Abstract
During embryonic development, DNA binding proteins help to specify and restrict the fates of pluripotent stem cells. In the developing kidney, Pax2 proteins are among the earliest markers for the renal epithelial cell lineage, with expression in the mesenchyme and in proliferating epithelia. The Pax2 protein is essential for interpreting inductive signals emanating from the ureter bud such that kidney mesenchyme can convert to epithelia. The biochemistry of Pax protein function is being studied in a variety of model systems. Through interactions with the adaptor PTIP, Pax proteins can recruit members of the Trithorax family of histone methyltransferases to imprint activating epigenetic marks on chromatin. However, the interactions with the co-repressor protein Grg4 can inhibit activation and instead recruit Polycomb repressor complexes to promote target gene silencing. We present a model whereby the regulated interactions of Pax proteins with available co-factors mediated activation or gene silencing at different stages of development. The implications for establishing and maintaining the epigenome are discussed.
Keywords: Kidney Development, Pax, PTIP, Grg4, Epigenetics, Trithorax, Polycomb
Introduction
How a single fertilized egg ultimately generates all of the highly specialized cell types in the body and incorporates those differentiated cells within a three-dimensional, architectural framework is one of the most amazing feats in biology. Although the environment can impact in utero development, the essential instructions for embryogenesis must be encoded in the genome. Given the complexity of the process, the human genome likely contains a large number of genes whose functions are limited to the developmental stages and whose expression may in fact be detrimental in adults. Many such developmental regulators have been identified in genetic screens of model organisms such as Drosophila, C. elegans, and Zebrafish. A surprisingly large number of genes that are essential for specifying cell types and tissues encode proteins with DNA binding domains, including homeoboxes, helix-loop-helix domains, zinc-fingers, HMG boxes, or paired-domains. As these genes and proteins were identified, it was assumed that the DNA binding proteins regulated transcription during development. Thus, the functions of these developmental DNA binding proteins were modeled based on the existing knowledge of known transcription factors or co-factors. However with our increased understanding of the chromatin and all of its epigenetic modifications, it is important to revisit some of our assumptions about DNA binding proteins in development and to consider alternative models for how DNA binding proteins impact the genome.
Despite sharing the identical genome, differentiated cells express unique sets of genes. These patterns of gene expression are in part imprinted through epigenetic modifications, such as histone methylation and DNA methylation, which are established during embryonic development [1]. The Polycomb and Trithorax family of epigenetic regulators function in development to maintain patterns of gene expression, yet how they recognize genes in a temporal and tissue specific manner remains unclear [2, 3]. In the developing kidney, many critical genes and DNA binding proteins have been identified that specify the renal stem and progenitor cells and their differentiated derivatives. How do these proteins impact the genome and the regulation of gene expression? Genetics and biochemistry have shed significant insight into the functions of early renal specification genes.
The adult kidney is composed of a large number of specialized epithelial, stromal and endothelial cells. Renal epithelial and stromal cells share a common lineage that is specified early in development, but diverges once kidney development proceeds [4]. In mammals, this lineage is apparent shortly after gastrulation in a region of mesoderm, called the intermediate mesoderm (IM) that lies between the paraxial mesoderm and the lateral plate mesoderm along the medio-lateral axis [5–7]. Moreover, the kidney develops along the anterior posterior axis in a temporal sequence and likely requires both medio-lateral and anterior-posterior patterning signals to determine the adult kidney field (Fig. 1). Early anterior kidney structures include the pro- and mesonephros, whose complexity, size, and duration varies greatly among vertebrate species. In the mouse, the pronephros is barely detectable, whereas mesonephric tubules are well developed with a proximal glomerulus and convoluted tubules that empty into the nephric duct (Fig. 1). The adult, or metanephric kidney, forms at the posterior end of this intermediate mesoderm when the ureteric bud grows out of the nephric duct and induces the metanephric mesenchyme to begin the process of nephrogenesis. Sequential branching morphogenesis of the ureteric bud induces new nephrons at the tips [8]. These nephrons arise form a population of stem cells, called the cap mesenchyme, that aggregate at the tips of each new branch point. Cell lineage tracing clearly shows that cap mesenchyme is capable of producing all the epithelial cells of the nephron [4, 9, 10]. Cap mesenchyme undergoes a mesenchymal-to-epithelial transition, generating first a primitive renal vesicle, followed by a comma-shaped body and an s-shaped body. Reiterative branching, induction, and renal vesicle formation along the radial axis continues such that older nephrons are located close to the medulla as young nephrons develop at the periphery.
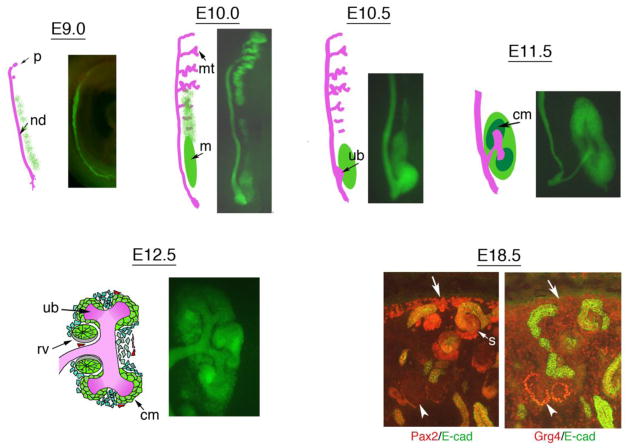
Figure 1. Pax2 Expression Marks the Intermediate Mesoderm and its Epithelial Derivatives.
Schematics of the developing kidney are shown from E9 to E12.5 in the mouse and compared to the expression of the Pax2 gene as shown by EGFP in the adjacent panel. The intermediate mesoderm begins to express Pax2 at 4 somite stage, approximately embryonic day 8 (E8, not shown). By E9, the nephric duct (nd) is clearly visible and extends towards the cloaca at the caudal end; the pronephros (p) is very rudimentary. By E10, the mesonephric tubules (mt) are well developed more anteriorly and the metanephric mesenchyme (m) is visible posteriorly. By E10.5, the ureteric bud begins to grow out of the nephric duct and invade the mesenchyme. By E11.5, the ureteric bud has bifurcated and the cap mesenchyme aggregates around the tips of the growing bud. By E12.5, the ureteric bud has branched again and renal vesicles, primitive epithelia derived from the cap mesenchyme, are visible. Sections through an E18.5 kidney show the complexity of the growing organ, with Pax2 positive cells (red, left panel) at the periphery (arrow), the cap mesenchyme and the comma and s-shaped derivatives (s), but not in the more developed tubules or in the glomeruli (arrowhead). An adjacent section shows the pattern of Grg4 (red, right panel) expression, which is low in the undifferentiated cells (arrow) and increases in podocyte precursors (arrowhead).
Pax Proteins in the developing kidney
Genetic analyses in mice clearly underscore the essential functions of the genes Lhx1, Osr1, and Pax2/8 in early specification of the IM. Lhx1 null mice lack the nephric duct, although Pax2 expression is observed in cells at the boundary between the paraxial and lateral plate mesoderm shortly after gastrulation [11]. Pax2 null mutants do develop a nephric duct [12], but the duct is completely absent in a Pax2/8 double mutant, suggesting that these Pax genes function redundantly in this early IM domain [13]. Pax2/8 double mutants also do not express Lhx1. Mice homozygous for an Osr1 null allele, the expression of which precedes that of Pax2/8, still exhibit nephric duct formation and Pax2 expression in the anterior IM, yet lack more developed mesonephric tubules and the metanephric mesenchyme in the posterior IM [14, 15]. Whether this anterior Pax2 expression in the Osr1 mutants is due to some partial rescue or redundancy by Osr2 or is completely cell autonomous and independent of Osr1 function remains to be determined.
At the time of metanephric mesenchyme induction, Pax2 is strongly expressed in the nephric duct, its outgrowth the ureteric bud, and in the metanephric mesenchyme prior to induction (Fig. 1). In homozygous Pax2 mutants, the ureteric bud fails to form, thus depriving the metanephric mesenchyme of the required inductive signals. This results in complete renal agenesis. However, attempts to replace the ureteric bud by exogenous inducers in vitro revealed that the Pax2 mutant mesenchyme was unable to respond to ectopic inductive signals in organ culture models [16]. We now know that WNT signals are essential for promoting the aggregation of the metanephric mesenchyme and for initiating the conversion of mesenchyme to renal epithelia [17]. Thus, Pax2 is essential for interpreting these inductive WNT signals into a biological response that includes cell aggregation, epithelial cell polarization, and proliferation of the primitive epithelial cells.
The question then remains as to how Pax proteins are able to transduce signals into a transcriptional response in a developing tissue. Several genes are known to require Pax2 for activation in the embryonic kidney, including GDNF [16] and GATA3 [18], yet the molecular mechanisms for Pax mediated activation are poorly characterized. Are Pax proteins transcription factors that promote polymerase II initiation or elongation, do they bind proximal promoters or distal enhancer sequences, do Pax proteins recruit or require multiple co-factors, do Pax proteins alter the structure or accessibility of chromatin? These are just some of the issues that require more detailed analyses before the developmental regulatory functions of Pax proteins can be fully understood.
Histone Methylation, Epigenetics, and Cell Lineage Decisions
As cells lose pluripotency and begin to differentiate towards specific lineages, this information must be imprinted somehow and then inherited such that the fates of all progeny cells are not altered during many rounds of cell division. It is becoming increasingly clear that cellular memory, as specified in development, is imprinted onto chromatin by epigenetic modifications. Chromatin consists of the genomic DNA and all the associated proteins, of which histones are the major component. The basic structural unit of chromatin is the nucleosome, which consists of approximately 1.6 turns of the double stranded DNA wrapped around a histone octamer. There are two copies of each of the four histones, H2A, H2B, H3 and H4 within the octamer. The crystal structure of the nucleosome revealed that the amino-terminal histone tails protrude out of the nucleosome thus making them accessible for post-translational modifications [19]. Post-translational modifications include acetylation or methylation of specific lysine or arginine residues, phosphorylation of serine residues, or ubiquitination of lysine residues. These types of histone modifications can directly impact gene expression and the structure of chromatin by providing a platform for additional proteins that recognize individual or combinations of modifications.
Among the most critical breakthroughs that helped define how chromatin modifications impact genome regulation was the cloning and characterization of enzymes that methylate histone tails. Genetic screens in Drosophila identified epigenetic modifier genes that established and maintained gene expression patterns in embryonic development [20, 21]. These epigenetic regulatory genes fell into two major groups, the Polycomb family of repressors and the Trithorax family of activators. However, the amino acid sequences of Polycomb and Trithorax proteins revealed little of their function. Not until the histone methyltransferases were cloned and sequenced from simple organisms like yeast and Tetrahymena did it become clear that many of the epigenetic regulators found in flies were histone methyltransferases or co-factors that were part of the methyltransferase complexes. Even more striking, the histone methyltransferases that were associated with compact or silent chromatin were found in the Polycomb family of epigenetic silencer genes, whereas the histone methyltransferases that were associated with expressed genes and accessible chromatin were among the Trithorax family of activators.
In mammals, the Polycomb and Trithorax complexes and their associated proteins have all been well described. The mammalian homologues of Drosophila Trithorax are the MLL (mixed lineage leukemia) genes that encode histone H3K4 methyltransferases, whereas the polycomb homologues Ezh1/2 and Suv39 encode H3K27 and H3K9 methyltransferases respectively. These enzymes are found in large protein complexes with multiple co-factors required for activity. However, biochemical purification of MLL complexes failed to find proteins that control DNA binding specificity. Thus how these histone methyltransferase complexes recognize specific genes in specific tissues remains to be determined.
Evidence that histone methylation is important for regulating developmental gene expression is not limited to Drosophila. In mammalian embryonic stem cells, many important lineage specific genes contain both positive and negative epigenetic marks. This pattern of low levels of H3K4 and H3K27 methylation was termed a bivalent chromatin domain and may be a general property of pluripotent stem cells [22, 23]. As stem cells differentiate towards a specific lineage, these bivalent domains are resolved into either fully activated H3K4 methylated or fully repressed H3K27 and H3K9 methylated domains. Clearly, during development the targeting of histone methyltransferase to specific genes must be controlled and is likely to involve many types of DNA binding proteins.
Linking Pax proteins to histone modifications
If particular histone modifications correlate with the loss of pluripotency and the restriction of cell fate, then how do the histone modifying complexes recognize specific genes at specific times in development? Strikingly, none of the biochemically purified mammalian H3K4 methyltransferase complexes contained sequence specific DNA binding proteins. Two possible explanations may account for the lack of DNA binding proteins in the MLL complexes. First, the complex may interact with many different DNA binding proteins such that they are not represented in stoichiometric quantities and thus are hidden among the background. Second, the MLL complex may only recognize DNA binding proteins when bound to chromatin and not in solution.
Recently, several members of the Pax family of DNA binding proteins have been linked to H3K4 methyltransferases and epigenetic control of gene expression. The Pax2 protein interacts with a ubiquitous nuclear factor, PTIP (Pax Transactivation-domain Interacting Protein), which is part of the MLL3/4 H3K4 methyltransferase complex [24]. Cell culture experiments demonstrate that Pax2 recruits the MLLs to a DNA binding site to increase levels of H3K4me3. In the absence of PTIP, Pax2 binds to chromatin but cannot recruit the MLL complex. Thus, PTIP links MLLs to a sequence specific DNA binding protein. The PTIP protein is essential for early development in mouse and in flies, as complete null mutants show significant decreases in global levels of H3K4me3 [25, 26]. The modular PTIP protein has 3 pairs of BRCT domains, which can recognize Phosphorylated serine residues [27]. Thus, it is likely that PTIP interacts with many other Phospho-Serine DNA binding proteins to link MLLs to specific DNA sequences.
Developing B cells require the Pax5 protein, which is nearly identical to Pax2 and binds the 3′ enhancer located on the immunoglobulin heavy chain (IgH) locus. This 3′ enhancer is thought to interact with more proximal IgH promoter sequences through long-range chromatin looping. Both PTIP and Pax5 are localized to proximal promoters whose levels of H3K4me3 greatly increase with signals that stimulate isotype class switch recombination [28]. However, PTIP mutant B cells do not recruit Pax5, show no evidence for chromatin looping, nor show increased H3K4me3 at proximal promoters in response to class switch stimulants [28, 29]. These data suggest a common role for PTIP in linking Pax proteins to MLL complexes in response to developmental or differentiation signals. Furthermore, the Pax-PTIP-MLL complex can orchestrate complex chromatin conformational changes, including long-range interactions between enhancers and promoters.
In muscle progenitor cells, the Pax7 protein is critical for the self-renewal and differentiation of satellite cells into myocytes [30]. At the promoters of activated genes, such as myf5 and myoD, Pax7 recruits the MLL2 histone methyltransferase complex to imprint high levels of H3K4me3 [31]. However, Pax3 and Pax7 appear to use a different adaptor protein, called Pax3/7BP (GCFC1), to link the DNA binding Pax proteins to the MLL/Wdr5/Ash2L complex [32].
The expression of Pax proteins in stem or progenitor cell populations, be it the metanephric mesenchyme, muscle satellite cells, or the pre B cells, suggest fundamental roles in cell lineage determination. Upon mutation of the corresponding Pax gene, such cell lineages are either ablated or arrested in differentiation. Thus, the recruitment of epigenetic complexes, such as the MLL family of H3K4 methyltransferases, is likely to imprint cell lineage specific modifications on chromatin such that the correct genes remain activated or accessible. Recent evidence also points to the capacity of Pax proteins to silence gene expression through mechanisms that require Polycomb complexes and repressive histone modifications.
Mechanisms of Pax mediated repression
Polycomb mediated gene silencing is essential for maintaining proper gene expression patterns in development. As stem cells differentiate they not only establish active histone marks but also repressive marks to generate heterochromatin for silencing parts of the genome. The most common repressive epigenetic marks are H3K27, H3K9 and H4R3 methylation and DNA methylation at CpG islands. These marks are thought to provide binding sites for proteins that function to compact chromatin into a more inaccessible state. This inaccessible chromatin limits the ability of common nuclear effectors to access target genes, essentially creating a novel epigenetic landscape for each cell type [33]. In Drosophila, Polycomb response elements guide the assembly of Polycomb repressor complexes (PRC1, PRC2) on DNA and act as nucleation centers to initiate gene silencing. However in mammalian organisms, Polycomb response elements are poorly defined and the mechanisms of DNA recognition by PRCs remains to be fully understood.
The Pax2/5/8 family of proteins has been linked to co-repressors of the Groucho family [34, 35], also known as Grg (Groucho related genes) or Tle (Transducin-like enhancer of split). In the developing kidney, Grg4 is expressed at low levels in the metanephric mesenchyme. However, expression of Grg4 increases in the primitive epithelia of the renal vesicle and is maximal in the podocyte precursors of the S-shaped body and in mature glomerular podocytes (Fig. 1). In cell culture assays, Grg4 inhibits Pax dependent gene activation [34]. Through the purification of Grg4 and associated proteins, the mechanism of Grg4 mediated repression at Pax DNA binding sites is becoming clear.
The Grg4 complex contains a phosphatase that can dephosphorylate the activation domain of Pax2 [34, 36]. Phosphorylation of the Pax2 carboxy-terminus increases its ability to activate gene expression and is likely to mediate its association with PTIP, via the P-serine binding BRCT domains [27]. Thus, Grg4 expression displaces PTIP from Pax2, consistent with the dephosphorylated state of Pax2. The Grg4 complex also contains an arginine methyltransferase PRMT5, capable of symmetric dimethylation at H4R3 [36], a common repressive epigenetic mark [37]. The symmetric dimethylation of H4R3 is necessary for the further recruitment of the PRC2 complex and subsequent H3K27 dimethylation. These data are consistent with the observation that Groucho proteins can drive chromatin compaction to limit accessibility of other DNA binding proteins at silenced loci [38].
Thus, our current model of Pax mediated gene regulation encompasses both activation and repression, depending at least in part on the availability of co-factors (Fig. 2). The ubiquitous adapter PTIP links Pax proteins to MLL complexes for maintaining gene activation, whereas the regulated co-repressor Grg4 can override this activation program to imprint repressive epigenetic marks. In the kidney, it is likely that Pax2/PTIP complexes predominate in mesenchyme, where Grg4 levels are low. As cap mesenchyme transitions to epithelia and Grg4 levels increase, the propensity for Pax2 to complex with Grg4 and PRC2 is likely to increase. Thus, we can envisage a situation whereby the same DNA binding Pax protein can maintain gene activation in undifferentiated precursor cells but then repress that same target in terminally differentiate cells. Confirmation of such a model still requires precise knowledge of chromatin modifications and structure at specific loci during the transition from stem cells to renal epithelia.
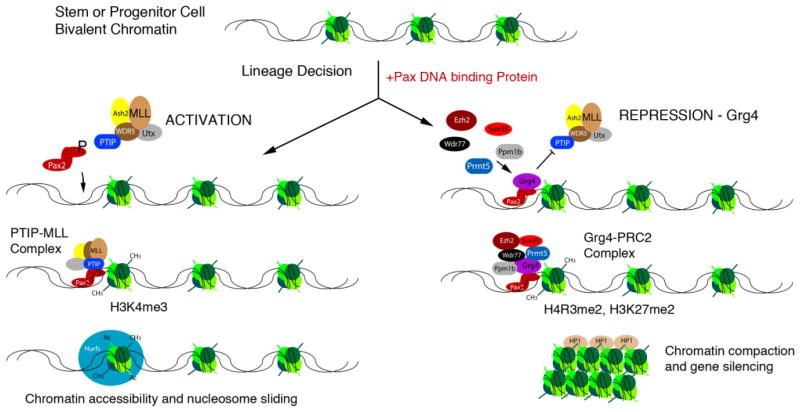
Figure 2. Model of Pax mediated Activation and Repression.
A gene in a progenitor or stem cell that has not yet been imprinted with a lineage specific epigenetic marks remains in a bivalent, or unspecified state. Upon expression of a Pax protein a lineage decision is initiated. Pax2 recruits the adapter PTIP and an MLL complex to imprint chromatin with active histone H3K4me marks. These marks prevent silencing and allow additional complex to access the gene, such as nucleosome remodeling factors (Nurfs) that are necessary for transcription. However in cells with high levels of Grg4, the PTIP/MLL complex is displaced by the Grg4/PRMT5/PPM1b complex that dephosphorylated Pax2 and imprints repressive epigenetic marks, such as H4R3me2 and H3K27me2. The recruitment of the Polycomb repressor complex 2 can lead to chromatin compaction and gene silencing.
Stability of the epigenetic state
If histone modifications are established as cells become specified and restricted in their fates, it is worth considering whether the proteins that establish patterns of histone methylation are needed in terminally differentiated cells and whether mutations in such pathways can lead to abnormalities. Once cells exit the mitotic cycle, it is possible that the pattern of histone modifications is stable. This question has been addressed in the kidney and the adult heart by deleting the adaptor protein PTIP in podocytes or cardiomyocytes respectively [39, 40]. In both cases, loss of histone H3K4me3 and alterations in gene expression patterns are observed. The resulting phenotypes suggest that MLL mediated H3K4me3 is still necessary even in non-dividing cells to maintain gene expression patterns. A decrease in H3K4me3 in podocytes leads to chronic glomerular disease, with foot process effacement and structural abnormalities evident even before the reduction in podocyte number. In the heart, changes in gene expression due to reduced H3K4me3 can lead to altered electrophysiology and arrhythmias.
Acute kidney injury through ischemia or nephrotoxins leads to cell death of proximal tubule epithelial cells and acute renal failure. However, proximal tubules can regenerate as surviving cells repopulate the damaged tubule [41]. Regeneration activates gene expression patterns reminiscent of developing tubules, including the re-expression of Pax2 [42]. This reactivation may be needed to reset the epigenetic landscape of the newly generated epithelial cells. Regeneration after injury also re-establishes the need for other DNA binding proteins, such as HNF1b, which regulates the PKD2 gene [43].
In conclusion, mechanisms of gene activation and repression by Pax proteins in the kidney may function to imprint both active and repressive epigenetic marks through interactions with histone modifying complexes. Such imprints can compartmentalize the genome into active and silent domains, unique for specific cell types. Maintaining such epigenetic marks throughout the life of a cell may be important to keep gene expression patterns stable. Thus, any variables that can alter the epigenetic landscape, such as environmental exposure, aging, and injury, may be critical factors driving the initiation and progression of chronic disease in the kidney.
Acknowledgments
This work was supported by National Institutes of Health grants DK073722 (G.R.D.) and DK082409 (S.R.P.). We thank the members of our labs for discussion and insights.
References
- 1.Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 3.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 7.Saxen L. In: Organogenesis of the Kidney. Barlow PW, Green PB, White CC, editors. Cambridge University Press; Cambridge, UK: 1987. (Developmental and Cell Biology Series 19). [Google Scholar]
- 8.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsang TE, Shawlot W, Kinder SJ, Kobayashi A, Kwan KM, Schughart K, Kania A, Jessell TM, Behringer RR, Tam PP. Lim1 activity is required for intermediate mesoderm differentiation in the mouse embryo. Dev Biol. 2000;223:77–90. doi: 10.1006/dbio.2000.9733. [DOI] [PubMed] [Google Scholar]
- 12.Soofi A, Levitan I, Dressler GR. Two novel EGFP insertion alleles reveal unique aspects of Pax2 function in embryonic and adult kidneys. Dev Biol. 2012;365:241–250. doi: 10.1016/j.ydbio.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Lan Y, Cho ES, Maltby KM, Jiang R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Dev Biol. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brophy PD, Ostrom L, Lang KM, Dressler GR. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development. 2001;128:4747–4756. doi: 10.1242/dev.128.23.4747. [DOI] [PubMed] [Google Scholar]
- 17.Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Grote D, Souabni A, Busslinger M, Bouchard M. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133:53–61. doi: 10.1242/dev.02184. [DOI] [PubMed] [Google Scholar]
- 19.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 21.Lanzuolo C, Orlando V. Memories from the polycomb group proteins. Annu Rev Genet. 2012;46:561–589. doi: 10.1146/annurev-genet-110711-155603. [DOI] [PubMed] [Google Scholar]
- 22.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 24.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol Cell Biol. 2003;23:1666–1673. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang M, Ren H, Liu J, Cadigan KM, Patel SR, Dressler GR. Drosophila ptip is essential for anterior/posterior patterning in development and interacts with the PcG and trxG pathways. Development. 2009;136:1929–1938. doi: 10.1242/dev.026559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 28.Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, Cho YW, Sun HW, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. PTIP Promotes Chromatin Changes Critical for Immunoglobulin Class Switch Recombination. Science. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwab KR, Patel SR, Dressler GR. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Mol Cell Biol. 2011;31:1503–1511. doi: 10.1128/MCB.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudnicki MA, Le Grand F, McKinnell I, Kuang S. The molecular regulation of muscle stem cell function. Cold Spring Harb Symp Quant Biol. 2008;73:323–331. doi: 10.1101/sqb.2008.73.064. [DOI] [PubMed] [Google Scholar]
- 31.McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell\Bbiol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diao Y, Guo X, Li Y, Sun K, Lu L, Jiang L, Fu X, Zhu H, Sun H, Wang H, Wu Z. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11:231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 33.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Y, Brophy PD, Levitan I, Stifani S, Dressler GR. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J. 2003;22:5522–5529. doi: 10.1093/emboj/cdg536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel SR, Bhumbra SS, Paknikar RS, Dressler GR. Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol Cell. 2012;45:185–195. doi: 10.1016/j.molcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Hoang S, Mayo MW, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics. 2011;11:396. doi: 10.1186/1471-2105-11-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lefevre GM, Patel SR, Kim D, Tessarollo L, Dressler GR. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet. 2010;6:e1001142. doi: 10.1371/journal.pgen.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, Dressler GR. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121:2641–2650. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphreys BD, Czerniak S, DiRocco DP, Hasnain W, Cheema R, Bonventre JV. Repair of injured proximal tubule does not involve specialized progenitors. Proc Natl Acad Sci U S A. 2011;108:9226–9231. doi: 10.1073/pnas.1100629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imgrund M, Grone E, Grone HJ, Kretzler M, Holzman L, Schlondorff D, Rothenpieler UW. Re-expression of the developmental gene Pax-2 during experimental acute tubular necrosis in mice 1. Kidney Int. 1999;56:1423–1431. doi: 10.1046/j.1523-1755.1999.00663.x. [DOI] [PubMed] [Google Scholar]
- 43.Verdeguer F, Le Corre S, Fischer E, Callens C, Garbay S, Doyen A, Igarashi P, Terzi F, Pontoglio M. A mitotic transcriptional switch in polycystic kidney disease. Nat Med. 2010;16:106–110. doi: 10.1038/nm.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]