Abstract
Background
Gene duplication is a source of evolutionary innovation and can contribute to the divergence of lineages; however, the relative importance of this process remains to be determined. The explosive divergence of the African cichlid adaptive radiations provides both a model for studying the general role of gene duplication in the divergence of lineages and also an exciting foray into the identification of genomic features that underlie the dramatic phenotypic and ecological diversification in this particular lineage. We present the first genome-wide study of gene duplication in African cichlid fishes, identifying gene duplicates in three species belonging to the Lake Malawi adaptive radiation (Metriaclima estherae, Protomelas similis, Rhamphochromis “chilingali”) and one closely related species from a non-radiated riverine lineage (Astatotilapia tweddlei).
Results
Using Astatotilapia burtoni as reference, microarray comparative genomic hybridization analysis of 5689 genes reveals 134 duplicated genes among the four cichlid species tested. Between 51 and 55 genes were identified as duplicated in each of the three species from the Lake Malawi radiation, representing a 38%–49% increase in number of duplicated genes relative to the non-radiated lineage (37 genes). Duplicated genes include several that are involved in immune response, ATP metabolism and detoxification.
Conclusions
These results contribute to our understanding of the abundance and type of gene duplicates present in cichlid fish lineages. The duplicated genes identified in this study provide candidates for the analysis of functional relevance with regard to phenotype and divergence. Comparative sequence analysis of gene duplicates can address the role of positive selection and adaptive evolution by gene duplication, while further study across the phylogenetic range of cichlid radiations (and more generally in other adaptive radiations) will determine whether the patterns of gene duplication seen in this study consistently accompany rapid radiation.
Background
Adaptive radiation, the evolution of genetic and ecological diversity leading to species proliferation in a lineage, is thought to be the result of divergent selection for resource specialization [1-3]. Differential selection in heterogeneous environments can result in adaptive radiation when there is a genetic basis for variability in organisms’ success in exploiting alternative resources [1-5]. Examples of such radiations include the Cambrian explosion of metazoans [6], the diversification of Darwin’s finches in the Galapagos [7], variations in amphipods and cottoid fishes in Lake Baikal [8], the Caribbean anoles [9], the Hawaiian Silverswords [10] and the explosive speciation of the cichlid fishes in the African Great Lakes [11].
The cichlid fishes are the product of an incredible series of adaptive radiations in response to the local physical, biological and social environment. While cichlids can be found on several continents [12], the most dramatic radiations are those of the haplochromine cichlids in the great lakes of East Africa. This speciose clade exhibits unprecedented diversity in morphological and behavioral characteristics [13] and accounts for ~10% of the world’s teleost fish. Interestingly, this clade also includes lineages that have remained in a riverine environment and have not radiated [14].
Classic work by Ohno [15] proposed a prominent role for gene duplication events in evolutionary expansion, despite their frequent loss due to drift [16]. Duplication makes extra gene copies available for dosage effects, subfunctionalization, or neofunctionaliztion [17], with the resultant phenotype potentially contributing to an organism’s fitness (for review see [18]). Current genomic research (e.g. primates: [19,20]) supports this, but the ability to compare closely related cichlid lineages that have and have not undergone an evolutionary radiation provides a critical tool for testing the association of gene duplication with adaptive radiation.
We used array-based comparative genomic hybridization (aCGH) to identify gene duplications among 5689 genes for three Lake Malawi radiation species, which began accumulating molecular diversity approximately 5 million years ago [21] (Metriaclima estherae, Protomelas similis, Rhamphochromis “chilingali”) and one closely related riverine species from a non-radiated lineage (Astatotilapia tweddlei). While previous mitochondrial data suggested a bifurcation that separated the Lake Malawi radiation from the riverine species (Figure 1), more recent data based on ALFP data and single nucleotide polymorphisms derived from low coverage whole genome sequence [22-24] suggest that the Malawi flock is not monophyletic and that some of the riverine lineages may have contributed to Malawi genomes. These insights further support the use of A. burtoni as a reference to the three approximately equidistant test species. This is the first genome-wide study of gene duplication among haplochromine cichlids.
Figure 1.
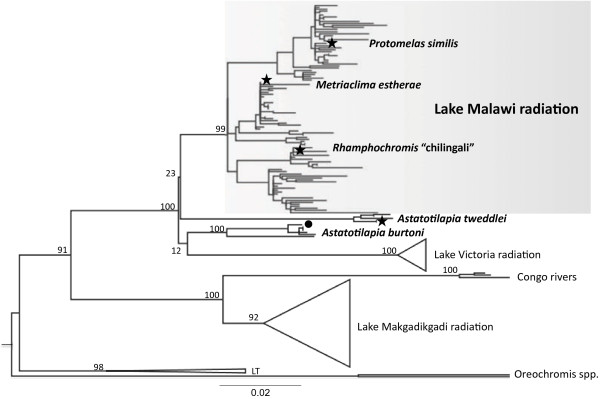
Maximum likelihood phylogeny illustrating the positions of experimental (stars) and reference (circle) taxa. The maximum likelihood tree is based on 1785 bp mitochondrial ND2. Nodes not supported by 50% maximum likelihood SH values collapsed and Lake Victoria, Lake Makgadikgadi, and Lake Tanganyika radiations are represented by triangles. The tree is rooted with Oreochromis and the scale bar indicates the mean number of nucleotide substitutions per site (DRYAD doi:10.5061/dryad.7vs2c).
Results
aCGH identification of duplicated genes
Microarray features, representing a total of 5689 genes, passed quality control measures in all four test species. Among these, 145 array features (representing 134 genes) were determined to have an increased genomic content (i.e. copy number) for one or more heterologous species relative to A. burtoni (P < 0.1 FDR corrected) (Tables 1, 2). This included duplications of 54 genes in M. estherae, 51 in P. similis, and 55 in R. “chilingali”, compared to only 37 in A. tweddlei, the species from the non-radiated lineage (Figure 2). The number of duplicated genes identified for the species from the radiated lineage represents a 38%–49% increase relative to the number of duplicated genes identified in A. tweddlei. Consistent with their shared evolutionary history, shared duplications were prevalent among the three Lake Malawi species, with 11 duplications shared among all three and 16 duplications shared between two of the three species (Figure 2). Five genes had greater gene copy number in all four species relative to A. burtoni. Genes found duplicated in only one of the four species were also identified. This included 27 genes in M. estherae, 20 in P. similis, 24 in R. “chilingali” and 27 in A. tweddlei.
Table 1.
Genes duplicated relative to A. burtoni with informative BLAST hits
| GenBank | Homology | A.twe | M.est | P.sim | R.chi | BitScore |
|---|---|---|---|---|---|---|
|
CN4688282/2 |
Adenine nucleotide translocator s598 |
ns |
ns |
ns |
0.60 |
567 |
| 0.0019 | ||||||
|
DY630000 |
Alcohol dehydrogenase Class VI |
ns |
ns |
0.73 |
ns |
379 |
| 0.0015 | ||||||
|
DY630424 |
Alkylated DNA repair protein alkB homolog 7 |
ns |
0.43 |
ns |
ns |
304 |
| 0.0002 | ||||||
|
DY629046 |
Arsenic (+3 oxidation state) methyltransferase |
ns |
ns |
ns |
1.06 |
150 |
| 0.0031 | ||||||
|
DY626788 |
ATPase, H + transporting, lysosomal V0 subunit E |
ns |
0.76 |
ns |
ns |
87.8 |
| 0.0028 | ||||||
|
DY628437 |
Claudin 29a (cldn29a) gene |
ns |
0.60 |
ns |
ns |
526 |
| 0.005 | ||||||
|
DY632040 |
Coiled-coil domain containing protein 80 |
ns |
ns |
1.19 |
2.13 |
434 |
| 3E-05 |
5E-07 |
|||||
|
DY629141 |
Crystallin gamma M2b |
ns |
ns |
ns |
0.43 |
829 |
| 0.0024 | ||||||
|
DY6262041/2 |
C-type lectin domain family 4 member C |
ns |
0.38 |
ns |
ns |
246 |
| 0.0039 | ||||||
|
DY631088 |
Cystatin-B |
0.45 |
ns |
ns |
ns |
150 |
| 0.0016 | ||||||
|
DY630353 |
Cytosolic sulfotransferase 3 |
ns |
ns |
0.62 |
0.64 |
713 |
| 0.0015 |
0.0013 |
|||||
|
CN470675 |
Dazl gene |
ns |
ns |
ns |
0.57 |
89.7 |
| 0.0040 | ||||||
|
DY6299674/8 |
Ferritin heavy subunit |
ns |
ns |
ns |
0.82 |
1160 |
| 8E-05 | ||||||
|
DY631817 |
Fish virus induced TRIM protein |
ns |
ns |
0.590 |
ns |
170 |
| 0.0005 | ||||||
|
DY626596 |
Fish virus induced TRIM protein |
ns |
ns |
0.41 |
0.44 |
145 |
| 0.0045 |
0.0030 |
|||||
|
DY628624 |
Gamma M7 crystallin |
ns |
ns |
ns |
0.42 |
169 |
| 0.0054 | ||||||
|
DY630388 |
Glutamyl-tRNA(Gln) amidotransferase |
0.48 |
ns |
ns |
ns |
347 |
| 0.0016 | ||||||
|
DY6261151/2 |
GTPase IMAP family member 7 |
ns |
ns |
1.14 |
ns |
370 |
| 0.0016 | ||||||
|
CN471284 |
High-mobility group 20B |
0.60 |
ns |
ns |
ns |
163 |
| 0.0004 | ||||||
|
CN469367 |
Hox gene cluster |
1.34 |
1.16 |
0.86 |
1.11 |
183 |
| 3E-05 |
9E-05 |
0.0006 |
0.0002 |
|||
|
DY627986 |
Hox gene cluster |
1.81 |
1.12 |
0.80 |
1.22 |
95.1 |
| 3E-07 |
1E-05 |
0.0001 |
5E-06 |
|||
|
DY629113 |
Immunoglobulin light chain |
ns |
ns |
0.65 |
ns |
482 |
| 0.0025 | ||||||
|
CN468953 |
Iron-sulfur cluster assembly enzyme ISCU |
ns |
ns |
ns |
0.86 |
610 |
| 0.0022 | ||||||
|
DY628151 |
Kallikrein-8 precursor |
1.02 |
ns |
ns |
ns |
102 |
| 0.0015 | ||||||
|
DY627800 |
Kinesin-like protein 2 (knsl2) |
ns |
0.86 |
1.84 |
1.14 |
398 |
| 0.0013 |
9E-06 |
0.0003 |
||||
|
CN469578 |
KLR1 gene |
1.04 |
ns |
ns |
ns |
154 |
| 4E-05 | ||||||
|
DY629760 |
LOC100150543, polyprotein |
1.35 |
ns |
0.65 |
0.79 |
141 |
| 8E-06 |
0.001 |
0.0003 |
||||
|
CN468718 |
LOC100151545, similar to Protein KIAA0284 |
0.72 0.0004 |
ns |
ns |
ns |
145 |
|
DY629780 |
MHC class I |
ns |
0.84 |
1.26 |
1.05 |
161 |
| 0.0053 |
0.0005 |
0.0016 |
||||
|
DY630620 |
MHC class IA antigen |
ns |
ns |
0.42 |
ns |
120 |
| 0.0026 | ||||||
|
DY630701 |
MHC class II alpha subunit |
ns |
ns |
0.49 |
ns |
764 |
| 0.0053 | ||||||
|
DY631898 |
MHC class II antigen alpha chain |
ns |
ns |
0.94 |
ns |
87.8 |
| 0.0004 | ||||||
|
DY631847 |
Mitotic spindle assembly checkpoint protein MAD2A |
0.60 |
ns |
ns |
ns |
374 |
| 0.0008 | ||||||
|
DY627079 |
Muscle-type creatine kinase CKM2 |
ns |
0.41 |
ns |
ns |
787 |
| 0.0049 | ||||||
|
CN4693752/3 |
Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 |
ns |
0.69 |
ns |
ns |
663 |
| 0.0003 | ||||||
|
DY628779 |
Post-GPI attachment to proteins factor 2 |
ns |
0.87 |
ns |
ns |
123 |
| 0.0002 | ||||||
|
DY6261142/2 |
Ras association domain-containing protein 4 |
ns |
0.82 |
ns |
ns |
1086 |
| 0.0001 | ||||||
|
DY630104 |
Ras-related C3 botulinum toxin substrate 2 |
1.44 |
0.83 |
1.47 |
1.90 |
331 |
| 8E-06 |
0.0003 |
7E-06 |
1E-06 |
|||
|
DY6305081/3 |
Replication factor C subunit 5 |
1.04 |
ns |
ns |
ns |
1234 |
| 2E-05 | ||||||
|
DY628495 |
Ribosomal protein, large P2 (60S) |
ns |
ns |
ns |
1.01 |
161 |
| 0.0001 | ||||||
|
DY6308322/3 |
Ribosomal protein S20 (40S) |
ns |
0.65 |
ns |
ns |
663 |
| 0.001 | ||||||
|
DY626643 |
Serine/threonine phosphatase gene |
ns |
0.57 |
0.57 |
0.54 |
87.8 |
| 0.0004 |
0.0004 |
0.0063 |
||||
|
CN470072 |
Sestrin 3 |
ns |
1.30 |
1.61 |
1.70 |
116 |
| 0.0007 |
0.0002 |
0.0002 |
||||
|
DY629126 |
Short coiled-coil protein |
ns |
ns |
ns |
0.59 |
242 |
| 0.0025 | ||||||
|
DY630540 |
Small inducible cytokine SCYA102 |
ns |
0.64 |
ns |
ns |
1204 |
| 0.0019 | ||||||
|
CN471492 |
Solute carrier family 9 (sodium/hydrogen exchanger) |
ns |
ns |
0.63 |
ns |
197 |
| 0.0021 | ||||||
|
CN4711031/3 |
Ubiquitin |
ns |
ns |
1.27 |
ns |
985 |
| 0.0042 | ||||||
|
DY629776 |
UDP glycosyltransferase 2 family, polypeptide A1 |
ns |
0.92 |
ns |
ns |
304 |
| 0.0006 | ||||||
|
CN469822 |
Vacuolar ATP synthase subunit G 1 |
0.79 |
ns |
ns |
ns |
277 |
| 0.0007 | ||||||
|
DY632057 |
Pituitary adenylate cyclase activating polypeptide receptor 1A |
ns |
1.73 |
1.98 |
1.79 |
170 |
|
DY626009 |
Non-LTR retrotransposon Rex1a |
0.70 |
ns |
ns |
ns |
82.4 |
| 0.002 | ||||||
|
DY629391 |
Non-LTR retrotransposon Rex3_Tet |
0.94 |
ns |
ns |
ns |
122 |
| 0.0028 | ||||||
| DY631649 | SINE sequence | ns | 0.78 |
ns | ns | 138 |
| 0.0002 |
Genomic Content Value: Log2 Hybridization coefficients for each species relative to A. burtoni as estimated by the linear model following background correction and normalization followed by the uncorrected p-value for those significantly different (P < FDR 0.1). BitScore: the quality of the alignment for the annotated homology. A.twe: A. tweddlei; M.est: M. estherae; P.sim: P. similis; R.chi: R. “chilingali”; “ns”: not significant; superscript numbers: when multiple array features represent a gene, these numbers indicate the number of significant array features and the total number of array features for that gene represented in the table by a single GenBank number and data row.
Table 2.
Genes duplicated relative to A. burtoni with no informative BLAST hit
| GenBank | A.twe | M.est | P.sim | R.chi |
|---|---|---|---|---|
|
CN4691252/2 |
1.32 |
ns |
ns |
ns |
| 2E-5 | ||||
|
CN469431 |
ns |
ns |
ns |
0.50 |
| 0.0017 | ||||
|
CN469460 |
ns |
ns |
0.64 |
ns |
| 0.0007 | ||||
|
CN469913 |
ns |
ns |
ns |
0.63 |
| 0.0008 | ||||
|
CN470216 |
ns |
0.39 |
ns |
ns |
| 0.0039 | ||||
|
CN470402 |
ns |
ns |
0.48 |
0.45 |
| 0.0034 |
0.0051 |
|||
|
CN470540 |
ns |
ns |
ns |
0.60 |
| 0.0051 | ||||
|
CN470597 |
0.65 |
ns |
ns |
ns |
| 0.0013 | ||||
|
CN470646 |
0.73 |
ns |
ns |
ns |
| 0.0003 | ||||
|
CN470701 |
ns |
ns |
ns |
0.65 |
| 0.0052 | ||||
|
CN470713 |
ns |
ns |
0.68 |
ns |
| 0.0009 | ||||
|
CN4707241/2 |
1.02 |
ns |
ns |
ns |
| 0.0027 | ||||
|
CN470781 |
0.58 |
ns |
ns |
ns |
| 0.0025 | ||||
|
CN470835 |
ns |
1.55 |
ns |
ns |
| 0.0002 | ||||
|
CN470857 |
ns |
0.67 |
ns |
0.96 |
| 0.005 |
0.0006 |
|||
|
CN470988 |
ns |
1.28 |
ns |
1.34 |
| 0.0007 |
0.0005 |
|||
|
CN471261 |
ns |
ns |
0.93 |
ns |
| 0.001 | ||||
|
CN471811 |
ns |
1.35 |
1.22 |
ns |
| 6E-6 |
1E-5 |
|||
|
CN471851 |
ns |
ns |
0.47 |
ns |
| 0.0018 | ||||
|
CN472050 |
ns |
ns |
ns |
0.70 |
| 0.0002 | ||||
|
DY625804 |
ns |
1.23 |
ns |
ns |
| 0.0001 | ||||
|
DY6258451/2 |
1.16 |
ns |
ns |
ns |
| 6E-6 | ||||
|
DY625884 |
0.49 |
ns |
ns |
ns |
| 0.0024 | ||||
|
DY625919 |
1.18 |
ns |
ns |
ns |
| 0.0001 | ||||
|
DY626122 |
1.50 |
ns |
ns |
ns |
| 2E-5 | ||||
|
DY626140 |
ns |
1.05 |
ns |
ns |
| 0.0036 | ||||
|
DY626192 |
ns |
0.75 |
ns |
ns |
| 0.0002 | ||||
|
DY626304 |
ns |
ns |
0.99 |
1.50 |
| 4E-5 |
2.2E-6 |
|||
|
DY626389 |
ns |
ns |
0.57 |
ns |
| 0.0022 | ||||
|
DY626428 |
ns |
ns |
ns |
0.39 |
| 0.0053 | ||||
|
DY626737 |
ns |
ns |
0.79 |
ns |
| 0.0001 | ||||
|
DY626766 |
ns |
0.81 |
0.98 |
0.68 |
| 0.0003 |
0.0001 |
0.0009 |
||
|
DY627085 |
ns |
0.44 |
ns |
ns |
| 0.0043 | ||||
|
DY627087 |
1.15 |
ns |
ns |
ns |
| 3E-5 | ||||
|
DY627338 |
0.72 |
ns |
ns |
ns |
| 0.0006 | ||||
|
DY627361 |
ns |
ns |
ns |
0.74 |
| 0.0001 | ||||
|
DY627641 |
ns |
0.76 |
0.85 |
ns |
| 0.0012 |
0.0006 |
|||
|
DY627780 |
ns |
1.51 |
ns |
ns |
| 3E-6 | ||||
|
DY627911 |
1.04 |
0.87 |
0.49 |
ns |
| 5E-5 |
0.0002 |
0.0046 |
||
|
DY628148 |
0.50 |
ns |
ns |
ns |
| 0.0012 | ||||
|
DY628172 |
1.38 |
ns |
ns |
ns |
| 4E-6 | ||||
|
DY628268 |
ns |
ns |
0.46 |
ns |
| 0.0017 | ||||
|
DY628316 |
ns |
0.64 |
ns |
ns |
| 0.0026 | ||||
|
DY628477 |
ns |
ns |
ns |
0.58 |
| 0.0052 | ||||
|
DY628517 |
0.76 |
ns |
ns |
ns |
| 0.0002 | ||||
|
DY628561 |
ns |
ns |
ns |
0.42 |
| 0.0052 | ||||
|
DY628642 |
ns |
ns |
1.62 |
1.13 |
| 9E-5 |
0.0087 |
|||
|
DY6287021/2 |
ns |
ns |
ns |
0.67 |
| 0.0004 | ||||
|
DY628714 |
ns |
ns |
ns |
0.48 |
| 0.0027 | ||||
|
DY629058 |
ns |
ns |
ns |
2.41 |
| 3.7E-5 | ||||
|
DY629123 |
ns |
0.87 |
0.83 |
1.41 |
| 0.0001 |
0.0001 |
3.8E-6 |
||
|
DY629387 |
ns |
ns |
1.18 |
ns |
| 4E-5 | ||||
|
DY629482 |
1.39 |
0.71 |
ns |
1.12 |
| 1E-5 |
0.0008 |
4.3E-5 |
||
|
DY629717 |
ns |
ns |
1.60 |
1.28 |
| 0.0002 |
0.0007 |
|||
|
DY6298821/2 |
ns |
ns |
ns |
0.77 |
| 0.0032 | ||||
|
DY629912 |
1.41 |
2.21 |
1.06 |
1.16 |
| 8E-6 |
0.0003 |
7E-6 |
1.1E-6 |
|
|
DY630229 |
ns |
0.88 |
0.83 |
ns |
| 0.0015 |
0.0021 |
|||
|
DY630284 |
ns |
0.54 |
ns |
ns |
| 0.0032 | ||||
|
DY6303731/2 |
ns |
0.89 |
1.00 |
1.23 |
| 0.0001 |
6E-5 |
1.5E-5 |
||
|
DY630867 |
0.97 |
0.64 |
ns |
ns |
| 0.0002 |
0.0024 |
|||
|
DY630964 |
ns |
ns |
ns |
0.95 |
| 0.0037 | ||||
|
DY630993 |
ns |
0.67 |
ns |
ns |
| 0.002 | ||||
|
DY631067 |
ns |
0.78 |
0.80 |
1.02 |
| 0.0008 |
0.0007 |
0.0002 |
||
|
DY631315 |
ns |
ns |
1.57 |
1.16 |
| 0.003 |
0.0019 |
|||
|
DY631408 |
ns |
ns |
0.50 |
ns |
| 0.0033 | ||||
|
DY631442 |
ns |
1.40 |
1.16 |
ns |
| 0.0014 |
0.004 |
|||
|
DY6315051/2 |
ns |
0.72 |
ns |
ns |
| 0.0029 | ||||
|
DY631507 |
0.67 |
0.69 |
0.78 |
0.61 |
| 0.0013 |
0.0011 |
0.0005 |
0.0022 |
|
|
DY631680 |
ns |
ns |
ns |
1.02 |
| 0.0019 | ||||
|
DY631698 |
ns |
ns |
1.03 |
ns |
| 0.0006 | ||||
|
DY631821 |
ns |
ns |
0.61 |
0.60 |
| 0.0038 |
0.0039 |
|||
|
DY631827 |
ns |
0.86 |
ns |
ns |
| 0.0046 | ||||
|
DY631850 |
0.85 |
ns |
ns |
ns |
| 0.0009 | ||||
|
DY631869 |
ns |
0.39 |
ns |
ns |
| 0.0038 | ||||
|
DY632007 |
0.99 |
ns |
ns |
ns |
| 0.0002 | ||||
|
DY6320583/3 |
ns |
0.90 |
0.72 |
0.71 |
| 0.0007 |
0.0026 |
0.0028 |
||
|
DY632092 |
ns |
1.09 |
ns |
ns |
| 0.0007 | ||||
|
DY632097 |
ns |
0.79 |
ns |
0.82 |
| 0.0031 |
0.0002 |
|||
|
DY6321342/2 |
0.94 |
0.86 |
ns |
0.84 |
| 0.0002 |
0.0003 |
0.0003 |
||
|
DY632256 |
ns |
ns |
1.56 |
ns |
| 0.0033 | ||||
| DY632294 | ns | 0.41 |
ns | ns |
| 0.0035 |
Genomic Content Value: Log2 Hybridization coefficients for each species relative to A. burtoni as estimated by the linear model following background correction and normalization followed by the uncorrected p-value for those significantly different. A.twe: A. tweddlei; M.est: M. estherae; P.sim: P. similis; R.chi: R. chilingali; “ns”: not significant; “*”: the GenBank number is a representative for multiple array features for that gene.
Figure 2.
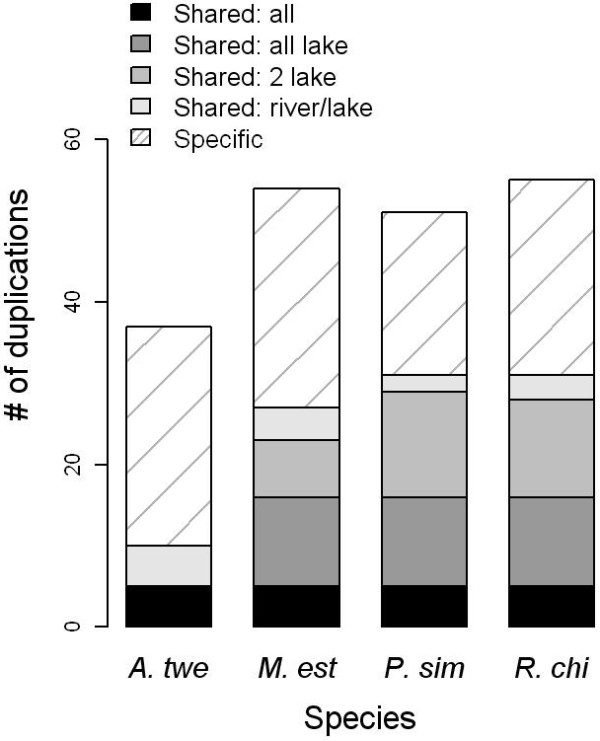
Genes identified as duplicated among test species (P < 0.1 FDR). A. twe: A. tweddlei; M. est: M. estherae; P. sim: P. similis; R. chi: R. “chilingali”. Shared: genes found duplicated in multiple species; Specific: genes found duplicated in only one species; lake: species belonging to the Lake Malawi radiation (M. estherae, P. similis, R. “chilingali”); river: the river species A. tweddlei.
In twenty cases, the gene identified as duplicated was represented on the array by multiple features. Five of these instances showed complete concordance among the two or three array features representing that gene such that all showed the same significant pattern across species. However, for some genes found to be duplicated, only one of the two (n = 8), one of the three (n = 4), two of three (n = 2) or in one case four of the eight array features representing that gene reached statistical significance. In most cases, those features that did not reach statistical significance followed a similar pattern (Additional file 1 Figure S1). However this was not always the case which may be due to different parts of the gene sequence being represented by the different features, high variance or poor quality for one of the features, miss-annotation of the array, or other technical reasons.
BLAST comparison of array feature sequence similarity to the nucleotide database allows annotation and predicted function for discussion of possible adaptive processes. Based on these annotations, several candidate genes were identified as duplicated in and among lineages. Repeated similarities of functional annotations were noticed, particularly for genes involved in immune response, ATP metabolism and detoxification.
Quantitative PCR verification
Four loci found to be duplicated in one or more test species according to aCGH were chosen for quantitative PCR (qPCR) validation for their observed duplication patterns- one duplicated in all species relative to A. burtoni, two duplicated in all three Lake Malawi radiation species and one species-specific duplication (Table 2). Primer pairs that were designed to A. burtoni sequence successfully amplified product with a similar or slightly reduced efficiency in each heterologous species tested (Table 2). We estimated the copy number relative to A. burtoni for these loci based on the array hybridization ratio, and compared that to the copy number estimated from the qPCR results. Each duplication of a given locus as identified by the microarray analysis also showed significantly increased copy number of that locus according to the qPCR analysis (Figure 3). Furthermore, the pattern of relative copy number among test species observed in the qPCR analysis, reflected, with few exceptions, the pattern of relative copy number observed in the microarray analysis. The only notable discrepancy was an increased genomic content for gene DY631898 detected for M. estherae that was not found by microarray analysis.
Figure 3.
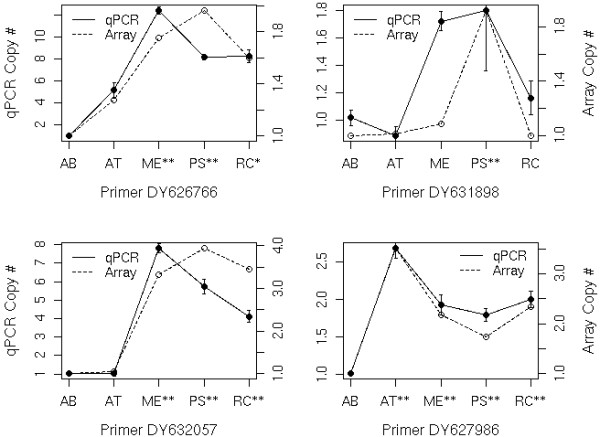
qPCR validates gene copy number determined by aCGH. Abbreviations are genus and species initials. Primer loci are named for the Genbank number of the A. burtoni array feature sequence. **P <0.1 FDR, *P <0.2 FDR found by array analysis.
Discussion
Gene duplication is an important source of functional novelty and has a demonstrated role in adaptive evolution [18]. Such adaptations can allow for niche diversification, as has been suggested for thermal adaptation (plants: [25], Antarctic ice fish: [26]) and for metabolic novelty (C–4 photosynthesis: [27]). The adaptive radiations of the African cichlid fishes exhibit remarkable niche exploitation in the presence of low levels of sequence divergence (reviewed by [13,21]). However, little is known regarding the relative number of duplicated genes, nor the identity of duplicated genes, within this group. If there is an increased rate of gene duplication or gene duplicate retention in radiated lineages, or if particular duplications are associated with these lineages, then their pattern and identity could provide insight into the processes facilitating the rapid expansion of the African cichlids. The patterns reported and validated here indicate shared and increased gene duplication within the Lake Malawi radiation compared to a close non-radiating lineage. While three of the identified gene duplicates were annotated as mobile elements (retrotransposons or SINE element), the majority of the genes could be assigned functional annotation based on a manually curated homology search to UniProtKB/Swiss-Prot for those genes found to be duplicated. Based on individual gene names and functional annotations, several candidate genes, including those that are involved in immune response, ATP metabolism and detoxification, are identified as duplicated in and among lineages (Table 1). Some of these gene duplicates may underlie adaptive phenotypic change.
Immune response
The evolution of immune response is a potent factor contributing to the divergence of lineages, resulting from strong selection on certain loci [28-30]. A greater number of genes associated with immune response (4–9) are found to be duplicated in the Lake Malawi lineage as compared to the riverine species (2). This list includes two finTRIM genes (one duplicated in P. similis and the other in both P. similis and R. “chilingali”), a gene family that is known to play a role in immunity against viral infection, and several finTRIM paralogs have been found in teleost fishes, resulting from duplication and positive selection (70 in trout, 84 in zebrafish) [31]. There are also five major histocompatibility complex (MHC) genes- two MHC class I, two MHC class II, and kinesin-like protein 2- found duplicated in one or more of the species from the radiated Lake Malawi lineage. The MHC gene family, in addition to being involved in immunity (salmon: [32]), has a history of expansion and contraction through duplication and deletion [33]. MHC gene families vary in size among teleosts, with particularly large families in cichlids [34-38]. Additional immune related genes duplicated in the Lake Malawi radiation include an immunoglobulin light chain, small inducible cytokine (associated with the MHC region in stickleback: [39]), and sestrin 3. In A. tweddlei, the test species from the non-radiated lineage, two immune genes, kallikrein-8 and natural killer cell lecin-type receptor, are also found to be duplicated. The identification of several duplicated immune function genes is consistent with previous work documenting size variability and rapid expansion of immune function gene families (Drosophila: [28], silkworm: [40]) that may allow species to invade new niches or better adapt to existing ones.
ATP metabolism
ATP metabolism and function is critical to many physiological processes. Two ATP synthases and one ATP transporter are found duplicated among the four species. Subunits G and E of vacuolar ATPases, which couple the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes, are duplicated in A. tweddlei and M. estherae, respectively. In R. “chilingali”, the adenine nucleotide translocator (ANT) s598 is found duplicated. This mitochondrial transmembrane protein is the most abundant mitochondrial protein and is integral in the exchange of ADP and ATP between the mitochondria and the cytoplasm. Increased expression of mitochondrial ATP synthase has been found in cold acclimated carp [41] and ANT genes are being studied for their potential adaptive role in thermal acclimation (fugu: [42]). Given that these ATP synthase and transport genes are found duplicated in all 4 species of this study rather than showing enrichment only within the Lake species, they may represent an ancestral duplication, or deletion in A. burtoni, nonetheless, their retention may be associated with adaptation to ecological conditions.
Detoxification
Selection on duplicated detoxification genes (those involved in the breakdown of toxic compounds) can determine survival in particular environments or can contribute to expansion into new niches. One example is seen in plant-herbivore interactions, where gene duplication has been implicated in the ability of herbivores to detoxify plant defense compounds and prevent exclusion of the herbivore from that food source [43,44]. We detect duplication of detoxification genes in all three species from the radiated lineage. In P. similis and R. “chilingali”, the sulfotransferase (SULT) gene cytosolic sulfotransferase 3 is found duplicated. SULT genes are detoxifying enzymes that catalyze the transfer sulfonate groups to endogenous compounds and xenobiotics. Once sulfated, compounds may become more easily excreted from the body. In zebrafish, ten SULT proteins have been cloned, two of which show strong activity towards environmental estrogens [45]. Zebrafish SULTs have also been found to act on other xenobiotics [46]. In Atlantic cod, a SULT gene was found to be upregulated in response to polluted water [47]. In R. “chilingali”, two other genes involved in detoxification, arsenic methyltransferase and ferritin (heavy subunit), are found duplicated. Arsenic methyltransferase converts inorganic arsenic into less harmful methylated species, and ferritin is an iron storage protein that is essential for iron homeostasis, keeping iron concentrations at non-toxic levels. Another iron-related protein, the iron-sulfur cluster assembly enzyme, was also duplicated in R. “chilingali”. It is possible that some of these gene duplicates have been retained due to a selective advantage for metabolic breakdown of environmental compounds and toxins. Such duplicates may allow novel physiological interactions with the chemical, physical and pathogenic environment that may play a role in adaptive divergence as a lineage radiates to inhabit new niches such as those associated with the African Great Lakes.
Gene family membership
Gene families by their very nature reveal a propensity for duplication and duplicate retention of certain genes. One study estimated that 38% of known human genes can be assigned to gene families, based on amino acid sequence similarity [48]. These gene families typically consist of two genes, but the largest gene families can have more than 100 members. In the present study, several of the genes found to be duplicated were members of large gene families, comprised of multiple known genes. These include 40 S and 60 S ribosomal proteins (duplicated in R. “chilingali” and M. estherae), claudin 29a (M. estherae), GTPase IMAP family member 7 (P. similis), C–type lectin domain family 4 (M. estherae), high-mobility group 20B (HMG20B) from HMG-box superfamily (A. tweddlei), and hox gene cluster genes (all species). Hox genes are important in the regulation of development, and have been found to be associated with differential jaw development in cichlid fishes [49,50]. An immunoglobulin light chain gene belonging to the largest gene family represented in this study was found duplicated in P. similis. Since large gene families are comprised of multiple paralogs and may possess a greater tendency for expansion, it is not surprising that large gene families are well represented in our list of duplicated regions.
qPCR verification
The robust validation of aCGH results using quantitative PCR not only verifies the increased genomic content for all four loci analyzed in test species relative to A. burtoni, it also provides a complementary approach that may prove to be a more efficient means to survey candidate loci in future population level analyses. For each locus except DY631898, the pattern of copy number among the four test species relative to A. burtoni is similar to that found by aCGH. However, the copy number estimated by qPCR differs from that estimated with array results. This is particularly true of the DY626766 and DY632057 loci, which showed greater qPCR copy number than predicted, despite the underestimation bias possible for those loci. Similarly, in M. estherae, the DY631898 locus appeared to be substantially higher in copy number than predicted by the array results. This discrepancy could result from three factors. First, it may be due to the fact that aCGH will produce an underestimate of true copy number when there is sequence divergence of the heterologous species relative to the platform provided the primers are in a conserved sequence region. Second, while qPCR and microarray analyses both provide relative rather than absolute measures, the scale of the relationship measurements may differ due to the difference in normalization techniques applied to the raw data. Finally, particularly for the case of the DY631898 locus in M. estherae, the micorray analysis includes only two replicates for each species and is thus sensitive to technical error where technical failure of qPCR is more easily replicated. Nonetheless, even for the two instances in which reduced primer efficiency in the tested heterologous species would have been expected to result in an underestimate rather than an overestimate of copy number, the pattern identified by aCGH was upheld. Regardless of discrepancies in magnitude, our quantitative PCR results demonstrate, with the exception of one data point, both qPCR,and aCGH are valid techniques for estimation of relative copy number in heterologous species. While aCGH allows one to survey a greater number of genes, the qPCR technique may provide an efficient means to assess copy number variation (CNV) of candidate loci within a larger population in order to illuminate the role of gene duplication on a microevolutionary scale.
Technical considerations
The use of aCGH was initially developed for cancer studies and has been applied to several within species studies, but has less frequently been used to assess between species patterns of gene duplication. Careful consideration of the technical biases and conservative interpretation of the results are warranted [51,52]. The array features analyzed represent only 5689 genes, a fraction (25-30% of a standard vertebrate genome) of predicted total gene content for these species. Furthermore, because genomic content for each gene has been assessed relative to the array platform species A. burtoni, any gene that has equivalent copy number (even if greater than 1) in both the platform and the heterologous species will go undetected. Similarly, those genes that appear to be duplicated in all heterologous species may actually represent a reduction in genomic content in A. burtoni due to gene deletion events. Furthermore, aCGH with spotted cDNA arrays does not allow quantification among different genes and it is therefore impossible to provide absolute copy numbers. We identify five such genes, two annotated as Hox gene cluster genes, one as a Ras-related C3 botulinum toxin substrate gene and two that lack annotation, that appear to be duplicated in all four test species, but which may in fact be deleted in A. burtoni. In our study we do not attempt to distinguish between these two scenarios.
The hybridization bias due to sequence divergence of the heterologous species from the platform species is another an important consideration for the interpretation of aCGH results. Diverged sequences will hybridize less well to the array feature than A. burtoni DNA. Therefore, it follows that duplicated genes for which the paralog is highly diverged will be less likely to be detected as duplicated than duplicated genes with paralogs that are less diverged from the platform species, as found by Machado and Renn [52]. Therefore, older gene duplication events, those with very little purifying selection pressure, and those with strong positive selection in the gene region represented on the array are less likely to be identified, while recent duplication events or highly conserved duplicates are more likely to be identified. Therefore, the results presented here represent a subset of the total gene duplicates that may differ from the subset of gene duplicates identified by other techniques such as sequence assembly or depth of coverage. Gene number and gene copy number identified by short read sequencing technology is prone to overestimation of copy number variation [53]. Nonetheless, the numbers reported here are clearly an under-representation of the total and may present a different phylogenetic pattern of retention than other subsets of gene duplicates.
In this study, we use a recent adaptive radiation so that, whilst strong positive selection on duplicates might be overlooked by the aCGH technique, the majority of very recent duplications are likely to be identified. We find a pattern of increased gene duplication in these Lake Malawi haplochromines, with 38-49% more genes duplicated than in the non-radiated lineage. Care must be taken in interpreting this increase in the context of adaptive radiation, with four primary considerations. First, only a subset of genes (i.e. those present on the array with available sequence) was tested. Second, gene duplicates may have become fixed in ancestral populations due to neutral processes such as founder events, genetic bottlenecks or drift during the relatively recent evolutionary past. Sequence data from multiple species will be necessary to distinguish neutral vs. adaptive evolutionary processes. Third, due to the shared evolutionary history of the three Lake Malawi species, they cannot be considered independent. Fourth, the ecology of the species, lake versus riverine, is confounded with the tendency to radiate. Therefore, as tantalizing as these results are, our single comparison of radiated versus non-radiated lineages requires further support before general patterns associated with adaptive radiation can be rigorously discussed. Fortunately, the African cichlids provide such a system with which to undertake this [14].
Conclusions
Only recently have studies begun to examine the patterns of gene duplication and copy number polymorphism across species in natural systems, beyond primates (e.g. [26,54-56]). While other studies have examined specific genes (e.g. [57-59]), we present the largest analysis thus far of genome wide patterns of gene duplication across lineages of the African cichlid radiations. We identify several candidate gene duplicates in four cichlid species and find a pattern of increased gene duplication within the Lake Malawi radiation. While our inference regarding the adaptive value of candidate gene duplicates must be tempered, the results of this study support the hypothesis that gene duplication, particularly of genes related to immune response, ATP metabolism and detoxification, is a characteristic of the Lake Malawi adaptive radiation. Assessment across a greater phylogenetic range of cichlid radiations will identify consistent patterns of gene duplication correlated with radiated and non-radiated lineages, and comparative sequence analysis will reveal the potential contribution of natural selection to gene duplicate evolution.
Methods
aCGH identification of duplicated genes
Genomic DNA, extracted from ethanol-preserved field tissue samples (n = 2 per species) by standard ProteinaseK/Phenol protocol, was size reduced by Hydroshear (Genome Solutions/Digilab) to 1–5 Kb. DNA (4 μg) and labeled with Alexa-Fluors (555 & 647) conjugated dCTP by Klenow polymerization (Invitrogen, BioPrime® Direct Array CGH Genomic Labeling System catalog# 18095–011). Each species was hybridized twice (once with each individual) (in dye swap) against a reference pool of A. burtoni genomic DNA using the A. burtoni cDNA PCR product spotted microarray which contains ~20,000 features, representing ~16,000 unique sequences of which ~65% have available EST sequence [60] (GEO platform GPL6416). After a 16 hour hybridization (67.5°C, 3.4× SSC, 0.15% SDS, 1 mM DTT, Cot-1DNA), arrays were washed and scanned (Axon 4100B, Genepix).
Microarray data (GEO series GSE19368) were filtered by omitting features with a lack of sequence information, known ribosomal content, or that had faint array signal (<2 SD above background). Only features that survived this quality control for all eight microarrays were analyzed. Data were corrected for background intensity (“minimum”) and were loess normalized within array using 250 conserved features [60]. This corrects for bias introduced by sequence divergence under standard normalization [61]. Duplicated genes were identified as those with increased fluorescence according to the “lmFit” statistical model with “eBayes” correction and FDR adjustment for P < 0.1 significance level [62]. The reported results are underestimates of duplication levels, due to the fact that diverged duplicates are less likely to be detected [52]. GEL50 measurements [63] indicated that experiments were of similar statistical power (M. estherae: 1.80, P. similis: 1.95, R. “chilingali”: 1.61, A. tweddlei: 1.89). The automated annotations available from DFCI were not used in this study because many proved to be uninformative. Instead, functional annotations for genes were gathered only for identified duplicates using BLASTn to compare EST sequences to the UniProtKB/Swiss-Prot database. The top 100 hits were returned in order to identify informative annotations and infer function based on homology. Bit scores are reported for these annotations. No filtering or masking was applied during the BLASTn thus annotations for repetitive sequences and transponsons are included.
Quantitative PCR
Genomic content was validated for four genes using qPCR (Table 3). gDNA concentration was quantified with 1.5× SYBR Green I (Roche Applied Science) on a Nanodrop 3300 (Thermosavant). Triplicate qPCR reactions (Opticon MJ Research) contained 0.75× SybrGreen, 1× Immomix (Biolabs), 200–500 nM primers and 0.2 ng sample DNA in 10 μl reactions (95°C– 10 min; 35 cycles of: 94°C– 2 min, 60°C- 20 sec, 72°C- 15 sec, and 2 min extension). Copy number relative to A. burtoni was calculated as CT, the cycle number at a set threshold relative to the A. burtoni standard curve, standardized to an A. burtoni copy number of 1. Primer efficiency was calculated with a dilution series for A. burtoni DNA and one test species (Additional file 2: Table S2).
Table 3.
Oligonucleotide primers used for qPCR designed against GenBank sequence available for microarray features
| GenBank | Primer sequence | Homology | |
Primer efficiency |
|
|---|---|---|---|---|---|
| Predicted length | A. burtoni | Test species | |||
|
DY626766 |
F: TCGGTCTCCTTAACCGGATG |
No Hit |
193 |
86 |
74 |
| |
R: CTGAGTTTGGCTGCCCGTAA |
|
|
|
(P. similis) |
|
DY627986 |
F: ACGAACACCCGAACGGAAAC |
Hox gene cluster |
222 |
100 |
104 |
| |
R: GGTGCACGCACATGAACTGT |
|
|
|
(M. estherae) |
|
DY631898 |
F: CGTCCCAGTGAGGATGAGGA |
MHC class II antigen |
161 |
82 |
82 |
| |
R: TGATGCTGATCGGTTGATGC |
|
|
|
(R. “chilingali”) |
|
DY632057 |
F: ATTACTGCGAGTGCCGTCCA |
Pituitary adenylate cyclase activating |
150 |
91 |
78 |
| R: CTGCGCCCTGAAAGAACAGA | polypeptide receptor 1A | (A. tweddlei) | |||
Primer Efficiency: percent is based on 4-fold template dilutions for A. burtoni and one heterologous test species indicated in parentheses.
Availability of supporting data
The data sets supporting the results of this article are available in the GEO repository, [GSE19368: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19368) and DRYAD (doi:10.5061/dryad.7vs2c http://datadryad.org/resource/doi:10.5061/dryad.7vs2c).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SCPR, DHL, DJ conceived of the project. HEM, CRLR, GJ performed the experiments. HEM conducted the analyses. SCPR, HEM, DHL prepared the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Genes Identified as duplicated that are represented on the array by more than one microarray feature. Those with perfect concordance for significance calls (n = 5) are not shown. In each plot, the y-axis represents Log2 hybridization ration for the heterologous species relative to A. burtoni. Each line indicates an individual microarray feature that is statistically significant (P < FDR 0.1, black), marginally significant (P < FDR 0.2, solid grey) or not significant (grey dashed) for genes represented by 2 microarray features (A-H), 3 microarray features (I-N) and 8 microarray features (O). AT: A. tweddlei; ME: M. estherae; PS: P. similis; RC: R. chilingali.
Oligonucleotide primers and efficiency used for qPCR designed against GenBank sequence available for microarray features.
Contributor Information
Heather E Machado, Email: machadoheather@gmail.com.
Ginger Jui, Email: ginger.jui@gmail.com.
Domino A Joyce, Email: d.joyce@hull.ac.uk.
Christian RL Reilly, III, Email: Christian.Reilly@santacatalina.org.
David H Lunt, Email: d.h.lunt@hull.ac.uk.
Suzy CP Renn, Email: renns@reed.edu.
Acknowledgements
Funded by Murdock Charitable Life Trust and NSF-OIS 0818957. Thanks to Martin J Genner for Rhamphochromis “chilingali” samples.
References
- Dobzhansky T. Genetics of the evolutionary process. New York: Columbia University Press; 1937. [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge, Massachusetts: Belknap Press of Harvard University Press; 1963. [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- Slatkin M. Ecological Character Displacement. Ecology. 1980;61:163–177. doi: 10.2307/1937166. [DOI] [Google Scholar]
- Smith JM. Sympatric speciation. Am Nat. 1966;100:637. doi: 10.1086/282457. [DOI] [Google Scholar]
- Gould SJ. Wonderful Life: The Burgess Shale and the Nature of History. New York: W.W. Norton & Company; 1989. [Google Scholar]
- Darwin C. The Origin of Species. New York: Bantam Books; 1859. [Google Scholar]
- Fryer G. Comparative aspects of adaptive radiation and speciation in Lake Baikal and the great rift lakes of Africa. Hydrobiologia. 1990;211:137–146. [Google Scholar]
- Losos JB, Jackman TR, Larson A, de Queiroz K, Rodriguez S. Contingency and determinism in replicated adaptive radiations of island lizards. Science. 1998;279:2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- Baldwin BG, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc Natl Acad Sci U S A. 1998;95(16):9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer G, Iles TD. The cichlid fishes of the Great Lakes of Africa: Their biology and evolution. Edinburgh: Oliver & Boyd; 1972. [Google Scholar]
- Farias IP, Orti G, Meyer A. Total evidence: molecules, morphology, and the phylogenetics of cichlid fishes. J Exp Zool. 2000;288(1):76–92. doi: 10.1002/(SICI)1097-010X(20000415)288:1<76::AID-JEZ8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5(4):288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Seehausen O. African cichlid fish: a model system in adaptive radiation research. Proc R Soc Lond B Biol Sci. 2006;273(1597):1987–1998. doi: 10.1098/rspb.2006.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, Karimpour-Fard A, Glueck D, McGavran L, Berry R, Pollack J, Sikela JM. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2(7):937–954. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang ZS, Baker C, Malfavon-Borja R, Fulton LA, Alkan C, Aksay G, Girirajan S, Siswara P, Chen L, Cardone MF, Navarro A, Mardis ER, Wilson RK, Eichler EE. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457(7231):877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, Turner GF. Age of cichlids: new dates for ancient lake fish radiations. Mol Biol Evol. 2007;24(5):1269–1282. doi: 10.1093/molbev/msm050. [DOI] [PubMed] [Google Scholar]
- Joyce DA, Lunt DH, Genner MJ, Turner GF, Bills R, Seehausen O. Repeated colonization and hybridization in Lake Malawi cichlids. Curr Biol. 2011;21(3):R108–R109. doi: 10.1016/j.cub.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Loh Y, Bezault E, Muenzel F, Roberts R, Swofford R, Barluenga M, Kidd C, Howe A, Di Palma F, Lindblad-Toh K, Hey J, Seehausen O, Salzburger W, Kocher TD, Streelman JT. Origins of Shared Genetic Variation in African Cichlids. Mol Biol Evol. 2013;30:906–917. doi: 10.1093/molbev/mss326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YHE, Katz LS, Mims MC, Kocher TD, Yi SV, Streelman JT. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9(7):R133. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve SR, Rudi H, Asp T, Rognli OA. Tracking the evolution of a cold stress associated gene family in cold tolerant grasses. BMC Evol Biol. 2008;8:245. doi: 10.1186/1471-2148-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZZ, Cheng CHC, Zhang JF, Cao LX, Chen L, Zhou LH, Jin YD, Ye H, Deng C, Dai ZH, Xu X, Hu P, Sun S, Shen Y, Chen L. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc Natl Acad Sci U S A. 2008;105(35):12944–12949. doi: 10.1073/pnas.0802432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson RK. Gene duplication, neofunctionalization, and the evolution of C-4 photosynthesis. Int J Plant Sci. 2003;164(3):S43–S54. [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39(12):1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11(1):17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Little TJ. Immunity in a variable world. Philos Trans R Soc Lond B Biol Sci. 2009;364(1513):15–26. doi: 10.1098/rstb.2008.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Aa LM, Levraud JP, Yahmi M, Lauret E, Briolat V, Herbomel P, Benmansour A, Boudinot P. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009;7:7. doi: 10.1186/1741-7007-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs MF, Harstad H, Grimholt U, Beetz-Sargent M, Cooper GA, Reid L, Bakke HG, Phillips RB, Miller KM, Davidson WS, Koop BF. Genomic organization of duplicated major histocompatibility complex class I regions in Atlantic salmon (Salmo salar) BMC Genomics. 2007;8:251. doi: 10.1186/1471-2164-8-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Kaukinen KH, Schulze AD. Expansion and contraction of major histocompatibility complex genes: a teleostean example. Immunogenetics. 2002;53(10–11):941–963. doi: 10.1007/s00251-001-0398-4. [DOI] [PubMed] [Google Scholar]
- Malaga-Trillo E, Zaleska-Rutczynska Z, McAndrew B, Vincek V, Figueroa F, Sultmann H, Klein J. Linkage relationships and haplotype polymorphism among cichlid Mhc class II B loci. Genetics. 1998;149(3):1527–1537. doi: 10.1093/genetics/149.3.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Withler RE. The salmonid class I MHC: limited diversity in a primitive teleost. Immunol Rev. 1998;166:279–293. doi: 10.1111/j.1600-065X.1998.tb01269.x. [DOI] [PubMed] [Google Scholar]
- Persson AC, Stet RJM, Pilstrom L. Characterization of MHC class I and beta(2)-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes. Immunogenetics. 1999;50(1–2):49–59. doi: 10.1007/s002510050685. [DOI] [PubMed] [Google Scholar]
- Sato A, Figueroa F, O’Huigin C, Steck N, Klein J. Cloning of major histocompatibility complex (Mhc) genes from threespine stickleback, Gusterosteus aculeatus. Mol Mar Biol Biotechnol. 1998;7(3):221–231. [PubMed] [Google Scholar]
- Sato A, Dongak R, Hao L, Shintani S, Sato T. Organization of Mhc class II A and B genes in the tilapiine fish Oreochromis. Immunogenetics. 2012;64(9):679–690. doi: 10.1007/s00251-012-0618-0. [DOI] [PubMed] [Google Scholar]
- Reusch TBH, Schaschl H, Wegner KM. Recent duplication and inter-locus gene conversion in major histocompatibility class II genes in a teleost, the three-spined stickleback. Immunogenetics. 2004;56(6):427–437. doi: 10.1007/s00251-004-0704-z. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1087–1110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Itoi S, Watabe S. Increased levels of mitochondrial ATP synthase beta-subunit in fast skeletal muscle of carp acclimated to cold temperature. Fish Sci. 1999;65(4):629–636. [Google Scholar]
- Itoi S, Misaki R, Hirayama M, Nakaniwa M, Liang CS, Kondo H, Watabe S. Identification of three isoforms for mitochondrial adenine nucleotide translocator in the pufferfish Takifugu rubripes. Mitochondrion. 2005;5(3):162–172. doi: 10.1016/j.mito.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Wen ZM, Rupasinghe S, Niu GD, Berenbaum MR, Schuler MA. CYP6B1 and CYP6B3 of the black swallowtail (Papilio polyxenes): adaptive evolution through subfunctionalization. Mol Biol Evol. 2006;23(12):2434–2443. doi: 10.1093/molbev/msl118. [DOI] [PubMed] [Google Scholar]
- Fischer HM, Wheat CW, Heckel DG, Vogel H. Evolutionary origins of a novel host plant detoxification gene in butterflies. Mol Biol Evol. 2008;25(5):809–820. doi: 10.1093/molbev/msn014. [DOI] [PubMed] [Google Scholar]
- Liu TA, Bhuiyan S, Snow R, Yasuda S, Yasuda T, Yang YS, Williams FE, Liu MY, Suiko M, Carter G, Liu MC. Identification and characterization of two novel cytosolic sulfotransferases, SULT1 ST7 and SULT1 ST8, from zebrafish. Aquat Toxicol. 2008;89(2):94–102. doi: 10.1016/j.aquatox.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Sugahara T, Yang YS, Liu CC, Pai TG, Liu MC. Sulphonation of dehydroepiandrosterone and neurosteroids: molecular cloning, expression, and functional characterization of a novel zebrafish SULT2 cytosolic sulphotransferase. Biochem J. 2003;375:785–791. doi: 10.1042/BJ20031050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie KK, Lanzen A, Breilid H, Olsvik PA. Gene expression profiling in Atlantic cod (Gadus morhua l.) from two contaminated sites using a custom-made cDNA microarray. Environ Toxicol Chem. 2009;28(8):1711–1721. doi: 10.1897/08-517.1. [DOI] [PubMed] [Google Scholar]
- Li WH, Gu ZL, Wang HD, Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409(6822):847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- le Pabic P, Stellwag EJ, Scemama JL. Embryonic Development and Skeletogenesis of the Pharyngeal Jaw Apparatus in the Cichlid Nile Tilapia (Oreochromis niloticus) Anat Rec (Hoboken) 2009;292(11):1780–1800. doi: 10.1002/ar.20960. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An Ancient Gene Network Is Co-opted for Teeth on Old and New Jaws. PLoS Biol. 2009;7(2):233–247. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SCP, Machado HE, Jones A, Soneji K, Kulathinal RJ, Hofmann HA. Using comparative genomic hybridization to survey genomic sequence divergence across species: a proof-of-concept from Drosophila. BMC Genomics. 2010;11:271. doi: 10.1186/1471-2164-11-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado HE, Renn SCP. A critical assessment of cross-species detection of gene duplicates using comparative genomic hybridization. BMC Genomics. 2010;11:304. doi: 10.1186/1471-2164-11-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating Gene Gain and Loss Rates in the Presence of Error in Genome Assembly and Annotation Using CAFE 3. Mol Biol Evol. 2013;30(8):1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104(50):19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A, Vidal O, Amills M. Copy number variation in the genomes of domestic animals. Anim Genet. 2012;43(5):503–517. doi: 10.1111/j.1365-2052.2012.02317.x. [DOI] [PubMed] [Google Scholar]
- Nicholas TJ, Baker C, Eichler EE, Akey JM. A high-resolution integrated map of copy number polymorphisms within and between breeds of the modern domesticated dog. BMC Genomics. 2011;12:414. doi: 10.1186/1471-2164-12-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz AJ, Gunter HM, Fischer JMF, Qiu H, Meyer A, Kuraku S. Ancestral and derived attributes of the dlx gene repertoire, cluster structure and expression patterns in an African cichlid fish. EvoDevo. 2011;2:1. doi: 10.1186/2041-9139-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah S, Laria RL, Gray SM, Hawryshyn CW. Functional diversity in the color vision of cichlid fishes. BMC Biol. 2010;8 doi: 10.1186/1741-7007-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Conte MA, Kocher TD. Circular DNA Intermediate in the Duplication of Nile Tilapia vasa Genes. PLoS One. 2011;6:e29477. doi: 10.1371/journal.pone.0029477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Renn SCP, Steinke D, Braasch I, Hofmann HA, Meyer A. Annotation of expressed sequence tags for the east African cichlid fish Astatotilapia burtoni and evolutionary analyses of cichlid ORFs. BMC Genomics. 2008;9(96):1–14. doi: 10.1186/1471-2164-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hijum S, Baerends RJS, Zomer AL, Karsens HA, Martin-Requena V, Trelles O, Kok J, Kuipers OP. Supervised Lowess normalization of comparative genome hybridization data - application to lactococcal strain comparisons. BMC Bioinforma. 2008;9:93. doi: 10.1186/1471-2105-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–26. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Townsend JP. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinforma. 2004;5:54. doi: 10.1186/1471-2105-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes Identified as duplicated that are represented on the array by more than one microarray feature. Those with perfect concordance for significance calls (n = 5) are not shown. In each plot, the y-axis represents Log2 hybridization ration for the heterologous species relative to A. burtoni. Each line indicates an individual microarray feature that is statistically significant (P < FDR 0.1, black), marginally significant (P < FDR 0.2, solid grey) or not significant (grey dashed) for genes represented by 2 microarray features (A-H), 3 microarray features (I-N) and 8 microarray features (O). AT: A. tweddlei; ME: M. estherae; PS: P. similis; RC: R. chilingali.
Oligonucleotide primers and efficiency used for qPCR designed against GenBank sequence available for microarray features.


