Abstract
Congenital anomalies of the kidney and urinary tract (CAKUT) are the leading cause of childhood chronic kidney disease (CKD). While mutations in several renal development genes have been identified as causes for CAKUT, most cases have not yet been linked to known mutations. Furthermore, the genotype-phenotype correlation is variable, suggesting that there are additional factors that impact the severity of CAKUT. MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression at the post-transcriptional level, and are involved in many developmental processes. Although little is known about the function of specific miRNAs in kidney development, several have recently been shown to regulate the expression of, and/or are regulated by, crucial renal development genes present in other organ systems. In this review, we discuss how miRNA regulation of common developmental signaling pathways may be applicable to renal development. We focus on genes that are known to contribute to CAKUT in humans, for which miRNA interactions in other contexts have been identified, with miRNAs that are present in the kidney. We hypothesize that miRNA-mediated processes play a role in kidney development through similar mechanisms, and speculate that genotypic variations in these small RNAs or their targets could be associated with CAKUT.
Keywords: microRNAs, kidney development, congenital anomalies, renal disease, epigenetics
Introduction
Congenital anomalies of the kidney and urinary tract (CAKUT) are amongst the most frequent birth defects in humans, and are the major cause of childhood chronic kidney disease and end-stage renal disease [1]. The underlying causes of CAKUT remain elusive for the most part, likely because these represent a heterogeneous group of disorders with many underlying etiologies, and a wide range of prognostic outcomes. Thus, molecular tools that may direct prevention strategies, predict functional outcomes or guide interventions would be a helpful addition to the clinical management of these children. This review will address emerging information regarding the role of miRNAs in regulating kidney development.
In humans, kidney development begins around the fifth week of gestation with the outgrowth of the ureteric bud (UB) from the Wolffian duct epithelium. This outgrowth is initiated via cues from a subset of the adjacent intermediate mesoderm, known as the metanephric mesenchyme (MM), which induces the UB to elongate and invade the MM [2]. The UB responds to signals from the MM to undergo iterative branching events to form the collecting duct system [2, 3]. The terminal collecting ducts, whose ampullae are referred to as ureteric buds, induce the formation of new nephrons with each successive round of branching [3]. In response to signals from the UB, MM cells are induced to either condense around the tips of the UB to form nephron progenitors (multipotent, self-renewing renal progenitors) or to become renal stromal cells [4].
Broadly speaking, CAKUTs occur due to abnormalities in kidney and urinary tract development. For example, abnormal UB induction can result in renal agenesis, duplicated collecting systems, vesicoureteral reflux and/or obstruction. Alternatively, if fewer nephrons are induced during kidney development, renal hypoplasia resulting in poor congenital nephron endowment occurs. Children with reduced nephron numbers are thought to have a poorer renal prognosis [5], and are at risk for developing primary hypertension [6]. In addition, impaired nephrogenesis can result in dysplastic kidneys or tumor formation (eg. Wilms tumors), related to abnormal cell fate specification, apoptosis or proliferation. For an in-depth review please refer to [7]. CAKUTs are a highly heterogeneous group of disorders, and the factors that influence the clinical presentations of CAKUTs are not fully understood. For example, the renal phenotype in families with known mutations affecting kidney development can be variable, suggesting that there are additional factors involved [8]. These factors could be mutational (i.e. genetic variation amongst individuals), epigenetic (i.e. DNA methlylation, histone modifications, miRNAs, etc.) and/or environmental (i.e. toxin exposure). Given the heterogeneity, it is likely that the genetic or molecular basis may be different from one subtype to another.
MiRNAs are small non-coding RNAs that act as regulators of gene expression through the repression of their target mRNAs (reviewed in [9]). miRNA genes are transcribed by RNA polymerase II, and are subsequently processed to their mature, 22-nucleotide form. They interact primarily with the 3′-untranslated regions (UTRs) of target mRNAs via an 8-nucleotide seed region to direct mRNA degradation or inhibit protein translation. A single miRNA can have hundreds of putative targets, and any given mRNA can be targeted by multiple miRNAs. Individual miRNAs are thought to be capable of targeting multiple proteins in a signaling pathway to maximize their effect. The majority of studies regarding miRNAs in kidney development have thus far blocked the miRNA biogenesis pathway in specific cell lineages. In renal tubules, loss of miRNAs impacted maturation and caused hydroureter, hydronephrosis, and tubular and glomerular cysts [10]. Without miRNAs in juxtaglomerular cells, they no longer expressed renin [11]. Inhibiting mature miRNAs in nephron progenitors increased apoptosis and severely impacted progenitor survival [12, 13]. Loss of miRNAs in podocytes resulted in marked proteinuria and tubular and glomerular injury including tuft collapse and foot process effacement [14]. Finally, miRNA biogenesis disruption in renal tubules along with part of the ureteric bud caused the CAKUT symptoms of low nephron endowment and ureteropelvic junction obstruction [15]. Please refer to several recent reviews for details [16-18].
Despite growing knowledge regarding miRNAs in the kidney, the roles of individual miRNAs in renal development remains largely obscure, and there is limited data regarding the function of miRNAs in human kidney development. Intriguingly, deep RNA-sequencing of murine embryonic kidneys revealed 51 miRNAs that are present in the developing kidney [19]. In this review, we will discuss recently published data on genes in which mutations have been defined in humans with CAKUT, and for which there is evidence of interaction in various experimental systems with miRNA(s) known to be expressed in the kidney (please see schematic 1 for a summary). We speculate that miRNAs may be responsible, at least in part, for the phenotypic variability observed in CAKUTs.
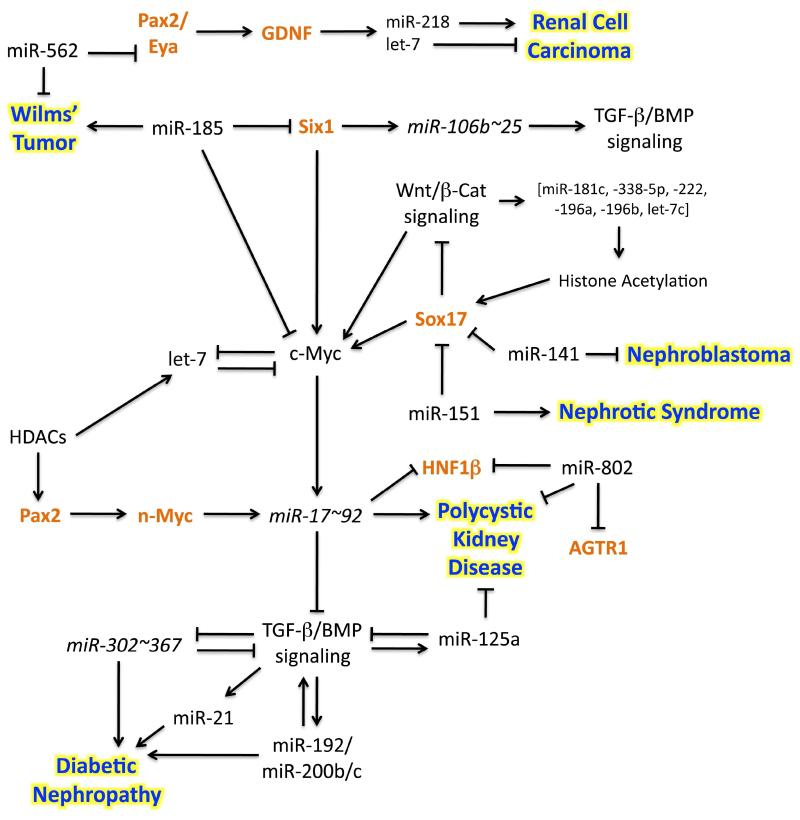
Schematic 1.
MiRNAs associated with renal disease can potentially regulate CAKUT related genes during kidney development. Several of the miRNAs mentioned in this review have already been linked to kidney disease. Presented are the hypothetical pathways in which these miRNAs can potentially interact with CAKUT linked renal development genes (orange text). Arrows indicate a positive interaction, where t-bars indicate inhibition.
A potential Pax2/n-Myc/miR-17~92 interaction in nephrogenesis
The Pax2 gene (paired box 2) codes for a transcription factor critical for the formation of tissues and organs during embryogenesis, including the kidney. Pax2 mutations are associated with Renal Coloboma Syndrome (RCS), which is associated with congenital anomalies of the kidneys [20, 21]. Null mutations in humans have not been described, presumably due to prenatal lethality; however, Pax2−/− mice completely lack ureters, kidneys and the entire genital tract [22]. In both mice and humans, heterozygous loss of Pax2 results in reduced kidney size [22, 23]. In contrast, transgenic mice that overexpress Pax2 globally had poorly developed podocyte foot processes, proteinaceous dilated tubules, abnormal renal function and died perinatally [24]. This suggests that the gene dosage of Pax2 is essential for normal kidney development, and implies that its levels must be tightly regulated.
One possible mechanism to regulate Pax2 levels is via an interaction with n-Myc and miRNAs. Pax2 and n-Myc can function concertedly to regulate cell proliferation in embryonic renal mesenchymal cells [25]. Pax2 and n-Myc levels appeared to positively regulate each other, where transfected Pax2 cDNA elevated n-Myc levels, and vice versa. In addition, the stimulation of Pax2 transcription by n-Myc was amplified under high glucose conditions, implying that this pathway could be modulated by stress [25]. n-Myc is required for kidney development, and hypomorphic mutations have been associated with fewer developing glomeruli and collecting ducts in the embryonic mouse kidney [26]. n-Myc is thought to transcriptionally activate miR-17~92 via several canonical E-box binding domains located in the 5′UTR of the miR-17~92 loci, and has been shown to do so in primary cerebellar granule neuron precursors [27].
miR-17~92 (homologous to human MIR17HG) codes for six miRNAs that have been shown via numerous studies to be linked to the regulation of cell cycle and proliferation in a variety of cellular contexts [28-30]. The observation that the hypomorphic n-Myc allele resulted in decreased cell division in the developing mouse kidney is interesting given the described role of miR-17~92 in the cell cycle [26, 31, 32]. Additionally, miR-17~92 expression is amplified in several cancers including Wilms’ tumor [27, 33], possibly in response to elevated n-Myc and/or c-Myc levels [34, 35]. In humans, miR-17~92 haploinsufficiency has been linked to Feingold Syndrome [28], a condition that is most frequently ascribed to mutations in n-Myc [36]. Renal defects have been reported in cases of Feingold Syndrome due to n-Myc mutations, including bilateral renal dysplasia and hypoplasia; however it remains unclear what role the miR-17~92 cluster plays during normal kidney development [36, 37]. Together, these data raise the question of whether a Pax2/n-Myc/miR-17~92 pathway plays an important role in kidney development.
miRNAs expressed in polycystic kidney disease can target HNF1β
Recent data showed that miR-17~92 was upregulated in the Ksp/cre; Kif3aflx/flx model of polycystic kidney disease (PKD) [38]. Interestingly, deletion of the miR-17~92 locus from developing renal tubules and ureters in this model ameliorated cyst growth [38]. Luciferase reporter assays subsequently demonstrated that the PKD1 and PKD2 genes can be targeted by miR-17 [32, 39, 40]. In renal epithelial cells, miR-92a (one of the miRNAs in the miR-17~92 cluster) was found to directly target the 3′UTR of hepatocyte nuclear factor-1β (HNF1β, mutations of which are associated with cystic renal hypodysplasia) [38, 41]. In addition, miR-17~92 expression in mouse kidneys was negatively correlated with HNF1β levels, which is consistent with HNF1β being a direct target of miR-92a [38].
Another miRNA that may target HNF1β is miR-802. In murine liver cells, upregulation of miR-802 was correlated with decreased HNF1β expression, and direct targeting of HNF1β by miR-802 was confirmed using a luciferase assay [42]. Levels of miR-802 may be responsive to stress, since miR-802 was increased in the livers of both obese mice and humans [42], as well as in response to a high potassium diet in the cortical collecting ducts of mice [43]. Interestingly, miRNA microarrays of adult PKD mice revealed that miR-802 is downregulated relative to controls in the kidney [38]. These studies suggest that miR-802 might target HNF1β in the kidney as well.
Potential HDAC/let-7/c-Myc/miR-17~92 pathway in kidney development
Chromatin modifications represent an epigenetic mechanism that allows genes to become more or less transcriptionally active. Examples include histone acetylation and deacetylation, and histone deacetylases (HDACs) have important roles in numerous cellular processes including cell cycle, proliferation, differentiation, and cell death [44]. For example, chromatin immunoprecipitation assays demonstrated that HDAC1 and HDAC2 regulate Pax2 in the developing mouse kidney [45]. Interestingly, about 40% of noncoding RNAs were either upregulated or down-regulated in response to HDAC inhibitors in cancer [46, 47]. In the kidney, HDAC inhibitors expanded the renal progenitor population in zebrafish [48], and enhanced renal recovery after acute kidney injury in both zebrafish and mice [49]. In addition to Pax2, other important kidney development genes regulated by HDACs include: Pax8, WT1, GDNF and Wnt9b [45]. Taken together, these data suggest that HDAC regulation of miRNAs and renal development genes could play an important role during kidney organogenesis.
HDAC inhibitors have been shown to downregulate several members of the let-7 miRNA family in human breast cancer and hepatic stellate cells [47, 50], as well as miR-17~92 levels in colorectal cancer cells [51]. In liver cells, let-7c could directly target the 3′UTR of c-Myc, subsequently altering the expression of miR-17~92 [52], a transcriptional target of c-Myc [27, 53]. Interestingly, recent work showed that c-Myc functions as a direct transcriptional repressor of miRNA genes in B cell lymphomas, including members of the let-7 family [54]. These data suggest that it is possible for c-Myc and let-7 miRNAs to reciprocally regulate each other possibly affecting at least the levels of miR-17~92 miRNAs. Let-7 member miRNAs are expressed fairly ubiquitously in the kidney [12, 55, 56], and their miss-expression was linked with renal cell carcinoma [57]. Since c-Myc, let-7g (at least), and miR-17~92 are all expressed in nephron progenitors [12, 45, 55, 56], it will be interesting to determine if an interaction between these signaling pathways exists during kidney development.
Regulation of TGF-β/BMP signaling via miRNAs
Bone morphogenic proteins (BMPs) are members of the transforming growth factor-β (TGF-β) super-family of growth factors. BMP signaling is required for normal ureteric budding and nephron induction during kidney development, and BMP4 mutations are associated with renal hypodysplasia [58, 59]. Numerous recent studies have provided evidence of BMP repression by miRNAs, and conversely, TGF-β and BMP signaling have the potential to regulate miRNAs. One intriguing mechanism by which TGF-β and BMP signaling might regulate miRNA levels is via an interaction of their downstream effector proteins, the SMADs, with a component of the DROSHA microprocessor complex (required for processing of mature miRNAs) [60]. A SMAD-DROSHA interaction was recently demonstrated to promote the processing of the miR-21 primary transcript to mature miR-21 in vascular smooth muscle cells [60]. However, it remains unknown exactly how this occurs or how many miRNAs are regulated via this mechanism. Though it was reported that BMP4 treatment increased miR-21 levels in vascular smooth muscle cells [61], it was also shown that BMP4 decreased miR-21 levels in epidermal keratinocytes, suggesting that there are additional factors that regulate the miR-21/BMP4 interaction [62]. Interestingly, upregulation of miR-21 in diabetic nephropathy mouse models correlated with increased expression of proteins that contribute to renal fibrosis [63].
Post-transcriptional regulation of miRNA biogenesis is not the only mechanism by which BMP signaling regulates miRNAs. miRNAs may also participate in regulatory loops for TGF-β/BMP signaling. For example, one member of the miR-17~92 cluster, miR-20a, was reported to target negative regulators of BMP signaling in human mesenchymal stem cells upon osteogenic differentiation [64]. BMP signaling was also associated with miR-17~92 in myocardial differentiation, where Bmp mutant mouse embryos had reduced levels of miR-17~92 miRNAs, and the BMP−/− phenotype worsened when one copy of miR-17~92 was removed [65].
Another example of the interaction between miRNAs and TGF-β/BMP signaling is miR-302~367. The miR-302~367 cluster was transcriptionally downregulated in human primary pulmonary artery smooth muscle cells in response to BMP4, likely to promote BMP signaling because miR-302 can directly target the type II BMP receptor (BMPRII) [66]. In the kidney, miR-302 is at least expressed in the glomerular mesangium, and its levels were increased in response to connective tissue growth factor (CTGF) [67]. Thus, increased levels of miR-302 might play a role in the progression of diabetic nephropathy as a mediator between CTGF and TGF-β signaling.
Yet another example was demonstrated when BMP4 was found to activate miR-125a transcription in a feedback loop where miR-125a targeted Dies1 (a protein associated with the BMP4 receptor complex), resulting in decreased BMP4 signaling [68]. In the kidney, miR-125a interacts with the RNA binding protein bicaudal-C homolog 1 (Bicc1) to silence at least the protein kinase inhibitor PKIα and the adenylate cyclase AC6 in a PKD mouse model implying a role for miRNAs in regulation of cystic growth [69].
Recent work suggests that a TGF-β1 regulatory loop is miRNA-dependent in diabetic nephropathy [70]. Increased TGF-β1 in mouse models of diabetic nephropathy were shown to result in increased miR-192 expression, which is thought to target the E-box repressor SIP1 [70, 71]. The increase in miR-192 is believed to be mediated by TGF-β1-induced Akt activation, which subsequently results in the release of repression of miR-192 via the transcription factor, Ets-1 [72]. In addition, transfection of mesangial cells with miR-192 and miR-200b/c induced TGF-β1 expression, presumably by down-regulating the E-box repressors, Zeb1/2 [70, 73]. However, the precise mechanism by which TGF-β regulates miR-192 expression may be cell type- or species-dependent. Expression profiling of biopsies from patients with diabetic nephropathy revealed lower miR-192 levels, in association with increased fibrosis and decreased glomerular filtration rate [73]. Additionally, treatment of cultured human proximal tubular cells with TGF-β1 resulted in decreased miR-192 levels [73]. Further studies will be required to reconcile these apparent conflicts. miR-192 also induced the expression of miR-200b/c, implying that miR-192 is upstream of miR-200b/c [70]. Both miR-200b/c and miR-192 enhanced the expression of collagens and are thought to play a role in fibrosis in diabetic nephropathy [17].
Additional miRNAs involved in TGF-β/BMP signaling are the highly conserved, non-homologous miR-143 and miR-145 [74]. TGF-β and BMP4 induced expression of miR-143 and miR-145 in pulmonary artery smooth muscle cells, presumably through transcriptional activation of an upstream CArG box [75]. This subsequently led to a decrease in their direct target, Krüppel-like factor-4 (KLF4) [75]; a transcription factor that defines pluripotency [76]. Both miR-143 and miR-145 are expressed in E15.5 mouse kidney by RNA-sequencing [19], and KLF4 is expressed in smooth muscle cells of the kidney where TGF-β signaling is active [77]. Interestingly, miR-145 overexpression decreased levels of BMP4 mRNA in esophageal adenocarcinoma cells [78]. Together, although multiple miRNAs have been implicated in TGF-β/BMP signaling, the function of these miRNAs remains largely undefined in the developing kidney.
Potential miRNA links to Eya1/GDNF
The invasion of the UB into the neighboring MM depends on glial-cell-line-derived neurotrophic factor (GDNF) signaling by the MM to its receptor tyrosine kinase, Ret, in the ureteric bud [79]. Mutations in both GDNF and Ret were demonstrated in stillborn fetuses with bilateral or unilateral renal agenesis [80]. The transcription factors, Pax2 and Eya1, function as part of a complex to activate expression of GDNF [81], and Eya1 is associated with Branchiootorenal (BOR) syndrome [82, 83]. Interestingly, miR-562 is expressed in the kidney and was shown to directly target Eya1 in Wilms’ tumors via luciferase reporter assays [84]. In addition, miR-562 is harbored in the genetic locus 2q37, whose deletion has been associated with Wilms’ tumor [84].
Notably, GDNF could regulate the expression of miR-21 and miR-24-2 precursors via an ERK1/2 MAPK signaling pathway in human BE(2)-C cells [85]. Another study showed GDNF induction of miR-2, −21, −218, and let-7f, −7g, −7i in human glioblastoma cells [86]. It is not clear to what extent miR-21, miR-24-2 or miR-2 are expressed in the kidney, but miR-218 and let-7 member miRNAs are [12, 55, 56]. Furthermore, the miss-expression of miR-218 has been linked with renal cell carcinoma [57, 87]. Whether these miRNAs play an important role in kidney development remains unknown.
Six1 and miRNAs
The transcription factor Six1 is required for UB invasion in renal development, and is also associated with BOR syndrome [88-90]. Six1 upregulated miR-106b~25, which was shown to activate TGF-β signaling via targeting Smad7 in human breast cancer [91]. The miR-106b~25 cluster is expressed in the developing kidney [12], and is paralogous to the miR-17~92 cluster; thus, having the potential to target similar mRNAs. Six1 was also shown to be a miRNA target of miR-185 in various cancers, including Wilms’ tumor [92]. Interestingly, Six1 has been suggested to regulate c-Myc, which can also be regulated by miR-185 [92, 93].
Sox17, Wnt signaling, and miRNAs
Mutations in the HMG-box transcription factor, Sox17, are associated with CAKUT, primarily vesicoureteric reflux (VUR) [94, 95]. Sox17 functions as an antagonist of the Wnt/β-Catenin pathway via a Wnt signaling repression domain [96-99]. In embryonic stem cells, Wnt signaling increased the expression of certain miRNAs (miR-181c/338-5p/222/196a/196b/let-7e) [100]. Furthermore, overexpression of a pool of these miRNAs recapitulated the effects of Wnt activation, including increased global histone acetylation, directly affecting the Sox17 promoter [100]. These data suggest a possible Sox17-Wnt signaling feedback loop regulated by miRNAs, mediated globally through chromatin modifications.
Sox17 has also been shown to be a direct miRNA target of miR-151 and miR-141 [101, 102]. miR-141 is expressed in the developing kidney, and its downregulation was reported in nephroblastomas along with miR-200c and miR-192 [103]. Interestingly, inhibition of Sox17 by miR-141 in cell culture resulted in upregulation of downstream genes of the Wnt signaling pathway including c-Myc [102]. MiR-151 was reported to be elevated in the serum of children with nephrotic syndrome, although the significance of this observation remains unclear [104].
The Renin-Angiotensin-Aldosterone system and miRNAs
The Renin-Angiotensin-Aldosterone system plays an important role in responding to physiological cues in regulating blood pressure, and is also causally linked to CAKUT. Evidence for this first emerged when ACE inhibitors or Angiotensin Receptor Type I (AGTR1) antagonists were found to cause fetal anuria [105, 106]. Subsequently mutations in AGT, REN, ACE, and AGTR1 are linked to renal tubular dysgenesis [107, 108]. Signaling through the Renin-Angiotensin-Aldosterone system can be altered upon stress, and in such instances, results in miss-expression of miRNAs [109-111].
In a spontaneous progressive nephropathy model in rats, miR-324-3p levels were increased in association with reduced levels of prolyl endopeptidase (Prep), an enzyme involved in angiotensin metabolism [112]. Accordingly, ACE inhibition down-regulated miR-324-3p, increased Prep levels, and alleviated renal fibrosis in that model [112]. Additionally, HEK293N cells over-expressing AGTR1 had increased levels of miR-29b, -129-3p, −132, -132-3p and −212 [113]. The RAAS can also be targeted by miRNAs, where miR-802 targeted AGTR1 in the gastrointestinal tract[114].
Conclusions
The precise molecular mechanisms behind many cases of CAKUT still remain elusive. Recent work on the functional roles of miRNAs suggests that miRNAs play a role in kidney development, and that their misexpression could contribute to kidney disease and progression. In that context, miRNAs are likely to be important in fine-tuning the expression of important renal development genes, and could be downstream effectors of developmental programs themselves.
While genetic mutations have clearly been linked to a subset of patients with CAKUT, there is significant phenotypic variation amongst family members. In these instances, one possibility is that there is an as yet undetermined gene interacting with the known mutated renal development gene. Another possibility that we would propose is that mutations in genes that code for miRNAs are potential candidates for disease modification. In addition, it is possible that single nucleotide polymorphisms (SNPs) in the 3′UTR of important renal development genes alter visibility to miRNA targeting. The idea of disease-related SNPs (dSNPs) in the 3′UTRs of crucial renal development genes provides a promising mechanism linking genetics, epigenetics, and disease phenotypes [115]. For example, a polymorphism in the miR-155 binding site of AGTR1 hinders the ability of miR-155 to downregulate AGTR1 in humans, and this polymorphic allele has been associated with hypertension [116]. Alternatively, the lack of genotype-phenotype correlation could be due to interactions with environmental or stress factors. Other work has shown that miss-expression of miRNAs in response to stress can surprisingly mimic the effects due to genetic mutations [117, 118]. Accordingly, some of the studies described in this review also integrate environmental stressors as a component of these regulatory pathways.
Ultimately, the emerging information regarding how miRNAs regulate developmental pathways (and are regulated by these pathways themselves) will provide a more comprehensive view of renal development, and has the potential to provide unique insights into the underlying causes of CAKUT. This may then, in turn, lead to novel molecular tools that may direct prevention strategies, predict functional outcomes or guide interventions would be a helpful addition to the clinical management of these children.
References
- 1.Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA. Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Pediatr Transplant. 2007;11:366–373. doi: 10.1111/j.1399-3046.2007.00704.x. [DOI] [PubMed] [Google Scholar]
- 2.Saxen L, Sariola H. Early organogenesis of the kidney. Pediatr Nephrol. 1987;1:385–392. doi: 10.1007/BF00849241. [DOI] [PubMed] [Google Scholar]
- 3.Piscione TD, Rosenblum ND. The molecular control of renal branching morphogenesis: current knowledge and emerging insights. Differentiation. 2002;70:227–246. doi: 10.1046/j.1432-0436.2002.700602.x. [DOI] [PubMed] [Google Scholar]
- 4.Aufderheide E, Chiquet-Ehrismann R, Ekblom P. Epithelial-mesenchymal interactions in the developing kidney lead to expression of tenascin in the mesenchyme. J Cell Biol. 1987;105:599–608. doi: 10.1083/jcb.105.1.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanna-Cherchi S, Ravani P, Corbani V, Parodi S, Haupt R, Piaggio G, Innocenti ML, Somenzi D, Trivelli A, Caridi G, Izzi C, Scolari F, Mattioli G, Allegri L, Ghiggeri GM. Renal outcome in patients with congenital anomalies of the kidney and urinary tract. Kidney Int. 2009;76:528–533. doi: 10.1038/ki.2009.220. [DOI] [PubMed] [Google Scholar]
- 6.Keller G, Zimmer G, Mall G, Ritz E, Amann K. Nephron number in patients with primary hypertension. N Engl J Med. 2003;348:101–108. doi: 10.1056/NEJMoa020549. [DOI] [PubMed] [Google Scholar]
- 7.Yosypiv IV. Congenital anomalies of the kidney and urinary tract: a genetic disorder? Int J Nephrol. 2012;2012:909083. doi: 10.1155/2012/909083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harambat J, van Stralen KJ, Kim JJ, Tizard EJ. Epidemiology of chronic kidney disease in children. Pediatr Nephrol. 2012;27:363–373. doi: 10.1007/s00467-011-1939-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel V, Hajarnis S, Williams D, Hunter R, Huynh D, Igarashi P. MicroRNAs Regulate Renal Tubule Maturation through Modulation of Pkd1. J Am Soc Nephrol. 2012;23:1941–1948. doi: 10.1681/ASN.2012030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sequeira-Lopez ML, Weatherford ET, Borges GR, Monteagudo MC, Pentz ES, Harfe BD, Carretero O, Sigmund CD, Gomez RA. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol. 2010;21:460–467. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho J, Pandey P, Schatton T, Sims-Lucas S, Khalid M, Frank MH, Hartwig S, Kreidberg JA. The pro-apoptotic protein Bim is a microRNA target in kidney progenitors. J Am Soc Nephrol. 2011;22:1053–1063. doi: 10.1681/ASN.2010080841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagalakshmi VK, Ren Q, Pugh MM, Valerius MT, McMahon AP, Yu J. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011;79:317–330. doi: 10.1038/ki.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartram MP, Hohne M, Dafinger C, Volker LA, Albersmeyer M, Heiss J, Gobel H, Bronneke H, Burst V, Liebau MC, Benzing T, Schermer B, Muller RU. Conditional loss of kidney microRNAs results in congenital anomalies of the kidney and urinary tract (CAKUT) J Mol Med (Berl) 2013;91:739–748. doi: 10.1007/s00109-013-1000-x. [DOI] [PubMed] [Google Scholar]
- 16.Ho JJ, Marsden PA. Dicer cuts the kidney. J Am Soc Nephrol. 2008;19:2043–2046. doi: 10.1681/ASN.2008090986. [DOI] [PubMed] [Google Scholar]
- 17.Kato M, Park JT, Natarajan R. MicroRNAs and the glomerulus. Exp Cell Res. 2012;318:993–1000. doi: 10.1016/j.yexcr.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho J, Kreidberg JA. MicroRNAs in renal development. Pediatr Nephrol. 2012;28:219–225. doi: 10.1007/s00467-012-2204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiagarajan RD, Cloonan N, Gardiner BB, Mercer TR, Kolle G, Nourbakhsh E, Wani S, Tang D, Krishnan K, Georgas KM, Rumballe BA, Chiu HS, Steen JA, Mattick JS, Little MH, Grimmond SM. Refining transcriptional programs in kidney development by integration of deep RNA-sequencing and array-based spatial profiling. BMC Genomics. 2011;12:441. doi: 10.1186/1471-2164-12-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanyanusin P, McNoe LA, Sullivan MJ, Weaver RG, Eccles MR. Mutation of PAX2 in two siblings with renal-coloboma syndrome. Hum Mol Genet. 1995;4:2183–2184. doi: 10.1093/hmg/4.11.2183. [DOI] [PubMed] [Google Scholar]
- 21.Sanyanusin P, Schimmenti LA, McNoe LA, Ward TA, Pierpont ME, Sullivan MJ, Dobyns WB, Eccles MR. Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet. 1995;9:358–364. doi: 10.1038/ng0495-358. [DOI] [PubMed] [Google Scholar]
- 22.Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- 23.Weber S, Moriniere V, Knuppel T, Charbit M, Dusek J, Ghiggeri GM, Jankauskiene A, Mir S, Montini G, Peco-Antic A, Wuhl E, Zurowska AM, Mehls O, Antignac C, Schaefer F, Salomon R. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol. 2006;17:2864–2870. doi: 10.1681/ASN.2006030277. [DOI] [PubMed] [Google Scholar]
- 24.Dressler GR, Wilkinson JE, Rothenpieler UW, Patterson LT, Williams-Simons L, Westphal H. Deregulation of Pax-2 expression in transgenic mice generates severe kidney abnormalities. Nature. 1993;362:65–67. doi: 10.1038/362065a0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang SL, Chen YW, Tran S, Liu F, Nestoridi E, Hebert MJ, Ingelfinger JR. Pax-2 and N-myc regulate epithelial cell proliferation and apoptosis in a positive autocrine feedback loop. Pediatr Nephrol. 2007;22:813–824. doi: 10.1007/s00467-007-0444-z. [DOI] [PubMed] [Google Scholar]
- 26.Bates CM, Kharzai S, Erwin T, Rossant J, Parada LF. Role of N-myc in the developing mouse kidney. Dev Biol. 2000;222:317–325. doi: 10.1006/dbio.2000.9716. [DOI] [PubMed] [Google Scholar]
- 27.Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Genevieve D, Goldenberg A, Oufadem M, Manouvrier S, Munnich A, Vidigal JA, Vekemans M, Lyonnet S, Henrion-Caude A, Ventura A, Amiel J. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin R, Bao W, Xing Y, Xi T, Gou S. MiR-19b-1 inhibits angiogenesis by blocking cell cycle progression of endothelial cells. Biochem Biophys Res Commun. 2012;417:771–776. doi: 10.1016/j.bbrc.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 32.Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, Kolle G, Gabrielli B, Grimmond SM. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kort EJ, Farber L, Tretiakova M, Petillo D, Furge KA, Yang XJ, Cornelius A, Teh BT. The E2F3-Oncomir-1 axis is activated in Wilms’ tumor. Cancer Res. 2008;68:4034–4038. doi: 10.1158/0008-5472.CAN-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodeur GM. Genetics of embryonal tumours of childhood: retinoblastoma, Wilms’ tumour and neuroblastoma. Cancer Surv. 1995;25:67–99. [PubMed] [Google Scholar]
- 35.Ariyaratana S, Loeb DM. The role of the Wilms tumour gene (WT1) in normal and malignant haematopoiesis. Expert Rev Mol Med. 2007;9:1–17. doi: 10.1017/S1462399407000336. [DOI] [PubMed] [Google Scholar]
- 36.Marcelis CL, Hol FA, Graham GE, Rieu PN, Kellermayer R, Meijer RP, Lugtenberg D, Scheffer H, van Bokhoven H, Brunner HG, de Brouwer AP. Genotype-phenotype correlations in MYCN-related Feingold syndrome. Hum Mutat. 2008;29:1125–1132. doi: 10.1002/humu.20750. [DOI] [PubMed] [Google Scholar]
- 37.Aslam M, van Bokhoven H, Taylor CM. End-stage renal failure, reflux nephropathy and Feingold’s syndrome. Pediatr Nephrol. 2008;23:159–161. doi: 10.1007/s00467-007-0602-3. [DOI] [PubMed] [Google Scholar]
- 38.Patel V, Williams D, Hajarnis S, Hunter R, Pontoglio M, Somlo S, Igarashi P. miR-17~92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc Natl Acad Sci U S A. 2013;110:10765–10770. doi: 10.1073/pnas.1301693110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H, Li QW, Lv XY, Ai JZ, Yang QT, Duan JJ, Bian GH, Xiao Y, Wang YD, Zhang Z, Liu YH, Tan RZ, Yang Y, Wei YQ, Zhou Q. MicroRNA-17 post-transcriptionally regulates polycystic kidney disease-2 gene and promotes cell proliferation. Mol Biol Rep. 2010;37:2951–2958. doi: 10.1007/s11033-009-9861-3. [DOI] [PubMed] [Google Scholar]
- 40.Tran U, Zakin L, Schweickert A, Agrawal R, Doger R, Blum M, De Robertis EM, Wessely O. The RNA-binding protein bicaudal C regulates polycystin 2 in the kidney by antagonizing miR-17 activity. Development. 2010;137:1107–1116. doi: 10.1242/dev.046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giglio S, Contini E, Toni S, Pela I. Growth hormone therapy-related hyperglycaemia in a boy with renal cystic hypodysplasia and a new mutation of the HNF1 beta gene. Nephrol Dial Transplant. 2010;25:3116–3119. doi: 10.1093/ndt/gfq315. [DOI] [PubMed] [Google Scholar]
- 42.Kornfeld JW, Baitzel C, Konner AC, Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM, Knippschild U, Seibler J, Cereghini S, Heeren J, Stoffel M, Bruning JC. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494:111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 43.Lin DH, Yue P, Pan C, Sun P, Wang WH. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol. 2011;22:1087–1098. doi: 10.1681/ASN.2010090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reichert N, Choukrallah MA, Matthias P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell Mol Life Sci. 2012;69:2173–2187. doi: 10.1007/s00018-012-0921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Bellew C, Yao X, Stefkova J, Dipp S, Saifudeen Z, Bachvarov D, El-Dahr SS. Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. J Biol Chem. 2011;286:32775–32789. doi: 10.1074/jbc.M111.248278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 48.de Groh ED, Swanhart LM, Cosentino CC, Jackson RL, Dai W, Kitchens CA, Day BW, Smithgall TE, Hukriede NA. Inhibition of histone deacetylase expands the renal progenitor cell population. J Am Soc Nephrol. 2010;21:794–802. doi: 10.1681/ASN.2009080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cianciolo Cosentino C, Skrypnyk NI, Brilli LL, Chiba T, Novitskaya T, Woods C, West J, Korotchenko VN, McDermott L, Day BW, Davidson AJ, Harris RC, de Caestecker MP, Hukriede NA. Histone Deacetylase Inhibitor Enhances Recovery after AKI. J Am Soc Nephrol. 2013;24:943–953. doi: 10.1681/ASN.2012111055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mannaerts I, Eysackers N, Onyema OO, Van Beneden K, Valente S, Mai A, Odenthal M, van Grunsven LA. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS One. 2013;8:e55786. doi: 10.1371/journal.pone.0055786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphreys KJ, Cobiac L, Le Leu RK, Van der Hoek MB, Michael MZ. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol Carcinog. 2013;52:459–474. doi: 10.1002/mc.21879. [DOI] [PubMed] [Google Scholar]
- 52.Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol Cell Biol. 2007;27:4238–4247. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 54.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harding SD, Armit C, Armstrong J, Brennan J, Cheng Y, Haggarty B, Houghton D, Lloyd-MacGilp S, Pi X, Roochun Y, Sharghi M, Tindal C, McMahon AP, Gottesman B, Little MH, Georgas K, Aronow BJ, Potter SS, Brunskill EW, Southard-Smith EM, Mendelsohn C, Baldock RA, Davies JA, Davidson D. The GUDMAP database--an online resource for genitourinary research. Development. 2011;138:2845–2853. doi: 10.1242/dev.063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 57.Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, Gajda MR, Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367–373. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 58.Miyazaki Y, Oshima K, Fogo A, Hogan BL, Ichikawa I. Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J Clin Invest. 2000;105:863–873. doi: 10.1172/JCI8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tabatabaeifar M, Schlingmann KP, Litwin M, Emre S, Bakkaloglu A, Mehls O, Antignac C, Schaefer F, Weber S. Functional analysis of BMP4 mutations identified in pediatric CAKUT patients. Pediatr Nephrol. 2009;24:2361–2368. doi: 10.1007/s00467-009-1287-6. [DOI] [PubMed] [Google Scholar]
- 60.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang H, Davis-Dusenbery BN, Nguyen PH, Lal A, Lieberman J, Van Aelst L, Lagna G, Hata A. Bone morphogenetic protein 4 promotes vascular smooth muscle contractility by activating microRNA-21 (miR-21), which down-regulates expression of family of dedicator of cytokinesis (DOCK) proteins. J Biol Chem. 2011;287:3976–3986. doi: 10.1074/jbc.M111.303156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed MI, Mardaryev AN, Lewis CJ, Sharov AA, Botchkareva NV. MicroRNA-21 is an important downstream component of BMP signalling in epidermal keratinocytes. J Cell Sci. 2011;124:3399–3404. doi: 10.1242/jcs.086710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Gao Y, Ma M, Li M, Zou D, Yang J, Zhu Z, Zhao X. Effect of miR-21 on Renal Fibrosis by Regulating MMP-9 and TIMP1 in kk-ay Diabetic Nephropathy Mice. Cell Biochem Biophys. 2013 doi: 10.1007/s12013-013-9539-2. DOI: 10.1007/s12013-013-9539-2. [DOI] [PubMed] [Google Scholar]
- 64.Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan G, Li G, Wang H, Lu G, Hu X, Jiang S, Li JN, Lin MC, Zhang YO, Kung HF. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011;8:829–838. doi: 10.4161/rna.8.5.16043. [DOI] [PubMed] [Google Scholar]
- 65.Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang H, Louie J, Weisman A, Sheu-Gruttadauria J, Davis-Dusenbery BN, Lagna G, Hata A. Inhibition of microRNA-302 (miR-302) by bone morphogenetic protein 4 (BMP4) facilitates the BMP signaling pathway. J Biol Chem. 2012;287:38656–38664. doi: 10.1074/jbc.M112.390898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faherty N, Curran SP, O’Donovan H, Martin F, Godson C, Brazil DP, Crean JK. CCN2/CTGF increases expression of miR-302 microRNAs, which target the TGFbeta type II receptor with implications for nephropathic cell phenotypes. J Cell Sci. 2012;125:5621–5629. doi: 10.1242/jcs.105528. [DOI] [PubMed] [Google Scholar]
- 68.Parisi S, Battista M, Musto A, Navarra A, Tarantino C, Russo T. A regulatory loop involving Dies1 and miR-125a controls BMP4 signaling in mouse embryonic stem cells. FASEB J. 2012;26:3957–3968. doi: 10.1096/fj.12-211607. [DOI] [PubMed] [Google Scholar]
- 69.Piazzon N, Maisonneuve C, Guilleret I, Rotman S, Constam DB. Bicc1 links the regulation of cAMP signaling in polycystic kidneys to microRNA-induced gene silencing. J Mol Cell Biol. 2012;4:398–408. doi: 10.1093/jmcb/mjs027. [DOI] [PubMed] [Google Scholar]
- 70.Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R. A microRNA circuit mediates transforming growth factor-beta1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011;80:358–368. doi: 10.1038/ki.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato M, Dang V, Wang M, Park JT, Deshpande S, Kadam S, Mardiros A, Zhan Y, Oettgen P, Putta S, Yuan H, Lanting L, Natarajan R. TGF-beta Induces Acetylation of Chromatin and of Ets-1 to Alleviate Repression of miR-192 in Diabetic Nephropathy. Sci Signal. 2013;6:ra43. doi: 10.1126/scisignal.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol. 2010;21:438–447. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, Lagna G, Hata A. down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J Biol Chem. 2011;286:28097–28110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song B, Niclis JC, Alikhan MA, Sakkal S, Sylvain A, Kerr PG, Laslett AL, Bernard CA, Ricardo SD. Generation of induced pluripotent stem cells from human kidney mesangial cells. J Am Soc Nephrol. 2011;22:1213–1220. doi: 10.1681/ASN.2010101022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boersema M, Katta K, Rienstra H, Molema G, Nguyen TQ, Goldschmeding R, Navis G, van den Born J, Popa ER, Hillebrands JL. Local medial microenvironment directs phenotypic modulation of smooth muscle cells after experimental renal transplantation. Am J Transplant. 2012;12:1429–1440. doi: 10.1111/j.1600-6143.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 78.van Baal JW, Verbeek RE, Bus P, Fassan M, Souza RF, Rugge M, Ten Kate FJ, Vleggaar FP, Siersema PD. microRNA-145 in Barrett’s oesophagus: regulating BMP4 signalling via GATA6. Gut. 2013;62:664–675. doi: 10.1136/gutjnl-2011-301061. [DOI] [PubMed] [Google Scholar]
- 79.Trupp M, Arenas E, Fainzilber M, Nilsson AS, Sieber BA, Grigoriou M, Kilkenny C, Salazar-Grueso E, Pachnis V, Arumae U. Functional receptor for GDNF encoded by the c-ret proto-oncogene. Nature. 1996;381:785–789. doi: 10.1038/381785a0. [DOI] [PubMed] [Google Scholar]
- 80.Skinner MA, Safford SD, Reeves JG, Jackson ME, Freemerman AJ. Renal aplasia in humans is associated with RET mutations. Am J Hum Genet. 2008;82:344–351. doi: 10.1016/j.ajhg.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;27:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 83.Vincent C, Kalatzis V, Abdelhak S, Chaib H, Compain S, Helias J, Vaneecloo FM, Petit C. BOR and BO syndromes are allelic defects of EYA1. Eur J Hum Genet. 1997;5:242–246. [PubMed] [Google Scholar]
- 84.Drake KM, Ruteshouser EC, Natrajan R, Harbor P, Wegert J, Gessler M, Pritchard-Jones K, Grundy P, Dome J, Huff V, Jones C, Aldred MA. Loss of heterozygosity at 2q37 in sporadic Wilms’ tumor: putative role for miR-562. Clin Cancer Res. 2009;15:5985–5992. doi: 10.1158/1078-0432.CCR-09-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoong LF, Wan G, Too HP. Glial cell-line derived neurotrophic factor and neurturin regulate the expressions of distinct miRNA precursors through the activation of GFRalpha2. J Neurochem. 2006;98:1149–1158. doi: 10.1111/j.1471-4159.2006.03959.x. [DOI] [PubMed] [Google Scholar]
- 86.Wan G, Lim QE, Too HP. High-performance quantification of mature microRNAs by real-time RT-PCR using deoxyuridine-incorporated oligonucleotides and hemi-nested primers. RNA. 2010;16:1436–1445. doi: 10.1261/rna.2001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamasaki T, Seki N, Yoshino H, Itesako T, Hidaka H, Yamada Y, Tatarano S, Yonezawa T, Kinoshita T, Nakagawa M, Enokida H. microRNA-218 inhibits cell migration and invasion in renal cell carcinoma through targeting caveolin-2 involved in focal adhesion pathway. J Urol. 2013 doi: 10.1016/j.juro.2013.02.089. DOI: 10.1016/j.juro.2013.02.089. [DOI] [PubMed] [Google Scholar]
- 88.Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kochhar A, Orten DJ, Sorensen JL, Fischer SM, Cremers CW, Kimberling WJ, Smith RJ. SIX1 mutation screening in 247 branchio-oto-renal syndrome families: a recurrent missense mutation associated with BOR. Hum Mutat. 2008;29:565. doi: 10.1002/humu.20714. [DOI] [PubMed] [Google Scholar]
- 90.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Jr., Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, Chen Y, Rao MK. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. 2010;29:4971–4979. doi: 10.1038/onc.2010.233. [DOI] [PubMed] [Google Scholar]
- 93.Liao JM, Lu H. Autoregulatory suppression of c-Myc by miR-185-3p. J Biol Chem. 2011;286:33901–33909. doi: 10.1074/jbc.M111.262030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Katoh M. Molecular cloning and characterization of human SOX17. Int J Mol Med. 2002;9:153–157. [PubMed] [Google Scholar]
- 95.Gimelli S, Caridi G, Beri S, McCracken K, Bocciardi R, Zordan P, Dagnino M, Fiorio P, Murer L, Benetti E, Zuffardi O, Giorda R, Wells JM, Gimelli G, Ghiggeri GM. Mutations in SOX17 are associated with congenital anomalies of the kidney and the urinary tract. Hum Mutat. 2010;31:1352–1359. doi: 10.1002/humu.21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M. SOX17 antagonizes WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Epigenetics. 2010;5:743–749. doi: 10.4161/epi.5.8.13104. [DOI] [PubMed] [Google Scholar]
- 97.Chen HL, Chew LJ, Packer RJ, Gallo V. Modulation of the Wnt/beta-catenin pathway in human oligodendroglioma cells by Sox17 regulates proliferation and differentiation. Cancer Lett. 2013;335:361–371. doi: 10.1016/j.canlet.2013.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yin D, Jia Y, Yu Y, Brock MV, Herman JG, Han C, Su X, Liu Y, Guo M. SOX17 methylation inhibits its antagonism of Wnt signaling pathway in lung cancer. Discov Med. 2012;14:33–40. [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang W, Glockner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, Ahuja N, Baylin SB. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu S, Fei Q, Jiang H, Chuai S, Shi S, Xiong W, Jiang L, Lu C, Atadja P, Li E, Shou J. Involvement of histone acetylation of Sox17 and Foxa2 promoters during mouse definitive endoderm differentiation revealed by microRNA profiling. PLoS One. 2011;6:e27965. doi: 10.1371/journal.pone.0027965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chiyomaru T, Yamamura S, Zaman MS, Majid S, Deng G, Shahryari V, Saini S, Hirata H, Ueno K, Chang I, Tanaka Y, Tabatabai ZL, Enokida H, Nakagawa M, Dahiya R. Genistein suppresses prostate cancer growth through inhibition of oncogenic microRNA-151. PLoS One. 2012;7:e43812. doi: 10.1371/journal.pone.0043812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jia Y, Yang Y, Zhan Q, Brock MV, Zheng X, Yu Y, Herman JG, Guo M. Inhibition of SOX17 by microRNA 141 and methylation activates the WNT signaling pathway in esophageal cancer. J Mol Diagn. 2012;14:577–585. doi: 10.1016/j.jmoldx.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 103.Senanayake U, Das S, Vesely P, Alzoughbi W, Frohlich LF, Chowdhury P, Leuschner I, Hoefler G, Guertl B. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis. 33:1014–1021. doi: 10.1093/carcin/bgs126. [DOI] [PubMed] [Google Scholar]
- 104.Luo Y, Wang C, Chen X, Zhong T, Cai X, Chen S, Shi Y, Hu J, Guan X, Xia Z, Wang J, Zen K, Zhang CY, Zhang C. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin Chem. 2013;59:658–666. doi: 10.1373/clinchem.2012.195297. [DOI] [PubMed] [Google Scholar]
- 105.Schaefer C. Angiotensin II-receptor-antagonists: further evidence of fetotoxicity but not teratogenicity. Birth Defects Res A Clin Mol Teratol. 2003;67:591–594. doi: 10.1002/bdra.10081. [DOI] [PubMed] [Google Scholar]
- 106.Tabacova S, Little R, Tsong Y, Vega A, Kimmel CA. Adverse pregnancy outcomes associated with maternal enalapril antihypertensive treatment. Pharmacoepidemiol Drug Saf. 2003;12:633–646. doi: 10.1002/pds.796. [DOI] [PubMed] [Google Scholar]
- 107.Weber S. Novel genetic aspects of congenital anomalies of kidney and urinary tract. Curr Opin Pediatr. 2012;24:212–218. doi: 10.1097/MOP.0b013e32834fdbd4. [DOI] [PubMed] [Google Scholar]
- 108.Gribouval O, Gonzales M, Neuhaus T, Aziza J, Bieth E, Laurent N, Bouton JM, Feuillet F, Makni S, Ben Amar H, Laube G, Delezoide AL, Bouvier R, Dijoud F, Ollagnon-Roman E, Roume J, Joubert M, Antignac C, Gubler MC. Mutations in genes in the renin-angiotensin system are associated with autosomal recessive renal tubular dysgenesis. Nat Genet. 2005;37:964–968. doi: 10.1038/ng1623. [DOI] [PubMed] [Google Scholar]
- 109.Goyal R, Lister R, Leitzke A, Goyal D, Gheorghe CP, Longo LD. Antenatal maternal hypoxic stress: adaptations of the placental renin-angiotensin system in the mouse. Placenta. 2011;32:134–139. doi: 10.1016/j.placenta.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goyal R, Leitzke A, Goyal D, Gheorghe CP, Longo LD. Antenatal maternal hypoxic stress: adaptations in fetal lung Renin-Angiotensin system. Reprod Sci. 2011;18:180–189. doi: 10.1177/1933719110385134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goyal R, Goyal D, Leitzke A, Gheorghe CP, Longo LD. Brain renin-angiotensin system: fetal epigenetic programming by maternal protein restriction during pregnancy. Reprod Sci. 2010;17:227–238. doi: 10.1177/1933719109351935. [DOI] [PubMed] [Google Scholar]
- 112.Macconi D, Tomasoni S, Romagnani P, Trionfini P, Sangalli F, Mazzinghi B, Rizzo P, Lazzeri E, Abbate M, Remuzzi G, Benigni A. MicroRNA-324-3p promotes renal fibrosis and is a target of ACE inhibition. J Am Soc Nephrol. 2012;23:1496–1505. doi: 10.1681/ASN.2011121144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jeppesen PL, Christensen GL, Schneider M, Nossent AY, Jensen HB, Andersen DC, Eskildsen T, Gammeltoft S, Hansen JL, Sheikh SP. Angiotensin II type 1 receptor signalling regulates microRNA differentially in cardiac fibroblasts and myocytes. Br J Pharmacol. 2011;164:394–404. doi: 10.1111/j.1476-5381.2011.01375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sansom SE, Nuovo GJ, Martin MM, Kotha SR, Parinandi NL, Elton TS. miR-802 regulates human angiotensin II type 1 receptor expression in intestinal epithelial C2BBe1 cells. Am J Physiol Gastrointest Liver Physiol. 2010;299:G632–642. doi: 10.1152/ajpgi.00120.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bruno AE, Li L, Kalabus JL, Pan Y, Yu A, Hu Z. miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC Genomics. 2012;13:44. doi: 10.1186/1471-2164-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marrone AK, Edeleva EV, Kucherenko MM, Hsiao NH, Shcherbata HR. Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol. 2012;13:26. doi: 10.1186/1471-2121-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kucherenko MM, Marrone AK, Rishko VM, Magliarelli Hde F, Shcherbata HR. Stress and muscular dystrophy: a genetic screen for dystroglycan and dystrophin interactors in Drosophila identifies cellular stress response components. Dev Biol. 2011;352:228–242. doi: 10.1016/j.ydbio.2011.01.013. [DOI] [PubMed] [Google Scholar]