkes et al., 2002). In yeast, mutations in any of the Elp genes lead to complete ablation of mcm5U34 (and ncm5U34) formation in tRNA (Huang et al., 2005). Together these observations are most remarkable considering that the same complex plays important roles in histone acetylation and transcription regulation (Svejstrup, 2007).
Despite apparent variance in mechanisms between Bacteria and Eukarya, the eight chemically distinct modifications occurring at C5 of wobble uridines (xm5U34, where x indicates any hypermodification of m5U, for example mnm5U, Figure 2A) all share a common 5-carboxymethyl structure at their core. As a reflection of the variegated chemistry involved, the enzymes involved require substrates such as S-adenosylmethionine (SAM), glycine, taurine, and folate, which are derived from different metabolic pathways. The modification pathway is thus part of a network of metabolic interactions, and beyond this, ties into components, like Elp, that because of their function in transcription, play critical roles in affecting gene expression.
Bacterial tRNAs also undergo further modifications at U34, including the thiolation reaction that replaces the exocyclix oxygen at C2 in tRNALysUUU, tRNAGlnUUG and tRNAGluUUC. Despite affecting the same nucleotide, s2U and xm5U (Figure 2A) formation are independent or each other. In terms of evolution, the enzymes responsible for s2U formation evolved independently in the eukaryotic and bacterial lineages. Both sets of enzymes exist in eukaryotic cells where the cytoplasmic pathway is uniquely eukaryotic, yet the mitochondrial pathway is still bacterial in nature. Cytoplasmic thiolation to s2U34 requires an enzymatic cascade performed by a series of ubiquitin ligase-like proteins (Leidel et al., 2009). In mitochondria, the complete pathway has not been elucidated but is known to include the MTU1 (TRMU) enzyme, a homolog of the mnmA enzyme from Bacteria (Umeda et al., 2005). The bacterial system also includes a series of small proteins that catalyze a modification cascade starting with transfer of the sulfur from cysteine and involvement of IscS, the same desulfurase enzyme essential for catalyzing sulfur incorporation during FeS-cluster assembly into many proteins, including many modification and metabolic enzymes (as discussed below) (Shigi et al., 2008).
When tRNAs are encoded with a guanosine at the wobble position (G34), in the context of a GUN anticodon (where N represents any nucleotide), this nucleotide is usually hypermodified to queuosine (yeast representing one of the few exceptions). This includes tRNAAsn, tRNAAsp, tRNAHis and tRNATyr, but tRNAAsp and tRNATyr are further glycosylated to mannosyl Q-tRNA (manQ34) and galactosyl Q-tRNA (galQ34) respectively (Chen et al., 2011) (Figure 4). Regardless of the tRNA species, in all cases Q plays critical roles in translation. Interestingly, the route to Q formation in tRNA differs markedly in Bacteria and Eukarya. In Bacteria, five enzymatic steps are necessary for the de novo biosynthesis of 7-aminomethyl-7-deazaguanine (preQ, Figure 4), which is then used to replace guanosine in the tRNA substrates by the enzyme tRNA-guanine transglycosylase (TGT) (Chen et al., 2011). Once on the tRNA, preQ is further modified to queuosine and eventually glycosylated, as described above, in tRNAAsp and tRNATyr but not tRNAAsn or tRNAHis. All eukaryotes, with the exception of yeast, where TGT is missing, contain Q in tRNA; however, these organisms do not synthesize preQ and can only incorporate queuine (the free base form of queuosine) through salvage from nutrients in the media, where queuine is preformed by the bacterial gut microflora (Iwata-Reuyl, 2003). These differences in substrate specificity led to the proposal that both pathways were the result of convergent evolution. However, recent crystallographic and biochemical evidence strongly support the view that in fact they are evolutionarily divergent. What is not clear is whether eukaryotes once had the preQ pathway and lost it or alternatively never had it. It stands to reason that the former is the likely explanation and the result of the intimate interaction between Bacteria and Eukarya in their crossing evolutionary paths (Chen et al., 2011).
In terms of cellular physiology, in Bacteria, lack of Q leads to issues of translational efficiency, best manifested in a reduction in virulence in some pathogens. This pathway has thus been proposed as an antibacterial target (Goodenough-Lashua and Garcia, 2003). In Eukarya, beyond translation, laboratory animals deficient in TGT grow normally but if the non-essential amino acid tyrosine is left out of their feed, they die within 18 days (Rakovich et al., 2011). The early symptoms of tyrosineless/Q-less diet easily disappear with the addition of tyrosine to growth media. The defect has been ascribed to an inability of Q-less mice to synthesize tyrosine from phenylalanine by phenylalanine hydrolase (PAH), presumably due to low levels of tetrahydrobiopterin (BH4), an essential co-factor for PAH. However, it is still not clear why this imbalance in BH4 levels only occurs in the absence of Q (Rakovich et al., 2011).
Also different between Bacteria and Eukarya is the nature of the TGT enzyme itself. The bacterial enzyme is either monomeric or homomultimeric depending on the study (Chen et al., 2011). The eukaryal enzyme, at least in mouse, requires two non-identical sub-units derived from alternative splicing of the TGT gene (Boland et al., 2009). Neither sub-unit by itself can catalyze the reaction, yet mixing together recombinant versions of the two, robustly reconstitutes activity in vitro. Presumably, one of the subunits is used in salvaging queuine from queuosine monophosphate resulting from tRNA turnover, while the second sub-unit is in charge of catalyzing the Q exchange on the tRNA. Just as complex is the localization of the two eukaryotic TGT subunits. Queuosine is found in both cytoplasmic and mitochondrial tRNAs in mice, yet both sub-units appear to localize to mitochondria (Boland et al., 2009). How or why this unusual localization, remains an open question.
The importance of good neighbors: hypermodifications of position 37
The only other position in the anticodon of tRNAs to undergo hyper-modification is position 37, neighboring the anticodon sequence (Figure 4). This position is always an encoded purine, which in all three domains of life is almost always modified. The evolutionary conservation of this modified position signals its importance in stabilizing anticodon-codon interaction during decoding. It also plays critical roles in reading-frame maintenance. In general modifications at position 37 help maintain and open loop conformation, preventing base pairing with neighboring nucleotides on the other side of the anticodon loop (U33) and helping formation of a canonical anticodon loop structure important for decoding (Cabello-Villegas et al., 2002).
In most tRNAs an encoded G37 is methylated at the base to form m1G, which by itself plays critical roles in aminoacylation and/or translational accuracy. Because m1G is found in tRNAs in all domains of life it was probably present in the last common ancestor, already serving important functions in establishment of the genetic code early in evolution (Bjork et al., 2001). However, m1G37, also serves as the chemical platform for additional group additions in the case of tRNAPhe of Archaea and Eukarya. In these organisms, m1G is the first step in formation of wybutosine (yW) in Eukarya; wyosine and derivatives in Archaea (de Crecy-Lagard et al., 2010; Noma and Suzuki, 2006). Synthesis of this nucleoside(s) requires a series of enzymatic steps starting with formation of a tricyclic ring on the methylated guanosine (Figure 3A). Interestingly, the eukaryotic TYW1 protein contains an FMN binding domain, which is presumably important for the reduction of the essential [4Fe-4S] cluster proposed to be critical for catalysis. Although this reaction has not been reconstituted in vitro in the eukaryotic system, it was shown that deletion of the TYW1 gene led to disappearance of yW with a concomitant appearance of m1G in tRNAPhe (Bjork et al., 2001).
In Eukarya following the TYW1-catalyzed reaction, TYW2 transfers the α-amino-α-carboxypropyl (acp) group from SAM, forming the intermediate yW-86, so called because it differs from fully formed wybutosine by 86 daltons (Noma et al., 2006) (Figure 2B). This intermediate is then methylated at the original purine ring N-3 position by TYW3 to form yW-72, the acp group is then methylated by TYW4 to yW-58 and subsequently methoxycarbonylated by the same enzyme to form wybutosine (yW) (Figure 2B). This last step is particularly interesting in that it involves CO2 fixation, which leads to the suggestion of a new role for SAM (Suzuki et al., 2009). Still however, all energetic details for this reaction have not been fully clarified.
An additional question is the origin of the different methylation and methoxycarbonylation activities that involve the synthesis of wybutosine. Curiously, at least in the case of TYW4, the crystal structure offers important clues. This enzyme contains a domain in common with protein phosphatase methyltransferase 1 (PPM1), which methylates protein phosphatase 2A (PP2A) in a SAM-dependent manner. In TYW4, a nine amino acid loop is present, which has been replaced in PPM1 (Suzuki et al., 2009). This loop contains key positively charged residues important for substrate binding and is proposed to have change specificity from a protein (PP2A) to a tRNAPhe substrate through evolution (Suzuki et al., 2009).
In Eukarya following the TYW1-catalyzed reaction, the C-7 side chain differs in the extent of modification. Unlike yeast, in mammals the final product is a hydroxylated wybutosine derivative, hydroxywybutosine (OHyW) (Urbonavicius, 2009) (Figure 2B). Although this difference was known for many years, it was only recently reported that a fifth enzyme in the pathway exists, appropriately named TYW5. This enzyme belongs to the Jumonji C (JmjC) domain-containing family, typical of histone demethylases (Kato et al., 2010). Recent biochemical data has shown that TYW5 catalyzes an Fe(II)/2-oxoglutarate hydroxylation reaction in the final step of wybutosine biosynthesis in humans. Additionally, analogous to the fusion of TYW2-3-4 in plants, in humans TYW4-5 are also fused suggesting coupling of the last two reactions (Kato et al., 2010).
The variability in the end product of wyosine and derivatives is even more complex in Archaea. Although the reaction usually ends after the TYW3 step, due to the lack of genes encoding TYW4 and 5, various combinations of wyosine derivatives exist depending on the organism. For example most euryarchaeota lack TYW3, yet others have the ACP side-change replaced by a methyl group (imG2). In these Archaea the observation of a gene duplication of the TRM5 methylase (responsible for m1G37) has led to the suggestion that the duplicated gene product is responsible for this additional methylation, but this has to be formally proven (de Crecy-Lagard et al., 2010).
Perhaps the biggest puzzle in wyosine synthesis in Archaea is the absence of the highly conserved FMN-binding domain found in eukaryotic TYW1 (de Crecy-Lagard et al., 2010). This raises the question of what provides the function needed to reduce the essential iron-sulfur clusters. It has been suggested that in Archaea an additional protein, perhaps thioredoxin reductase, serves this function in trans. In addition the nature of the 2-carbon compound needed to form the third ring had precluded reconstitution of the TYW1 reaction until recently, when the use of dithionite as the source of reducing power in vitro, established that pyruvate is the key substrates needed for ring closure (Young and Bandarian, 2011). Further studies then showed that TYW1 from Archaea contains, not one, but two iron-sulfur clusters, whereby cluster I is proposed to bind SAM and mediate formation of a SAM radical that abstracts a proton from the methyl group of m1G, creating the substrate radical that attacks pyruvate, forms the tricyclic wyosine and releases CO2 (Perche-Letuvee et al., 2012). Although pyruvate has not been validated as the 2-C donor in eukaryotes, a similar mechanism is expected.
Presence of wyosine and derivatives is suggested to lead to a single functional outcome, hypermodification may provide base-stacking interactions that play a key function in reading-frame maintenance (Urbonavicius et al., 2003). Why then are wyosine and derivatives only found at position 37 of tRNAPhe? It may well be due to the nature of the phenylalanine codons, UUU and UUC codons, and their propensity for ribosome “slippage” (Atkins and Bjork, 2009). These observations then lead to the possibility that the extent of tRNAPhe modification at position 37 may correlate with the frequency of poly-uridine slippery sequences in genomes. Why then use the wybutosine strategy when m1G alone can prevent unwanted frameshifting at most codons? The answer may rest in the idea of “frameshifting potential” (Waas et al., 2007), whereby, like in many viruses, frameshifting is utilized in a programmed fashion perhaps to increase coding diversity. This then explains the variability of tRNAPhe modification at G37 ranging from no wybutosine in Drosophila to the hypermodification in mammals. “frameshifting potential” implies that cells as part of their normal translation programs may exploit frameshifting as a regulatory mechanism and not just avoid it, at all cost, as a source of translational infidelity.
The only other anticodon loop hypermodifications occur when position 37 is an encoded adenosine. In all domains of life this position is usually isopentenylated to form isopentenyl adenosine (i6A), while in Bacteria i6A can be further hypermodified to ms2i6A or ms2io6A depending on the tRNA and the organism. The i6A system of Bacteria targets tRNAs for all codons with a U at the first position while in Eukarya the tRNA substrates vary but are generally restricted to a smaller set of tRNAs. In terms of substrate specificity the bacterial enzymes recognize A36A37A38 as a major determinant for i6A formation while in Eukarya what determines a good substrate depends once again on the organism, revealing a remarkable plasticity in substrate recognition by this family of enzymes (Lamichhane et al., 2011).
Whether the end product is i6A or the methylthiolated counterpart, the isopentenyl addition reaction is catalyzed by the product of the miaA gene in Bacteria (MOD5 in Eukarya), the enzyme catalyzes the transfer of the dimethylallyl moiety from dimethyallyl pyrophosphate to A37 in various tRNAs, using magnesium as the only required co-factor in vitro. In Bacteria, i6A can be further modified to ms2i6A or ms2io6A by a bi-functional enzyme encoded by miaB. Coincidentally, this enzyme also contains two iron-sulfur clusters analogous to the TYW1 enzyme of Archaea. Despite i6A and derivatives constituting bulky modifications, their function differ from that of the previously described wybutosine/wyosine derivatives in that rather than playing roles in frame maintenance, A37 modifications are key determinants for codon-specific translation and therefore translation efficiency (Lamichhane et al., 2011).
For tRNAs decoding the ANN codons, it has been established for a long time that A37 is universally modified to form N6-threonylcarbamoyladenosine (t6A) and can be further methylated at the base (e.g. m6t6A); this modification is also crucial to translational accuracy. In all systems, the t6A biosynthetic pathway requires threonine, ATP and carbonate as substrates and in Bacteria it involves the Sua5/YrdC family of proteins while in Eukarya the Kae1/YgjD/Qri7 family (El Yacoubi et al., 2009a). Despite the fact that each system has recruited different evolutionarily unrelated proteins to catalyze the reaction, minimally Sua5 and Qri (a Kae1 homolog) are required for t6A formation as shown in the mitochondrial system. Significantly, mutants in these families of proteins lead to pleiotropic effects in cells including defects in transcription and genome stability (El Yacoubi et al., 2009b).
The t6A story besides its biological significance also offers a cautionary tale when studying modifications. Despite its existence being accepted for over 20 years, recent work showed that in fact in bacteria (and perhaps in all organisms) the native modification is cyclic t6A (ct6A), a dehydrated form of t6A. It turns out that ct6A is so reactive that at neutral pH the cyclic side chain is destroyed. In line with this observation, an E1-like enzyme with ATPase activity (TcdA) has been identified as the dehydratase responsible for cyclization. Notably, ct6A is so reactive that it can also readily form adducts with the amines in Tris or ethanolamine buffers. Biologically lack of TcdA leads to respiratory defects in yeast suggesting its particular importance in mitochondrial decoding (Miyauchi et al., 2013).
Environmental sensors and metabolic integrators
Efforts in the last few years have focused on the identification and characterization of the enzymes responsible for various modifications. New technologies have considerably accelerated the process and in certain organisms, such as S. cerevisiae and E. coli almost all of the tRNA modifications and corresponding enzymes have been cataloged. Many of the enzymatic reactions have also been established in vitro and much progress has been made toward understanding different modification enzyme mechanisms. Even in cases where reactions have not been recapitulated in vitro, strong genetic evidence has linked a number of distinct proteins to a given modification. This knowledge has also benefited from progress in the structure determination of modification enzymes, which has provided further insights into their specificity and modes of substrate recognition. The next major challenge in the modification field will be to establish how environmental changes affect modification sets and how these in turn get transduced into various cellular signals. The idea of a connection between tRNA modifications and environmental states dates back to several reports in the 1970s where changes in tRNA modification content were correlated to how cells respond to various environmental stimuli, including exposure to UV light, challenge by phage infections, growth rate and changes in growth temperature (Emilsson et al., 1992). Back then, the technical challenges in quantitatively assigning modifications to specific nucleotides and specific positions in a tRNA sequence, forced these studies to remain mostly descriptive and could not push the ideas of modifications as environmental sensors much further.
Several subsequent reports have measured the impact of modification in response to temperature changes. A key observation here was the induction of a set of tRNA modifications at elevated temperatures in thermophiles (Kowalak et al., 1994) - These studies followed the line of reasoning that given the important roles played by modifications in maintaining the stability of the folded L-shape tRNA, these may play particularly important roles in organisms that grow at high temperatures. Not surprisingly, the lack of some modifications, for example s2T, led to temperature sensitivity in several bacteria. Likewise, lack of pseudouridine 55 (Ψ55) leads to a decrease in the levels of Gm18 and lack of both modifications lead to temperature sensitivity in Thermus thermophilus (Ishida et al., 2011). Since these modifications are important for necessary tertiary contacts between the D-loop and TΨC loops and maintaining the tertiary fold of many tRNAs, their role in coping with elevated temperatures makes logical sense and therefore represent a modification-mediated adaptation to an extreme environment.
The questions still remain how, more globally, modifications play roles in dealing with environmental extremes. To this end, while applying different stressors to yeast cells in the laboratory, liquid-chromatography tandem mass spectrometry (LC-MS/MS) approaches revealed that the global modification set of tRNAs changed in a dynamic way and depending on the specific stress (Chan et al., 2010). For example, the levels of m5C, m22G, Cm and t6 A changed in response to H2O2-induced oxidative stress. If an alkylating agent, like MMS (methylmethanesulphonate) was used instead, a different set of modifications changed (including, m1A, m3C and m7G). Although these studies could not tell if these changes were specific to tRNA sets, affected all tRNAs or a single tRNA to different levels at a specific location, some of the modifications like t6A and m3C are significant in that these affect subsets of tRNAs and only occur in the anticodon loop, perhaps more directly implicating modification dynamics to translational changes. Such effects on translation have indeed been reported by studies showing that tampering with TRM9 has significant effects on how cells cope with ionizing radiation or MMS (Chan et al., 2012). Since TRM9 is key in the formation of xm5U (Figure 2A), the observed phenotypes directly point to changes in translational efficiency of mRNAs rich in codons for which the modification is needed for decoding.
Along the same lines it was shown that, more specifically, increases in m5C methylation in tRNALeuCAA were crucial to the cells response to oxidative stress. Remarkably, an increase in m5C levels induced by oxidative stress is coupled to translation of a TTG-rich mRNA encoding the ribosomal protein RPL22A, but not of an mRNA for a paralogous protein which has a lower usage of the TTG codon (Chan et al., 2012). This observation is significant in that the ribosomal protein RPL22A has an established role in oxidative stress response. This observation speaks volumes for the dynamic nature of modifications in response to environmental stress and in at least one respect, suggested by the authors, it implies a codon-specific remodeling of translational efficiency imparted by modifications.
Current work on the connection between modifications and environmental signals has concentrated on areas with a vast body of knowledge on how cells respond to certain well-known stresses, for example oxidative stress. However, response to stress caused by environmental extremes may just be one side of cellular modification dynamics. A careful look at the few hypermodification pathways discussed in this review quickly points to the systemic nature of modifications (Figure 5). Many modifications require co-factors and substrates that belong to seemingly disparate yet intricately interconnected metabolic pathways and as such, far and beyond stress, modification dynamics may also play a role in the adjustment and coupling of metabolic rates to tRNA function and protein synthesis as means of overall cell homeostasis and during normal growth conditions. Clearly many modifications borrow metabolites from other biosynthetic pathways in cells and we view these as an additional level of metabolic integration. For example, formation of t6A (and derivatives) utilizes threonine which itself is made from aspartic acid in Bacteria and salvaged from the media in humans (as an essential amino acid). In fact, in humans a potential connection between threonine and SAM has been reported, whereby Thr and SAM metabolism are clearly coupled in determining the pluripotency of stem cells (Shyh-Chang et al., 2012). Thr-derived glycine, but not Ser-derived, contributes to 5-m-tetrahydrofolate levels and SAM generation. When Thr is limiting there is an observed decrease in acetyl CoA pools, which in turn leads to a decrease in NADH. Thus changes in Thr levels can affect production of reducing equivalents (Shyh-Chang et al., 2012). All these factors combined decrease embryonic-stem cell growths and increase differentiation. Although these represent the result of cellular nutrient stress, clearly changing levels of SAM, Thr and glycine indirectly affect several of the key hypermodifications discussed in the previous sections of this review. As such, nutrient response should involve the integration of metabolic signals and protein synthesis via tRNA modifications during normal cell growth, far and beyond the realm of nutritional stress. The same argument can be made for the biosynthesis of I6A, which again requires the substrate DMAPP (dimethylallyl pyrophosphate). DMAPP itself is derived from acetyl-CoA (Figure 5), which in turn may come from glycolysis via pyruvate or fatty acid beta oxidiation. Again this begs the question of how small physiological changes in acetyl-CoA levels may affect I6A biosynthesis and thus lead to reduction in the translation of certain mRNAs rich in codons that require I6A-containing tRNAs. Along these lines, a genetic screen in S. cerevisiae involving the MOD5 enzyme (responsible for I6A biosynthesis), revealed a competition between sterol and I6A biosynthesis; two pathways that require DMAPP, which is in limited supply in yeast (Benko et al., 2000). Similar arguments have been made for the possible connection between thiolation and iron-sulfur cluster assembly pathways in bacteria and eukarya, which may extend to metabolic connections beyond tRNA maturation and may again involve seemingly unrelated pathways.
CONCLUSIONS
Exploring the limits of RNA natural product synthesis: modifications yet to be found
The preceding pages have highlighted the integrative nature of RNA modifications, in further support of the idea that these do not act alone but rather operate in a tangled web of interconnections, which in the context of the four canonical nucleotides determine both their chemical diversity and modification enzyme specificity. This level of chemical integration has led us to suggest that the occurrence of subtle phenotypes, when approaching modification function genetically, has unfairly led to the conclusion that the impact of modification on cell function is minor, if any. However, it is possible that to a great degree lack of a single modification may be partially or fully compensated by the appearance of additional modifications in the tRNA, which in turn may satisfy the function provided by the normally occurring but now missing modification. Thus tackling to what extent modifications affect RNA function at a cellular scale is indeed a difficult business. Reductionism dictates system minimalization and the study of modifications as single entities, yet their effects cannot be fully realized without thinking about modifications in the context of the ever-changing cellular environment.
The recent identification of geranylated uridines (Dumelin et al., 2012) will not certainly be the last in a long line of discoveries that started in the 1950s, and produced surprises with remarkable steadiness, as illustrated by the discovery dates given in Figure 1. What may mark a new trend is the approach taken by the Liu laboratory: the search for new modifications by anticipation of what could be out there, while taking into consideration where and how one had to look for it, in chemical terms, rather than in what organism. Because geranylation makes RNA more lipophilic, geranylated RNAs were identified in a search for unusually hydrophobic RNAs. Similar approaches based on the hydrophobicity of wybutosine led earlier to the isolation of large amounts of tRNAPhe necessary for crystallography, eventually leading to the first X-ray structure of a tRNA (Kim et al., 1974; Robertus et al., 1974)(Hoskinson and Khorana, 1965; Rajbhandary et al., 1967). With this in mind, future search strategies for new modifications may be tailored towards efficient affinity purification methods. For these there is ample inspiration in the literature for example the retention of sulfur containing modifications in mercury gels (Igloi, 1988) and vicinal diols by boronic acids (Igloi and Kossel, 1985).
The few modifications mentioned here are not meant as a conclusive list, but rather to highlight a few cases where the connections may be more obvious yet widely understudied. In the end the next challenge will be to integrate the fields of modomics and metabolomics to establish where the lines are drawn. It is therefore, the use of multiple approaches that will lead to the elucidation of the connectivity between modifications and other cellular metabolic programs that will place RNA modifications at the heart of key regulatory loops controlling cellular systems.
HIGHLIGHTS.
- RNA modifications can occur in a combinatorial fashion within single sites
-Some modifications occur in cascades
-Modifications may coordinate metabolic and protein synthesis rates
-Modifications may serve as cellular sensors
ACKNOWLEDGEMENTS
We thank all members of the Helm and Alfonzo laboratories for useful discussions. This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (HE 3397/4) and the Volkswagen Foundation to MH and an NIH grant GM084065-05 to JDA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Armengod ME, Moukadiri I, Prado S, Ruiz-Partida R, Benitez-Paez A, Villarroya M, Lomas R, Garzon MJ, Martinez-Zamora A, Meseguer S, Navarro-Gonzalez C. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie. 2012;94:1510–1520. doi: 10.1016/j.biochi.2012.02.019. [DOI] [PubMed] [Google Scholar]
- Atkins JF, Bjork GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko AL, Vaduva G, Martin NC, Hopper A. Competition between a sterol byosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense supression. Proc Natl Acad Sci U S A. 2000;97:61–66. doi: 10.1073/pnas.97.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? Embo J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland C, Hayes P, Santa-Maria I, Nishimura S, Kelly VP. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J Biol Chem. 2009;284:18218–18227. doi: 10.1074/jbc.M109.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregeon D, Colot V, Radman M, Taddei F. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Villegas J, Winkler ME, Nikonowicz EP. Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe). J Mol Biol. 2002;319:1015–1034. doi: 10.1016/S0022-2836(02)00382-0. [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, why?. Mol Cell. 2013;5:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Brooks AF, Goodenough-Lashua DM, Kittendorf JD, Showalter HD, Garcia GA. Evolution of eukaryal tRNA-guanine transglycosylase: insight gained from the heterocyclic substrate recognition by the wild-type and mutant human and Escherichia coli tRNA-guanine transglycosylases. Nucleic Acids Res. 2011;39:2834–2844. doi: 10.1093/nar/gkq1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crecy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, Grosjean H. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol Biol Evol. 2010;27:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dumelin CE, Chen Y, Leconte AM, Chen YG, Liu DR. Discovery and biological characterization of geranylated RNA in bacteria. Nat Chem Biol. 2012;8:913–919. doi: 10.1038/nchembio.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crecy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. Embo J. 2009a;30:882–893. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009b;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Naslund AK, Kurland CG. Thiolation of transfer RNA in Escherichia coli varies with growth rate. Nucleic Acids Res. 1992;20:4499–4505. doi: 10.1093/nar/20.17.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough-Lashua DM, Garcia GA. tRNA-guanine transglycosylase from E. coli: a ping-pong kinetic mechanism is consistent with nucleophilic catalysis. Bioorg Chem. 2003;31:331–344. doi: 10.1016/S0045-2068(03)00069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, Krappmann D, Scheidereit C, Thomas CL, Schiavo G, Erdjument-Bromage H, et al. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- Hoskinson RM, Khorana HG. Studies on Polynucleotides. Xli. Purification of Phenylalanine-Specific Transfer Ribonucleic Acid from Yeast by Countercurrent Distribution. J Biol Chem. 1965;240:2129–2134. [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. Rna. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry. 1988;27:3842–3849. doi: 10.1021/bi00410a048. [DOI] [PubMed] [Google Scholar]
- Igloi GL, Kossel H. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res. 1985;13:6881–6898. doi: 10.1093/nar/13.19.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2011;39:2304–2318. doi: 10.1093/nar/gkq1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Reuyl D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg Chem. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- Kato M, Araiso Y, Noma A, Nagao A, Suzuki T, Ishitani R, Nureki O. Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res. 2010;39:1576–1585. doi: 10.1093/nar/gkq919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. Rna. 2011;17:1846–1857. doi: 10.1261/rna.2628611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- Li S, Limbach PA. Method for comparative analysis of ribonucleic acids using isotope labeling and mass spectrometry. Anal Chem. 2012;84:8607–8613. doi: 10.1021/ac301638c. [DOI] [PubMed] [Google Scholar]
- Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013;41:D262–267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jager G, Gong Z, Bystrom AS, Schaffrath R, Breunig KD. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol. 2010;76:1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Bohme S, Kruger A, Steinhoff HJ, Klare JP, Wittinghofer A. Kissing G domains of MnmE monitored by X-ray crystallography and pulse electron paramagnetic resonance spectroscopy. PLoS Biol. 2009;7:e1000212. doi: 10.1371/journal.pbio.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kimura S, Suzuki A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem. 2013;9:105–113. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- Moukadiri I, Prado S, Piera J, Velazquez-Campoy A, Bjork GR, Armengod ME. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 2009;37:7177–7193. doi: 10.1093/nar/gkp762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. Embo J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A, Suzuki T. Ribonucleome analysis identified enzyme genes responsible for wybutosine synthesis. Nucleic Acids Symp Ser (Oxf) 2006:65–66. doi: 10.1093/nass/nrl032. [DOI] [PubMed] [Google Scholar]
- Perche-Letuvee P, Kathirvelu V, Berggren G, Clemancey M, Latour JM, Maurel V, Douki T, Armengaud J, Mulliez E, Fontecave M, et al. 4-Demethylwyosine synthase from Pyrococcus abyssi is a radical-S-adenosyl-L-methionine enzyme with an additional [4Fe-4S](+2) cluster that interacts with the pyruvate co-substrate. J Biol Chem. 2012;287:41174–41185. doi: 10.1074/jbc.M112.405019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado S, Villarroya M, Medina M, Armengod ME. The tRNA-modifying function of MnmE is controlled by post-hydrolysis steps of its GTPase cycle. Nucleic Acids Res. 2013;41:6190–6208. doi: 10.1093/nar/gkt320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajbhandary UL, Chang SH, Stuart A, Faulkner RD, Hoskinson RM, Khorana HG. Studies on polynucleotides, lxviii the primary structure of yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1967;57:751–758. doi: 10.1073/pnas.57.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovich T, Boland C, Bernstein I, Chikwana VM, Iwata-Reuyl D, Kelly VP. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J Biol Chem. 2011;286:19354–19363. doi: 10.1074/jbc.M111.219576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat Chem Biol. 2010;6:733–740. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- Schiesser S, Hackner B, Pfaffeneder T, Muller M, Hagemeier C, Truss M, Carell T. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew Chem Int Ed Engl. 2012;51:6516–6520. doi: 10.1002/anie.201202583. [DOI] [PubMed] [Google Scholar]
- Scrima A, Vetter IR, Armengod ME, Wittinghofer A. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. Embo J. 2005;24:23–33. doi: 10.1038/sj.emboj.7600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N, Sakaguchi Y, Asai S, Suzuki T, Watanabe K. Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. Embo J. 2008;27:3267–3278. doi: 10.1038/emboj.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Wada T, Saigo K, Watanabe K. Novel taurine-containing uridine derivatives and mitochondrial human diseases. Nucleic Acids Res Suppl. 2001:257–258. doi: 10.1093/nass/1.1.257. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res. 2009;37:2910–2925. doi: 10.1093/nar/gkp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ. Elongator complex: how many roles does it play? Curr Opin Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2012;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J Biol Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Droogmans L, Armengaud J, Grosjean H. Deciphering the Complex Enzymatic Pathway for Biosynthesis of Wyosine Derivatives in Anticodon of tRNAPhe. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution (Landes Biosciences) 2009:423–435. [Google Scholar]
- Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Bjork GR. Transfer RNA modifications that alter +1 frameshifting in general fail to affect -1 frameshifting. Rna. 2003;9:760–768. doi: 10.1261/rna.5210803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AP, Bandarian V. Pyruvate is the source of the two carbons that are required for formation of the imidazoline ring of 4-demethylwyosine. Biochemistry. 2011;50:10573–10575. doi: 10.1021/bi2015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waas WF, Druzina Z, Hanan M, Schimmel P. Role of a tRNA modification and its precursors in frameshifting in eukaryotes. J Biol Chem. 2007;36:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]