Abstract
Tivantinib has been described as a potent and highly selective inhibitor of the receptor tyrosine kinase c-MET and is currently in advanced clinical development for several cancers including non-small cell lung cancer (NSCLC). However, recent studies suggest that tivantinib’s anticancer properties are unrelated to c-MET inhibition. Consistently, in determining tivantinib’s activity profile in a broad panel of NSCLC cell lines, we found that, in contrast to several more potent c-MET inhibitors, tivantinib reduces cell viability across most of these cell lines. Applying an unbiased, mass-spectrometry-based, chemical proteomics approach, we identified glycogen synthase kinase 3 (GSK3) alpha and beta as novel tivantinib targets. Subsequent validation showed that tivantinib displayed higher potency for GSK3α than for GSK3β and that pharmacological inhibition or simultaneous siRNA-mediated loss of GSK3α and GSK3β caused apoptosis. In summary, GSK3α and GSK3β are new kinase targets of tivantinib that play an important role in its cellular mechanism-of-action in NSCLC.
Keywords: Kinase inhibitor, tivantinib, c-MET, non-small cell lung cancer, chemical proteomics, off-target, GSK3
The receptor tyrosine kinase c-MET is involved in cell migration and metastasis of various malignancies.(1) c-MET amplification and activity provides an important mechanism by which cancer cells develop resistance to targeted drugs, such as EGFR inhibitors in non-small cell lung cancer (NSCLC).(2) Thus, c-MET is an attractive therapeutic target and several inhibitors are currently in clinical development. One of the most advanced c-MET inhibitors, tivantinib (ARQ197),(3) has progressed into phase III trials.(4–6) In phase II studies, tivantinib displayed clinical activity in NSCLC patients in combination with the EGFR inhibitor erlotinib, particularly in patients with KRAS mutations.(5) This was unexpected as the primary rationale for testing tivantinib in NSCLC was to prevent emergence of resistance to erlotinib due to compensatory c-MET signaling in patients with EGFR mutations, which are mutually exclusive with KRAS mutations.(7) Moreover, although described to be highly selective for c-MET, reportedly due to its unique ATP-independent binding mode,(3, 8) tivantinib showed anticancer activity in various cell lines across diverse tumor types, many of which are not driven by c-MET signaling.(3) We therefore hypothesized that tivantinib inhibits a wider range of targets than appreciated and that some of these are functionally relevant for its activity. Further supporting this hypothesis, two recent studies suggest that tivantinib’s anticancer activity in different tumor types may be related to modulation of microtubule dynamics rather than c-MET inhibition.(9–11) Here, we report tivantinib’s antiproliferative activity in a large panel of lung cancer cell lines showing that tivantinib action in NSCLC is indeed independent of inhibition of c-MET activity, but furthermore also of KRAS status. Subsequent cellular target profiling by chemical proteomics identified glycogen synthase kinase (GSK) 3α and GSK3β as novel tivantinib targets, both being more potently inhibited than c-MET. Loss of function studies suggest that inhibition of these kinases plays an important role for the antiproliferative activity of tivantinib in NSCLC cells.
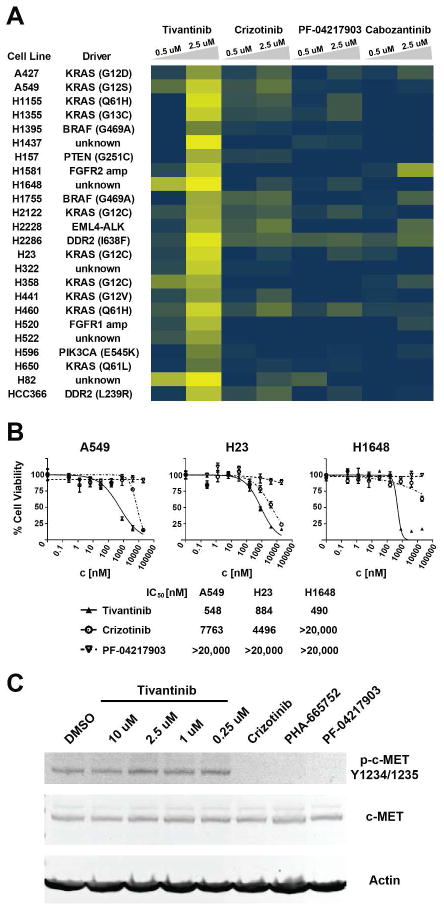
To obtain a broader view of tivantinib’s activity in lung cancer, we screened a panel of 24 KRAS-mutant and wild-type lung cancer cell lines (Figure 1A). Tivantinib inhibited the viability of the majority of these whereas crizotinib (PF-02341066), PF-04217903 and cabozantinib (XL-184), which are much more potent c-MET kinase inhibitors than tivantinib, had no significant effects. There was no obvious link between tivantinib sensitivity and KRAS mutation status. Determination of the IC50 values for inhibition of cellular viability confirmed the differential activity of these compounds with tivantinib displaying an IC50 of about 500 nM for the most sensitive NSCLC cell lines. In comparison, the highly selective c-MET inhibitor PF-04217903 and the less selective crizotinib had no measurable or only weak activity, respectively (Figure 1B). Confirming the functional integrity of these compounds, though, c-MET autophosphorylation in A549 cells was effectively inhibited by crizotinib, PF-04217903 and another widely used c-MET inhibitor, PHA-665752, whereas tivantinib showed essentially no effect (Figure 1C). Considering the reported maximum plasma concentration of 5–7 μM from phase I clinical trials,(12, 13) tivantinib’s activity against several of these cell lines was well within physiologically relevant concentrations. In summary, tivantinib displayed potent activity against a broad panel of lung cancer cell lines, which is unrelated to inhibition of c-MET kinase activity and KRAS mutation status.
Figure 1. Cellular activity of tivantinib and various c-MET inhibitors in lung cancer cell lines.
(A) Effects of tivantinib, crizotinib, PF-04217903 and cabozantinib at 0.5 and 2.5 μM on cellular viability across the indicated panel of KRAS WT and mutant lung cancer cell lines. Relative cellular viability is displayed as a gradient from 0% (yellow) to 100% (blue) compared to DMSO control. (B) Dose-response curves and IC50 values for inhibition of viability by tivantinib, crizotinib and PF-04217903 in the A549 and H23 (both KRAS-mutant) and the H1648 (KRAS-wild-type) NSCLC cell lines. (C) Effect on c-MET autophosphorylation in A549 cells following 30 minute treatment at the indicated concentrations of tivantinib and 250 nM each of the c-MET inhibitors crizotinib, PHA-665752 and PF-04217903.
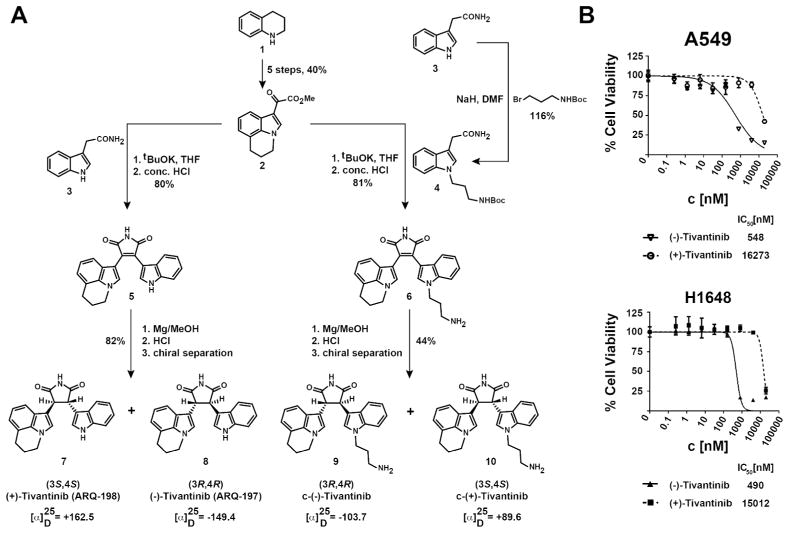
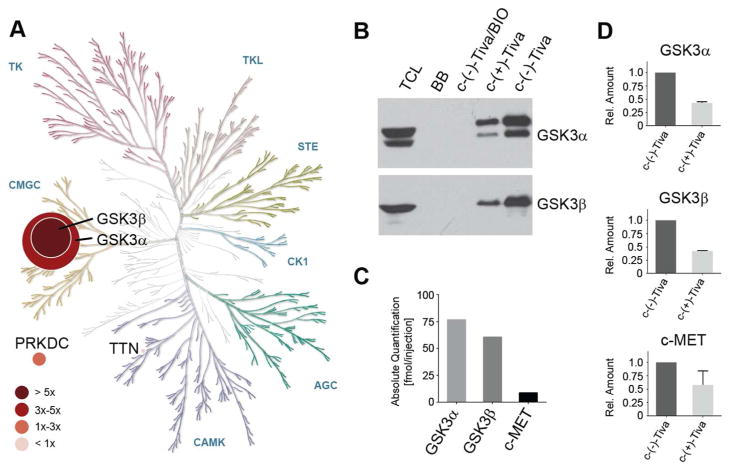
Tivantinib’s in vitro inhibition profile was originally determined against a panel of 230 kinases, based on which it was considered a selective c-MET inhibitor.(3) In light of our data, however, we hypothesized that one or more of the remaining almost 300 kinases in the human kinome could be previously unrecognized tivantinib targets responsible for the cellular activity in NSCLC cells. We therefore applied a mass spectrometry-based chemical proteomics strategy to characterize tivantinib’s target profile in NSCLC cells in a proteome-wide and unbiased fashion.(14) To this end, we designed the tivantinib analogue c-(−)-tivantinib (9, Figure 2A) based on the reported co-crystal structure of tivantinib with c-MET, which suggests that the indole moiety is solvent accessible.(8) According to our previous experience performing chemical proteomics with various kinase inhibitors, similar structure-activity relationships are likely maintained across the majority of targets.(15, 16) c-(−)-Tivantinib (9) was synthesized by modifying the published synthetic route to tivantinib (8) and its enantiomer (+)-3S,4S-tivantinib (7), which has been reported to be inactive against c-MET (Figures 2A and S1A).(8, 17) Interestingly, 7 also showed substantially weaker activity in (−)-tivantinib-sensitive cell lines making it an excellent control compound (Figure 2B). Separation of the racemic mixture resulting from reduction of the maleimide intermediate 6 by preparative chiral HPLC yielded the optically pure enantiomers c-(−)-tivantinib (9) and c-(+)-tivantinib (10), which retained differential cellular activity, albeit somewhat reduced possibly due to altered cell permeability (Figure S1B). Chemical proteomics with A549 total cell lysates using analogues 9 and 10 (the latter as control) identified several protein kinases not previously implicated as tivantinib targets (Figure 3A, Tables S1,S2). Applying stringent filters for reproducibility between replicates and enrichment by 9 over 10, GSK3α and GSK3β were found to be the highest confidence target candidates that interacted with tivantinib. Subsequent immunoblotting confirmed the interaction between c-(−)-tivantinib (9) and GSK3α and GSK3β and suggested selectivity of (−)-tivantinib over both (+)-tivantinib and blocked beads (no immobilized drug, ruling out non-specific binding to the matrix) for these kinases (Figure 3B). Furthermore, competition with the potent ATP-competitive pan-GSK3 inhibitor BIO suggested that tivantinib interacts directly with GSK3α and GSK3β by binding to their ATP-binding pockets. In comparison, c-MET was only weakly detectable by LC-MS/MS analysis in A549 cells and did not pass our stringent filtering criteria. Furthermore, while a more sensitive targeted proteomics analysis using multiple reaction monitoring (MRM) with stable isotope-labeled standard (SIS) peptides also detected c-MET as a tivantinib binder, absolute quantification demonstrated that GSK3α and GSK3β were much more strongly enriched than c-MET (Figure 3C). All three kinases were selectively recovered by c-(−)-tivantinib (9) over c-(+)-tivantinib (10) (Figure 3D). Chemical proteomics using H1648 cells, which are known to overexpress c-MET, again prominently enriched for GSK3α and GSK3β, but now also for c-MET (Figure S2, Tables S3,S4). Consistent with in vitro kinase assays (Figure S3), these results suggest that c-MET is only a weak tivantinib target. Instead, our data demonstrate that tivantinib is prominently binding to GSK3α and GSK3β.
Figure 2. Synthesis of tivantinib enantiomers and analogues.
(A) Synthetic strategy and chemical structures of (−)-tivantinib (8), its enantiomer (+)-tivantinib (7) and their coupleable analogues, (9) and (10), respectively. For the full synthetic route see Figure S1A. (B) Differential inhibition of cellular viability of A549 and H1648 cells by (−)-tivantinib and (+)-tivantinib.
Figure 3. Determination of tivantinib’s kinase target interaction profile in A549 cells by chemical proteomics.
(A) Phylogenetic tree of the human protein kinome displaying targets identified by affinity chromatography experiments with coupled tivantinib analogues in A549 cells. Node size indicates the average ratio of the number of unique spectra to the molecular weight of the proteins identified with the c-(−)-tivantinib affinity matrix. Node color corresponds to the ratio of the number of unique spectra identified by c-(−)-tivantinib to c-(+)-tivantinib affinity chromatography experiments. Displayed are all protein kinases that have been observed in both biological replicates of the (−)-tivantinib affinity purifications with more than 2 unique spectra across both replicates. (B) Immunoblot analysis of GSK3α and GSK3β purification by c-(−)-tivantinib and c-(+)-tivantinib affinity chromatography. Competition experiments were conducted by 2-hour treatment of A549 total cell lysate with the pan-GSK3 inhibitor BIO prior to affinity chromatography. (C) Absolute quantification of GSK3α, GSK3β and c-MET peptides identified by LC-MS/MS analysis of c-(−)-tivantinib affinity purifications using LC-MRM with stable isotope labeled peptides. (D) Relative abundance ratios of GSK3α, GSK3β and c-MET peptides between c-(−)-tivantinib and c-(+)-tivantinib affinity purifications. The Human Kinome Map was adapted with permission from Cell Signaling Technology (www.cellsignal.com). TCL: total cell lysate; BB: blocked beads (no immobilized compound); Tiva: tivantinib.
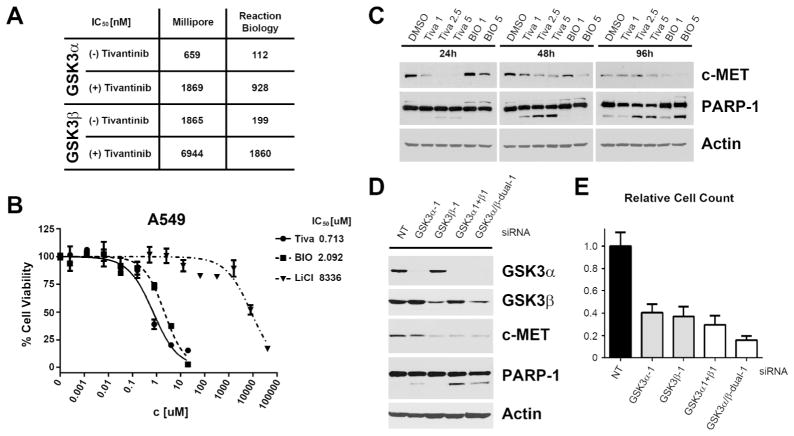
To evaluate the functional consequences of tivantinib binding to GSK3α and GSK3β, enzyme inhibition was determined by in vitro kinase assays. These experiments demonstrated that (−)-tivantinib potently inhibits GSK3α and GSK3β with IC50 values in the upper nanomolar range, whereas (+)-tivantinib is a significantly weaker inhibitor of both kinases (Figure 4A). Thus, GSK3α and GSK3β are more potently inhibited than c-MET by (−)-tivantinib and the differential activity between the tivantinib enantiomers is supported by the difference observed in their cellular activities. Also consistent with GSK3 inhibition, an increase in total β-catenin levels, which is degraded upon phosphorylation by GSK3,(18) was detected following tivantinib treatment of A549 cells (Figure S4). Notably, in contrast to the vast majority of GSK3 inhibitors, which either affect GSK3α and GSK3β with equal potency or display selectivity for GSK3β,(19) tivantinib inhibits GSK3α 2–3-fold more strongly than GSK3β. Although recently, there has been significant progress towards GSK3α-specific inhibitors,(20, 21) tivantinib constitutes to the best of our knowledge the first GSK3 inhibitor in clinical development that has selectivity for GSK3α. Furthermore, although other targets with different structure-activity relationships cannot be completely ruled out, tivantinib appears to be much more selective for GSK3α and GSK3β on a kinome-wide level than established GSK3 inhibitors. This relative GSK3α vs. GSK3β specificity, as well as the kinome-wide target selectivity, may harbor novel therapeutic opportunities for tivantinib in malignant diseases, such as acute myeloid leukemia or pancreatic cancer, in which GSK3α has recently been described to be a promising new target.(22, 23) Furthermore, GSK3 inhibitors may have utility in Alzheimer’s disease, diabetes and bipolar disorder.(24, 25) Considering the narrow therapeutic index of the FDA-approved GSK3 inhibitor LiCl, the clinical safety and remarkable target selectivity of tivantinib may offer an interesting alternative.
Figure 4. Functional analysis of GSK3α and GSK3β inhibition.
(A) IC50 values for inhibition of GSK3α and GSK3β in vitro kinase activity by (−)-tivantinib and (+)-tivantinib on Millipore and Reaction Biology assay platforms (mean values of independent experimental duplicate analyses). (B) Effect of tivantinib and the known pan-GSK3 inhibitors BIO and LiCl on cell viability of A549 NSCLC cells. (C) Effects of tivantinib and BIO (in μM) on A549 cells regarding PARP-1 cleavage and total c-MET levels. (D) Effects of individual or combined siRNA-mediated knockdowns of GSK3α and GSK3β in A549 cells on PARP-1 cleavage and c-MET levels 72 hours post-transfection. GSK3α1+β1: combination of gene-specific siRNAs GSK3α-1 and GSK3β-1; GSK3α/β-dual-1: single siRNA targeting both genes. NT: non-targeting siRNA. (E) Effects of individual or combined GSK3α and GSK3β knockdowns on cell number of A549 cells 96 hours post-transfection.
In the context of cancer, GSK3 is generally considered a tumor suppressor and thus, inhibition of GSK3 by tivantinib as a potential mechanism-of-action in NSCLC is initially counterintuitive. However, in addition to the aforementioned studies, several recent reports propose GSK3 to be a promising anticancer target, e.g. in glioblastoma multiforme and MLL-rearranged leukemia,(26, 27) suggesting that GSK3 function is context-dependent. Consistently, tivantinib also potently inhibited viability of MV-4–11 and RS4;11 MLL-rearranged leukemia cells, stabilized β-catenin and induced G1 cell cycle arrest, as has been described previously for GSK3 inhibitors in these cells (Figure S5).(27) To evaluate the functional relevance of GSK3 inhibition by tivantinib in NSCLC cells, we compared tivantinib activity with the known GSK3 inhibitors BIO and LiCl (Figure 4B). Both compounds showed antiproliferative activity in A549 cells. Although initially it may seem high, the low millimolar activity of LiCl is in excellent agreement with GSK3-dependent growth inhibition in leukemia (Figure S5A).(22, 27) Like tivantinib, BIO was able to induce apoptosis in A549 cells, albeit slightly delayed, as indicated by PARP-1 cleavage (Figure 4C). Curiously, we also noted decreased c-MET levels upon treatment with either compound. Regulation of c-MET levels may offer an explanation for why tivantinib has clinical activity in tumors driven by mutant or overexpressed c-MET. Interestingly, siRNA-mediated knockdown of GSK3α and GSK3β suggest that these kinases have non-redundant functions in A549 cells (Figure 4D). Loss of GSK3β was primarily responsible for reducing c-MET levels, whereas GSK3α knockdown had a stronger effect on PARP-1 cleavage and closely resembled tivantinib’s characteristic cell-rounding phenotype, which is possibly due to the observed G2/M cell cycle arrest elicited by tivantinib (Figures S6,S7).(9, 10) The combination of GSK3α- and GSK3β-specific siRNAs or using single siRNAs that cause simultaneous knockdown of both genes, however, was yet more effective in causing apoptosis (Figures 4D, S8A). Dual knockdown of GSK3α and GSK3β furthermore caused a significant reduction of viability in A549 cells compared to non-targeting siRNA (Figures 4E, S8B), which is consistent with the essential functions of GSK3α and GSK3β in many cells.
In summary, applying an unbiased, integrated chemical biology approach, we have identified GSK3α and GSK3β as novel targets of the clinical kinase inhibitor tivantinib in NSCLC cells. Importantly, tivantinib is a more potent inhibitor of GSK3α and GSK3β than of its intended target c-MET, which is only weakly inhibited. Furthermore, simultaneous loss of function of GSK3α and GSK3β causes apoptosis in lung cancer cells suggesting that inhibition of these kinases rather than c-MET plays an important role in tivantinib’s mechanism-of-action in NSCLC cells. Considering the large number of substrates and pathways that are modulated by GSK3, many of which also affect microtubules,(18) further global and unbiased studies are required to elucidate the exact downstream mechanism, by which tivantinib impairs cell viability. We anticipate that this knowledge will provide the basis for identification of relevant biomarkers that will greatly benefit tivantinib’s further clinical development, which is hampered by the erroneous assumption that tivantinib is a potent c-MET inhibitor. In addition, identification of GSK3α/β as tivantinib targets may pave the way for novel clinical studies with tivantinib, for instance in MLL-rearranged leukemia.
Methods
Cell culture and reagents, Flow cytometry, Immunoblotting, RNA interference
For details see Supplementary Methods.
Viability assays
Cell viability assays were conducted using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). For details see Supplementary Methods.
Chemical synthesis
(+)-Tivantinib (7) and (−)-tivantinib (8) were synthesized by the Moffitt Chemistry Core as described in the patent literature(17) followed by an adapted method for chiral separation. For details see Supplementary Methods.
Kinase assays
In vitro kinase inhibition assays and IC50 determinations were performed on the Reaction Biology Kinase HotspotSM and Millipore KinaseProfiler™platforms. For details see Supplementary Methods.
Affinity purification and mass spectrometry
Affinity chromatography experiments with c-(−)-tivantinib (9) and c-(+)-tivantinib (10) were performed in duplicate essentially as described previously.(28) For details see Supplementary Methods.
Supplementary Material
Acknowledgments
This work was supported by the American Cancer Society Institutional Research Grant (Award No. IRG-93-032-16), the Moffitt NIH/NCI SPORE in Lung Cancer (Award No. P50 CA119997), Moffitt Pinellas Partners and the H. Lee Moffitt Cancer Center and Research Institute. We wish to acknowledge the Moffitt Lung Cancer Center of Excellence and the Moffitt Chemical Biology (Chemistry Unit), Proteomics, Flow Cytometry and Microscopy Core Facilities. Moffitt Core Facilities are supported by the National Cancer Institute (Award No. P30-CA076292) as a Cancer Center Support Grant. Proteomics is also supported by the US Army Medical Research and Material Command (Award No. W81XWH-08-2-0101) for a National Functional Genomics Center, the Moffitt Foundation, and the Bankhead-Coley Cancer Research program of the Florida Department of Health (09BE-04). We also thank J. Kufs for skillful technical assistance.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Additional data and methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 2.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 3.Munshi N, Jeay S, Li Y, Chen CR, France DS, Ashwell MA, Hill J, Moussa MM, Leggett DS, Li CJ. ARQ 197, a novel and selective inhibitor of the human c-Met receptor tyrosine kinase with antitumor activity. Mol Cancer Ther. 2010;9:1544–1553. doi: 10.1158/1535-7163.MCT-09-1173. [DOI] [PubMed] [Google Scholar]
- 4.Scagliotti GV, Novello S, Schiller JH, Hirsh V, Sequist LV, Soria JC, von Pawel J, Schwartz B, Von Roemeling R, Sandler AB. Rationale and Design of MARQUEE: A Phase III, Randomized, Double-Blind Study of Tivantinib Plus Erlotinib Versus Placebo Plus Erlotinib in Previously Treated Patients With Locally Advanced or Metastatic, Nonsquamous, Non-Small-Cell Lung Cancer. Clin Lung Cancer. 2012 doi: 10.1016/j.cllc.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Sequist LV, von Pawel J, Garmey EG, Akerley WL, Brugger W, Ferrari D, Chen Y, Costa DB, Gerber DE, Orlov S, Ramlau R, Arthur S, Gorbachevsky I, Schwartz B, Schiller JH. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–3315. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]
- 6.Santoro A, Rimassa L, Borbath I, Daniele B, Salvagni S, Van Laethem JL, Van Vlierberghe H, Trojan J, Kolligs FT, Weiss A, Miles S, Gasbarrini A, Lencioni M, Cicalese L, Sherman M, Gridelli C, Buggisch P, Gerken G, Schmid RM, Boni C, Personeni N, Hassoun Z, Abbadessa G, Schwartz B, Von Roemeling R, Lamar ME, Chen Y, Porta C. Tivantinib for second-line treatment of advanced hepatocellular carcinoma: a randomised, placebo-controlled phase 2 study. Lancet Oncol. 2013;14:55–63. doi: 10.1016/S1470-2045(12)70490-4. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 8.Eathiraj S, Palma R, Volckova E, Hirschi M, France DS, Ashwell MA, Chan TC. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J Biol Chem. 2011;286:20666–20676. doi: 10.1074/jbc.M110.213801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basilico C, Pennacchietti S, Vigna E, Chiriaco C, Arena S, Bardelli A, Valdembri D, Serini G, Michieli P. Tivantinib (ARQ197) Displays Cytotoxic Activity That Is Independent of Its Ability to Bind MET. Clin Cancer Res. 2013;19:2381–2392. doi: 10.1158/1078-0432.CCR-12-3459. [DOI] [PubMed] [Google Scholar]
- 10.Katayama R, Aoyama A, Yamori T, Qi J, Oh-Hara T, Song Y, Engelman JA, Fujita N. Cytotoxic Activity of Tivantinib (ARQ 197) Is Not Due Solely to c-MET Inhibition. Cancer Res. 2013;73:3087–3096. doi: 10.1158/0008-5472.CAN-12-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michieli P, Di Nicolantonio F. Targeted therapies: Tivantinib--a cytotoxic drug in MET inhibitor’s clothes? Nat Rev Clin Oncol. 2013;10:372–374. doi: 10.1038/nrclinonc.2013.86. [DOI] [PubMed] [Google Scholar]
- 12.Adjei AA, Schwartz B, Garmey E. Early clinical development of ARQ 197, a selective, non-ATP-competitive inhibitor targeting MET tyrosine kinase for the treatment of advanced cancers. Oncologist. 2011;16:788–799. doi: 10.1634/theoncologist.2010-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yap TA, Olmos D, Brunetto AT, Tunariu N, Barriuso J, Riisnaes R, Pope L, Clark J, Futreal A, Germuska M, Collins D, deSouza NM, Leach MO, Savage RE, Waghorne C, Chai F, Garmey E, Schwartz B, Kaye SB, de Bono JS. Phase I trial of a selective c-MET inhibitor ARQ 197 incorporating proof of mechanism pharmacodynamic studies. J Clin Oncol. 2011;29:1271–1279. doi: 10.1200/JCO.2010.31.0367. [DOI] [PubMed] [Google Scholar]
- 14.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 15.Remsing Rix LL, Rix U, Colinge J, Hantschel O, Bennett KL, Stranzl T, Muller A, Baumgartner C, Valent P, Augustin M, Till JH, Superti-Furga G. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia. 2009;23:477–485. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 16.Winter GE, Rix U, Carlson SM, Gleixner KV, Grebien F, Gridling M, Muller AC, Breitwieser FP, Bilban M, Colinge J, Valent P, Bennett KL, White FM, Superti-Furga G. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nat Chem Biol. 2012 doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li CJ, Ashwell MA, Hill J, Moussa MM, Munshi N. Meleimide derivatives, pharmaceutical compositions and methods for treatment of cancer. ArQule, Inc; USA: 2006. p. 133. [Google Scholar]
- 18.Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends in biochemical sciences. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Kramer T, Schmidt B, Lo Monte F. Small-Molecule Inhibitors of GSK-3: Structural Insights and Their Application to Alzheimer’s Disease Models. International journal of Alzheimer’s disease. 2012;2012:381029. doi: 10.1155/2012/381029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atilla-Gokcumen GE, Williams DS, Bregman H, Pagano N, Meggers E. Organometallic compounds with biological activity: a very selective and highly potent cellular inhibitor for glycogen synthase kinase 3. Chembiochem: a European journal of chemical biology. 2006;7:1443–1450. doi: 10.1002/cbic.200600117. [DOI] [PubMed] [Google Scholar]
- 21.Lo Monte F, Kramer T, Gu J, Anumala UR, Marinelli L, La Pietra V, Novellino E, Franco B, Demedts D, Van Leuven F, Fuertes A, Dominguez JM, Plotkin B, Eldar-Finkelman H, Schmidt B. Identification of glycogen synthase kinase-3 inhibitors with a selective sting for glycogen synthase kinase-3alpha. J Med Chem. 2012;55:4407–4424. doi: 10.1021/jm300309a. [DOI] [PubMed] [Google Scholar]
- 22.Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, Hahn CK, Kakoza RM, Chow KT, Ross L, Alexe G, Tolliday N, Inguilizian H, Galinsky I, Stone RM, DeAngelo DJ, Roti G, Aster JC, Hahn WC, Kung AL, Stegmaier K. The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. The Journal of clinical investigation. 2012;122:935–947. doi: 10.1172/JCI46465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bang D, Wilson W, Ryan M, Yeh JJ, Baldwin AS. GSK-3alpha Promotes Oncogenic KRAS Function in Pancreatic Cancer via TAK1-TAB Stabilization and Regulation of Noncanonical NF-kappaB. Cancer Discov. 2013;3:690–703. doi: 10.1158/2159-8290.CD-12-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen P, Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat Rev Drug Discov. 2004;3:479–487. doi: 10.1038/nrd1415. [DOI] [PubMed] [Google Scholar]
- 25.Hooper C, Killick R, Lovestone S. The GSK3 hypothesis of Alzheimer’s disease. Journal of neurochemistry. 2008;104:1433–1439. doi: 10.1111/j.1471-4159.2007.05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotliarova S, Pastorino S, Kovell LC, Kotliarov Y, Song H, Zhang W, Bailey R, Maric D, Zenklusen JC, Lee J, Fine HA. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res. 2008;68:6643–6651. doi: 10.1158/0008-5472.CAN-08-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Kocher T, Superti-Furga G. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.