Abstract
Misincorporation of genomic uracil and formation of DNA double strand breaks (DSBs) are known consequences of exposure to TS inhibitors such as pemetrexed. Uracil DNA glycosylase (UNG) catalyzes the excision of uracil from DNA and initiates DNA base excision repair (BER). To better define the relationship between UNG activity and pemetrexed anticancer activity, we have investigated DNA damage, DSB formation, DSB repair capacity, and replication fork stability in UNG+/+ and UNG−/− cells. We report that despite identical growth rates and DSB repair capacities, UNG−/− cells accumulated significantly greater uracil and DSBs compared with UNG+/+ cells when exposed to pemetrexed. ChIP-seq analysis of γ-H2AX enrichment confirmed fewer DSBs in UNG+/+ cells. Furthermore, DSBs in UNG+/+ and UNG−/− cells occur at distinct genomic loci, supporting differential mechanisms of DSB formation in UNG-competent and UNG-deficient cells. UNG−/− cells also showed increased evidence of replication fork instability (PCNA dispersal) when exposed to pemetrexed. Thymidine co-treatment rescues S-phase arrest in both UNG+/+ and UNG−/− cells treated with IC50-level pemetrexed. However, following pemetrexed exposure, UNG−/− but not UNG+/+ cells are refractory to thymidine rescue, suggesting that deficient uracil excision rather than dTTP depletion is the barrier to cell cycle progression in UNG−/− cells. Based on these findings we propose that pemetrexed-induced uracil misincorporation is genotoxic, contributing to replication fork instability, DSB formation and ultimately cell death.
Keywords: base excision repair, uracil DNA glycosylase, double strand breaks, pemetrexed, uracil, thymineless death
Pemetrexed is a multi-target antifolate that inhibits several folate-dependent enzymes, leading to a global reduction in nucleotide synthesis and cell death.1 In addition to reduced nucleotide biosynthesis, inhibition of thymidylate synthase (TS) – the major target for pemetrexed activity – directly limits the methyl reduction of deoxyuridine monophosphate (dUMP) to form deoxythymidine monophosphate (dTMP).2, 3, 4 The resulting imbalance in deoxyuridine and deoxythimidine nucleotide pools favors the utilization of dUTP in DNA replication and the accumulation of uracil in DNA.2
Uracil DNA glycosylase (UNG) is the major glycosylase for the removal of uracil from DNA and the initiation of base excision repair (BER).2, 5, 6, 7, 8, 9 As a mechanism of TS-inhibitor-induced cell death (or so called thymine-less death), it has been hypothesized that futile cycles of uracil excision by UNG, BER pathway repair activity, and dUTP re-insertion contribute to the fragmentation of genomic DNA.4, 10 Though uracil accumulation and UNG excision activity have been observed in cells treated with TS-inhibitors,11 little evidence supports futile cycles of BER as effectors of thymine-less death. If TS-inhibitor-induced genomic instability were reliant upon UNG excision of uracil, one would anticipate increased TS-inhibitor sensitivity when UNG expression is elevated. However, pemetrexed-resistant subtypes of human cancer have statistically higher UNG expression than pemetrexed sensitive subtypes.12 Additionally, overexpression of UNG does not sensitize cells to raltitrexed or plevitrexed, a similar agent.13
Recently, we have reported that UNG is predictive of pemetrexed response in experimental human lung cancer models and that loss of UNG expression in human cancer sensitizes cells to pemetrexed.12 These data imply that pemetrexed-induced genomic instability is mediated by accumulation rather than removal of uracil. The objective of the present study was to further investigate the pemetrexed hypersensitivity and DNA double strand break (DSB) formation observed in UNG−/− cells in order to better understand the consequences of deficient uracil excision during pemetrexed exposure.
Herein, we show that despite equivalent rates of proliferation, DLD1 human colon cancer cells with stable knockout of UNG (UNG−/−) are hypersensitive to pemetrexed-induced DNA DSB formation and cell death. Using γ-H2AX chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) we observed significantly greater number of γ-H2AX-associated genomic loci (DSB) in pemetrexed UNG−/− cells compared with UNG+/+ cells. Additionally, ChIP-Seq data reveal distinct patterns of DSB susceptible loci in UNG+/+ and UNG−/− cells, suggesting disparate mechanisms of DSB formation in UNG-proficient and -deficient cells. Using the complimentary approaches of CldU/IdU DNA labeling and PCNA dispersal experiments, we show evidence of replication fork instability in pemetrexed-treated cells. UNG+/+ and UNG−/− have similar basal level DSB repair protein expression and DSB repair capacities, yet UNG−/− cells show delayed recovery from pemetrexed-induced DNA damage, including DSBs. Based on these findings we propose that the enhanced pemetrexed cytotoxicity in UNG−/− cells is indicative of the genotoxicity of misincorporated uracil, which persists at sufficient quantities to stall DNA replication triggering cell death.
Results
Loss of UNG expression hyper-sensitizes human cancer cells to pemetrexed
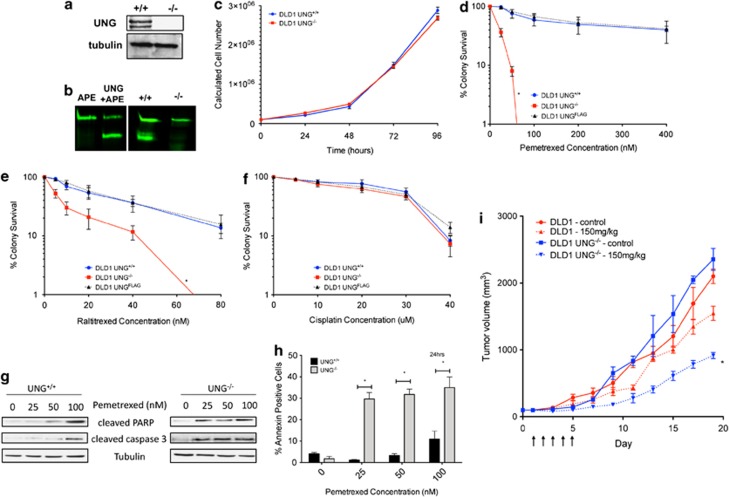
Previously, we showed that UNG expression predicts sensitivity to pemetrexed in experimental models of human lung cancer.12 As we were interested in the impact of UNG expression on pemetrexed response in human cancer cells, we evaluated sensitivity to pemetrexed in UNG+/+ and UNG−/− colon cancer cells. These UNG−/− cells were engineered as described14 and lack expression of both nuclear and mitochondrial UNG isoforms (Figure 1a). Loss of expression of UNG abolished in vitro cutting activity in UNG/APE oligonucleotide cutting assay (Figure 1b), suggesting that in the absence of UNG, uracil is not efficiently removed from DNA.
Figure 1.
Loss of UNG enhances pemetrexed sensitivity in DLD1 human colon cancer cells. (a) Western blot of UNG nuclear (top band, 39 kDa) and mitochondrial (bottom band, ∼36 kDa) in UNG+/+ and UNG−/− DLD1 human colon cancer cells. (b) UNG cutting activity assay using either purified enzyme (APE and UNG+APE lanes, 1 U of each) or 2.5 μg of whole-cell extract from UNG+/+ and UNG−/− cells. (c) Proliferation rates were determined by cell counting with trypan blue exclusion of non-viable cells over a period of 0–96 h using a hemocytometer. Colony survival assays for UNG+/+ and UNG−/− cells treated with pemetrexed (d), raltitrexed (e) and cisplatin (f) for 10-days. UNG−/− cells are 10-fold more sensitive to pemetrexed, P<0.0001 and 4.3-fold more sensitive to raltitrexed, P<0.0001. Sensitivity to cisplatin was not significantly different for the two cell lines, P=0.17. UNGFLAG cells are UNG−/− cells that were transfected with a FLAG-tagged UNG vector to induce re-expression of UNG. UNGFLAG cells showed resistance to pemetrexed and raltitrexed similar to UNG+/+ cells (g) Western blot of apoptotic markers cleaved PARP and cleaved caspase 3 in UNG+/+ and UNG−/− cells treated for 24 h with 0–100 nM pemetrexed. (h) Percentage of Annexin V-positive pemetrexed-treated UNG+/+ and UNG−/− cells after 24-h exposure to 0–100 nM pemetrexed. (i) Tumor volumes of xenograft tumors grown in NODSCID mice that were treated for 5 consecutive days with pemetrexed (150 mg/kg, intraperitoneal injection)
UNG expression is coordinated with DNA replication.7, 8, 15, 16, 17 This relationship prompted us to evaluate the impact of UNG loss on cellular proliferation rates for UNG+/+ and UNG−/− cells (Figure 1c). An identical number of cells (1 × 105) were seeded in 60-mm culture dishes and incubated for 24–96 h. Following incubation, cells were stained with trypan blue and the total number of viable cells per sample was determined using a hemocytometer. This simple experiment yielded identical proliferation rates for UNG+/+ and UNG−/− cells (P=0.292; Figure 1c). Clonogenic survival assays revealed that UNG−/− cells were 10-fold more sensitive to pemetrexed (IC50 UNG+/+=205.8 nM and IC50 UNG−/−=22.0 nM, P<0.0001, Figure 1d). UNG−/− cells displayed cross-sensitivity to the TS inhibitor, raltitrexed (P<0.0001; Figure 1e), but not the cross-linking agent, cisplatin (P=0.17; Figure 1f). The impact of pemetrexed on cell killing was further analyzed by cleaved PARP measurements using western blot (Figure 1g) and Annexin V measurements using flow cytometry (Figure 1h) at 24-hours in cell treated with 0–100 nM pemetrexed. Results indicated that UNG−/− cells were significantly more sensitive to pemetrexed-induced apoptosis compared with UNG+/+ cells. Re-expression of UNG (UNGFLAG) rescued pemetrexed and raltitrexed sensitivity in UNG−/− cells (Figures 1d–f), confirming that TS-inhibitor sensitivity in UNG−/− cells is due to UNG loss.
To evaluate the impact of UNG loss on tumor cell growth in vivo, UNG+/+ and UNG−/− tumors were xenografted subcutaneously into NODSCID mice (Figure 1i). Treatment with five daily consecutive intraperitoneal (IP) injections of pemetrexed (150 mg/kg) resulted in tumor quadrupling time of 7.83±0.88 days in UNG+/+ tumors compared with 12.54±0.42 days in UNG−/− tumors, P<0.001. At day 20 pemetrexed-treated UNG−/− tumors were 61% smaller than untreated UNG−/− tumors while UNG+/+ tumors were only 26% smaller with pemetrexed treatment compared with untreated controls (Figure 1i). Together these data indicate a significant increase in the antitumor effect of pemetrexed in UNG-deficient tumor cells in vitro and in vivo.
Increased DNA DSB formation in UNG−/− cells during pemetrexed exposure
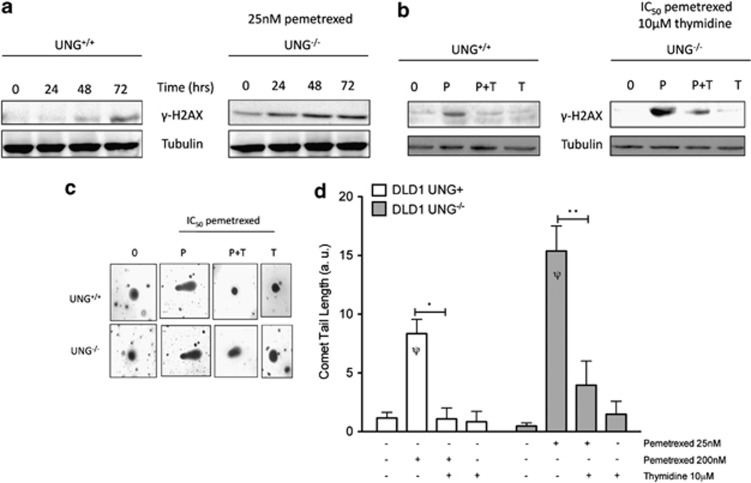
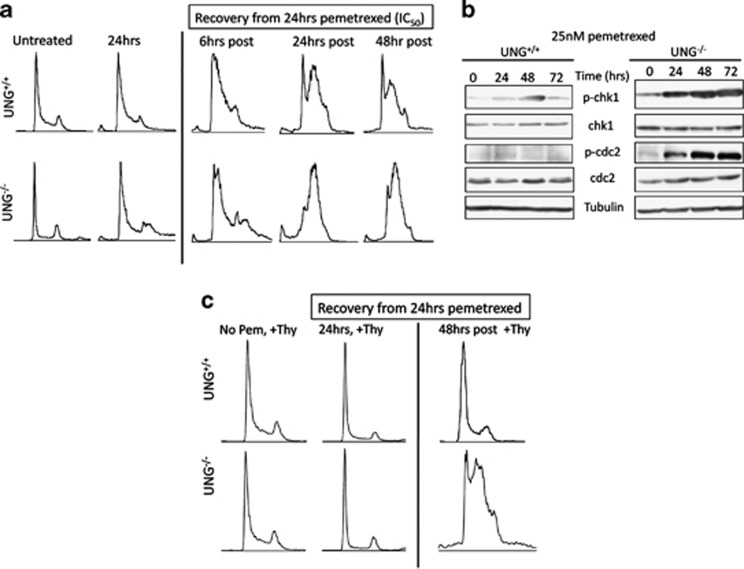
The formation of DNA DSBs is a long observed consequence of TS inhibitor exposure and thymine-less death,10, 18, 19 yet the mechanism of DSB formation has remained ill defined. It has been hypothesized that futile cycles of uracil excision, BER and dUTP reinsertion contribute to DNA fragmentation and DSB formation in cells treated with TS-inhibitors.4, 10 However, this hypothesis lacks experimental support13 and fails to explain the pemetrexed hypersensitivity observed in UNG−/− cells (Figure 1). We therefore evaluated DNA DSB formation in our isogenic UNG+/+ and UNG−/− cells to better understand the role of UNG in mediating DSB formation and thymine-less death during pemetrexed exposure. Western blots for the DNA DSB surrogate marker, γ-H2AX, show significant induction in UNG−/− cells compared with UNG+/+ cells (Figure 2a). Importantly, even when the two cell lines are treated with equally cytotoxic concentrations of pemetrexed (IC50, UNG−/− 25 nM and UNG+/+ 200 nM), induction of γ-H2AX protein expression appears greater in UNG−/− cells (Figure 2b, ‘P' lanes). As a complimentary experiment, we measured DNA DSBs using neutral single-cell gel electrophoreses (comet assay). Pemetrexed treatment increased comet tail lengths in UNG−/− cells by 1.85-fold compared with UNG+/+ cells, P<0.05 (Figures 2c and d). By providing a salvage pathway for dTTP synthesis, supplemental thymidine should limit the effects of TS inhibition. Thymidine supplementation rescued IC50 pemetrexed-induced γ-H2AX induction (Figure 2b) and comet tail formation (Figure 2c) in both DLD1 UNG−/− and UNG+/+ cells, suggesting that in both cell lines dUTP/dTTP nucleotide pool ratio drives DNA DSB formation. Of note, the increased formation of DNA DSBs in UNG−/− cells occurs despite equivalent expression of DNA DSB repair proteins and DSB repair capacities in UNG+/+ and UNG−/− cells (Figure 3).
Figure 2.
Increased DNA DSB formation in UNG−/− cells treated with pemetrexed. (a) Western blot for γ-H2AX was performed using whole-cell extracts from UNG+/+ and UNG−/− cells treated with 25 nM pemetrexed for 0–72 h to compare sensitivity of the two cell lines to pemetrexed-induced DSB formation. (b) Western blot for γ-H2AX was performed using whole-cell extracts from UNG+/+ and UNG−/− cells treated with IC50-level pemetrexed alone or in combination with 10 μM supplemental thymidine for 24 h. UNG+/+ cells were treated with 200 nM pemetrexed and UNG−/− cells were treated with 25 nM pemetrexed. (c and d) Single-cell gel electrophoresis (comet) assay was performed under neutral conditions for UNG+/+ and UNG−/− cells treated with IC50-level pemetrexed alone or in combination with supplemental thymidine. At least 50 comet tail lengths were measured using NIH Image J software from two independent experiments for a total of ≥100 cells per drug treatment. Representative images are shown in (c) and the mean and S.E.M. for this experiment is plotted in (d). Pemetrexed treatment caused 1.85-fold increase in comet tail length in UNG−/− compared with UNG+/+ cells, Ψ and P<0.05. Thymidine rescued sensitivity to pemetrexed in UNG+/+ (*P<0.001) and UNG−/− (**P<0.004) cells
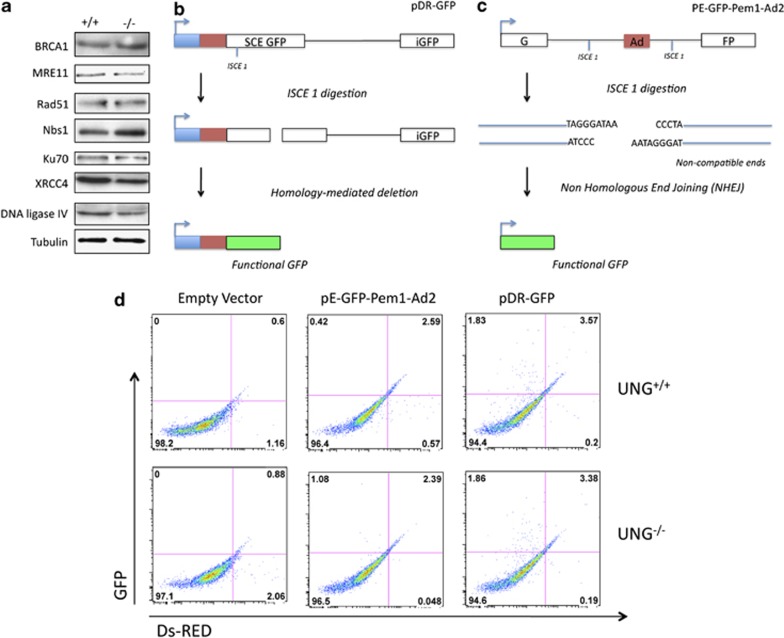
Figure 3.
Comparison of DNA DSB repair capacity in UNG+/+ and UNG−/− cells. (a) Western blots comparing basal expression of proteins involved in DNA DSB repair in UNG+/+ and UNG−/− cells. (b) Schematic for HR repair assay. Briefly, transient transfection of ISCE1 linearized pDR-GFP was used to measure homologous recombination efficiency. pDR-GFP contains two tandem but inactive GFP repeats, one of which contains an ISCE restriction enzyme cutting site. GFP fluorescence is activated by homology-mediated deletion between the two repeats during functional HR to generate a functional GFP. (c) Schematic for NHEJ repair assay. Briefly, pE-GFP-Pem1-Ad2 has a non-functional GFP interrupted by Pem 1 intron into which there is inserted an adenovirus exon sequenced flanked by ISCE1 restriction enzyme cleavage sites. ISCE1 plasmid linearization generates non-complimentary ended DSB and removes the AD2 sequence. NHEJ allows juxtaposition of the two GFP fragments to reconstitute fluorescence. (d) Representative flow cytometry dot blots from untreated UNG+/+ and UNG−/− cells transfected with an empty vector, ISCE1 linearized pDR-GFP or ISCE1 linearized pE-GFP-Pem1-Ad2. pdSRed was co-transfected in each instance as a transfection efficiency control. Similar percentages of GFP+/dSRed+ cells were observed for UNG+/+ and UNG−/− cells for each transfection suggesting similar capacity for BER
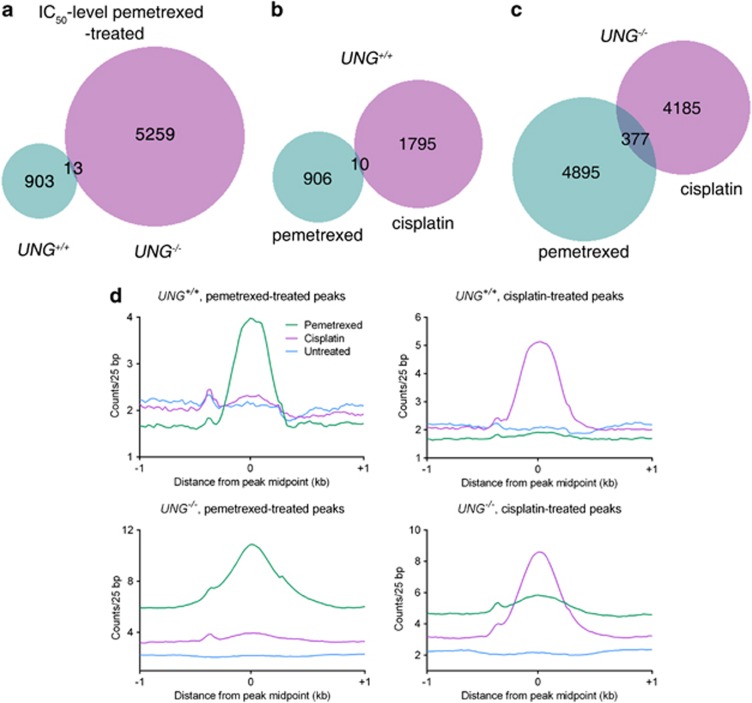
To further compare the mechanism of DSB formation and thus cell death in UNG+/+ and UNG−/− cells, we performed γ-H2AX ChIP-seq to compare the genomic distributions of DSBs in pemetrexed-treated UNG+/+ and UNG−/− cells. Pemetrexed treatment resulted in a 5.75-fold increase in distinct γ-H2AX-bound genomic regions in UNG−/− cells compared with UNG+/+ cells. Moreover, of the γ-H2AX-enriched sequences identified in UNG−/− cells, only 0.0025% of them overlapped with γ-H2AX bound sites in pemetrexed-treated UNG+/+ cells (Figure 4a). Aggregate plots of γ-H2AX peaks confirm that enrichment is significantly elevated in treated compared with untreated samples and reaffirm a more robust enrichment of γ-H2AX in UNG−/− cells (Figure 4d). When a detailed analysis of genomic features of γ-H2AX-enriched sequences was performed, DSBs were observed to map more frequently to transcription start sites (TSS), CpG islands, DNAse-hypersenstive clusters and putative origins of replication in pemetrexed-treated UNG−/− cells compared with UNG+/+ cells (Table 1). In all, our data indicate that UNG−/− cells accumulate significantly greater number of DSBs in response to pemetrexed and differential chromosomal distribution of γ-H2AX enrichment in the genome suggests disparate mechanism of DSB formation in UNG-proficient and -deficient cells.
Figure 4.
γ-H2AX ChIP-seq in UNG+/+ and UNG−/− cells. Cells were treated with IC50-level pemetrexed (UNG+/+, 200 nM and UNG−/−, 25 nM) or with 20 μM cisplatin for 24 h and subsequently processed for ChIP-seq as described in the Materials and Methods section. Cisplatin was utilized as a control DNA-damaging agent that forms DNA double-strand breaks. (a–c) Venn diagrams of the ChIP-seq peaks generated comparing signal overlap for pemetrexed treated UNG+/+ and UNG−/− cells (a); pemetrexed treated and cisplatin treated UNG+/+ cells (b) and pemetrexed-treated and cisplatin-treated UNG−/− cells (c). (d) Midpoint coordinates for each γ-H2AX peak were used to obtained signals ±1 kb for the coordinate (within a 25-bp window). The average signal in each window for each peak shown for UNG+/+ pemetrexed treated (top left); UNG+/+ cisplatin-treated (top right); UNG−/− pemetrexed-treated (bottom left) and UNG−/− cisplatin-treated cells (bottom right)
Table 1. Characteristics of γ-H2AX-enriched sequences.
| DNA feature analyzed for overlap | WT (%) | KO (%) |
|---|---|---|
| Encode clustered TFBS | 35.1 | 55.8* |
| DNAse clusters | 45.5 | 62.8* |
| CpG islands | 4.5 | 11.4* |
| Lamin-associated domains | 33.9 | 1.4* |
| Caco-2 CTCF peaks | 6.3 | 11.0* |
| HCT116 methyl sequencing peaks | 5.1 | 23.1* |
| K562 and MCF7 identified origins of replication | 12.7 | 36.7* |
*P<0.001
Delayed recovery from S-phase arrest in pemetrexed-treated UNG−/− cells
Pemetrexed-treated UNG−/− cells accumulate 40-fold more genomic uracil compared with UNG+/+ cells.20 Additionally, there are data to suggest that heavily uracilated DNA alters DNA–protein interactions due to perturbations in basal genomic methylation, which may negatively impact replication and transcription.21, 22 Putative origins of replication in human cancer cells were identified as common sites pooled from published nascent DNA sequencing of K562 and MCF7 cells.23, 24 In ChIP-seq experiments, we observed more significant association of γ-H2AX enrichment with putative origins of replication (Table 1). We therefore surmised a mechanistic link between UNG loss, DNA DSB formation and replication instability.
Both UNG+/+ and UNG−/− cells accumulate in S-phase in response to 24-h exposure to IC50-level pemetrexed (Figure 5a). Phosphorylation of chk1 (ser345) and cdc2 (tyr15) also suggested S-phase arrest (Figure 5b). Interestingly, UNG−/− but not UNG+/+ cells show delayed recovery from S-phase arrest when pemetrexed is withdrawn, compared with UNG+/+ cells (Figure 5a). Expression of replication-activated checkpoint proteins and arrest in S phase with delayed recovery suggest pemetrexed-induced replication instability. Supplemental thymidine co-treatment blocked S-phase accumulation in pemetrexed-treated UNG+/+ and UNG−/− cells (Figure 5c), suggesting S-phase arrest is related to dUTP/dTTP pool imbalance. However, culturing cells in medium containing supplemental thymidine after pemetrexed exposure accelerates recovery from S-phase arrest in UNG+/+ cells but not UNG−/− cells (Figure 5d). The inability of thymidine to restore cell cycle progression in UNG−/− cells after pemetrexed is withdrawn suggests that DNA damage caused by misincorporated dUTP is irreversible in the context of deficient uracil removal.
Figure 5.
Delayed recovery from cell cycle arrest in pemetrexed-treated UNG−/− cells. (a) Cell cycle progression was monitored using PI staining and flow cytometry. UNG+/+ and UNG−/− cells treated with IC50 level pemetrexed (UNG+/+, 200 nM; UNG−/− 25 nM) both show evidence of S-phase accumulation after 25 h of exposure. When allowed to recover in drug-free media, UNG+/+ cells are capable of resuming cell cycle (return of G1 peak in +48 h sample) while UNG−/− cells are not. (b) Western blot from UNG+/+ and UNG−/− cells treated for 0–72 h with 25 nM pemetrexed showing increased sensitivity in UNG−/− cells to pemetrexed-induced phosphorylation of chk1 and cdc2, S-phase checkpoint kinases. (c) Cell cycle histograms for cells treated with supplemental thymidine. Cells were treated with 10 μM thymidine for 24 h (No Pem, +Thy), co-treated with IC50 level pemetrexed and thymidine for 24 h (24 h, +Thy) or treated with IC50 level pemetrexed for 24 h and allowed to recover for 48 h in media containing 10 μM thymidine (48 h post, +Thy)
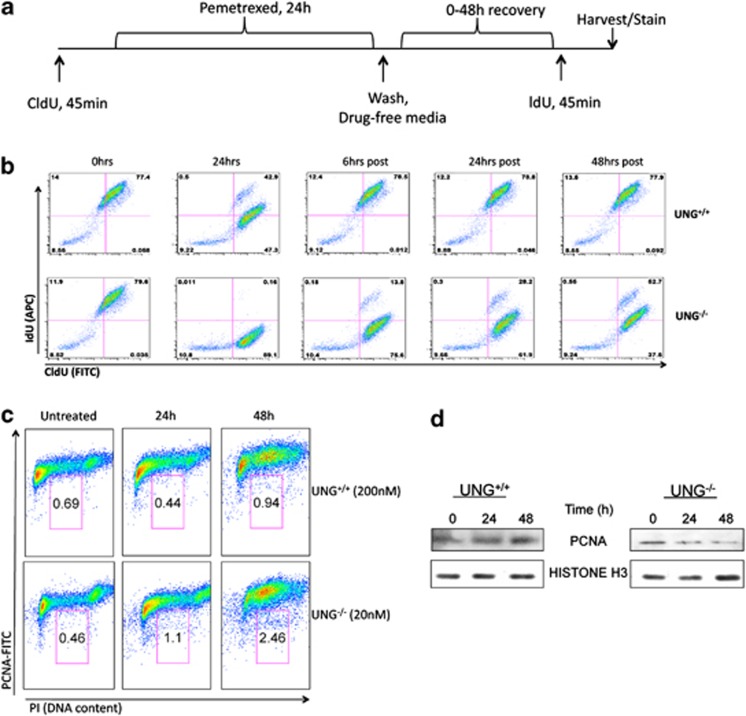
The association of H2AX phosphorylation, S-phase arrest and replication fork collapse has previously been studied in cells treated with ionizing radiation, nucleoside analogs and antimetabolites,25, 26, 27 suggesting that H2AX is phosphorylated during inhibition of DNA synthesis. We therefore evaluated the stability of DNA replication fork in UNG−/− cells. Intracellular replication fork progression is typically studied using nucleotide incorporation assays (Figure 6a). Using this approach in a flow cytometry-based pulse chase assay, we observed a decrease in post-pemetrexed treatment IdU incorporation in UNG−/− cells (Figure 6b) While this is consistent with decreased replication fork progression in pemetrexed-treated UNG−/− cells, these data alone could simply indicate that misincorporated uracil impedes IdU incorporation because they compete for nucleotide insertion sites. Therefore, as a complimentary experiment, we assessed the dispersal of the replication processivity factor, PCNA, from chromatin using both flow cytometry (Figure 6c) and western blot (Figure 6d). Similar experiments have been used to analyze collapsing replication forks in etoposide and hydroxyurea-treated cells.28, 29 In these assays, decreased PCNA staining of S-phase-gated cells or chromatin is indicative of replication fork instability or fork collapse. Our data show increased PCNA dispersal in pemetrexed-treated UNG−/− cells compared with UNG+/+ cells, suggesting that pemetrexed causes replication fork instability in UNG-deficient cells (Figures 6c and d).
Figure 6.
Intracellular replication fork instability in UNG−/− cells. (a) Schematic for CldU and IdU pulse labeling of pemetrexed-treated cells. Briefly, cells were incubated for 45 min with CldU (50 μM) prior to treatment with IC50-level pemetrexed (UNG+/+, 200 nM, UNG−/− 25 nM) for 24 h. Cells were then allowed to recover in drug-free media for 0–48 h prior to incubation for 45 min with IdU (50 μM). Cells were then stained with fluorescent antibodies that recognize CldU (rat-antiBrdU-FITC) and IdU (mouse-antiBrdU-APC). Representative data from flow cytometry sorting of CldU/IdU-labeled cells is shown in (b) where the x axis is CldU (FITC) and the y axis is IdU (APC). (c) Cells were treated with IC50-level pemetrexed for 24 and 48 h and subsequently stained with PI (x axis) to and FITC-labeled PCNA antibody (y axis). (d) Cells were treated as in (c) and subsequently incubated in 1% formaldehyde for crosslinking and chromatin extraction for western blots of chromatin-bound PCNA and histone H3 (loading control)
Discussion
We have previously shown that loss of UNG expression sensitizes human cancer cells to pemetrexed and that UNG expression predicts pemetrexed sensitivity in experimental models of human cancer.12, 20 Here, we analyzed the DNA damage response to pemetrexed in UNG+/+ and UNG−/− DLD1 human colon cancer cells to better understand the role of UNG in the mechanism of pemetrexed-induced DNA DSB formation and cell death. We observed that loss of UNG hyper-sensitized DLD1 cells to pemetrexed in vitro and in vivo despite equivalent proliferation rates. This hypersensitivity is associated with increased replication fork instability and DNA DSB formation, despite an equivalent capacity for DNA DSB repair in the two cells.
The formation of DNA DSBs in cells treated with TS inhibitors has been studied for decades,10, 30, 31, 32, 33, 34, 35 yet the precise role of uracil misincorporation and UNG excision of uracil in the mechanism of DNA DSB formation and cell death is not clear. The dominant futile cycle hypothesis proposes that DSBs arise as a result of continuous cycles of uracil excision, BER and uracil re-insertion. Experimental evidence from other labs13, 32, 36 and the data presented herein indicate that futile cycles of BER do not adequately explain thymine-less death. Overexpression of UNG, which should exacerbate futile cycles of UNG, does not enhance TS-inhibitor sensitivity.13 UNG loss does not cause compensatory upregulation of the other DNA glycosylases capable of uracil excision,12 so it is unlikely that futile cycles of BER are initiated in UNG−/− cells treated with pemetrexed. The relatively fewer DNA DSBs observed in UNG+/+ cells compared with UNG−/− cells treated with equally toxic concentrations of pemetrexed implies that, at least in the models examined, UNG activity limits rather than promotes pemetrexed-mediated DNA DSB formation and cell death. Based on these data, we bring forth a novel hypothesis for the mechanism of thymine-less death in UNG-deficient cancer cells. In our model, uracil accumulates at a critical level near replication origins, stalling DNA replication fork progression and leading to fork collapse, DNA DSB formation and cell death (Table 2).
Table 2. Possible pathways to DSB formation and cell death in pemetrexed-treated cells.
| Possible route to cell death | Problem with hypothesis |
|---|---|
| UNG+/+ cells | |
| Stalled replication due to nucleotide pool aberrations caused by reduced dTTP | Does not explain differential sensitivity in UNG+/+ and UNG−/− cells. |
| Endonuclease cleavage of uracil and/or AP sites | No endonucleases with uracil activity have been studied in pemetrexed treated cells. Products of APE incision are rapidly targeted by activities of Polβ and downstream BER. |
| Futile cycles of BER | UNG+/+ cells are more resistant than UNG−/− cells, suggesting excision of uracil protects from pemetrexed cytotoxicity. |
| UNG−/− cells | |
| Uracil-mediated replication fork stall and collapse | Currently there is no available method to directly interrogate various degrees of uracil base substitution within living cells. |
| Uracil-mediated endonuclease cleavage | No endonucleases with uracil activity have been studied in this context. |
Several levels of experimental evidence support this model. First, we have observed substantial accumulation of genomic uracil,20 coordinated with replication fork instability and DNA DSB formation in UNG−/− cells. Otherwise isogenic UNG+/+ cells are largely protected from these effects, suggesting uracil is intrinsically cytotoxic. The altered methylation status and secondary structure of heavily uracilated DNA may explain this toxicity.21, 22, 37 An alternative explanation for uracil-mediated DSB formation in UNG−/− cells is increased endonuclease cleavage in uracil-containing DNA,38, 39 yet the activities of endonucleases with this activity have not been assessed in pemetrexed-treated cells.
Secondly, we have noted persistence of S-phase arrest in pemetrexed-treated UNG−/− cells with concomitant activation of intra-S phase checkpoint proteins. UNG+/+ cells still undergo S-phase arrest and cell death, albeit at significantly higher pemetrexed concentrations than UNG−/− cells. While genomic uracil does not accumulate20 in UNG competent cells, dTTP levels are still aberrantly low during TS-inhibitor exposure. Low dTTP may limit allosteric regulation of ribonucleotide reductase, resulting in elevated dATP and decreased dGTP nucleotide pools.40 In UNG+/+ cells, S-phase arrest and cell death in response to pemetrexed may be a consequence of global nucleotide pool imbalance.
Thymidine salvage prevents S-phase arrest but has limited capacity to reverse it in pemetrexed-treated UNG−/− cells. Correction of dUTP/dTTP ratios is therefore ineffective after uracil has been misincorporated in cells with reduced uracil excision activity. Literature suggests that replication fork reversal is limited during thymine starvation, while fork reversal readily occurs during dNTP depletion or ribonucleotide reductase inactivation.41 Lack of replication fork reversal during thymine deprivation may explain the failure to recover from cell cycle arrest in UNG−/− cells.
Lastly, distinct patterns of genomic γ-H2AX enrichment (DSBs) identified in this study suggest disparate mechanisms of DSB formation in UNG-competent and -deficient cells. In UNG−/− cells, enrichment of γ-H2AX occurred more frequently in accessible chromatin; TSS, DNAse-hypersensitivity regions, and origins of replication tend to be nucleosome-depleted and more readily accessible to proteins. This feature is consistent with the negative impact uracilated DNA has on DNA–protein interactions.21, 22 Of interest, our analysis indicates that 36.7% of UNG−/− and 12.67% of UNG+/+ γ-H2AX enriched sites overlap with putative human origins of replication,23, 24 suggesting that UNG excision of uracil protects origins of replication from pemetrexed-induced DSB formation. It has been suggested that continued replication in thymine-starved cells triggers destabilization of origins of replication resulting in DSBs at sites of origin firing.32, 36 The fact that UNG-deficient cells appear to be more sensitive to this mode of DSB formation is consistent with prior observations of coordination of UNG-initiated BER and the observation of UNG complexed at replication foci.6, 7, 8, 17
ChIP-seq of γ-H2AX to localize DSBs has been previously performed.42 It has been suggested that only a small fraction of γ-H2AX foci represent actual DSBs.43, 44 Given this caveat, we have offered comet assay as corroborating evidence of DSB formation. Additionally, over-expression of UNG and H2AX in cancer and co-expression of these genes in pemetrexed-resistant cancers (Supplementary Figures 1 and 2)45, 46, 47 reflect the association of UNG and H2AX with DNA replication. The deposition of endogenous unphosphorylated H2AX at replication origins, sub-telomeres, and TFS coincides with sites of endogenous DNA damage and γ-H2AX enrichment.42 We took steps in our experimental design to confirm that the γ-H2AX enrichment observed was not due to altered replication rates or endogenous DNA damage. DSBs were not increased in untreated UNG−/− cells, which had identical proliferation rate as UNG+/+ cells. Moreover, <1% overlap between UNG+/+ and UNG−/− γ-H2AX-enriched sites suggests different mechanisms of direct DSB formation in the two cell lines.
Combined, the observation of fewer DSBs, reduced predilection of DSBs for origins of replication, lack of observed replication fork instability, and S-phase arrest that is adequately rescued with supplemental thymidine in UNG+/+ cells suggests pemetrexed-induced toxicity is more related to dTTP depletion than to dUTP misincorporation. In contrast, UNG−/− cells, which accumulate genomic uracil,20 have increased DSBs that localize to replication origins, show a significant degree of replication instability and have irreversible S-phase arrest; pemetrexed toxicity is more related to dUTP misincorporation (Table 2).
Together these findings highlight the importance of genomic uracil accumulation in mediating cellular response to pemetrexed. Significant enhancement of pemetrexed cell killing in UNG−/− cells suggests that targeting UNG may have therapeutic impact in human cancer. Additionally, our data suggest in pemetrexed-treated cancer cells, cell death is mediated by direct toxicity of uracil accumulation and by nucleotide perturbations induced by dTTP depletion, with genotoxic uracil accumulation becoming the dominant mode of cell killing as UNG activity declines. Apart from our studies using pemetrexed, studies of changes in UNG expression have reported little impact on cellular sensitivity to TS-inhibitor-induced cell killing.13, 48 In contrast, one study has reported diminished DNA replication rates and increased apoptosis in UNG-deficient cells even in the absence of drug treatment.49 We speculate that variations in sensitivity among previously studied UNG deficient cells may be explained by differences in induced genomic uracil accumulation in the TS/inhibitor models investigated. Indeed, the sensitization index for UNG−/− cells to raltitrexed was far smaller than the sensitization index for UNG−/− cell to pemetrexed (Figure 1), suggesting that UNG expression and activity has a greater role in pemetrexed versus raltitrexed response. To address this, we propose future quantitative studies that compare genomic uracil and nucleotide pool levels in UNG−/− and UNG+/+ cells treated with equally toxic concentrations of various TS inhibitors.
Materials and Methods
Cell lines and reagents
Pemetrexed was purchased from LC Laboratories (Woburn, MA, USA). Thymidine, raltitrexed and cisplatin were purchased from Sigma Aldrich (St. Louis, MO, USA). Cells were maintained in complete DMEM (10% FBS, 2 mM L-glutamine) at 37 °C in a 5% CO2 incubator. DLD1 cells were purchased from ATCC and expanded upon delivery into numerous vials of low passage cells for cryopreservation. Cell line characterization by ATCC is conducted through short tandem repeat (STR) typing. Re-authentication was not performed. DLD1 UNG−/− cells14 were a gift from Sanford Markowitz.
Western blots
Protein extracts (30 μg) were resolved by SDS-PAGE, transferred to PVDF membrane (Millipore, Billerica, MA, USA) and detected by incubations in primary antibodies followed by HRP-linked secondary antibodies at manufacturer's specifications. Chromatin-bound proteins were extracted from formaldehyde cross-linked (1%) cells using the Pierce Chromatin Prep module (Thermo Pierce, Rockford, IL, USA). Antibody sources: UNG-23936 (39 kDa band, nuclear UNG), Nbs1, DNA ligase IV, Ku70, BRCA1, MRE11, and PCNA (Abcam, Cambridge, MA, USA); Tubulin (Calbiochem, via Millipore, Billerica, MA, USA); γ-H2AX, cleaved caspase 3, and Histone-H3 (Millipore); cleaved PARP, XRCC4, and secondary antibodies (Cell Signaling, Danvers, MA, USA); and Rad51, p-chk1, chk1, p-cdc2, cdc2 (Santa Cruz, Dallas, TX, USA).
UNG activity assay
UNG activity was measured using 40-mer oligodeoxynucleotide duplex containing a single U:A mispair as described.12 Products were resolved by denaturing polyacrylamide gels (20%), prior to visualization with a Typhoon 9200 fluorescence imager (Amersham Bioscience via GE Healthcare, Pittsburgh, PA, USA) and UNG activity was calculated (percentage of cut band relative to total oligonucleotide).
Determination of cell proliferation rate/double time
Cell counting with trypan blue exclusion determined cellular proliferation rates. Briefly, 1 × 105 cells were seeded in 60-mm cell culture dishes and allowed to grow in typical cell culture medium for 24–96 h. At the time of the harvest, cells were stained with trypan blue, counted using a hemocytometer and the number of viable cells was recorded to generate a growth curve.
Colony survival
Colony survival was assessed by methylene blue staining of colonies formed after 10-day exposure to the drugs indicated. Only colonies containing ≥50 cells were counted.
Annexin V staining
Cells were stained with Annexin-V FITC and propidium iodide (PI) for evaluation of apoptosis by flow cytometry according to the manufacturer's protocol (BD Pharmingen, San Diego, CA, USA). At the time of harvest, cells were stained with 5 μl of Annexin V FITC reagent and 10 μl of PI (5 μg/ml) in 1 × binding buffer (10 mM HEPES, 140 mM NaOH, 2.5 mM CaCl2, pH 7.4) for 30 min in the dark. Apoptotic cells were determined using a Coulter flow cytometer (EPICS-XL-MCL). Cell death calculations are the sum of early apoptotic (Annexin V positive, PI-negative) and late (Annexin V-positive, PI-positive) cells.
Cell cycle and PCNA dispersal assays
PI staining of methanol-fixed cells was used for cell cycle determinations. For PCNA dispersal assays, FITC-labeled PCNA antibody (PCNA-FITC, Abcam) was added for PCNA detection. Both uni-parameter (PI-only) and dual-parameter (PI+PCNA-FITC) analyses were performed on a Beckman Coulter flow cytometer (Brea, CA, USA) (EPICS-XL-MCL). Cell cycle histograms (PI) and PCNA dot plots (PCNA-FITC) were deconvoluted from ≥20 000 events using Flow Jo software (TreeStar, INC, Ashland, OR, USA).
Tumor growth assays in xenograft mice
Tumor cells (5 × 106) in early passage were injected into bilateral flanks of female NODSCID mice (∼5 weeks old). When tumor volumes reached 100 mm3, mice were divided into control and treatment cohorts (n=6). Mice bearing tumors were treated with pemetrexed (150 mg/kg) or 100 μl sterile PBS (control) by daily IP injection for 5 consecutive days. Tumor measurements were taken every 2 days and the response was quantified by calculating the tumor volume.
Comet assay under neutral conditions
Cells were processed for comet tail formation under neutral comet assay conditions according to the manufacturer's protocol for cell lysis and single-cell gel electrophoresis (Trevigen, Gaithersburg, MD, USA). Tail lengths were recorded for at least 50 comets on two separate slides (∼100 cells per treatment) using ImageJ software.
CldU/IdU pulse labeling and flow cytometry
Cells were incubated for 45 min in media containing 50 μM chlorodeoxyuridine (CldU) prior to drug treatments. Following treatment, cells were incubated for 45 min with iododeoxyuridine (IdU, 50 μM). For analysis, fixed cells were incubated in DNA denaturation solution (4 N HCl, 0.1% Triton X-100 in PBS) for 15 min, followed by neutralization washes with 0.1 M Na2B4O7 buffer. Cells were then incubated for 30 min at 37 °C in 1 mg/ml RNAseA in PBS. Each sample was labeled using rat anti-BrdU-FITC (for detecting CldU) and mouse anti-BrdU-APC (for detecting IdU) antibodies and counterstained with PI. Uptake of CldU and IdU was determined using a BD LSR II flow cytometer. Dot plots of cycling cells (cells in G1/S/G2M) were deconvoluted from univariate analysis of PI staining. Bivariate dot plots of CldU/IdU content were obtained for cycling cells using Flow Jo software.
DNA DSB repair assays
DNA DSB assays were performed as described.50 Briefly, cells were transiently transfected with either circular pDRGFP or ISCE1-linearized pDRGFR for homologous recombination (HR) assays or with circular pE_GFP-PEM1-Ad2 or ISCE1-linearized PE-GFP-PEM1-Ad2 for non-homologous end joining (NHEJ) assays. DNA repair plasmids were obtained from Elisa Tichy (University of Cincinnati) and plasmid stocks were expanded using bacterial transformation. Plasmid pDsRed express (Clontech, Mountainview, CA, USA) was co-transfected in each instance as a transfection efficiency marker. Transfection of 2.5 × 105 cells in six-well culture dishes was carried out using Opti-MEM media and oligofectamine according to standard laboratory procedures. In all, 5 μg of pDsRed Express N1 and 40 μg of construct DNA were utilized for transfection. Seventy two hours post transfection, cells were trypsinized and suspended in cold PBS containing 0.1% BSA for analysis by flow cytometry. Samples were analyzed using a BD LSRII (BD Biosciences, San Jose, CA, USA). Dot plots for DsRed and GFP were acquired for ≥20 000 events using Flow Jo software.
γ-H2AX ChIP sequencing
ChIP was performed as described,51 with 10 μg of rabbit anti γ-H2AX, p-S139 (Abcam, ab2893) per ChIP. Sequencing libraries were prepared as described,51 with the exception of the library amplification step. Instead of Phusion DNA polymerase, Expand High Fidelity Plus (Roche Diagnostics, Indianapolis, IN, USA) was used. The following parameters were used for thermal cycling: 5 min at 94 °C, 30 s at 65 °C and 60 s at 72 °C followed by a 7 min final elongation at 72 °C. Libraries were sequenced at the Case Western Reserve University Genomics Core Facility. The following unique read numbers were obtained: WT untreated γH2AX ChIP, 28 089 742; WT cisplatin-treated γH2AX ChIP, 22 505 752; WT pemetrexed-treated γH2AX ChIP 30 494 966; UNG−/− untreated γH2AX ChIP 36 823 368; UNG−/− cisplatin-treated γH2AX ChIP 38 155 972; UNG−/− pemetrexed-treated γH2AX ChIP 52 013 817. ChIP-seq data generated in this study were deposited into the Gene Expression Omnibus under accession number GSE50692. After removal of duplicate reads, ChIP-seq data were aligned to the hg18 human genome assembly with Bowtie,52 discarding reads with >1 reportable alignment. Peaks were called using MACS.53 For MACS analysis, the pemetrexed- or cisplatin-treated ChIP was used as the treatment file and the untreated ChIP was used as the control file. Fold enrichment cutoffs used were 4 for WT cells and 6 for UNG−/− cells. Peak coordinates are given in Supplementary Data 1.
To determine the overlap of γH2AX with putative replication origins, peaks were converted to the hg19 human genome assembly with liftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Lists of putative origins of replication determined by nascent DNA sequencing in K562 and MCF7 cells23, 24 were combined and regions <10 bp apart were collapsed into a single putative origin. Lists of peaks were intersected with the condensed list of origins with the GFF-overlap feature of the ChIP-seq Tool Set (http://havoc.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi), with peaks ≤200 bp apart considered overlapping. The UCSC table browser was used to overlap γH2AX peaks with various regulatory features. The proportions of γH2AX peaks overlapping putative origins and other regulatory features in pemetrexed- and cisplatin-treated WT and UNG−/− cells were compared by χ2 contingency test.
Statistical analysis
Data are presented as the mean±S.E.M. of at least three independent experiments. Significance, assigned for P-values<0.05, was determined by unpaired two-tailed Student's t-test with standard software (GraphPad Prism, San Diego, CA, USA).
Acknowledgments
We thank Sanford Markowitz for generously supplying the DLD1 UNG−/− cells. We thank Alex Almasan, Clive Hamlin, Ruth Keri, Shigemi Matsuyama, and George Stark for their careful review of this manuscript. This study was supported by grants from the NIH (CA86357 and CA 430703) to SL Gerson. LD Weeks is the recipient of an F31 NRSA training grant from the NCI (CA159614) and was also supported by an NIGHMS MSTP training grant T32-GM007250.
Glossary
- BER
base excision repair
- ChIP-Seq
chromatin immunoprecipitation high-throughput sequencing
- CldU
chlorodeoxyuridine
- DNA
deoxyribonucleic acid
- DSB
double strand break
- dTMP
deoxythymidine monophosphate
- dUMP
deoxyuridine monophosphate
- IdU
iododeoxyuridine
- TS
thymidylate synthase
- TSS
transcription start sites
- UNG
uracil DNA glycosylase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by H-U Simon
Supplementary Material
References
- Curtin NJ, Hughes AN. Pemetrexed disodium, a novel antifolate with multiple targets. Lancet Oncol. 2001;2:298–306. doi: 10.1016/s1470-2045(00)00325-9. [DOI] [PubMed] [Google Scholar]
- Grogan BC, Parker JB, Guminski AF, Stivers JT. Effect of the thymidylate synthase inhibitors on dUTP and TTP pool levels and the activities of DNA repair glycosylases on uracil and 5-fluorouracil in DNA. Biochemistry. 2011;50:618–627. doi: 10.1021/bi102046h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SH, Pittman DL, Wyatt MD. Uracil in DNA: consequences for carcinogenesis and chemotherapy. Biochem Pharmacol. 2008;76:697–706. doi: 10.1016/j.bcp.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner RD. The role of dUTPase and uracil-DNA repair in cancer chemotherapy. Curr Protein Pept Sci. 2001;2:361–370. doi: 10.2174/1389203013380991. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Drablos F, Slupphaug G. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, et al. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnes T, Akbari M, Hagen L, Slupphaug G, Krokan HE. The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair (Amst) 2008;7:1869–1881. doi: 10.1016/j.dnarep.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Dickey L, Goulian M. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry. 1986;25:3225–3230. doi: 10.1021/bi00359a022. [DOI] [PubMed] [Google Scholar]
- Andersen S, Heine T, Sneve R, Konig I, Krokan HE, Epe B, et al. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- Weeks LD, Fu P, Gerson SL. Uracil DNA glycosylase expression determines human lung cancer cell sensitivity to pemetrexed. Mol Cancer Ther. 2013;12:2248–2260. doi: 10.1158/1535-7163.MCT-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh SJ, Hobbs S, Aherne GW. Expression of uracil DNA glycosylase (UDG) does not affect cellular sensitivity to thymidylate synthase (TS) inhibition. Eur J Cancer. 2003;39:378–387. doi: 10.1016/s0959-8049(02)00610-x. [DOI] [PubMed] [Google Scholar]
- Guo C, Zhang X, Fink SP, Platzer P, Wilson K, Willson JK, et al. Ugene, a newly identified protein that is commonly overexpressed in cancer and binds uracil DNA glycosylase. Cancer Res. 2008;68:6118–6126. doi: 10.1158/0008-5472.CAN-08-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Olsen LC, Helland D, Aasland R, Krokan HE. Cell cycle regulation and in vitro hybrid arrest analysis of the major human uracil-DNA glycosylase. Nucleic Acids Res. 1991;19:5131–5137. doi: 10.1093/nar/19.19.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen L, Kavli B, Sousa MM, Torseth K, Liabakk NB, Sundheim O, et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T-a, Ayusawa D, Shimizu K, Koyama H, Seno T. Chromosome breakage induced by thymidylate stress in thymidylate synthase-negative mutants of mouse FM3A cells. Cancer Res. 1984;44:703–709. [PubMed] [Google Scholar]
- Yoshioka A, Tanaka S, Hiraoka O, Koyama Y, Hirota Y, Ayusawa D, et al. Deoxyribonucleoside triphosphate imbalance. 5-Fluorodeoxyuridine-induced DNA double strand breaks in mouse FM3A cells and the mechanism of cell death. J Biol Chem. 1987;262:8235–8241. [PubMed] [Google Scholar]
- Bulgar AD, Weeks LD, Miao Y, Yang S, Xu Y, Guo C, et al. Removal of uracil by uracil DNA glycosylase limits pemetrexed cytotoxicity: overriding the limit with methoxyamine to inhibit base excision repair. Cell Death Dis. 2012;3:e252. doi: 10.1038/cddis.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Riggs AD. Lac repressor binding to operator analogues: comparison of poly(d(A-T)), poly(d(A-BrU)), and poly(d(A-U)) Biochem Biophys Res Commun. 1971;45:1542–1547. doi: 10.1016/0006-291x(71)90195-1. [DOI] [PubMed] [Google Scholar]
- Ivarie R. Thymine methyls and DNA-protein interactions. Nucleic Acids Res. 1987;15:9975–9983. doi: 10.1093/nar/15.23.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, Ryan M, Kim R, Zakas AL, Fu H, Lin CM, et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela MS, Chen Y, Davis S, Yang F, Walker RL, Bilke S, et al. Preferential localization of human origins of DNA replication at the 5'-ends of expressed genes and at evolutionarily conserved DNA sequences. PLoS One. 2011;6:e17308. doi: 10.1371/journal.pone.0017308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald B, Sampath D, Plunkett W. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 2007;6:1239–1248. doi: 10.1158/1535-7163.MCT-06-0633. [DOI] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB. Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell. 2005;16:2566–2576. doi: 10.1091/mbc.E04-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Ferrari G, Scovassi AI, Prosperi E, Biamonti G. Etoposide induces the dispersal of DNA ligase I from replication factories. Mol Biol Cell. 2001;12:2109–2118. doi: 10.1091/mbc.12.7.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayusawa D, Shimizu K, Koyama H, Takeishi K, Seno T. Accumulation of DNA strand breaks during thymineless death in thymidylate synthase-negative mutants of mouse FM3A cells. J Biol Chem. 1983;258:12448–12454. [PubMed] [Google Scholar]
- Hori T, Ayusawa D, Shimizu K, Koyama H, Seno T. Chromosome breakage induced by thymidylate stress in thymidylate synthase-negative mutants of mouse FM3A cells. Cancer Res. 1984;44:703–709. [PubMed] [Google Scholar]
- Kuong KJ, Kuzminov A. Disintegration of nascent replication bubbles during thymine starvation triggers RecA- and RecBCD-dependent replication origin destruction. J Biol Chem. 2012;287:23958–23970. doi: 10.1074/jbc.M112.359687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaloe O, Hanawalt PC, Guarino E, Salguero I, Jimenez-Sanchez A, Guzman EC, et al. Thymine deficiency and the normal DNA replication cycle. J Mol Biol. 1961;3:144–155. doi: 10.1016/s0022-2836(61)80041-7. [DOI] [PubMed] [Google Scholar]
- Seno T, Ayusawa D, Shimizu K, Koyama H, Takeishi K, Hori T. Thymineless death and genetic events in mammalian cells. Basic Life Sci. 1985;31:241–263. doi: 10.1007/978-1-4613-2449-2_15. [DOI] [PubMed] [Google Scholar]
- Waldman BC, Wang Y, Kilaru K, Yang Z, Bhasin A, Wyatt MD, et al. Induction of intrachromosomal homologous recombination in human cells by raltitrexed, an inhibitor of thymidylate synthase. DNA Repair (Amst) 2008;7:1624–1635. doi: 10.1016/j.dnarep.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuong KJ, Kuzminov A. Stalled replication fork repair and misrepair during thymineless death in Escherichia coli. Genes Cells. 2010;15:619–634. doi: 10.1111/j.1365-2443.2010.01405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Ali MM, Hazra TK, Hong D, Kow YW. Action of human endonucleases III and VIII upon DNA-containing tandem dihydrouracil. DNA Repair (Amst) 2005;4:679–686. doi: 10.1016/j.dnarep.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Gates FT, 3rd, Linn S. Endonuclease V of Escherichia coli. J Biol Chem. 1977;252:1647–1653. [PubMed] [Google Scholar]
- Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, McIntosh EM, Reidy JA. International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Guarino E, Salguero I, Jimenez-Sanchez A, Guzman EC. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J Bacteriol. 2007;189:5782–5786. doi: 10.1128/JB.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Kim SC, Lee HS, Kim JK, Shon HJ, Salleh NL, et al. Genome-wide profiles of H2AX and gamma-H2AX differentiate endogenous and exogenous DNA damage hotspots in human cells. Nucleic Acids Res. 2012;40:5965–5974. doi: 10.1093/nar/gks287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE, Feeney L, Revet I. Phosphorylated H2Ax is not an unambiguous marker for DNA double-strand breaks. Cell Cycle. 2011;10:3223–3224. doi: 10.4161/cc.10.19.17448. [DOI] [PubMed] [Google Scholar]
- de Feraudy S, Revet I, Bezrookove V, Feeney L, Cleaver JE. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc Natl Acad Sci USA. 2010;107:6870–6875. doi: 10.1073/pnas.1002175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer. 2009;63:32–38. doi: 10.1016/j.lungcan.2008.03.033. [DOI] [PubMed] [Google Scholar]
- Luo Y, Walla M, Wyatt MD. Uracil incorporation into genomic DNA does not predict toxicity caused by chemotherapeutic inhibition of thymidylate synthase. DNA Repair (Amst) 2008;7:162–169. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulukuri SM, Knost JA, Estes N, Rao JS. Small interfering RNA-directed knockdown of uracil DNA glycosylase induces apoptosis and sensitizes human prostate cancer cells to genotoxic stress. Mol Cancer Res. 2009;7:1285–1293. doi: 10.1158/1541-7786.MCR-08-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy ED, Pillai R, Deng L, Liang L, Tischfield J, Schwemberger SJ, et al. Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 2010;19:1699–1711. doi: 10.1089/scd.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.