Abstract
Mutations in the olfactomedin domain of myocilin (myoc-OLF) are the strongest link to inherited primary open angle glaucoma. In this recently-identified protein misfolding disorder, aggregation-prone disease variants of myocilin hasten glaucoma-associated elevation of intraocular pressure, leading to vision loss. In spite of its well-documented pathogenic role, myocilin remains a domain of unknown structure or function. Here we report the first small-molecule ligands that bind to the native state of myoc-OLF. To discover these molecules, we designed a general label-free, mix-and-measure, high throughput chemical assay for restabilization (CARS), which is likely readily adaptable to discover ligands for other proteins. Of the 14 hit molecules identified from screening myoc-OLF against the Sigma-Aldrich Library of Pharmacologically Active Compounds using CARS, surface plasmon resonance binding studies reveal three are stoichiometric ligand scaffolds with low micromolar affinity. Two compounds, GW5074 and apigenin, inhibit myoc-OLF amyloid formation in vitro. Structure-activity-relationship-based soluble derivatives reduce aggregation in vitro as well as enhance secretion of full-length mutant myocilin in a cell culture model. Our compounds set the stage for a new chemical probe approach to clarify the biological function of wild-type myocilin, and represent lead therapeutic compounds for diminishing intracellular sequestration of toxic mutant myocilin.
Keywords: ligand binding, olfactomedin, glaucoma, amyloid, myocilin, high throughput assay
INTRODUCTION
In spite of the 15-year-old knowledge of its genetic linkage to the prevalent ocular disorder glaucoma (1), myocilin remains poorly understood. Inherited mutations localized to the myocilin olfactomedin domain (myoc-OLF, ~30 kDa) are the strongest link to early-onset primary open angle glaucoma (2, 3). Mutant myocilin aggregates within trabecular meshwork (TM) cells (2, 3), and recent evidence indicates that these deposits exhibit hallmarks of amyloid (4). Myocilin misfolding appears at least in part due to an aberrant interaction with endoplasmic-reticulum (ER) resident chaperones (5) that then is cytotoxic, leading to apoptosis (6, 7). Wildtype myocilin is also causative in steroid-induced glaucoma (8). How myocilin behavior leads to the major glaucoma-associated risk factor of elevated intraocular pressure has not yet been clarified, however. Likewise, the biological function of myocilin is still enigmatic. Myocilin has been proposed to be a molecular chaperone (9), play structural or signaling role in the TM extracellular matrix, but few binding partners have been identified (3). To complicate matters, myocilin appears to have additional intracellular functions (10, 11) and is expressed in tissues throughout the body, where it does not cause disease (3).
Chemical probes would be excellent tools to address these outstanding functional issues for myocilin, and could lead to targeted therapeutic molecules to inhibit mutant myocilin aggregation. However, like many gene products associated with disease and/or discovered through genome sequencing efforts that lack homology to other well-characterized proteins or functional inferences from bioinformatics, high throughput (HT) screening methods to identify lead compounds are highly limited. Biophysical methods to measure ligand binding such as calorimetry or surface plasmon resonance (SPR) require peripheral automation for HT application and suffer from slow rates of data collection and complex data analysis (12, 13). Similarly, methods to detect changes in stability as a function of temperature, a proxy for binding affinity, are not easily converted to HT (12). While such methods have been reported (13–17), they typically require substantial set-up or workup, specialized equipment, elevated or increasing temperatures, or long incubation times. We were unable to adapt any existing target-independent assay to discover ligands for myocilin in our HT screening facility.
Here we report the discovery of the first ligands for myoc-OLF. To enable HT compound library screening, we first developed a pragmatic chemical assay for re-stabilization (CARS). Under ambient conditions, CARS measures the apparent extent of stabilization upon ligand binding using a visible-wavelength sensor of hydrophobic environments. As detailed below, CARS has a number of positive attributes including the retention of near-native protein structure, fast readout, straightforward data analysis, and requirement for only basic laboratory instrumentation. Three scaffolds that bind stoichiometrically to folded myoc-OLF were identified by CARS and evaluated by secondary assays. These compounds set the stage for an exciting chemical biology approach to probe the biological function of wild-type myocilin ex vivo. Further, derivatives tested as part of a structure-activity relationship (SAR) effort represent novel lead therapeutic compounds for diminishing intracellular sequestration of toxic mutant myocilin. More generally, on the basis of ease of implementation and our success with myoc-OLF, we believe CARS to be readily adaptable as a primary assay to discover ligands for other proteins in which structure or function is not known.
RESULTS
Assay principle and development
Our assay (Figure 1Supporting Information (SI), Table S1) derives from the physico-chemical interplay between ligand binding, equilibrium dissociation constant (Kd), and protein stability, namely, the apparent increase in free energy (IG0) of protein unfolding in the presence of a ligand, L (18)
and the consequence of such apparent stabilization in reducing exposure of interior hydrophobic regions of a protein. The assay takes advantage of the visible wavelength fluorescence properties of Sypro Orange (SO, λex = 470 nm, λem = 580 nm), an environmentally sensitive extrinsic probe. A primary asset of CARS is that no prior knowledge of protein structure and/or function is required to identify binders to a receptor. Conversely, since CARS has no direct functional readout, it is intended for cases where a functional assay is not available. The assay is compatible with routine laboratory equipment kept under ambient conditions as well as HT robotics found in screening facilities.
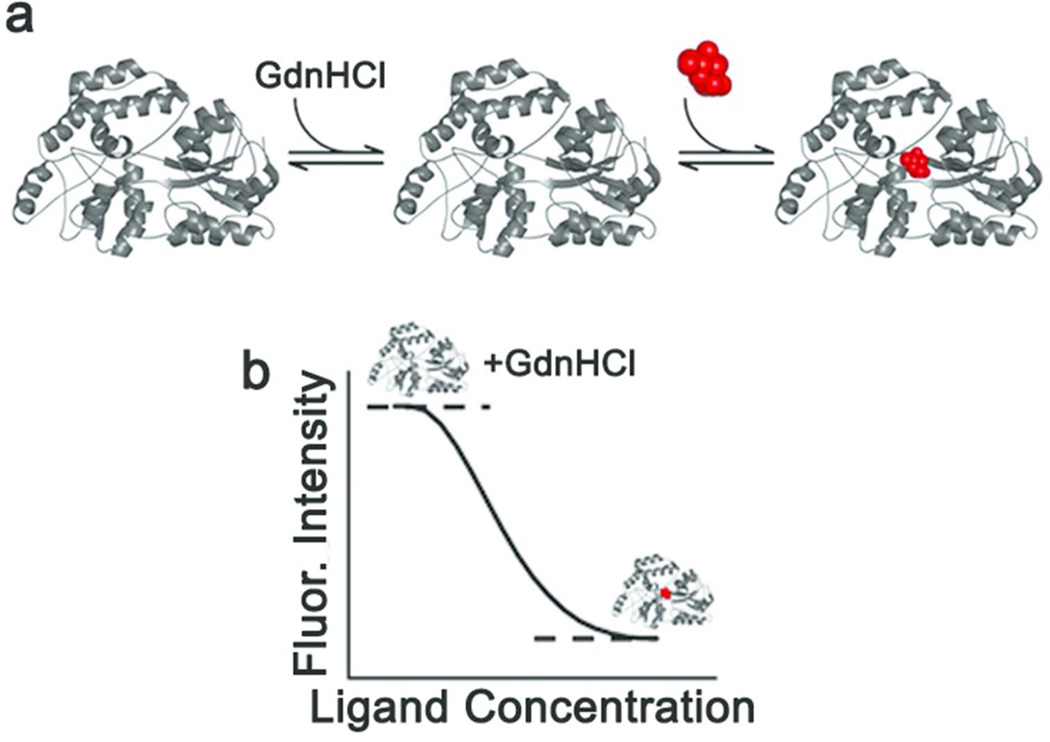
Figure 1. Schematic of CARS.
(a) Native protein (left) is slightly destabilized by the addition of low levels of guanidine hydrochloride (GdnHCl, middle) where it is still folded but less stable, and is restabilized by binding of the ligand (red, right). (b) Sypro Orange (SO) fluorescence signal expected upon the addition of increasing amount of ligand in the CARS setup. High signal is observed for the initially destabilized protein. Upon ligand binding, protein stabilization results in a reduction of hydrophobic exposure, leading to a decrease in SO fluorescence. This creates the signal window.
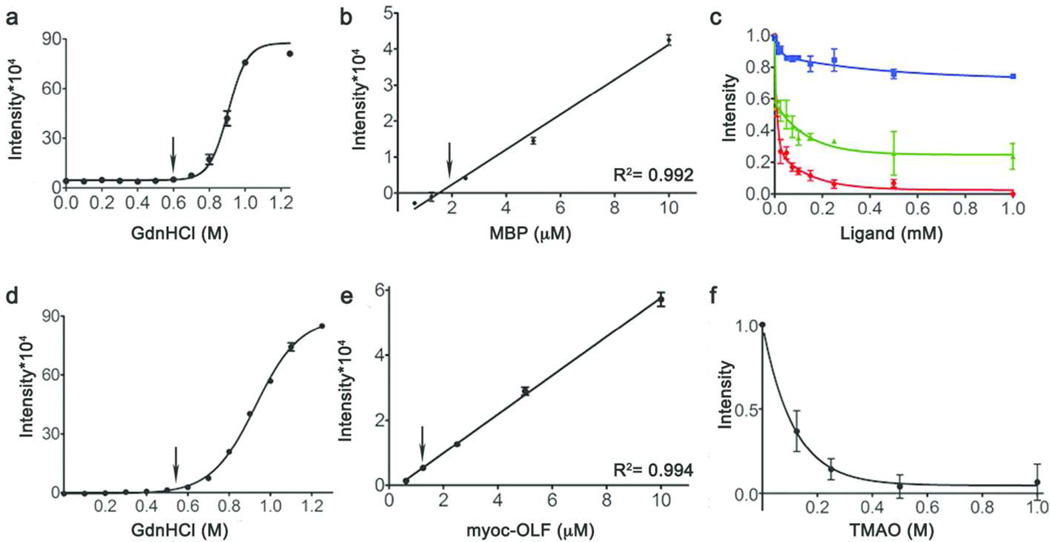
E. coli MBP was used as a proof of concept model protein for CARS. MBP binds the disaccharide maltose as well as longer linear and some circular maltodextrins with Kd values in the low micromolar range (19), within the typical potency range of 100 nM to 5 µM for HT compound library screening (20). The CARS method uses low levels of a chemical denaturant such as guanidinium (GdnHCl) to bring the target protein to a native-like state that has an initial high SO signal. To find the proper concentration of GdnHCl, a chemical melt was conducted with MBP while SO fluorescence was monitored (Figure 2a). In the presence of 0.6 M GdnHCl, a value well below the unfolding transition, MBP was destabilized enough to yield strong SO fluorescence. While higher levels of GdnHCl would further increase the SO fluorescence signal, the native binding site(s) must remain intact for the assay. In addition, for many proteins like myoc-OLF, higher GdnHCl levels would likely lead to irreversible aggregation.
Figure 2. Assay development and application in 96-well format.
(a–c) Development using MBP. (d–f) Application to myoc-OLF. (a, d) Chemical melt with addition of GdnHCl monitoring SO fluorescence. (b, e) Serial dilution to optimize protein concentration for subsequent assays. (c, f) Dose-dependent stabilization by ligands for MBP (red, maltose; green, maltotetraose; blue, maltitol) (c) or TMAO for myoc-OLF (f) as monitored by SO fluorescence. Error bars denote standard deviation, arrows at selected concentrations.
Serial dilutions of MBP in 0.6 M GdnHCl indicated that 2 µM MBP provided a sufficient signal in 96-well format (Figure 2b). To test whether SO fluorescence could detect stabilization in a dose-dependent manner, MBP re-stabilization upon binding of three known ligands – maltose, maltotetraose, and maltitol – was monitored in 96-well format (Figure 2c). Maltose, the highest affinity ligand (Kd = 1 µM) (19), decreased SO fluorescence to the greatest extent, with a 50% decrease in intensity at low micromolar concentrations (Figure 2c). Addition of maltitol had the weakest effect (Figure 2c), consistent with its lower affinity (Kd = 50 µM) (19), but a decrease in SO fluorescence was likewise seen by ~10 µM maltitol. Thus, this setup provides a fluorescence readout that is sufficiently sensitive within the low micromolar concentration range of compounds tested with compound libraries (20) and the binding site of MBP remains recognizable to known ligands under the assay conditions.
CARS applied to MBP exhibits excellent reproducibility and statistics (SI Figure S1). Upon binding to MBP, all three sugars decrease SO fluorescence reproducibly, day-to-day, and plate-to-plate (SI Figure S1a). Neither of the two negative controls tested, PMSF, a known protease inhibitor, nor iodoacetamide, a thiol-modifying reagent (MBP lacks cysteine residues), elicited a change in SO fluorescence in the presence of MBP (SI Figure S1b). Similarly, the assay is compatible with DMSO (SI Figure S1c). The combination of a signal-to-background (S/B) = 2, a Z′ factor of 0.76 (SI Figure S1d), and coefficient of variation (CV) of 4.0% indicates a good HT assay with a large separation between signal and background populations (21–23). The corresponding changes in thermal stability were evaluated by differential scanning fluorimetry (DSF), a medium-throughput thermal assay used to evaluate protein stability using an RT-PCR instrument (24). Using 1 µM MBP, 1mM ligand, the change in melting temperature (ITm) of is 10 K with maltose and maltotetraose, but just 0.9K for the weaker maltitol ligand (SI Table S2).
Assay adaptation to myoc-OLF
For myoc-OLF, chemical melts in the presence of SO also revealed a suitable concentration of 0.5 – 0.6 M GdnHCl for high starting fluorescence signal prior to the onset of unfolding (Figure 2d), and serial dilutions indicated that in the 96-well format, 1 µM myoc-OLF provides measurable signal (Figure 2e). Because no ligands for myoc-OLF were known prior to this assay, our strategy for creating a signal window was to mimic the effect of ligand-binding on myoc-OLF using TMAO, a compound previously shown to stabilize myoc-OLF (25). TMAO is an osmolyte, and thus exerts its stabilizing effect by altering the hydration state of protein surfaces (26). Experiments with TMAO were conducted in the appropriate concentration range for osmolytes, leading to a decrease in SO fluorescence as a function of increasing TMAO, which levels off by 1 M TMAO (Figure 2f). The CARS signal window for myoc-OLF was therefore defined by the difference in SO fluorescence of myoc-OLF in 0.6 M GdnHCl with and without 1 M TMAO. This corresponds to a change in melting temperature (ITm) of ~ K, similar to the ~ K seen for myoc-OLF in the presence of 1 M TMAO without GdnHCl (SI Table S3).
The application of CARS to myoc-OLF is robust (SI Figure S2). Stabilization measured for myoc-OLF exhibits a S/B = 7.7, 4-fold higher than that observed for MBP. The signal window persists for an hour at room temperature or up to three days of incubation in the dark at room temperature with only a slight baseline drift (SI Figure S2a). In addition, there is little plate-to-plate variability (SI Figure S2b) and 5% DMSO is tolerated (SI Figure S2c). Statistical analysis yields a Z′ factor of 0.72 (SI Figure S2d) and a calculated CV of 3.3%, values well above acceptable thresholds (21–23).
Biophysical characterization of myoc-OLF under assay conditions
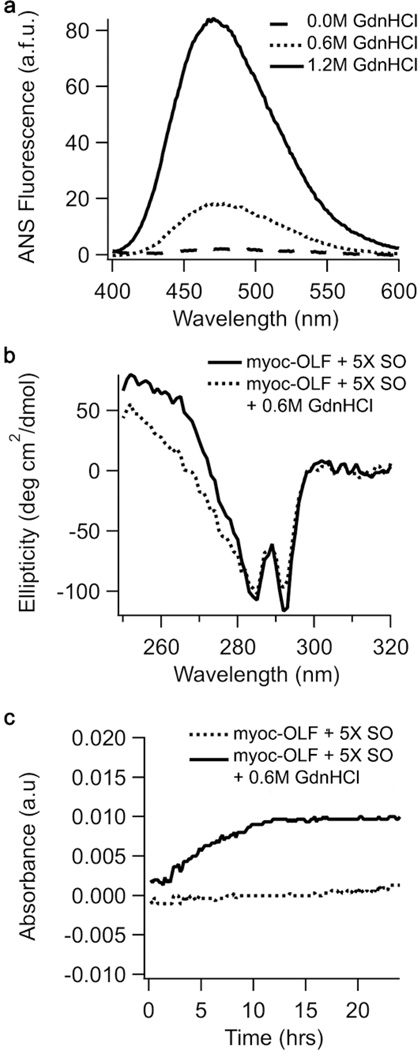
An important aspect of our assay design is to preserve native-like binding sites. Biophysical characterization using 8-anilinonaphthalene-1-sulfonic acid (ANS) fluorescence as an independent measure of exposed hydrophobicity, combined with near-UV circular dichroism (CD) spectra to probe tertiary protein structure, reveals that CARS experimental conditions do not appreciably perturb myoc-OLF. In the presence of 0.6 M GdnHCl, ANS fluorescence is ~25% of fully denatured myoc-OLF (Figure 3a) and there is little difference in tertiary structure between myoc-OLF under experimental conditions when compared to native (Figure 3b). We also wanted to ensure that myoc-OLF did not aggregate under assay conditions. Scatter measurements reveal no appreciable aggregation over a time frame well beyond that required for CARS (Figure 3c). Notably, our optimized experimental conditions are consistent with those of classical biophysical investigations of protein unfolding, in which optical rotation changes were measured upon the addition of compounds to a protein in its partially denatured state (see for example, refs (27, 28)).
Figure 3. Biophysical characterization of myoc-OLF under assay conditions.
(a) ANS fluorescence (b) CD spectra in near-UV region (c) Time-dependent aggregation monitored at 620 nm.
HT screening
Next, CARS was automated at the Emory Chemical Biology Discovery Center and used to screen myoc-OLF against the Sigma-Aldrich Library of Pharmacologically Active Compounds (LOPAC 1280), with a total assay time of ~ 2h. Prior to screening, the assay was readily adapted to a 384-well format (see Methods, SI Table S1). More than adequate signal was measured using 4 µM (0.12 mg/ml) myoc-OLF (S/B = 8) with 0.54 M GdnHCl and restabilized with 0.8 M TMAO. At higher protein concentrations, no appreciable increase in S/B was observed.
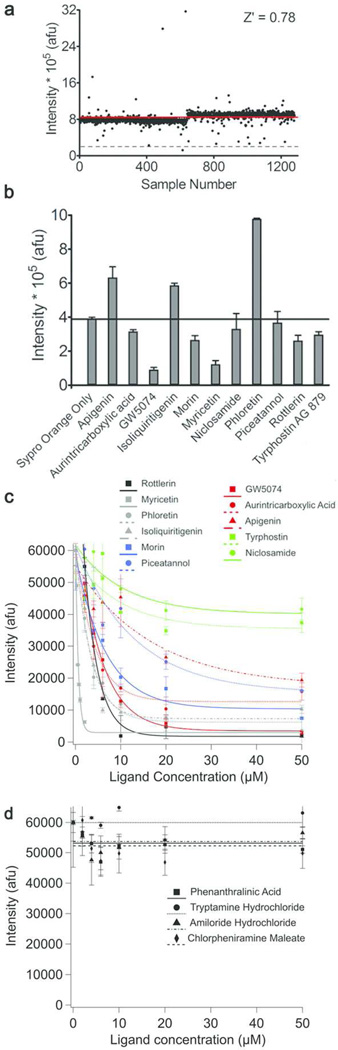
Each compound was added at a single concentration of 16.7 µM. Two wells in each plate were reserved for GdnHCl-destabilized myoc-OLF without any compound (blank) and with 0.8 M TMAO (positive signal), respectively, to define the signal window, and allow for calculation of plate-to-plate statistics to detect any irregularities in the assay. The average Z′ value = 0.78 (0.76 – 0.83) over the 384-well plates used indicates a high-quality HT screening assay (Figure 4a). Similar to values obtained in 96-well format, the S/B ranged between 4.2 and 5.5 and the CV value between 4.3% and 6.1%.
Figure 4. Initial CARS hits and analysis.
(a) Assay readout for myoc-OLF tested against LOPAC library conducted in 384-well format. Red horizontal line: average value; grey dashed horizontal line: fluorescence decrease with TMAO. Hit compounds selected for further analysis were identified by a decrease in fluorescence of 50% or greater compared to that of TMAO. (b) Effect of hits on SO fluorescence in absence of myoc-OLF. (c) Positive hit (d) representative negative hit dose-dependent response. Fluorescence is reported as a function of compound concentration under CARS experimental conditions.
From the pilot screen, hit compounds (hit rate 1.25%) resulted in a 50% or better decrease in SO fluorescence compared to that observed with 0.8 M TMAO (Table 1SI Table S4). Two compounds led to an increase in fluorescence, likely due to interference with SO or highly destabilizing effects. We proceeded with further evaluation of 11 hit compounds based on feasibility of obtaining enough material for downstream experiments. Several compounds that did not elicit a change in fluorescence were selected at random from the screen as negative controls.
Table 1.
Summary of compounds identified in myoc-OLF pilot screen by CARS and binding by SPR
| Compound | ΔTm(°C) | Kd (µM) from SPR |
|---|---|---|
| Apigenin | −0.3 ± 0.2 | 41.6 ± 39.6c |
| Aurintricarboxilic acid | 0.3 ± 0.2 | 17.1 ± 9.6 |
| GW5074 | 0.2 ± 0.0 | 33.5 ± 20.9d |
| Tyrphostin AG 879 | −0.8 ± 0.0 | 38.6 ± 12.7c |
| Myricetin | 1.7 ± 0.0 | linear dose-response |
| Isoliquiritigenin | 1.3 ± 0.1 | linear dose-response |
| Rottlerin | N/Af | 30.1 ± 0.6e |
| Phloretin | 1.2 ± 0.0 | linear dose-response |
| Piceatannol | −0.4 ± 0.1 | sigmoidal dose-response |
| Morin | −0.4 ± 0.4 | weak sigmoidal responsec,d |
| Niclosamide | 0.0 ± 0.2 | no detectable bindingd |
| Quercetin dihydratea | N/A | N/A |
| Reactive Blue 2a | N/A | N/A |
| (R,R)-cis-diethyl tetrahydro-2,8-chrysenediola | N/A | N/A |
Not tested.
Result obtained with surfactant P-20 in buffer solution.
Poor solubility.
Non-stoichiometric, promiscuous binder.
N/A = not available for further analysis.
Evaluation of binding modes of hits by SPR
We first examined the extent of the decrease in modulation of SO fluorescence in the absence of myoc-OLF (Figure 4b); compound library resources precluded testing all compounds in the library for their effects on SO fluorescence without the target protein. GW5074 and myricetin significantly quench SO fluorescence in the absence of myoc-OLF, while three compounds, apigenin, phloretin, and isoliquiritigenin, increase the fluorescence of SO (Figure 4b). The corresponding changes in thermal stability of myoc-OLF in the presence of hits at concentrations used for screening are statistically insignificant (13) (−1 K < ITm < K, Table 1), raising the possibility of false positives, but further evaluation revealed more nuanced results.
Although positive hits exhibit a dose-dependent SO signal decrease when tested in 96- well format (Figure 4c), and in principle a Kd could be estimated this way, traditional secondary assays are required to remove non lead-like compounds and identify the stoichiometric ligands (29). Due to the lack of a functional assay for myoc-OLF, we used SPR (Figure 5, Table 1, and SI Figure S3). The standard immobilization procedure, conducted using buffers in which myoc-OLF is folded and stable (30), provides a random orientation of folded myoc-OLF on the SPR chip for ligand binding. Several compounds, including GW5074, morin, niclosamide, and tyrphostin exhibited poor solubility behavior under standard experimental conditions of SPR, in which the compounds are dissolved in 5% DMSO. As noted below, the binding properties of some compounds benefitted from the addition of surfactant P-20 to the buffer solution, whereas others did not. The only compound that failed to elicit an interpretable response in SPR under all conditions tested was niclosamide, suggesting this compound may be a very weak binder with poor solubility or a false-positive hit. Negative controls produced no dose response in CARS and were well-behaved in SPR (see Figure 4d and SI Figure S3).
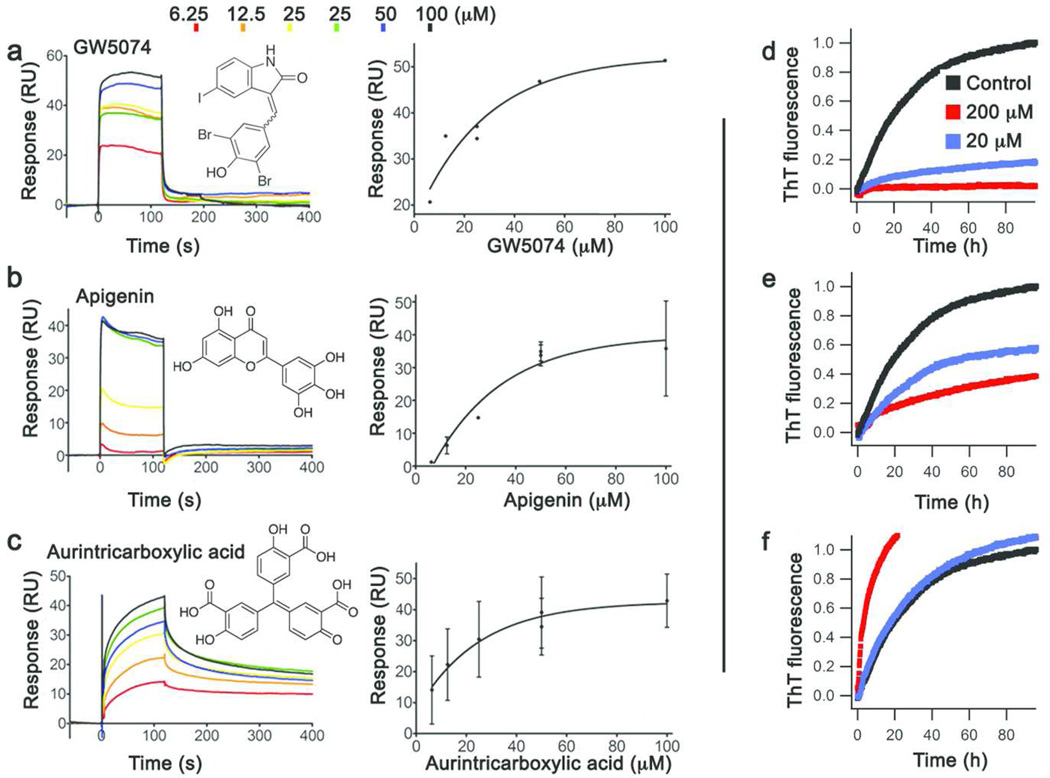
Figure 5. Stoichiometric binders evaluated by SPR and effect on myoc-OLF amyloidogenesis.
Sensorgrams and binding curves for (a) GW5074 (b) apigenin (c) aurintricarboxylic acid. (See also Table 1). (d–f) corresponding inhibition of fibril formation.
Inspection of the SPR data reveals a few promising lead compounds: scaffolds that exhibit saturated binding, with Kd values of ~20–50 µM (Figure 5 and Table 1). GW5074 [3-(3,5-dibromo-4-hydroxybenzylidine-5-iodo-1,3-dihydro-indol-2-one)] elicits a binding curve with a maximal response in SPR corresponding to a stoichiometry of 1:1 (Figure 5a). Apigenin [4’,5,7- trihydroxyflavone] also binds to myoc-OLF with a stoichiometry of approximately 0.5:1, when surfactant P-20 was present in the SPR buffer (Figure 5b). Aurintricarboxylic acid is also an apparent 1:1 stoichiometric binder. Although dissociation from myoc-OLF is not complete (Figure 5c) and aurintricarboxylic acid is known to self-polymerize (31), we elected to continue to consider this compound because carboxylic acid groups were proposed previously to be of functional relevance given the buffer preferences of myoc-OLF (30). Similarly, tyrphostin may also be a ligand, as stoichiometric binding is observed upon the addition of surfactant P-20, but like aurintricarboxylic acid, sensorgrams indicate poor compound behavior in equilibrium response and dissociation (SI Figure S3j). Among the remaining hits, rottlerin is a promiscuous binder (SI Figure S3d), while myricetin, phloretin and isoliquiritigenin bind non-specifically (SI Figure S3a, e, f). We note that myricetin quenched SO fluorescence without myoc-OLF (Figure 4b) with strong dose-dependence, and even appeared to be the best thermal stabilizer (Table 1), but is not a veritable ligand for myoc-OLF. Piceatannol and morin are also non-stoichiometric binders, and their sensorgrams are in line with concentration-dependent compound aggregation (SI Figure S3g,k). Only GW5074, apigenin and aurintricarboxylic acid were considered further; poorly behaved, non-stoichiometric, and/or promiscuous binders, commonly identified from LOPAC, are not good hits for further lead compound development (32, 33).
Evaluation of ligands for inhibition of myoc-OLF fibrilization
Next, with the goal of an anti-aggregation therapeutic agent for myocilin, we developed a fluorescence plate reader-based assay combined with atomic force microscopy (AFM) to evaluate the extent of myoc-OLF amyloid fibrillization upon the addition of compound. The assay monitors amyloid content by thioflavin T (ThT) fluorescence over the course of ~ 3 d in 96-well format (see Methods). GW5074 and apigenin decreased myoc-OLF fibrillization in a dose-dependent manner (Figure 5d, e) with no fibrils apparent by AFM (SI Figure S4); GW5074 exhibited potent inhibition in line with its micromolar affinity measured for myoc-OLF. By contrast, the addition of aurintricarboxylic acid to myoc-OLF led to an increase in ThT fluorescence (Figure 5f) and high levels of deposits similar to vehicle control seen by AFM (SI Figure S4), demonstrating an enhancement of fibril propensity. Considering this result, combined with its known polymerization propensity(31) and problematic behavior in SPR, aurintricarboxylic acid was abandoned at this point in the study.
SAR
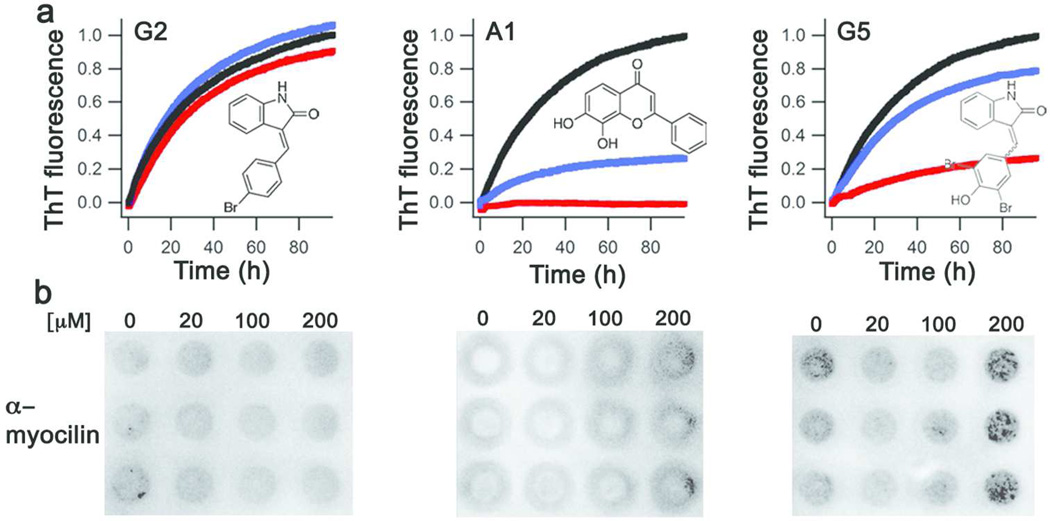
On the basis of results from SPR and the fibrillization assay, we synthesized or purchased several derivatives of GW5074 (SI Figure S5a, SI Methods) with improved potency and water solubility as potential lead compounds. Of the GW5074 derivatives, potent fibril inhibition was only retained in G5 (Figure 6a), in which the hydrophobic sphere has been altered by removal of the iodine from the indolinone ring. Modifications to the benzylidene substructure had either a weak or no effect on myoc-OLF fibrillization (see, for example, G2 in Figure 6a). The water-soluble apigenin surrogate, flavone A1 (SI Figure S5a), in which the two hydroxyl groups are located at the 7- and 8- positions, also elicited a dose-dependent decrease in myoc-OLF fibrilization (Figure 6a).
Figure 6. Assays to evaluate compounds for myocilin glaucoma therapeutics.
Evaluation of selected derivatives for (a) fibril inhibition and (b) enhanced cellular secretion of mutant myocilin.
Evaluation of validated stoichiometric ligands using mutant myocilin cellular secretion assay
Finally, the compounds were tested for their ability to rescue mutant myocilin secretion and prevent the glaucoma-associated HTM cell death. We used a known secretion assay (34) and a model HEK-cell line stably expressing I477N-mutant myocilin (35). This mutant represents one of the least stable glaucoma-causing OLF variants (36). Several compounds, including the original hits GW5074 and apigenin, were not suitable because they precipitated upon addition to the cell media. However, G2, G5 and A1 exhibited adequate solubility as well as cellular toxicity profile (SI Figure S5b) and could be tested for dose dependent secretion enhancement of I477N-mutant myocilin, up to 200 µM (Figure 6bSI Figure S5b). Consistent with results from in vitro fibrillization assays, G2 did not enhance secretion, whereas G5 and A1 increased I477N-mutant myocilin secretion in a dose-dependent manner, with G5 exhibiting the stronger increase (SI Figure S5c).
DISCUSSION
In contrast to other misfolding disorders that require crossing the blood brain barrier (37), the anterior segment of the eye is a suitable target for localized drug delivery (38). Currently, however, the only options for glaucoma patients are to manage ocular hypertension with drugs that lower aqueous humour production or increase outflow, or through a surgical procedure called trabeculectomy (39). A better understanding of myocilin biology and pathology could lead to new therapeutics for patients with myocilin-associated glaucoma, but the protein has thus far eluded requisite molecular structural and functional characterization.
The small molecule ligands for myocilin reported here are critical advance in the development of chemical probes of function/pathology and therapeutics. Apigenin and GW5074, which are only modestly similar to each other in structure and functional groups, bind to myoc-OLF stoichiometrically with micromolar affinity, and apparently shield the myoc-OLF amyloidogenic region. (30)After further improving hits for binding affinity, selectivity for myoc-OLF, solubility and cell permeability, we anticipate that it will be possible to configure derivatives of molecules featured in our study to identify myocilin binding partners and probe function ex vivo, and that these compounds are potentially adaptable for the study of the biology other poorly understood OLF domains (10) linked to a variety of (non-ocular) cancers (40–42). Furthermore, second generation molecules G5 and A1 show promise for their development into targeted molecules to diminish intracellular mutant myocilin aggregation, akin to therapies under development for other protein misfolding disorders (43–45).
With the intention for use as an early ligand discovery assay for myoc-OLF, CARS was motivated by the requirement to be fully compatible with routine robotic instrumentation and compound resources available at a HT screening facility, and due to the lack of alternative functional assay, is based on the thermodynamic underpinnings of ligand-induced protein stabilization. Measurements recorded immediately using a fluorescence plate reader at a single temperature required little sample work-up, and changes in fluorescence were trivial to compute and analyze. The assay was robust and reproducible in our own laboratory in 96-well format, and miniaturization and automation in 384-well format was straightforward; we anticipate no major hurdles for further miniaturization to 1536-well plates for screening larger compound libraries. The use of an osmolyte to define a signal window for ligand-induced protein stability is another novel aspect of the assay, especially as a proxy for a positive control ligand. CARS is reminiscent of, but is different from, DSF (24) and isothermal denaturation calorimetry (ITD) (46, 47), both of which also use SO fluorescence for assay readout. DSF has experienced some popularity in identifying new ligands for selected targets (13, 48–51). DSF can be conducted in 384-well setup with a specialized instrument called ThermoFluor(52, 53), which was not available to us. In addition to issues of myoc-OLF aggregation at elevated temperatures that are not apparent upon chemical destabilization in CARS, analogous screening of myoc-OLF likely would have required the use of significantly higher levels of compound (SI Figure S6) to obtain statistically relevant hits (ITm ± ~1.5°C (13)). For ITD, the elevated temperatures required are again incompatible with myoc-OLF due to its fibrillization.
Given the focus of the LOPAC library on GPCRs and kinases involved in cell signaling and cell cycle, it is encouraging that valid hits emerged for myoc-OLF using our assay. On the basis of ease of implementation and results for myocilin, we are optimistic that CARS could be adaptable for other protein systems where ligands are desired but no functional assay yet exists or is suitable. SO, originally designed for high sensitivity protein detection after denaturing gel electrophoresis (54), continues to prove compatible with numerous unrelated proteins, but in principle the CARS method should be also applicable using other dyes with properties suitable for membrane proteins, protein-protein interactions, or complex mixtures.
METHODS
Molecular Biology, Protein Expression, and Purification
MBP was expressed as described in Supporting Methods. The myoc-OLF protein was expressed as a Factor Xa-cleavable MBP fusion and purified as described previously (25).
Chemical Stability Assay Development
Denaturant concentration was first determined by unfolding myoc-OLF with GdnHCl as described in Supporting Methods. Protein concentration was optimized by serial dilutions of the protein (0.625−10 µM) in PBS (10 mM sodium phosphate dibasic, 10 mM potassium phosphate monobasic, 200 mM NaCl pH 7.2), SO, and 0.6 M GdnHCl. Fluorescence data was acquired and analyzed as for GdnHCl concentration determination. Next, maltose, maltotetraose, and maltitol were added at a final concentration of 0−1 mM to MBP in PBS, 0.6 M GdnHCl, and SO. PMSF and iodoacetamide were tested (0–1 mM) as negative controls for MBP. For myoc-OLF, TMAO (0−1 M) was used to assess dosedependent SO fluorescence, and 0 M and either 1 M or 0.8 M TMAO were included as negative and positive controls, in subsequent screening in 96- or 384-well formats, respectively. The protein was added last and prior to fluorescence readings.
DSF
DSF was conducted and analyzed as described previously (25).
CD, AFM imaging, ANS fluorescence, Absorbance assay to detect aggregation, and amyloid aggregation
are described in Supporting Methods.
HT Screening of Small Molecule Library
Details of optimization and screening are presented in Supporting Information Table S1 and Methods.
SPR
SPR was conducted using a BIAcore T200 (GE Healthcare) in the Georgia Tech Institute for Electronics and Nanotechnology as described in Supporting Methods.
Compound synthesis
Compounds were either purchased (GW5074, Sigma Aldrich; 7,8-dihydroxyflavone (A1), TCI chemicals, or synthesized. For G2 and G5, 2-Indolinone (1.0 equiv), aldehyde (1.0−1.2 equiv of either 4-bromobenzaldehyde (G2) or p-toluenesulfonic acid monohydrate (G5), piperidine (0.2−2 equiv), and absolute ethanol (10 mL) were charged to a flask equipped with a condenser and stir bar. The mixture was heated to a reflux for 16 hours. The resulting solid was filtered, washed repeatedly with low boiling petroleum ether, collected, and dried in vacuo overnight. Samples were characterized, examined for purity by NMR, and compared to the literature (56). Other details are described in Supporting Methods.
Cell secretion assay
The assay was conducted as described previously (5) except that DMSO vehicle or compound at varying concentrations (20, 100, 200 µM compound dissolved in DMSO) were added 3 days after induction. Media were collected by centrifugation after cells were incubated for 24 h with compounds and detected by immunoblotting as described previously (5).
Supplementary Material
ACKNOWLEDGMENT
We thank T. Sulchek (Georgia Tech) for use of the AFM instrument, L. Stafford for assistance in developing the ThT aggregation assay and M.G. Finn for helpful discussions.
Funding
This work was funded by grants from NIH (R01EY021205) and Pew Scholar in Biomedical Sciences program to R. L. L., U. S. Department of Education (Graduate Assistance in Areas of National Need P200A060188) to S.D.O, NSF Graduate Research Fellowship (DGE-1148903) and Georgia Tech Presidential Fellowship to M. A. C., and a Georgia Tech (Presidential Undergraduate Research Award) and Merck Fellowship to P. V. C.
Abbreviations
- HT
high throughput
- CARS
chemical assay for restabilization
- myoc-OLF
myocilin olfactomedin domain
- TM
trabecular meshwork
- SPR
surface plasmon resonance
- DSF
differential scanning fluorimetry
- Tm
melting temperature
- SO
Sypro Orange
- TMAO
trimethylamine-N-oxide
- GdnHCl
guanidine hydrochloride
- MBP
maltose binding protein
- CD
circular dichroism
- ANS
8-Anilinonaphthalene-1-sulfonic acid
- CV
coefficient of variation
- S/B
signal-to-background
- ITD
isothermal denaturation calorimetry
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 2.Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. N. EnglJMed. 2009;360:1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Resch Z, Fautsch M. Glaucoma-associated myocilin: A better understanding but much more to learn. Exp. Eye Res. 2009;88:704–712. doi: 10.1016/j.exer.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orwig SD, Perry CW, Kim LY, Turnage KC, Zhang R, Vollrath D, Schmidt-Krey I, Lieberman RL. Amyloid fibril formation by the glaucoma-associated olfactomedin domain of myocilin. J. Mol. Biol. 2012;421:242–255. doi: 10.1016/j.jmb.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suntharalingam A, Abisambra JF, O'Leary JC, 3rd, Koren J, 3rd, Zhang B, Joe MK, Blair LJ, Hill SE, Jinwal UK, Cockman M, Duerfeldt AS, Tomarev S, Blagg BS, Lieberman RL, Dickey CA. Glucose-regulated protein 94 triage of mutant myocilin through endoplasmic reticulum-associated degradation subverts a more efficient autophagic clearance mechanism. J. Biol. Chem. 2012;287:40661–40669. doi: 10.1074/jbc.M112.384800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joe MK, Tomarev SI. Expression of myocilin mutants sensitizes cells to oxidative stress-induced apoptosis: implication for glaucoma pathogenesis. Am. J. Pathol. 2010;176:2880–2890. doi: 10.2353/ajpath.2010.090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yam GH-F, Gaplovska-Kysela K, Zuber C, Roth J. Aggregated myocilin induces russell bodies and causes apoptosis: implications for the pathogenesis of myocilin-caused primary open-angle glaucoma. Am. J. Pathol. 2007;170:100–109. doi: 10.2353/ajpath.2007.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polansky JR, Fauss DJ, Chen P, Chen H, Lutjen-Drecoll E, Johnson D, Kurtz RM, Ma ZD, Bloom E, Nguyen TD. Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica. 1997;211:126–139. doi: 10.1159/000310780. [DOI] [PubMed] [Google Scholar]
- 9.Anderssohn AM, Cox K, O'Malley K, Dees S, Hosseini M, Boren L, Wagner A, Bradley JM, Kelley MJ, Acott TS. Molecular chaperone function for myocilin. Invest. Ophthalmol. Vis. Sci. 2011;52:7548–7555. doi: 10.1167/iovs.11-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol. Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joe MK, Kee C, Tomarev SI. Myocilin Interacts with Syntrophins and Is Member of Dystrophin-associated Protein Complex. J. Biol. Chem. 2012;287:13216–13227. doi: 10.1074/jbc.M111.224063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magliery TJ, Lavinder JJ, Sullivan BJ. Protein stability by number: highthroughput and statistical approaches to one of protein science's most difficult problems. Curr. Opin. Chem. Biol. 2011;15:443–451. doi: 10.1016/j.cbpa.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holdgate GA, Anderson M, Edfeldt F, Geschwindner S. Affinity-based, biophysical methods to detect and analyze ligand binding to recombinant proteins: matching high information content with high throughput. J. Struct. Biol. 2010;172:142–157. doi: 10.1016/j.jsb.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 14.Comess KM, Schurdak ME. Affinity-based screening techniques for enhancing lead discovery. Curr. Opin. Drug. Discov. Devel. 2004;7:411–416. [PubMed] [Google Scholar]
- 15.Senisterra GA, Finerty PJ., Jr High throughput methods of assessing protein stability and aggregation. Mol. Biosyst. 2009;5:217–223. doi: 10.1039/b814377c. [DOI] [PubMed] [Google Scholar]
- 16.Jonker N, Kool J, Irth H, Niessen WM. Recent developments in protein-ligand affinity mass spectrometry. Anal. Bioanal. Chem. 2011;399:2669–2681. doi: 10.1007/s00216-010-4350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 2009;131:3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pace CN, McGrath T. Substrate stabilization of lysozyme to thermal and guanidine hydrochloride denaturation. J. Biol. Chem. 1980;255:3862–3865. [PubMed] [Google Scholar]
- 19.Ferenci T, Muir M, Lee KS, Maris D. Substrate specificity of the Escherichia coli maltodextrin transport system and its component proteins. Biochim. Biophys. Acta. 1986;860:44–50. doi: 10.1016/0005-2736(86)90496-7. [DOI] [PubMed] [Google Scholar]
- 20.Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. BrJPharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 22.Macarron R, Hertzberg RP. Design and implementation of high throughput screening assays. Mol. Biotechnol. 2011;47:270–285. doi: 10.1007/s12033-010-9335-9. [DOI] [PubMed] [Google Scholar]
- 23.Achyuthan KE, Whitten DG. Design considerations for high-throughput screening and in vitro diagnostic assays. Comb. Chem. High Throughput Screen. 2007;10:399–412. doi: 10.2174/138620707781996475. [DOI] [PubMed] [Google Scholar]
- 24.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 25.Burns JN, Orwig SD, Harris JL, Watkins JD, Vollrath D, Lieberman RL. Rescue of glaucoma-causing mutant myocilin thermal stability by chemical chaperones. ACS Chem. Biol. 2010;5:477–487. doi: 10.1021/cb900282e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu P, Bolen DW. Osmolyte-induced protein folding free energy changes. Proteins. 2006;63:290–296. doi: 10.1002/prot.20868. [DOI] [PubMed] [Google Scholar]
- 27.Gordon JA, Jencks WP. The relationship of structure to the effectiveness of denaturing agents for proteins. Biochemistry. 1963;2:47–57. doi: 10.1021/bi00901a011. [DOI] [PubMed] [Google Scholar]
- 28.Pace CN, Marshall HF., Jr A comparison of the effectiveness of protein denaturants for beta-lactoglobulin and ribonuclease. Arch. Biochem. Biophys. 1980;199:270–276. doi: 10.1016/0003-9861(80)90281-7. [DOI] [PubMed] [Google Scholar]
- 29.Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov. Today. 2003;8:86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- 30.Orwig SD, Lieberman RL. Biophysical characterization of the olfactomedin domain of myocilin, an extracellular matrix protein implicated in inherited forms of glaucoma. PLoS One. 2011;6:e16347. doi: 10.1371/journal.pone.0016347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez RG, Haxo RS, Schleich T. Mechanism of action of polymeric aurintricarboxylic acid, a potent inhibitor of protein--nucleic acid interactions. Biochemistry. 1980;19:4299–4303. doi: 10.1021/bi00559a023. [DOI] [PubMed] [Google Scholar]
- 32.Giannetti AM, Koch BD, Browner MF. Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J. Med. Chem. 2008;51:574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- 33.McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J. Med. Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, Vollrath D. A cellular assay distinguishes normal and mutant TIGR/myocilin protein. Hum. Mol. Genet. 1999;8:2221–2228. doi: 10.1093/hmg/8.12.2221. [DOI] [PubMed] [Google Scholar]
- 35.Joe MK, Sohn S, Hur W, Moon Y, Choi YR, Kee C. Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem. Biophys. Res. Commun. 2003;312:592–600. doi: 10.1016/j.bbrc.2003.10.162. [DOI] [PubMed] [Google Scholar]
- 36.Burns JN, Turnage KC, Walker CA, Lieberman RL. The stability of myocilin olfactomedin domain variants provides new insight into glaucoma as a protein misfolding disorder. Biochemistry. 2011;50:5824–5833. doi: 10.1021/bi200231x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pardridge WM. Blood-brain barrier delivery. Drug Discov. Today. 2007;12:54–61. doi: 10.1016/j.drudis.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Lang JC. Ocular Drug-Delivery Conventional Ocular Formulations. Adv. Drug Deliver. Rev. 1995;16:39–43. [Google Scholar]
- 39.Donegan RK, Lieberman RL. New direction for glaucoma therapeutics: focus on the olfactomedin domain of myocilin. Future Med. Chem. 2012;4:2131–2134. doi: 10.4155/fmc.12.132. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi D, Koshida S, Moriai R, Tsuji N, Watanabe N. Olfactomedin 4 promotes S-phase transition in proliferation of pancreatic cancer cells. Cancer Sci. 2007;98:334–340. doi: 10.1111/j.1349-7006.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu RH, Yang MH, Xiang H, Bao LM, Yang HA, Yue LW, Jiang X, Ang N, Wu LY, Huang Y. Depletion of OLFM4 gene inhibits cell growth and increases sensitization to hydrogen peroxide and tumor necrosis factor-alpha induced-apoptosis in gastric cancer cells. J. Biomed. Sci. 2012;19:38. doi: 10.1186/1423-0127-19-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu L, Wang L, Chen S. Olfactomedin 4, a novel marker for the differentiation and progression of gastrointestinal cancers. Neoplasma. 2011;58:9–13. doi: 10.4149/neo_2011_01_9. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhuri TK, Paul S. Protein-misfolding diseases and chaperone-based therapeutic approaches. FEBS J. 2006;273:1331–1349. doi: 10.1111/j.1742-4658.2006.05181.x. [DOI] [PubMed] [Google Scholar]
- 44.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 45.Ulloa-Aguirre A, Janovick JA, Brothers SP, Conn PM. Pharmacologic rescue of conformationally-defective proteins: implications for the treatment of human disease. Traffic. 2004;5:821–837. doi: 10.1111/j.1600-0854.2004.00232.x. [DOI] [PubMed] [Google Scholar]
- 46.Sarver RW, Rogers JM, Epps DE. Determination of ligand-MurB interactions by isothermal denaturation: application as a secondary assay to complement high throughput screening. J. Biomol. Screen. 2002;7:21–28. doi: 10.1177/108705710200700104. [DOI] [PubMed] [Google Scholar]
- 47.Senisterra GA, Soo Hong B, Park HW, Vedadi M. Application of highthroughput isothermal denaturation to assess protein stability and screen for ligands. J. Biomol. Screen. 2008;13:337–342. doi: 10.1177/1087057108317825. [DOI] [PubMed] [Google Scholar]
- 48.Abad MC, Askari H, O'Neill J, Klinger AL, Milligan C, Lewandowski F, Springer B, Spurlino J, Rentzeperis D. Structural determination of estrogen-related receptor gamma in the presence of phenol derivative compounds. The J. Steroid Biochem. Mol. Biol. 2008;108:44–54. doi: 10.1016/j.jsbmb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Carver TE, Bordeau B, Cummings MD, Petrella EC, Pucci MJ, Zawadzke LE, Dougherty BA, Tredup JA, Bryson JW, Yanchunas J, Jr, Doyle ML, Witmer MR, Nelen MI, DesJarlais RL, Jaeger EP, Devine H, Asel ED, Springer BA, Bone R, Salemme FR, Todd MJ. Decrypting the biochemical function of an essential gene from Streptococcus pneumoniae using ThermoFluor technology. J. Biol. Chem. 2005;280:11704–11712. doi: 10.1074/jbc.M413278200. [DOI] [PubMed] [Google Scholar]
- 50.Clemente JC, Nulton E, Nelen M, Todd MJ, Maguire D, Schalk-Hihi C, Kuo LC, Zhang SP, Flores CM, Kranz JK. Screening and characterization of human monoglyceride lipase active site inhibitors using orthogonal binding and functional assays. J. Biomol. Screen. 2012;17:629–640. doi: 10.1177/1087057112441012. [DOI] [PubMed] [Google Scholar]
- 51.Parks DJ, Lafrance LV, Calvo RR, Milkiewicz KL, Gupta V, Lattanze J, Ramachandren K, Carver TE, Petrella EC, Cummings MD, Maguire D, Grasberger BL, Lu T. 1,4-Benzodiazepine-2,5-diones as small molecule antagonists of the HDM2-p53 interaction: discovery and SAR. Bioorg. Med. Chem. Lett. 2005;15:765–770. doi: 10.1016/j.bmcl.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 53.Cummings MD, Farnum MA, Nelen MI. Universal screening methods and applications of ThermoFluor. J. Biomol. Screen. 2006;11:854–863. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 54.Steinberg TH, Jones LJ, Haugland RP, Singer VL. SYPRO orange and SYPRO red protein gel stains: one-step fluorescent staining of denaturing gels for detection of nanogram levels of protein. Anal. Biochem. 1996;239:223–237. doi: 10.1006/abio.1996.0319. [DOI] [PubMed] [Google Scholar]
- 55.Johnsson B, Lofas S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 56.Alam M, Du Bois DJ, Hawley RC, Kennedy-Smith J, Minatti AE, Palmer WS, Silva T, Wilhelm RS. Indole derivatives as CRAC modulators. US2011/71150. US Patent. 2011
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.