Abstract
Tailed bacteriophages and herpesviruses consist of a structurally well conserved dodecameric portal at a special five-fold vertex of the capsid. The portal plays critical roles in head assembly, genome packaging, neck/tail attachment, and genome ejection. Although the structures of portals from phages φ29, SPP1 and P22 have been determined, their mechanistic roles have not been well understood. Structural analysis of phage T4 portal (gp20) has been hampered because of its unusual interaction with the E. coli inner membrane. Here, we predict atomic models for the T4 portal monomer and dodecamer, and fit the dodecamer into the cryoEM density of the phage portal vertex. The core structure, like that from other phages, is cone-shaped with the wider end containing the “wing” and “crown” domains inside the phage head. A long “stem” encloses a central channel, and a narrow “stalk” protrudes outside the capsid. A biochemical approach was developed to analyze portal function by incorporating plasmid-expressed portal protein into phage heads and determining the effect of mutations on head assembly, DNA translocation, and virion production. We found that the protruding loops of the stalk domain are involved in assembling the DNA packaging motor. A loop that connects the stalk to the channel might be required for communication between the motor and portal. The “tunnel” loops that project into the channel are essential for sealing the packaged head. These studies established that the portal is required throughout the DNA packaging process, with different domains participating at different stages of genome packaging.
Keywords: portal vertex, structural modeling, DNA packaging motor, head assembly, headful packaging
Introduction
Tailed bacteriophages and herpesviruses use powerful molecular machines to package their genomes into a head or capsid. The packaging machine consists of two basic components: a portal through which DNA genome enters the capsid and a motor that drives DNA translocation fueled by ATP (Fig. 1).1–3
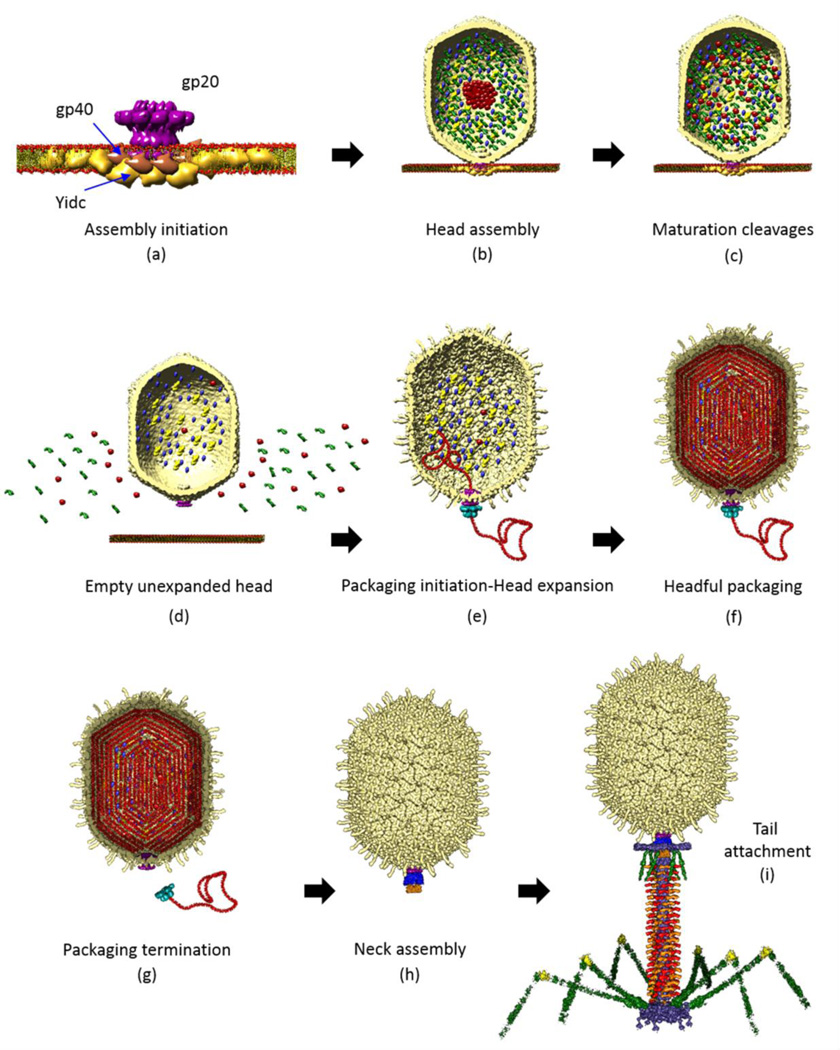
Fig. 1.
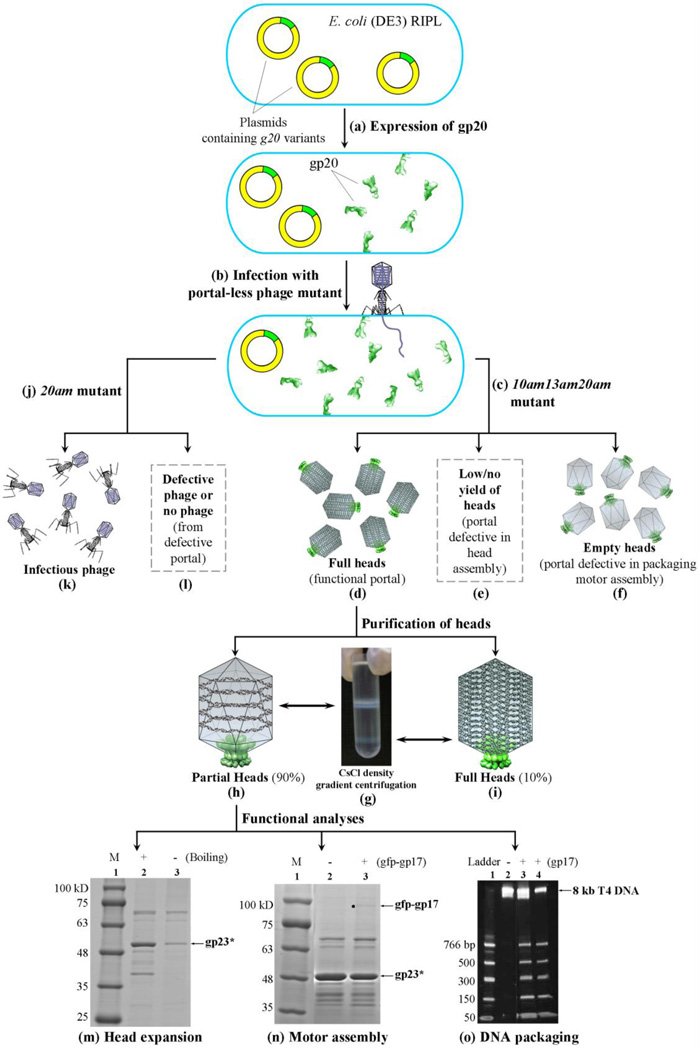
Schematic of phage T4 assembly showing the functional roles of portal. (a) A dodecameric portal (magenta) is assembled on the inner membrane of E. coli with the assistance of the phage-coded chaperone gp40 (brown) and the E. coli chaperone YidC (yellow). The portal assembly acts as an initiator for head assembly, leading to co-polymerization of the major capsid protein gp23 and the scaffolding proteins gp21 (protease), gp22, gp67, gp68, IPI, IPII, and IPIII, and gpAlt (b). A symmetry mismatch is created between the five-fold capsid and dodecameric portal. Following maturation cleavages by gp21 protease (c), the cleaved prohead is released from the membrane and the scaffold proteins degraded to small peptides that diffuse out of the capsid (d). A pentameric gp17 motor assembles on the portal and packaging is initiated. The proheads expand after about 25% of the genome is packaged (e). Packaging continues until the head is filled with the 171 kb genome (headful packaging) (f). The packaging motor dissociates (g) and neck proteins (gp13, gp14, and gp15) assemble on the portal (h). Tail and tail fibers assemble to produce an infectious virion (i).
The heads of phage T4 are assembled on the membrane. The portal of T4 (gp20) and of other phages is a dodecamer.4,5 It is the first structure assembled in the head assembly pathway (Fig. 1a). The portal nucleates the assembly of the hexameric capsomers each composed of six copies of the major capsid protein (gp23) into capsids. The portal also nucleates the assembly of the major scaffolding protein (gp22). Together, these interactions lead to the formation of the first five-fold vertex of the icosahedral capsid.6–8 It also creates a symmetry mismatch between the dodecameric portal and the five-fold capsid, a feature strictly conserved in all wellcharacterized tailed phages and herpesviruses. Head assembly continues by co-polymerization of the capsid protein and the scaffolding proteins (gp21, gp22, gp67, gp68, IPI, IPII, and IPIII, gpAlt) to form a “prehead” (Fig. 1b). A unique feature of phage T4 is that its portal assembles on the E. coli inner membrane, assisted by the membrane-bound phage chaperone gp40 and the E. coli membrane insertase protein YidC (Fig. 1a).9–11 If the portal protein function is missing (e.g., chain termination mutants under non-permissive conditions), gp23 polymerizes in the cytosol producing cylindrical tubes known as polyheads, which sometimes span the entire length of the E. coli cell.8 The preheads undergo maturation cleavage reactions catalyzed by a scaffoldassociated protease (gp21) that cleaves off the N-terminal ~8 kDa domain of the major capsid protein.12,13 The scaffolding proteins, for the most part, are degraded to small peptides and probably diffuse out of the head during head maturation and DNA packaging (Fig. 1c). The cleaved unexpanded empty “proheads” are then released into the cytosol (Fig. 1d).
Parallel to head assembly, the phage T4 large terminase protein gp17 and the small terminase protein gp16 make an endonucleolytic cut in the newly synthesized concatemeric viral DNA genome, generating a free end.14 Five molecules of gp17 and the cut end of the DNA attach to the prohead by interacting with the portal.15,16 A pentameric motor is thus assembled and genome packaging is initiated.17 gp17 contains an ATPase activity that provides energy for DNA translocation.18,19 After about 10% of the genome is packaged, gp23 undergoes a major conformational change causing expansion of the head in all dimensions by ~15% and increase in the capsid volume by ~50% (Fig. 1e).20 We have determined the X-ray structures of gp17, as well as the cryo-EM structure of the T4 prohead-motor complex.17,21,22 Based on these structures we proposed an electrostatic force driven mechanism in which the packaging motor alternates between a relaxed and a tensed state. The hydrolysis of one ATP molecule was predicted to translocate 2 bp of DNA into the capsid.17 After encapsidating ~171 kb of the viral genome, equivalent to 1.03 genome lengths (one headful), gp17 makes a termination cut (Fig. 1f).22,23 Subsequently, the motor dissociates (Fig. 1g) and the neck proteins (gp13, gp14, and gp15) assemble on the portal (Fig. 1h) followed by assembly of the tail and tail fibers to produce an infectious virion (Fig. 1i).24
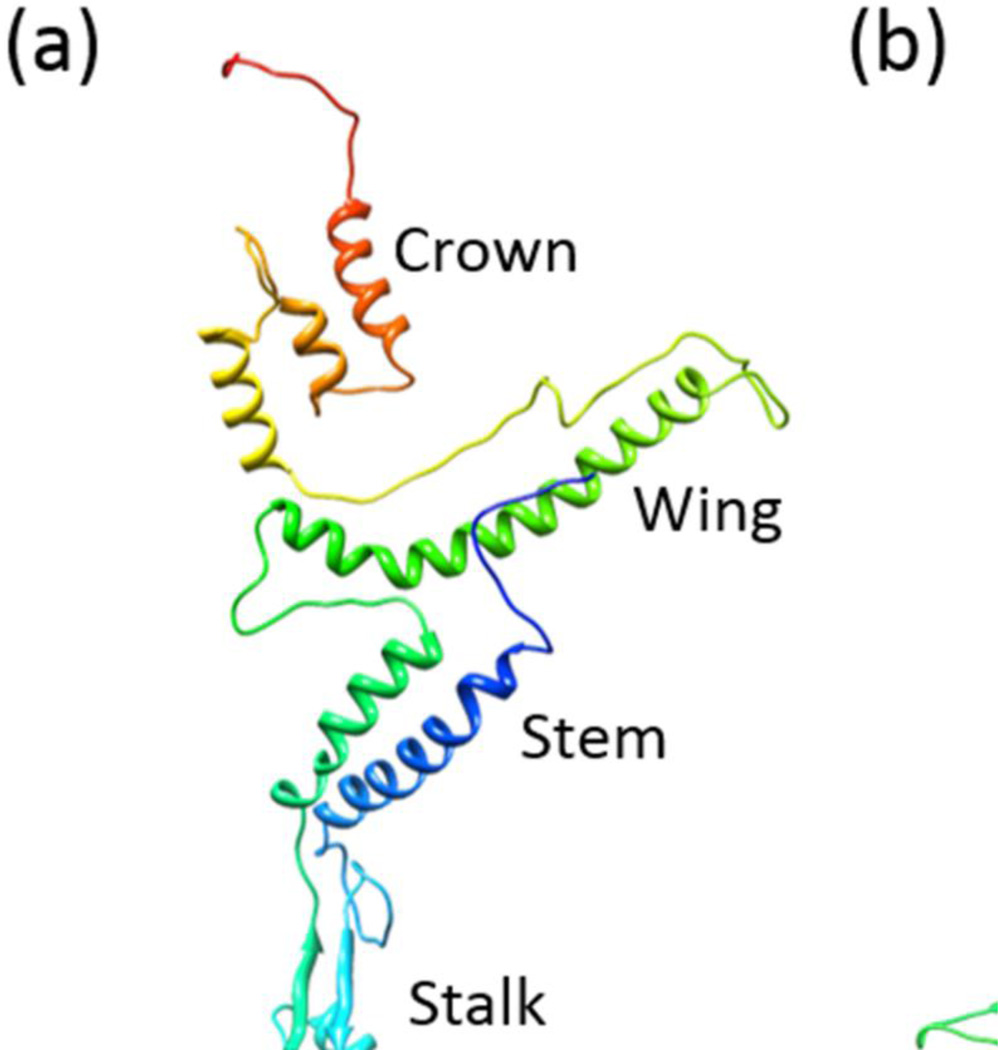
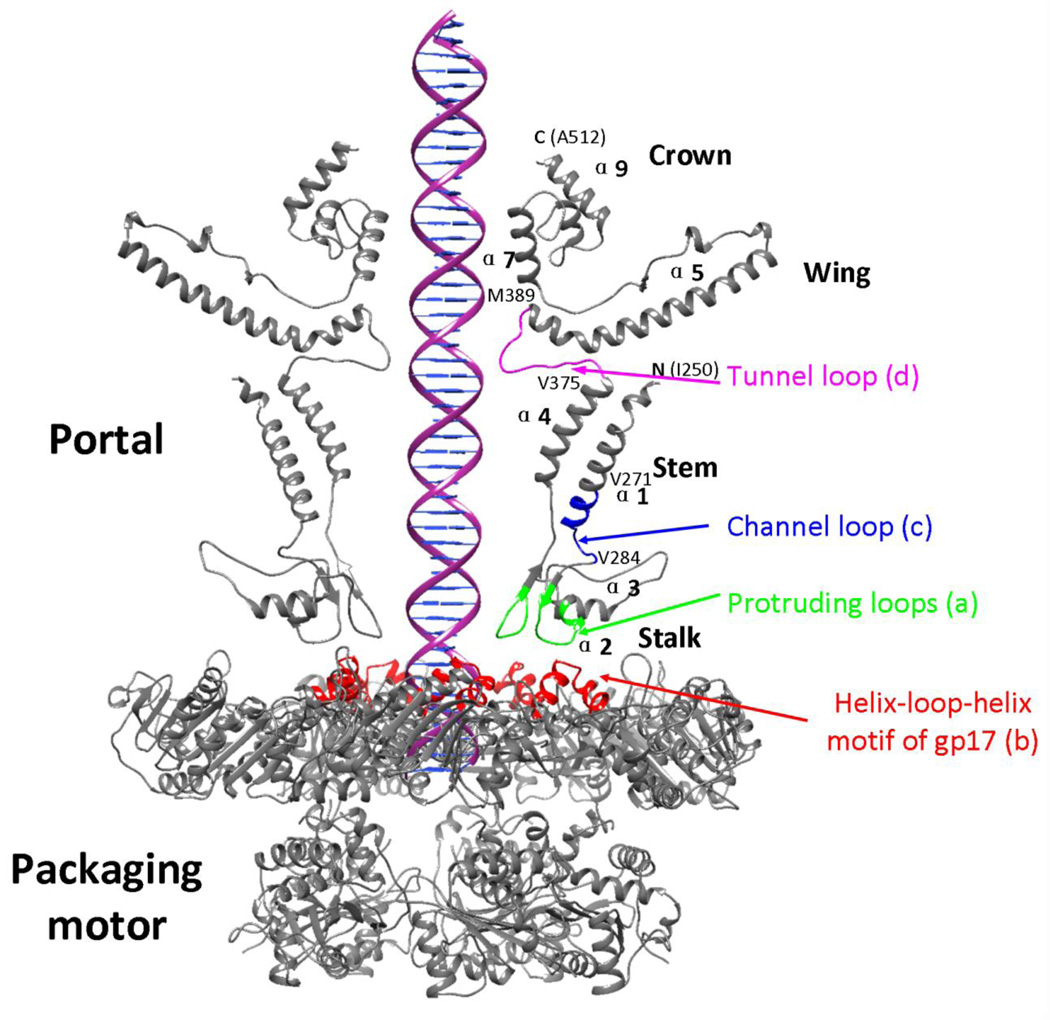
The crystal structures of phages φ29, SPP1, and P22 portals have been determined.25–27 A conserved feature of these structures is the 75–110Å long cone-shaped central domain, also referred to as the “core” structure. It consists of a wider opening inside the head formed by “wing” and “crown” domains, a stem enclosing a ~35–60Å wide central channel that traverses the capsid wall, and a stalk that protrudes out of the capsid (also referred to as “clip”). The channel is formed by twenty four helices, two from each subunit angled at ~40° from the central axis. The stalk, consisting of an α,β domain, binds the packaging motor while the genome is being pumped into the head, and to the neck proteins after the motor has dissociated from the portal following packaging termination. In addition to the core structure, the portal protein of phage P22 consists of an additional 200Å-long glutamine-rich α-helical barrel domain inside the head that extends from the crown domain of the portal into the interior of the capsid.25–27
The dodecameric portal is critical for head assembly, DNA translocation, and neck-tail attachment.3,5 For initiation of head assembly, the portal interacts with the major capsid protein and the scaffolding proteins. For DNA translocation, the portal interacts with the packaging motor protein, and for tail attachment, it interacts with the neck proteins. Conformational changes exposing different interaction sites of the portal assembly might be responsible for these transitions. Recent evidence28 suggests that some of these conformational changes might be reversible because the fully matured phage head can re-assemble the motor and permit a second round of packaging into an emptied head. Of special interest is the mechanistic role(s) the portal plays in DNA translocation. Several roles have been proposed: rotation coupled to DNA movement;25,26,29 DNA compression and release generating a power stroke;30 and one-way valve to stop the escape of DNA during the hand-over from one gp17 to a neighboring gp17 of pentameric motor.31,32 However, there has been no direct evidence for any of these functions and recent evidence shows that portal rotation is unlikely.33,34
Here, we report on the prediction of atomic models for the T4 portal protein monomer and dodecamer and the fit of the latter into the portal vertex of the 16Å cryoEM structure of phage T4. A biochemical approach was developed to dissect the portal functions, in the context of the predicted models. Mutant portal proteins expressed from a plasmid were incorporated into phage heads, or phage, in order to determine the importance of a given amino acid or motif for head assembly, DNA packaging, or virion production. We found that the loops in the stalk domain are essential for the assembly of the packaging motor at the external end of the portal. The loops that connect the stalk domain to channel helices are essential for DNA translocation. The “tunnel loops” that protrude into the channel are involved in stabilizing the last packaged genome and sealing the packaged head. These results suggest that the portal is required throughout the packaging process, with different segments of the portal protein participating at different stages of DNA packaging.
Results
Structural modeling of gp20 portal
We aimed at constructing computational models of T4 gp20 portal monomer using the known portal structures of SPP1 gp6 (PDB ID: 2JES-A), φ29 gp10 (1FOU-A), and P22 gp1 (3LJ5-A) as templates. However, even though these structures are very similar, they do not share sufficiently high sequence identity with each other, or with the T4 portal protein gp20. The sequence identity between gp20 and each of the above structures is 18.31%, 12.23%, and 12.7%, respectively. Since it would not be possible to construct a reliable model based on such low sequence identity, we built multiple models using different computational methods to identify common features (Fig. 2). Models a-e were constructed using combinations of a comparative modeling method, Modeller35, and various structure refinement methods. Model f was constructed by comparative modeling using the Swiss Model method (Fig. 2; see Materials and Methods, and supplementary section for details on model constructions).
Fig. 2.
Atomic models of T4 gp20 portal protein. Models (a) to (e) were computed by combinations of a threading method, a comparative modeling, followed by residue-based and atom-based refinements. Model f was generated by a comparative modeling method using SwissModel server. The colors indicate the position of residues in the polypeptide chain, transitioning from blue at the N-terminus (aa 250) to red at the C-terminus (aa 512). The N- and C-termini of model (a) are marked with blue and red dots respectively, and the amino acid numbers of the termini are also shown. The same applies to all other models (b) to (f) shown in the figure. See Materials and Methods for details of model building.
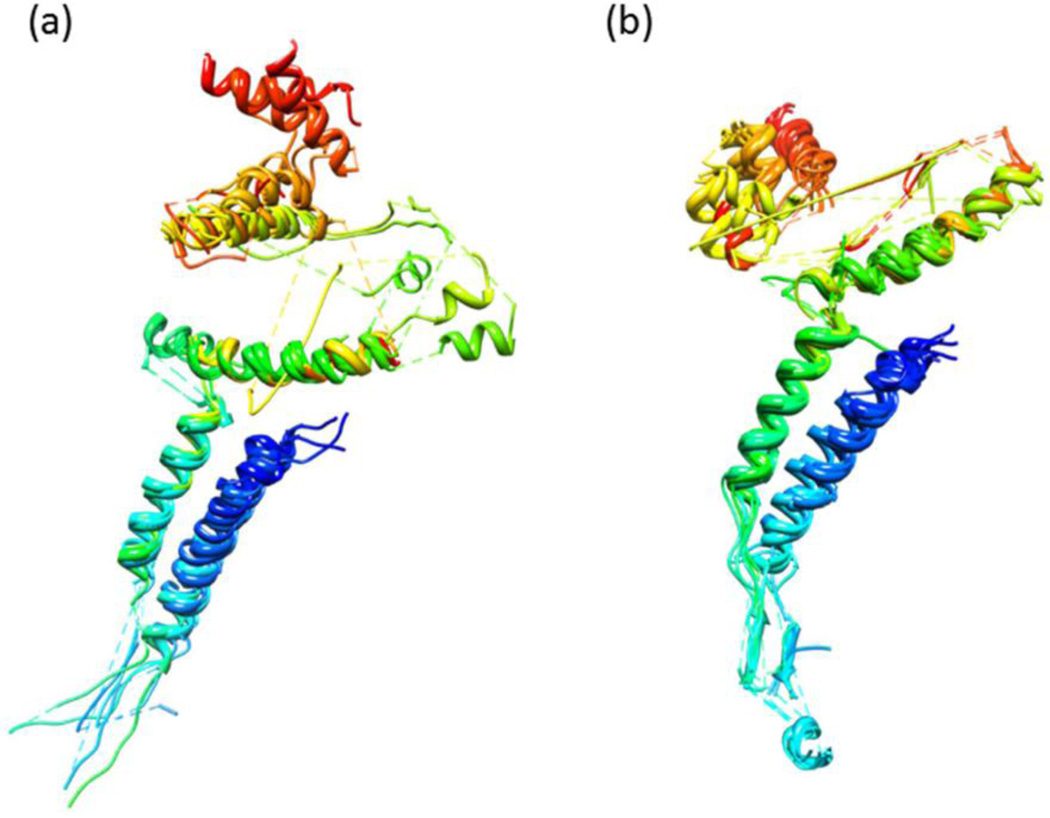
All the models predicted a common core structure for the gp20 portal monomer, which spans from aa 250 to aa 512, covering approximately 50% of the sequence. It consists of two long channel helices, namely α1 and α4, that line the portal channel (the helices are labeled in Fig. 2f); a “kinked” helix, α5; and the tunnel loop between α4 and α5 (Fig. 2a–f). These motifs superimpose with each other (Fig. 3a), as well as with those from the three known phage portal protein structures25–27 (Fig. 3b), and are consistent with the previously predicted secondary structure alignments26,36. The root mean square deviation (RMSD) between pairs of models for the common regions ranged between 2.59 to 3.89 Å. These values are in the same range as the RMSD values between pairs of the known portal protein structures, which is 3.56 to 4.45, and between model f and the known portal protein structures, which is 1.15 to 4.35. However, not unexpectedly, the models differed from each other in some respects. The actual amino acid sequences of some of the motifs are not the same among different models. For instance, the amino acid sequence of helix α1 in model b (Fig. 2b) is different from the rest of the models. Although all the models predicted a stalk domain in the same region of the gp20 sequence, its secondary and tertiary structural details varied depending on the computational method used (Fig. 2). Nevertheless, it can be concluded that the phage T4 portal monomer contains a core structure containing stalk domain, channel helices, tunnel loop, kinked helix, wing and crown domains, which are similar to portal protein structures from phages SPP1, φ29, and P22 (Figs. 2 and 3).
Fig. 3.
(a) Co-aligned parts of six structures of Figure 2 using flexible structure alignment, POSA17. (b) In addition to the six structures shown in (a), three core parts extracted from the three templates (2JES-A, 3LJ5-A, 1FOU-A) were aligned together. The regions where more than half of the input structures were co-aligned are presented.
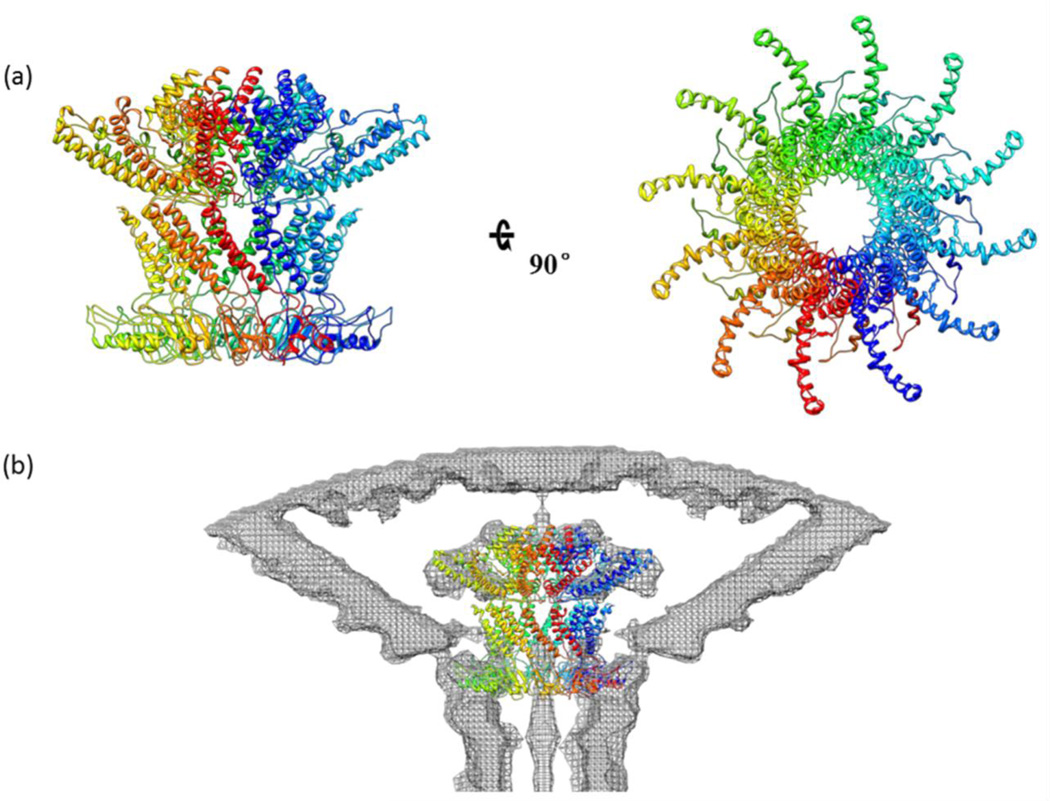
The homology model f, which is very similar to model a, also exhibited a secondary structure that aligned most closely with the SPP1 portal structure (Figs. S1, S2, and S3). The previous genetic and biochemical data are also consistent with this model (see below).15 Therefore, a dodecamer model of T4 portal vertex was constructed by aligning model f with each subunit of the SPP1 portal dodecamer structural model (Fig. 4a). The dodecamer model was then fitted into the cryo-EM density of phage T4 portal vertex24 (EMD ID: 1086) using the UCSF Chimera software37 (Fig. 4b). As observed in other phage portal models, the fitting aligned the wider end of the T4 portal dodecamer to the density inside the capsid and the narrower end to the density protruding out of the five-fold vertex. Thus, the basic architecture of the dodecameric portal appears to be well conserved in phage T4.
Fig. 4.
Structural model of phage T4 portal dodecamer. (a) A dodecamer model was constructed by aligning the gp20 portal monomer model (Fig. 2f) to each subunit of the SPP1 portal dodecamer model26. Side (left) and top (right) views are shown. (b) The dodecamer model was fitted into the cryo-EM density of phage T4 portal vertex (EMD ID: 1086) using the UCSF Chimera.
A biochemical approach to dissect portal function
A mutagenesis-coupled biochemical approach was developed to analyze portal function (Fig. 5). The DNAs corresponding to the wild-type (WT) portal protein and various mutants constructed based on model f were cloned into an IPTG-inducible plasmid (pET28b) and transformed into the expression strain E. coli BL21 (DE3) RIPL. Following brief (20 min) induction to express gp20 (Fig. 5a), cells were infected with a portal-less phage mutant [20amE481(W12am) or 20amN50(Q325am)] (Fig. 5b). Only the plasmids expressing the WT portal, or the mutant portals that are functional, would be able to complement the 20am mutation.33 Mutants that are defective will be unable to complement, producing various phenotypes. For instance, when infected with the 10amB255(W430am)13amE609(Q39am) 20amE481/20amN50 phage mutant (Fig. 5c), this mutant, since it lacks neck (13am) and tail (10am) but packages normally, will accumulate packaged heads in cells expressing a functional portal28 (Fig. 5d) [in tailed phages, the function of the neck protein(s) is exerted after packaging is completed].38 If a portal mutant is defective for nucleation of head assembly, no heads would be produced (Fig. 5e). If the head assembly is normal but the portal is unable to assemble the packaging motor, empty heads might accumulate (Fig. 5f).
Fig. 5.
A biochemical approach to dissect portal function. (a) The plasmids containing g20 variants (WT or mutants) were transformed into E. coli BL21 (DE3) RIPL strain for IPTG induced expression of the respective gp20 protein (green). (b) The E. coli cells were then infected with portal-less mutant phage. (c) With 10am13am20am phage infection, head particles can be produced only if the E. coli-expressed gp20 protein is functional for initiation of head assembly. (d) Fully-functional gp20 produces full heads. (e) gp20 mutants that are defective for initiation of head assembly lead to low to no yield of heads. (f) Packaging-defective gp20 mutants produce empty heads. (g-i) CsCl density gradient ultracentrifugation (g) separates partial heads (h) and full heads (i). With 20am phage infection (j), phage would be produced if the E. coli-expressed gp20 protein is functional (k) whereas defective gp20 produces defective phage or no phage (l). (m-o) Phage heads were tested for head expansion (m), packaging motor assembly (n), and DNA packaging (o). (m) Approximately 1010 head particles were tested for resistance to SDS at room temperature. Coomassie blue stained SDS polyacrylamide gel (10% w/v) showing the unboiled (−; gp23* not dissociated) and boiled for 5 min (+; gp23* dissociated) samples. The position of the 49 kDa gp23* band is marked with an arrow. M represents the molecular weight standards. (n) Purified heads were tested for gp17 binding. 5×1010 head particles were incubated either alone (−) or with gfp-gp17 (+). (o) Approximately 2 × 1010 head particles and 2 µM gp17 were used to package the 50- to 766-bp ladder DNA. In lanes 2 and 3, heads assembled with WT gp20 were used. Lane 2 is a negative control lacking gp17. Lane 4 is a positive control in which WT heads assembled using phage-produced portal (10am13am infection) were used for comparison. The position of the 8 kb T4 DNA retained in the partial heads is indicated. See Materials and Methods and Results for additional details.
Consistent with the above predictions and our previous data28, about 90% of the packaged heads produced in 10am13am20am infections using the plasmid-expressed WT portal were unstable, ejecting the DNA spontaneously during purification (Fig. 5d; see Materials and Methods). The released DNA was digested with DNAse I to produce “partial” phage heads that were essentially empty except for some residual ~8 kb DNA (Fig. 5h). The remaining ~10% of the heads retained the packaged genome (~170 kb DNA), presumably because the portal channel was closed, or the end of the DNA packaged last might have slipped away from the portal channel. In either case, DNA release would be blocked (Fig. 5i). These “full” heads were separated from the partial heads by CsCl density gradient centrifugation (Fig. 5g). Presence of 170 kb or 8 kb DNA, respectively, in these heads indicated that the portal mutant retained the ability to package DNA.28
If, in the above scheme, the infecting phage is the 20am mutant instead of the 10am13am 20am mutant (Fig. 5j), infectious phage would be produced in cells expressing functional portal (Fig. 5k), and either no phage or defective phage would be produced if the portal is defective (Fig. 5l). Consistent with these predictions, the WT gp20 construct gave a yield of ~15–30 partial heads per cell when the infecting phage was 10am13am20am mutant. This yield was similar to that obtained in the positive control in which the heads were assembled using the WT portal produced from phage genome using 10am13am mutant as the infecting phage. Approximately the same number of phage (measured in terms of plaque forming units, or pfu) per cell were produced if the infecting phage was 20am mutant. In a negative control lacking the plasmid for gp20, the yield was 0.02–0.04 phage heads or pfu per cell, respectively.
A series of biochemical assays were performed based on our previous studies,15,20,39 which further demonstrated that the plasmid portal-derived heads exhibited the same properties as the phage portal-derived heads. These include: i) the heads were of expanded type as it required boiling in the presence of SDS to dissociate the heads (unexpanded heads are less stable and dissociate at room temperature)20 [Fig. 5m; note the appearance of the major capsid protein (gp23*; “*” represents the cleaved form) band after boiling in lane 2]; ii) the heads bound gp17, which means that the portal can assemble the packaging motor15 (Fig. 5n, lane 3), Gfp-gp17 was used for these binding assays because purified gp17 contains three species, the full-length (70 kDa) and two shorter forms (~68 kDa and 63 kDa) generated by nonspecific protease(s) during purification. These co-migrated with gpAlt (70 kDa) or gp20 (61 kDa) bands making it difficult to determine the binding of gp17 to the head. Therefore, gfp-gp17 fusion protein was constructed,15 which increased the molecular weight of gp17 by 29 kDa and separated the gp17 band from the overlapping gp20 and gpAlt bands. Ggfp-gp17 showed similar functional behavior as the WT gp17 with respect to gp16-stimulated ATPase, nuclease, and DNA packaging activities. This approach allowed unambiguous quantification of gp17 binding to WT or mutants heads (see below); and iii) the heads packaged DNA at similar efficiency as the WT (Fig. 5o).
The external loops are essential for packaging motor assembly
The homology model predicts two loops (TGNMPARKAAEHM, aa 289–301; RDGKAVTEV, aa 339–347) in the stalk domain which are located at the protruded end of the portal dodecamer (Fig. 6a, shown in green). Genetic and biochemical evidence15,40 suggests that these loops interact with the helix-loop-helix (HLH) motif of gp17 (WQWSIQTINGSSLAQFRQEH, aa 333–352) (Fig. 6b; shown in red). For instance, peptides corresponding to either the protruding loop sequences40 or the HLH motif of gp1715, inhibited DNA packaging in vitro. Furthermore, the gp17 crystal structure shows that the HLH motif is exposed on the surface of the N-terminal domain. Fitting of the gp17 structure into the 32Å cryo-EM reconstruction of the pentameric prohead-motor complex indicates that the portal loops and the HLH motif might be in close proximity in the assembled motor (Fig. 6).17 However, a higher resolution reconstruction is needed to precisely position the HLH residues relative to the portal loops since an alternative orientation with the the C-terminal domain facing the portal has also been suggested for gp17 and gp2 of phage SPP1 based on FRET and biochemical evidence.41,42 On the other hand, a reconstruction of negatively stained phage T7 portal-large terminase complex is consistent with the T4 cryoEM reconstruction.43
Fig. 6.
Functional importance of phage T4 portal domains predicted by the homology model. Three linker (loop) regions of the model, which connect different domains of portal and packaging motor have been mutated and their effects on portal function have been determined. These include: the protruding loops of the stalk domain (a) that might interact with the helix-loop-helix motif of gp17 (b), the channel loop (c), and the tunnel loop (d).
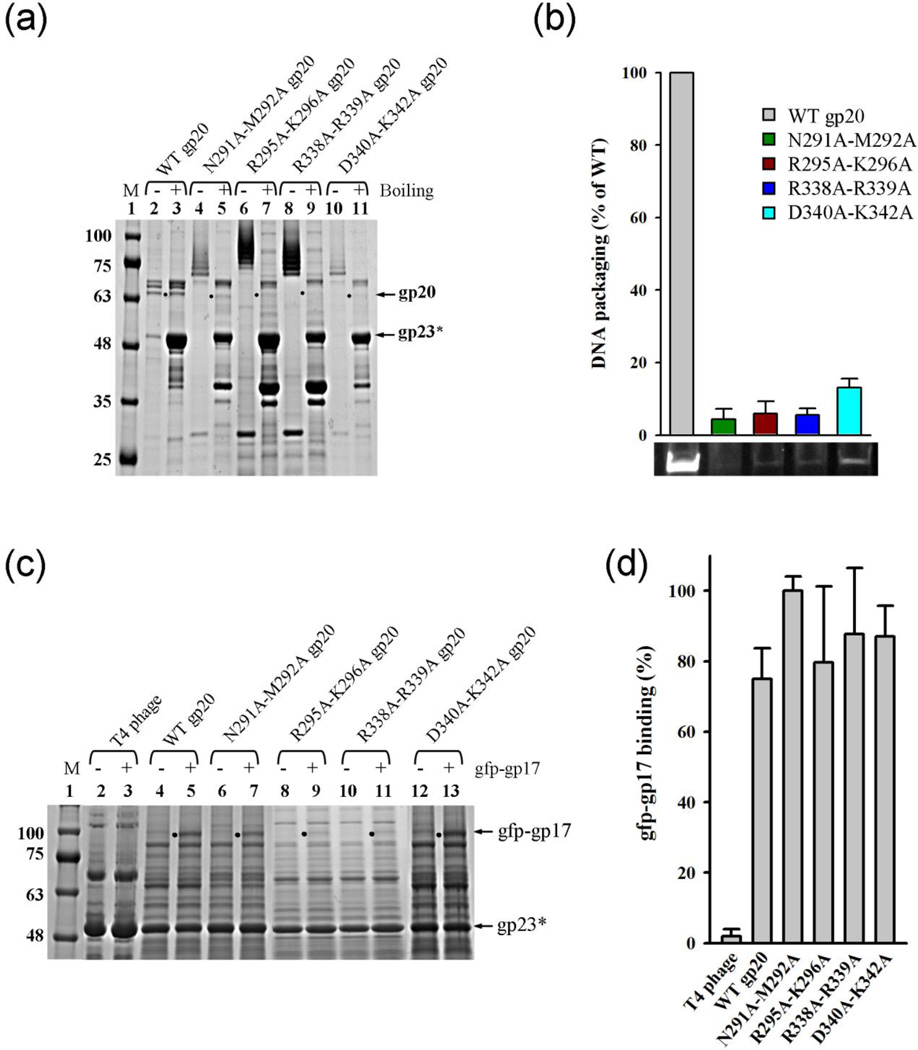
To test the functional importance of the stalk domain loops, the following mutations were introduced: N291A–M292A and R295A–K296A (loop 1 mutants); and R338A–R339A and D340A–K342A (loop 2 mutants). Charged residues were mutated because in previous genetic studies15,44–46 charge-charge interactions were implicated in portal-motor interactions. All the mutants showed a similar phenotype, producing expanded heads, at comparable yields to the WT portal (Fig. 7a; see Materials and Methods for additional details). The gp23* of mutant heads, like that of the WT heads, did not dissociate in the unboiled samples. That these are heads, but not nonspecific aggregates of gp23*, was evident from CsCl density gradient centrifugation which showed that the mutant heads banded at a similar position as the WT heads. Moreover, these heads further purified by Q-column chromatography showed similar protein patterns as the WT heads (Fig. S4).
Fig. 7.
The external loops of stalk domain are essential for packaging. (a) 4–20% (w/v) gradient SDS-PAGE showing the unboiled (−) and 5-min boiled (+) samples of the loop mutant head particles. M represents the molecular weight standards. The position of gp23* band is marked with an arrow. The position of gp20 in the boiled lanes is indicated with a black dot. (b) Histogram showing the comparison of the DNA packaging activity of the loop mutant heads with the heads assembled with WT gp20. Approximately 0.5–2 × 1010 head particles and 1–3 µM gp17 were used to package 50–766 bp ladder DNA, or linearized 4 kb pVAX1 recombinant plasmid DNA, or linearized 6 kb pUC18-luciferase plasmid DNA. Each type of heads was tested in 3 to 5 independent experiments. The DNA packaging activity was presented as the percentage of the activity of WT gp20 heads under the same conditions. The error bars represent standard error. A representative agarose gel image of DNA packaging by various gp20 constructs is shown at the bottom. (c) gp17 binding assays showing the binding ability of the loop mutant heads to gfp-gp17. Approximately 5 × 1010 head particles were incubated either alone (−) or with gfp-gp17 (+) according to the procedure described in Materials and Methods. T4 phage particles were included as the negative control (lanes 2 and 3) as the mature phage does nt bind gp17. “.” indicates the bound gfp-gp17 band positions. (d) Histogram showing the percent binding activity of the loop mutant heads to gfp-gp17 when compared to the WT head (the highest binding is taken as 100%). Bound gfp-gp17 was quantified using gp23 band in the same lane as the internal standard for the number of head particles. Error bars represent the standard error from 3 independent experiments for each type of heads.
The stalk domain mutant heads did not package DNA in vitro. Their packaging activity in the bulk assay was less than 10% of the WT heads (Fig. 7b). These data are consistent with the predictions of the model and the genetic observations described above in that the protruding loop mutants might be defective for the assembly of the packaging motor.
However, unexpectedly, gp17 binding assays showed that the mutant heads bound gp17 as well as the heads containing WT portals (Fig. 7c, d). Single molecule optical tweezers experiments were conducted to analyze the packaging defect of individual mutant motors.47 Preliminary results showed that, unlike the WT portals, the loop mutant portals formed fewer tethers between the motor and DNA, and the tethers broke after only a few seconds at a minimal applied force of 5 pN. In contrast, the tethers in the WT motors were intact even at an applied force of 50–60 pN. Furthermore, no DNA translocation was observed with the mutant motors, which agreed with the results from bulk assays. This behavior is consistent with the hypothesis that although the mutant portal can bind the packaging motor, the interaction was unstable and failed to initiate DNA packaging.
The channel loops are essential for DNA packaging
The stalk domain is linked to the channel helices through short loops (Fig. 6c). These loops might be important for movement of the channel helices during DNA translocation. For instance, the helices, in coordination with the motor, might alternately bind and release the DNA during the translocation process. Therefore, these loops might be a part of communication mechanism between the external domain of the portal that binds gp17 motor and the internal domains that interact with the DNA. Previous studies showed that interfering with the movement of channel helices by disulphide crosslinking of the helices of adjacent monomers resulted in loss of DNA packaging.48
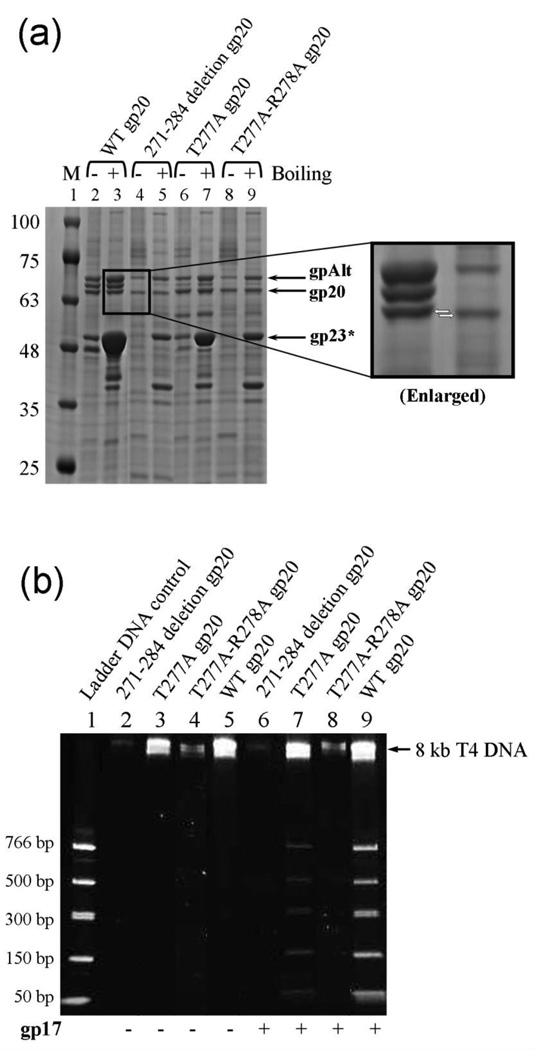
To test the functional importance of the channel loops, the sequence corresponding to the loop, VVIYRITRAPDRRV271–284, was deleted. This deletion mutant produced cleaved expanded proheads, but the yields were 2–5 fold lower than that of the WT. These heads contained a slightly shorter gp20 band (confirmed by Western blotting using gp20 antibodies) (Fig. 8a, compare lanes 4 and 5), evidence that the mutant portal expressed from the plasmid was incorporated into the heads. Furthermore, the channel loop mutant heads lacked the 8 kb DNA (Fig. 8b, lane 2), suggesting that these heads did not package DNA in vivo. This property was further confirmed by in vitro packaging assays, which showed that the mutant heads lacked significant DNA packaging activity (lane 6). To ensure that the lack of activity was not due to a structural perturbation caused by deleting the loop, two additional point mutants were constructed by mutating the T277 and R278 residues present in the loop sequence (T277A and T277A-R278A). Both the mutants showed a similar phenotype as the deletion mutant; decreased yield of heads; expanded heads (Fig. 8a, lanes 6–9); and very low DNA packaging activity (Fig. 8b, lanes 7 and 8).
Fig. 8.
The channel loop is essential for DNA packaging. (a) 4–20% gradient (w/v) SDS-PAGE showing the unboiled (−) and 5-min boiled (+) samples of the channel loop mutant head particles. M represents the molecular weight standards. The positions of gpAlt, gp20 and gp23* bands are marked with arrows. The gp20 band of the deletion mutant (lanes 4 and 5) migrated slightly faster than the WT or the T277A and T277A-R278A mutants. (b) 4–20% gradient PAGE showing the DNA packaging results of the channel loop mutant heads. Approximately 2 × 1010 head particles and 2 µM gp17 were used to package the 50–766 bp ladder DNA. Heads assembled with WT gp20 were used as the positive control (lane 9). The negative controls lacked gp17 (lanes 2–5). The position of the 8 kb T4 DNA retained in the heads is indicated with an arrow.
The tunnel loops are not essential for DNA translocation
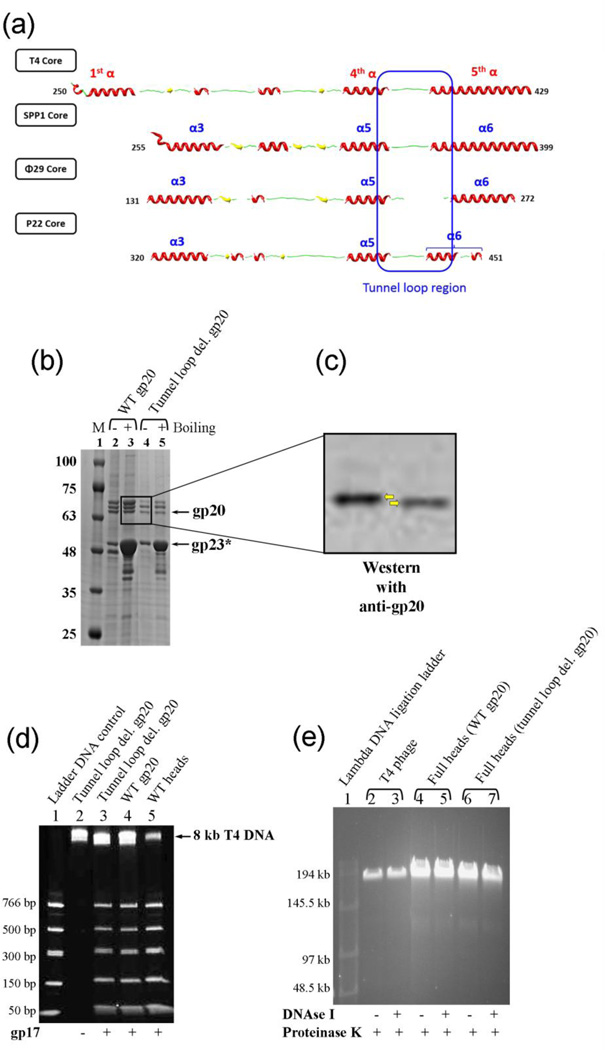
The models (Fig. 2) and the secondary structure alignments of phage portal sequences (Fig. 9a)26,36 predicted a 15 amino acid tunnel loop between the channel helix α4 and the kinked helix α5 of the T4 portal protein (VPLSRIPQDQQGGVM375–389), which is also well-conserved in its size and location, but not sequence, among all the known portal protein structures (Fig. 6d) (the loop was disordered in the φ29 structure).25–27 These twelve relatively long loops, one from each of the 12 portal subunits, that are projected into the channel would constrict the available space in the channel. Hence, some of the loop residues must come into contact with the DNA during translocation.
Fig. 9.
The tunnel loops are not essential for DNA translocation. (a) Secondary structure alignment of the tunnel loop regions of the portal proteins from different phages. The solved structures of different portal proteins are aligned with the predicted secondary structure of the T4 gp20 homology model f (Fig. 2). The presence of the tunnel loop between the inner helix α4 of portal channel and the kinked helix α5 of wing domain is a conserved feature among all the phage portals. (b) 4–20% gradient (w/v) SDS-PAGE showing the unboiled (−) and 5-min boiled (+) samples of the tunnel loop deletion head particles. M represents the molecular weight standards. The positions of gp20 and gp23* bands are marked with arrows. The gp20 band of the deletion mutant (lanes 4 and 5) migrated slightly faster than the WT gp20 (lanes 2 and 3). (c) The same gel was subjected to Western blotting using gp20 antiserum to confirm the positions of gp20 bands (indicated by the yellow arrows). (d) 4–20% PAGE showing the DNA packaging results of the tunnel loop deletion mutant heads. Approximately 2 × 1010 head particles and 2 µM gp17 were used to package the 50–766 bp ladder DNA. Heads assembled with WT gp20 and WT heads prepared from phage-produced portal were used as positive controls (lanes 4 and 5). The negative control lacked gp17 (lane 2). The position of the 8 kb T4 DNA retained in the heads is indicated. (e) T4 phage (5 × 108 particles per lane; lanes 2 and 3) or full heads (2 × 109 particles per lane; lanes 4–7) were treated with DNAse I (37°C, 30 min) followed by proteinase K (65°C, 30 min) or proteinase K only, as shown by “+” or “−” under the figure, and subjected to pulse field (field inversion) agarose gel (0.8% w/v) electrophoresis. The phage λ DNA ligation ladder (lane 1) was used to determine the size of DNA present in phage or heads.
A deletion mutant was constructed by deleting twelve of the tunnel loop residues, LSRIPQDQQGGV377–388, to determine its functional importance. This mutant produced cleaved expanded heads with a similar yield as the WT (Fig. 9b). As expected, the mutant gp20 band was slightly shorter than the WT protein (Fig. 9b), which was also confirmed by Western blotting (Fig. 9c). The tunnel loop deletion did not significantly affect DNA packaging per se in vivo, as shown by the production at similar yields of partial heads with 8 kb DNA (Fig. 9d, lane 2) and full heads with 170 kb DNA as the WT portal (Fig. 9e, lanes 6 and 7) (see Fig. 5g, h, and i for details). The phage heads lacking the tunnel loops packaged DNA (Fig. 9d, lane 3) with similar efficiency as the WT heads in vitro (Fig. 9d, lanes 4 and 5). Single molecule optical tweezers experiments further demonstrated that the rate of packaging of the tunnel loop deleted portal mutant was in the same range as the WT portal.
Tunnel loop mutant portal produced noninfectious virus particles
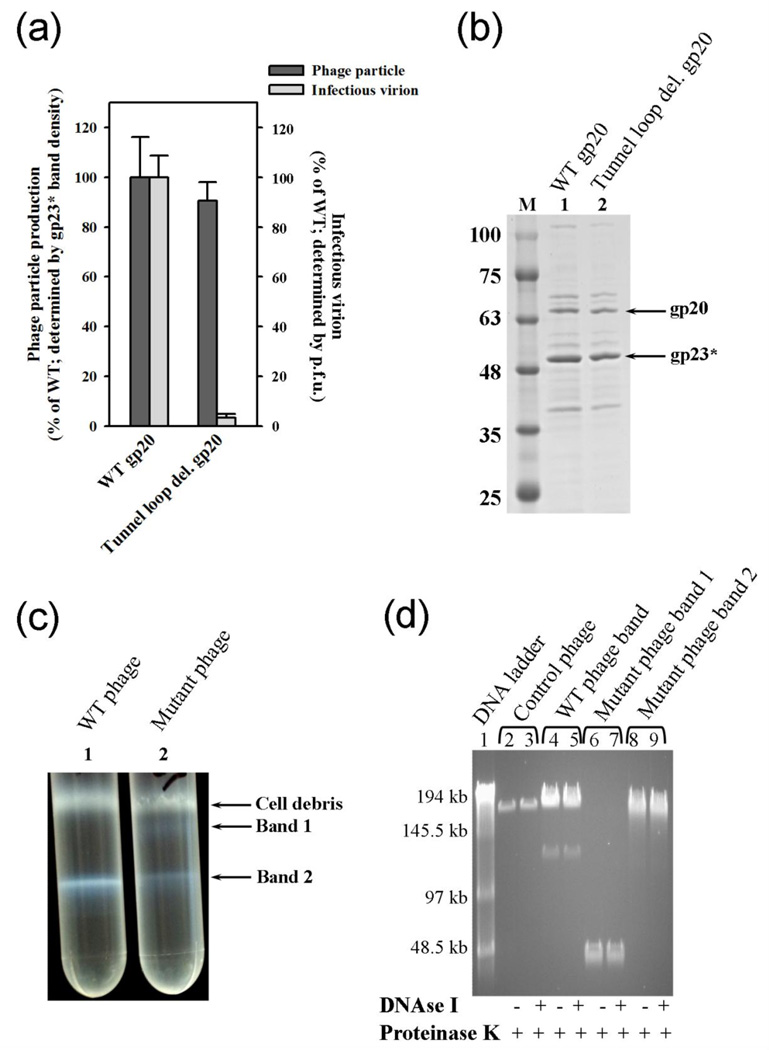
Since the tunnel loop mutant was not defective for DNA translocation per se, it was expected to produce infectious virions upon infection with 20am phage, as per the scheme shown in Fig. 5 (j). Surprisingly, the infectious titer of the progeny phage produced by this mutant was about 30-fold lower than the WT (Fig. 10a). On the other hand, the protein pattern (Fig. 10b) and yield of the progeny phage (Fig. 10a) were similar to that of the WT phage.
Fig. 10.
The tunnel loop mutant produces non-infectious virus particles containing shorter genomes. (a) Histogram showing the infectious titer of the phage particles produced. The particle number was determined by quantifying the gp23* band intensity of lanes 1 and 2 in (b) (black). The infectious titer was determined by plaque assay (grey). Error bars represent standard error. (b) 4–20% gradient (w/v) PAGE showing the protein pattern of phage samples produced from plasmid-expressed gp20. Approximately 109 phage particles (4 × 108 pfu) produced from WT gp20 and 5 × 108 phage particles (6 × 106 pfu) produced from tunnel loop deletion mutant gp20 were loaded. M represents the molecular weight standards. (c) The phage particles produced by the WT heads and tunnel loop mutant heads were subjected to CsCl gradient centrifugation. (d) Phage particles isolated from (c) were treated with DNAse I followed by proteinase K, or proteinase K only, as shown by “+” or “−” rows under the figure. The samples were then subjected to pulse field agarose gel (0.8% w/v) electrophoresis to separate the large DNAs. The phage λ DNA ligation ladder (lane 1) was used to determine the size of DNA present in the particles. Lanes 2 and 3 are the DNA isolated from control WT T4 phage. Lanes 4–9 are the DNA isolated from phage fractions from the CsCl gradient centrifugation (c).
Further analysis showed that the tunnel loop deleted phage, unlike the WT phage, was separated into two bands by CsCl density gradient centrifugation (Fig. 10c). In contrast, the WT phage, as expected, showed only one band corresponding to the mature virion. The lower band of the mutant phage (band 2) migrated slightly slower than the WT phage, which means that its density is lower than the WT phage. Indeed, pulse field agarose gel electrophoresis of the phage DNA isolated from this band showed a DNA smear that is shorter than that of the DNA isolated from the WT phage band (Fig. 10d, compare lanes 8 and 9 of the mutant to lanes 4 and 5 of the WT). The top mutant phage band (band 1), which has much lower density than the bottom band, contained only a ~45 kb piece of DNA (lanes 6 and 7). The plaque forming activity of mutant phage from the lower and top bands, normalized to the amount of the major capsid protein gp23*, was about 4% and 0.001% of the WT, respectively. These data demonstrate that the tunnel loop deleted portals, although proficient for DNA translocation, are defective to package the full-length genome. Consequently, the progeny phage cannot productively infect the E. coli cell.
Discussion
The portal and motor are the two principal components of the phage DNA packaging machine.1–3 Although the functions of the ATPase motor have been well established,17,21,22,47,49,50 the portal functions, in particular its role in DNA packaging is poorly understood. There has been considerable debate on whether the portal is mechanistically involved in translocation or merely serves as a channel to transport DNA into and out of the capsid. The widely considered rotation model coupling portal rotation to mechanical movement of DNA has not been proven,25,26,29 and evidence strongly argues against a rotary mechanism.33,34 Here, we analyzed the structure and function of the phage T4 portal protein gp20. Although gp20 was the first portal protein purified and determined to be a dodecameric ring4, further structural analyses have not been possible because the protein aggregates into insoluble inclusion bodies when expressed in E. coli10 Using a variety of computational methods, we generated atomic models of phage T4 portal monomer that were based on the crystal structures of SPP1, φ29, and P22 portal proteins.25–27 The models showed that the T4 portal protein, like the known structures, consists of two long helices that presumably line a central channel, but the actual amino acid sequence of these helices varied in different models. Helix α1 is the least variant, consisting of a common stretch of amino acids in 5 of 6 models, whereas the helix α4 is more variant. But the basic architecture of the core consisting of stalk domain, channel helices, tunnel loop, wing and crown domains, appears to be preserved in gp20, despite lacking significant amino acid sequence identity between any of these portal protein sequences. The structural features are also consistent with the previous biochemical and genetic data.15,40 For instance, the amino acid sequence that was reported to bind the packaging motor is part of the loop regions of the external stalk domain, well-positioned to interact with the gp17 subunits.
A mutagenesis-coupled biochemical approach was developed to dissect portal functions in the context of the structural models. By expressing gp20 in E. coli cells that also express other head proteins through phage infection, the plasmid-expressed portal protein could be efficiently incorporated into the phage head, rather than partitioning into inclusion bodies. This allowed testing the importance of a structural motif or an amino acid for portal function, such as initiation of head assembly, DNA translocation, and virion production. Mutations in stalk domain, channel loops, and tunnel loops did not significantly affect head assembly or head expansion since the gp23* band appeared only after boiling the heads in the presence of SDS. That the gp23* was assembled into heads but not into nonspecific aggregates was demonstrated by CsCl density gradient centrifugation which banded the mutant particles at a similar position as the WT heads. Moreover, the mutant heads further purified by column chromatography showed similar protein pattern as the WT heads (Fig. S4). On the other hand, deletions in the N-terminal region of gp20 greatly reduced the production of heads (data not shown), suggesting that this region might be important for head assembly.
Our results showed that the portal function is required throughout the packaging process, with different domains participating at different stages of packaging. The stalk domain is critical at the pre-translocation stage, to assemble a functional gp17 motor and initiate DNA packaging. Loops that connect the stalk domain and the associated motor to the channel are defective for DNA packaging. These residues might be required for interaction with the DNA as it translocates through the ~4 nm channel, either directly or through other residues lining the channel. The tunnel loops on the other hand are essential in the post-packaging stage, for stabilizing the last packaged genome and sealing the packaged head. However, other explanations are possible. For instance, some of the mutations might be affecting the interactions between the portal subunits or altered the stoichiometry of the portal. But since the dodecamer stoichiometry of the portal in the phage head is strictly conserved, such portals may affect initiation of the head assembly but not DNA packaging.
Tunnel loops were previously thought to be involved in pushing the DNA into the capsid.26 This model was proposed, in part, because these loops are present in two, “up” or “down”, conformations in the SPP1 portal structure. A “molecular lever” model was proposed in which the loops “embrace” the DNA by matching its shape through nonspecific van der Waals interactions. Portal rotation driven by the motor and powered by ATP hydrolysis causes sliding and repositioning of the loops in sequence, like a “Mexican wave”, resulting in directional translocation of DNA into the head. Our results are inconsistent with such a model because DNA translocation was unaffected in a tunnel loop deletion mutant that lacked 12 of the 15 predicted tunnel loop residues. Similar observations were made with the tunnel loop deletion mutants of the phage φ29 portal.51
Although the tunnel loop deletion mutant is not defective for DNA translocation per se, the progeny virus particles produced with this mutant contained shorter genomes and did not produce plaques. Majority of the genomes are in the range of 150–160 kb, approximately 90–95% the size of the full-length genome (~170 kb). These data lead to the hypothesis that the tunnel loop might be essential for stabilizing the packaged genome at the late stage of head filling. A conformational change must occur after head filling, presumably signaled by the internal pressure reaching a threshold,52,53 which leads to dissociation of the motor and termination of DNA packaging. Although the details of the mechanism are unknown, several studies reported structural changes in the portal at the time of packaging termination, which might involve constriction of the portal channel.25,53–55 Consequently, the tunnel loops would tightly bind the last packaged DNA, preventing its release as well as positioning it for delivery. In the portal mutant lacking tunnel loops, the DNA would be “loose”, forced to leak out a portion of the last packaged DNA due to the internal pressure and be degraded by nucleases before the neck proteins, gp13, gp14, and gp15 assemble and seal the portal channel. This explains why the tunnel loop deletion phages have a shorter genome and low infectivity. An alternative hypothesis is that the tunnel loops might be part of the headful sensing mechanism that signals the motor to dissociate and terminate packaging after the head is full. Portal mutations that are defective in this sensing mechanism have been reported in phages P22 and SPP1. These either over-package or under-package the head.54,56 The gp20 tunnel loop mutants might belong to the underpackaging phenotype, causing premature signaling and termination. This hypothesis could also explain why a fraction of the phage produced by the tunnel loop deletion mutant contained very short genomes, on the order of 45–50 kb, which is equivalent to 25–30% of the headful length. Coincidentally, this size correlates with the size of packaged genome when the head transitions into an expanded state.3,57 Head expansion in phage λ, which also occurs when 25–30% genome is packaged, is associated with a 4 pN increase in internal pressure and 50% dip in packaging rate.58 Ray et al57 proposed that a structural change in gp20 portal dodecamer is associated with head expansion. These observations lead to the speculation that the tunnel loops might also be involved in stabilizing the DNA during the expansion transition. Mutants lacking the loops, being unable to stablilize the genome at the time of expansion, might cause premature termination when only 25–30% of genome is packaged.
In conclusion, our studies suggest that the portal dodecamer is actively involved throughout the packaging process, not merely providing a DNA translocation channel. Although the motor provides the energy as well as, most likely, the “power stroke” for translocating DNA, coordination of this process requires active participation of the portal through its interactions with motor and DNA. In particular, the tunnel loops may be part of a headful sensing and packaging termination mechanism in which translocation of the last piece of DNA, motor dissociation, channel closure, engagement of DNA for delivery, and neck/tail attachment must be precisely orchestrated to produce a virion that can efficiently deliver a full complement of virus genome into a new host cell.
Materials and Methods
Structural modeling of gp20
Models a-c in Fig. 2 were constructed using the comparative modeling method, Modeller35, followed by two-steps of structural refinements, refinement of main-chain conformation and a subsequent atom-level refinement. The model building procedure started with computing alignments between the gp20 sequence with each of the three known structures, SPP1 gp6, φ29 gp10, and P22 gp1. We used a threading method, FUGUE59, for computing the alignments because it considers structural features of proteins (e.g. the secondary structures and solvent accessibility) as well as amino acid similarity in computing alignments and thus can construct more accurate alignments than regular pairwise sequence alignment methods particularly when the sequence similarity between them is low. Then, each of the three alignments were used as input of comparative modeling method, Modeller, obtaining three structural models of gp20. Next, a protein main-chain structure modeling method, CABS60 was used to refine the intital models from Modeller. CABS places a protein structure onto a 0.5Å 3D grid and move the mainchain of the structure on the grid with a Monte Carlo procedure so that the energy of the structure is improved. In the refinement by CABS, the secondary structure of the models are essentially kept intact. The secondary structure information of gp20 was predicted by Jpred61, PredictProtein62, and PsiPred63. CABS generates over a thousand structures. Of these, three best structures were selected using our in-house residue-based scoring function (Hyung-Rae Kim & Daisuke Kihara, in preparation) that examines propensities of neighboring residues in structures. Finally, a short run of molecular dynamics (MD) with the NAMD program64 was used to refine the atomic details of the models.
Models d and e were constructed using a slightly different procedure from the models a to c. Five structures, 3LJ5-A, 2JES-A, 1FOU-A, 1H5W-A, and 3LJ4-A, which were identified by FUGUE from the Protein Data Bank were used as templates of modeling [includes three structures used to construct models a-c, 3LJ5-A (P22 gp1 1–725 aa); 2JES-A (SPP1 gp10); and 1FOU-A (φ29, 3.2 Å resolution) plus two additional structures 1H5W-A (φ29 gp6 2.1 Å resolution) and 3LJ4-A (P22 gp1 1–602 aa). In the same way used for constructing models a-c, alignments between the gp20 sequence and each template was computed with FUGUE and structures were modeled with Modeller using the alginments as inputs. The resulting strucures were used as input for CABS, which refines the stereochemistry of the main-chain of the models. Over one thousand structures by CABS were clustered and centroid structures of ten largest clusters were obtained. Subsequently, the ten structures were further refined by a short MD run to avoid steric clashes. Finally, two structures were selected based on the residue-based scoring function (Fig. 2d, e).
The last model (Fig. 2f) was constructed using a comparative modeling procedure (Fig. 2f and S1). The procedure consists of identifying the template from the database that aligns best with the T4 gp20 sequence by HHpred server. This process identified the SPP1 portal protein gp6 (PDB: 2JES-A) as the best match for T4 gp20 and generated a secondary structure alignment (Fig. S2). This alignment was manually adjusted to improve matching of secondary structure elements (Fig. S3). Structural modeling was done using the the Swiss Model homology modeling server65. For this modeling only gp6 was used as template because it has the largest sequence identity to gp20 and also because it was identified as the closest homolog by HHpred66, a sequence-based database search method (Fig. S2). The final alignment (Fig. S3) was entered into the Swiss Model homology modeling server65 to generate a raw model. The model was then refined by Kobamin server67, which uses a refinement protocol that minimizes a knowledgebased potential that reflect statistics of structural features of proteins in PDB. The quality of the model was assessed using the SIB assessment server68, which assesses quality of structures based on a composite scoring function (QMEAN) and is able to derive both global and local error estimates of the submitted model (Tables S1 and S2). SIB gave a Z-score of 6.14, which indicates that the geometric features of the model are reasonable.
Bacteria, phage, and plasmids
Escherichia coli XL10 Gold-ultra competent cells (Stratagene, La Jolla, CA) were used for maintaining the recombinant plasmids. The plasmid clones were then transformed into E. coli BL21 (DE3) RIPL cells (Stratagene) for expression of recombinant proteins. E. coli P301 was used as a suppressor-negative strain and E. coli B40 (sup1) was used for preparation of T4 phage amber mutant stocks. The 10amB255 (W430am) 13amE609 (Q39am) 20amE481 (W12am) and 10amB255 (W430am) 13amE609 (Q39am) 20amN50 (Q325am) phage T4 mutants were constructed by crossing the 10am 13am and 20am mutant phages. The T7 expression plasmid vector pET28-b was used to construct all the gp20 WT and mutant clones.
Construction of recombinant plasmids
The gp20 mutant clones constructed in this study included a series of single amino acid and multiple amino acid substitutions, and deletions. These were constructed by the PCR-directed splicing by overlap extension strategy.69,70 Two WT end primers and two overlapping mutant primers containing the desired mutations were used for amplification of g20 DNA. Phage T4 DNA was used as a template. The amplified DNA fragments were digested with appropriate restriction enzymes, ligated with the linearized vector plasmid DNA, and transformed into XL10 Gold-ultra competent cells. In-frame insertion of these fragments into the vector resulted in the fusion of a 23 amino acid sequence containing hexa-histidine tag (His-tag) to the N-terminus of each gp20 construct. Thus, the WT and all the mutant gp20 constructs used in this study contain His-tag at the N-terminus of the portal protein. The accuracy of the cloned DNA was confirmed by DNA sequencing (Retrogen, Inc., San Diego, CA).
Incorporation of E. coli expressed gp20 into phage heads
E. coli BL21 (DE3) RIPL cells containing the gp20 recombinant plasmids was induced with isopropyl-β-D-thio-galactoside (IPTG; 1 mM) for 20 min at 37°C. The cells were infected with the 10am13am20am or 20am mutant phage [multiplicity of infection (MOI) = 4) and superinfected at the same MOI 7 min post-initial infection and incubated in a 37°C shaker for 28 min. The infected cells (500 ml) were then centrifuged at 8,000g for 12 min and lysed in 40 ml of Pi-Mg buffer (26 mM Na2HPO4, 68 mM NaCl, 22 mM KH2PO4 and 1mM MgSO4, pH 7.5) containing 10 µg/ml DNAse I and chloroform (1 ml). The sample was incubated at 37°C for 30 min to release the heads and digest the DNA. The lysate was centrifuged at 4,300g for 10 min and the supernatant was centrifuged at 34,500g for 45 min. The sample was then resuspended in 2.5 ml of Pi-Mg buffer containing 200 mM NaCl and subjected to another round of low- and high-speed centrifugations. The heads were then resuspended in 500 µl of Pi-Mg buffer. To compare the yields of the heads produced by portal mutants with the yields produced by the WT portal, heads were isolated by the above procedure from the same volume of the infected E. coli culture and the same volume of the final samples was loaded on the SDS gel. The gp23* band was quantified by densitometry (Personal Densitometer; GE Healthcare, Piscataway, NJ). In most cases, the mutant heads were further purified by CsCl density gradient ultracentrifugation.28 The mutant heads banded at a similar position as the WT head. The head bands were extracted and dialyzed overnight against 10 mM Tris-HCl pH 7.5, 50 mM NaCl and 5 mM MgCl2. The heads were concentrated by Amicon Ultra-15 centrifugal filters (Millipore, Temecula, CA) and stored at 4°C. For some experiments, the heads were further purified by Q-column (GE Healthcare) or DEAE-Sephacel column chromatography.28
Analysis of phage heads
Unexpanded heads dissociate in the presence of SDS at room temperature but the expanded heads are resistant. Therefore, the major capsid protein (gp23*) subunits of the expanded head do not migrate into the gel unless the sample is boiled.20 Thus, electrophoresis of the heads samples with and without boiling allows the determination of the extent of head expansion. The head particles were quantified by densitometry of the Coomassie blue stained gp23* band of the boiled and unboiled samples following SDS-PAGE on 4–20% (w/v) gradient gels (Personal Densitometer; GE Healthcare, Piscataway, NJ).
Two criteria were used to determine the size differences between the WT gp20 and the deletion mutants of gp20 lacking the tunnel loop or channel loop. First, comparisons were made by electrophoresing the samples on the same gel. The position of gp20 was compared with that of gpAlt, an internal standard which migrates to the same position in all the samples. Thus, the small changes in the distance between gp20 and gpAlt reflect the differences in the size of the gp20 mutant protein. Second, gp20 was stained using a polyclonal rabbit gp20 antiserum and the WesternBreeze Immunodetection System (Life Technologies, Grand Island, NY), to confirm that the protein bands identified by Coomassie blue staining were indeed the gp20 protein. In addition, these data further confirmed the differences in the molecular weights of WT and deletion mutant gp20 bands.
DNA packaging assay
In vitro DNA packaging assays were performed by the procedure described earlier.39 The main difference was that the reaction mixture contained purified heads containing either the WT portal or the mutant portal expressed from the plasmid (0.5–2 × 1010 particles). As a positive control, heads prepared from 10am13am phage infection (phage-produced portal) was used. The reaction mixture in addition contained purified full-length gp17 (1–3 µM), DNA (600 ng of 50–766 bp ladder DNA [New England Biolabs, Ipswich, MA], or linearized 4 kb pVAX1 recombinant plasmid DNA, or linearized 6 kb pUC18-luciferase plasmid DNA), and a buffer containing 30 mM Tris-HCl pH 7.5, 100 mM NaCl, 3 mM MgCl2 and 1 mM ATP. Packaging was terminated by the addition of DNAse I, and the encapsidated DNAse I-resistant DNA was released by treatment with proteinase K and analyzed by polyacrylamide (4–20% gradient) or agarose (0.8%) gel electrophoresis. The negative controls lacked gp17 (or ATP). A standard lane containing a small amount of the DNA substrate used in the packaging reaction allowed for quantification of packaged DNA using the Gel DOC XR imaging system (Bio-Rad, Hercules, CA).
gp17 binding assay
Purified heads (0.5–2 × 1011 particles) were incubated with gfp-gp17 (gfp gene was fused to the N-terminus of gp17 to increase its molecular mass by 29 kDa, which clearly separated the gp17 band from the gp20 band15) at a gfp-gp17 to gp20 ratio of 50:1 in 500 µl binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 5 mM MgCl2) for 30 min at room temperature. The heads were sedimented by centrifugation at 32,000g for 45 min at 4°C and the pellet was washed twice with 1 ml binding buffer and resuspended in 20 µl of bind buffer for SDS-PAGE analysis. The bound gfp-gp17 was quantified by ImageQuant software (GE Healthcare) using gp23* band in the same lane as the internal standard.
Supplementary Material
Highlights.
Portal of tailed bacteriophages is critical for head assembly and DNA packaging.
Atomic models have been constructed for phage T4 portal monomer and dodecamer.
A biochemical approach has been established to dissect portal functions.
Different portal domains participate at different stages of DNA packaging.
Tunnel loops of portal are essential for stabilizing the last packaged genome.
Acknowledgements
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R01AI081726) to M.G.R. and V.B. R., and the National Science Foundation (MCB-0923873) to V.B.R., and in part by grants from the National Institute of General Medical Sciences (GM097528) and the National Science Foundation (EF0850009, IOS1127027, DBI1262189) to D.K.
Abbreviations used
- aa
amino acids
- am
amber mutation
- EM
electron microscopy
- gp
gene product
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MD
molecular dynamics
- MOI
multiplicity of infection
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- PDB
Protein Data Bank
- pfu
plaque forming units
- RMSD
root mean square deviation
- SDS
sodium dodecyl sulfate
- WT
wild type
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contribution. V. P.S, S.G., H.R.K., D.K., L.S., M.G.R., and V.B.R. designed research. V. P.S, S.G., H.R.K., L.S. performed research. D.K., M.G.R., and V.B.R. wrote the paper.
Conflict of interest. The authors declare that they have no conflict of interest.
References
- 1.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 2.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 3.Black LW, Rao VB. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv Virus Res. 2012;82:119–153. doi: 10.1016/B978-0-12-394621-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driedonks RA, Engel A, tenHeggeler B, van D. Gene 20 product of bacteriophage T4 its purification and structure. J Mol Biol. 1981;152:641–662. doi: 10.1016/0022-2836(81)90121-2. [DOI] [PubMed] [Google Scholar]
- 5.Valpuesta JM, Carrascosa JL. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q Rev Biophys. 1994;27:107–155. doi: 10.1017/s0033583500004510. [DOI] [PubMed] [Google Scholar]
- 6.Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao VB, Black LW. Structure and assembly of bacteriophage T4 head. Virol J. 2010;7:356. doi: 10.1186/1743-422X-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eiserling FA, Black LW. Pathways in T4 morphogenesis. In: Molecular Biology of Bacteriophage T4. In: Karam JD, editor. Washington, D.C: ASM Press; 1994. [Google Scholar]
- 9.Hsiao CL, Black LW. Head morphogenesis of bacteriophage T4. II. The role of gene 40 in initiating prehead assembly. Virology. 1978;91:15–25. doi: 10.1016/0042-6822(78)90351-3. [DOI] [PubMed] [Google Scholar]
- 10.Michaud G, Zachary A, Rao VB, Black LW. Membrane-associated assembly of a phage T4 DNA entrance vertex structure studied with expression vectors. J Mol Biol. 1989;209:667–681. doi: 10.1016/0022-2836(89)90599-8. [DOI] [PubMed] [Google Scholar]
- 11.Quinten TA, Kuhn A. Membrane interaction of the portal protein gp20 of bacteriophage T4. J Virol. 2012;86:11107–11114. doi: 10.1128/JVI.01284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Showe MK, Isobe E, Onorato L. Bacteriophage T4 prehead proteinase. I. Purification and properties of a bacteriophage enzyme which cleaves the capsid precursor proteins. J Mol Biol. 1976;107:35–54. doi: 10.1016/s0022-2836(76)80016-2. [DOI] [PubMed] [Google Scholar]
- 14.Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci U S A. 2012;109:817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegde S, Padilla-Sanchez V, Draper B, Rao VB. Portal-large terminase interactions of the bacteriophage t4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. J Virol. 2012;86:4046–4057. doi: 10.1128/JVI.07197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliveira L, Cuervo A, Tavares P. Direct interaction of the bacteriophage SPP1 packaging ATPase with the portal protein. J Biol Chem. 2010;285:7366–7373. doi: 10.1074/jbc.M109.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J Biol Chem. 2000;275:37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 19.Goetzinger KR, Rao VB. Defining the ATPase center of bacteriophage T4 DNA packaging machine: requirement for a catalytic glutamate residue in the large terminase protein gp17. J Mol Biol. 2003;331:139–154. doi: 10.1016/s0022-2836(03)00636-3. [DOI] [PubMed] [Google Scholar]
- 20.Rao VB, Black LW. DNA packaging of bacteriophage T4 proheads in vitro. Evidence that prohead expansion is not coupled to DNA packaging. J Mol Biol. 1985;185:565–578. doi: 10.1016/0022-2836(85)90072-5. [DOI] [PubMed] [Google Scholar]
- 21.Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Alam TI, Draper B, Kondabagil K, Rentas FJ, Ghosh-Kumar M, Sun S, Rossmann MG, Rao VB. The headful packaging nuclease of bacteriophage T4. Mol Microbiol. 2008;69:1180–1190. doi: 10.1111/j.1365-2958.2008.06344.x. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh-Kumar M, Alam TI, Draper B, Stack JD, Rao VB. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 2010;39:2742–2755. doi: 10.1093/nar/gkq1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fokine A, Zhang Z, Kanamaru S, Bowman VD, Aksyuk AA, Arisaka F, Rao VB, Rossmann MG. The molecular architecture of the bacteriophage t4 neck. J Mol Biol. 2013;425:1731–1744. doi: 10.1016/j.jmb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Kottadiel VI, Vafabakhsh R, Dai L, Chemla YR, Ha T, Rao VB. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 2011;9:e1000592. doi: 10.1371/journal.pbio.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing P, Haque F, Shu D, Montemagno C, Guo P. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 2010;10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang H, Jing P, Haque F, Guo P. Role of channel lysines and the “push through a one-way valve” mechanism of the viral DNA packaging motor. Biophys J. 2012;102:127–135. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 34.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Tavares P, Zinn-Justin S, Orlova EV. Genome gating in tailed bacteriophage capsids. Adv Exp Med Biol. 2012;726:585–600. doi: 10.1007/978-1-4614-0980-9_25. [DOI] [PubMed] [Google Scholar]
- 39.Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 40.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J Mol Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 41.Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor: endonuclease VII resolvase release of arrested Y-DNA substrates. J Biol Chem. 2011;286:18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornilleau C, Atmane N, Jacquet E, Smits C, Alonso JC, Tavares P, Oliveira L. The nuclease domain of the SPP1 packaging motor coordinates DNA cleavage and encapsidation. Nucleic Acids Res. 2013;41:340–354. doi: 10.1093/nar/gks974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL. Large terminase conformational change induced by connector binding in bacteriophage T7. J Biol Chem. 2013;288:16998–17007. doi: 10.1074/jbc.M112.448951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30:4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J Mol Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 46.Yeo A, Feiss M. Mutational analysis of the prohead binding domain of the large subunit of terminase, the bacteriophage lambda DNA packaging enzyme. J Mol Biol. 1995;245:126–140. [PubMed] [Google Scholar]
- 47.Kottadiel VI, Rao VB, Chemla YR. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. Proc Natl Acad Sci U S A. 2012;109:20000–20005. doi: 10.1073/pnas.1209214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J Biol Chem. 2007;282:18907–18913. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- 49.Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage t4 terminase. J Biol Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 50.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grimes S, Ma S, Gao J, Atz R, Jardine PJ. Role of phi29 connector channel loops in late-stage DNA packaging. J Mol Biol. 2011;410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 53.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 54.Orlova EV, Dube P, Beckmann E, Zemlin F, Lurz R, Trautner TA, Tavares P, van Heel M. Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat Struct Biol. 1999;6:842–846. doi: 10.1038/12303. [DOI] [PubMed] [Google Scholar]
- 55.Zheng H, Olia AS, Gonen M, Andrews S, Cingolani G, Gonen T. A conformational switch in bacteriophage p22 portal protein primes genome injection. Mol Cell. 2008;29:376–383. doi: 10.1016/j.molcel.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- 57.Ray K, Oram M, Ma J, Black LW. Portal control of viral prohead expansion and DNA packaging. Virology. 2009;391:44–50. doi: 10.1016/j.virol.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 60.Kolinski A. Protein modeling and structure prediction with a reduced representation. Acta Biochim Pol. 2004;51:349–371. [PubMed] [Google Scholar]
- 61.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 64.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 66.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chopra G, Kalisman N, Levitt M. Consistent refinement of submitted models at CASP using a knowledge-based potential. Proteins. 2010;78:2668–2678. doi: 10.1002/prot.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rao VB, Mitchell MS. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J Mol Biol. 2001;314:401–411. doi: 10.1006/jmbi.2001.5169. [DOI] [PubMed] [Google Scholar]
- 70.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 71.Ye Y, Godzik A. Multiple flexible structure alignment using partial order graphs. Bioinformatics. 2005;21:2362–2369. doi: 10.1093/bioinformatics/bti353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.