Abstract
The discovery of underlying mechanisms of drug resistance, and the development of novel agents to target these pathways, is a priority for patients with advanced colorectal cancer (CRC). We previously undertook a systems biology approach to design a functional genomic screen and identified fibroblast growth factor receptor 4 (FGFR4) as a potential mediator of drug resistance. The aim of this study was to examine the role of FGFR4 in drug resistance using RNAi and the small-molecule inhibitor BGJ398 (Novartis). We found that FGFR4 is highly expressed at the RNA and protein levels in colon cancer tumour tissue compared with normal colonic mucosa and other tumours. Silencing of FGFR4 reduced cell viability in a panel of colon cancer cell lines and increased caspase-dependent apoptosis. A synergistic interaction was also observed between FGFR4 silencing and 5-fluorouracil (5-FU) and oxaliplatin chemotherapy in colon cancer cell lines. Mechanistically, FGFR4 silencing decreased activity of the pro-survival STAT3 transcription factor and expression of the anti-apoptotic protein c-FLIP. Furthermore, silencing of STAT3 resulted in downregulation of c-FLIP protein expression, suggesting that FGFR4 may regulate c-FLIP expression via STAT3. A similar phenotype and downstream pathway changes were observed following FGFR4 silencing in cell lines resistant to 5-FU, oxaliplatin and SN38 and upon exposure of parental cells to the FGFR small-molecule inhibitor BGJ398. Our results indicate that FGFR4 is a targetable regulator of chemo-resistance in CRC, and hence inhibiting FGFR4 in combination with 5-FU and oxaliplatin is a potential therapeutic strategy for this disease.
Keywords: FGFR4, drug resistance, 5-fluorouracil, oxaliplatin, colorectal
Colorectal cancer (CRC) is the second commonest cause of cancer-related deaths in the Western World, and resistance to chemotherapy remains the primary reason for treatment failure in advanced CRC.1 The exposure of increasing numbers of metastatic CRC patients to oxaliplatin, as well as its introduction into the adjuvant therapy of early stage CRC, has led to efforts to identify and target mechanisms of drug resistance to oxaliplatin. In a previous study, we used a systems biology approach to reveal novel genes and pathways mediating drug resistance to 5-fluorouracil (5-FU) and oxaliplatin in CRC and identified fibroblast growth factor receptor 4 (FGFR4) as a potential druggable mediator of chemo-resistance.2
FGFR4 is one of a family of highly conserved tyrosine kinase receptors that regulate cellular pathways involved in proliferation, differentiation and survival.3 Four of the receptors (FGFR1–4) possess tyrosine kinase domains and are activated by the binding of one of 18 secreted glycoprotein FGFs to their extracellular immunoglobulin domains (Ig I–III).4 Ligand binding results in FGFR dimerisation, auto-phosphorylation of the intracellular split tyrosine kinase domain and activation of downstream signalling pathways, such as the Ras-dependent mitogen-activated protein kinase (MAPK) pathway, AKT-dependent anti-apoptotic pathway and signal transducer and activator of transcription (STAT) signalling.3, 5 FGFR4 is expressed as a single FGFR4-IIIc isoform resulting in increased selectivity in relation to its FGF binding partners such that, in the absence of the single transmembrane containing co-factor β-Klotho, only FGF19 can bind to FGFR4 and initiate its downstream signal transduction pathways.4, 6, 7
FGFR4 has been found to be of importance in a number of tumour types, with overexpression occurring in prostate, breast, pancreatic, pituitary, hepatocellular and gynaecological tumours.8, 9, 10, 11, 12, 13, 14, 15, 16 Initial observations of an increased rate of hepatocellular carcinoma formation in FGF19 transgenic mice and the predominant expression of FGFR4 in the liver provided a link between FGF19-FGFR4 signalling and tumourigenesis.11, 17, 18 Recently, FGFR4 has been recognised as an oncogene in breast cancer cell lines and activating mutations occur in up to 7% of rhabdomyosarcoma (RMS) tumours during tumourigenesis.19, 20 In other tumour types, FGFR4 mutations are infrequent, but the single-nucleotide polymorphism FGFR4 G388R increases stability, prolongs activation of the receptor, and is associated with a poor prognosis in melanoma, breast, prostate and head and neck cancers.21, 22, 23, 24 In colon cancer, FGFR4 has an important role in tumour–stroma interaction and the presence of the FGFR4 G388R substitution correlates significantly with advanced tumour stage and lymph node metastases.21, 25 Inhibition of FGF19-FGFR4 signalling in colon cancer using an FGF19 blocking antibody (IA6) has been shown to disrupt FGF19 binding to FGFR4, inhibiting the growth of HCT116 and Colo201 xenograft tumours.26 In relation to chemo-resistance, a role for FGFR4 has recently been described, with upregulation of FGFR4 in response to the DNA-damaging agent Doxorubicin.27
Considering the role of FGFR4 in resistance to DNA-damaging agents and the efficacy of disrupting FGF19-FGFR4 signalling in colon cancer, both in vitro and in vivo, we hypothesised that inhibition of FGFR4 represents a novel strategy for overcoming drug resistance in CRC. In this study, we demonstrate that silencing of FGFR4 induces caspase-dependent apoptosis in a panel of parental and drug-resistant colon cancer cell lines. Mechanistically, we discovered a role for FGFR4 as a regulator of signal transducer and activator of transcription 3 (STAT3) activity and apoptosis via its regulation of the anti-apoptotic proteins cellular FLICE-inhibitory protein (c-FLIP) and B-cell lymphoma-2 (Bcl-2). Our results indicate the influential role played by FGFR4 in CRC and the potential importance of targeting this receptor as a strategy to overcome drug resistance.
Results
Altered FGF expression in parental and drug-resistant CRC models
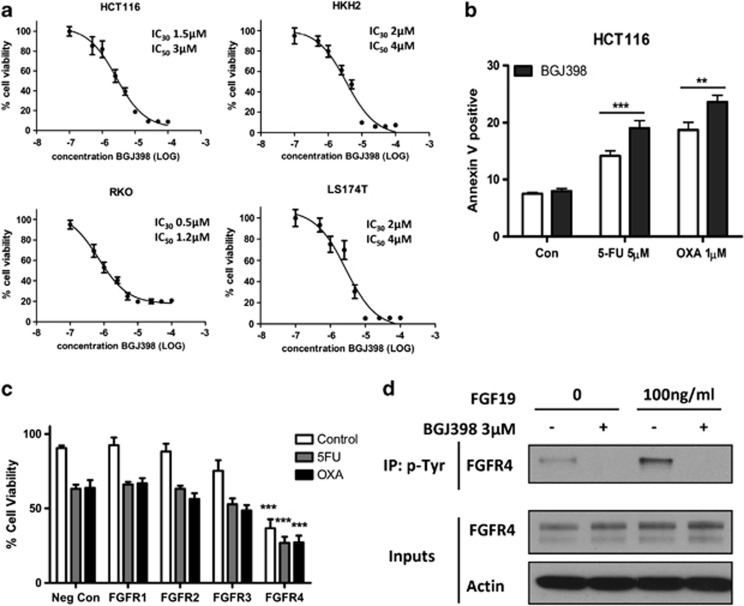
Transcriptional profiling of parental, 5-FU- and oxaliplatin-resistant HCT116 cells was carried out as previously described.2 Analysis of the microarray data revealed that gene expression of a number of FGFR ligands was acutely altered in response to chemotherapy treatment or constitutively altered in the 5-FU- or oxaliplatin-resistant cells (Supplementary Table 1). Of note, only FGFR4 is capable of binding all of the ligands significantly altered in our data set. We next evaluated pharmacological blockade of FGFR signalling using BGJ398 (Novartis, Basel, Switzerland), a small-molecule inhibitor of FGFR1-4 (in vitro kinase assay IC50 values of 0.9, 1.4, 1 and 60 nM for FGFR1–4, respectively).28 BGJ398 inhibited cell viability in a dose-dependent manner in HCT116, HKH2, RKO and LS174T colon cancer cells with IC50 doses in the low micromolar range (Figure 1a) and significant increases in apoptosis were observed in HCT116 cells treated with BGJ398 in combination with 5-FU or oxaliplatin (P<0.001 and P<0.01, respectively) (Figure 1b). We subsequently included FGFR1-4 in a functional genomic small interfering RNA (siRNA) screen and identified FGFR4 (but not FGFR1, FGFR2 or FGFR3) as a potential mediator of chemo-resistance (Figure 1c). Inhibition of FGFR4 activity (as monitored by its phosphorylation status) following stimulation with FGF19 and BGJ398 treatment was confirmed at an IC50 dose (Figure 1d). Taken together, these results indicate that disruption of FGFR signalling occurs in response to chemotherapy treatment and that inhibition of FGFR4 activity sensitises colon cancer cells to chemotherapy.
Figure 1.
Small-molecule inhibition of the FGFRs inhibits cell growth and induces apoptosis. (a) MTT cell viability assays of HCT116, HKH2, RKO and LS174T cells treated for 48 h with increasing concentrations of BGJ398. Absorbance was measured at 570 nM. (b) Annexin V/PI flow cytometric analysis was used to measure the levels of apoptosis in the HCT116 cell line following treatment with IC30 (48 h) doses of 5-FU or oxaliplatin for 48 h and co-treatment with IC30 (48 h) doses of BGJ398 for a further 24 h. Significance was determined by two-way analysis of variance. Results are presented as mean values±S.E.M. for three replicate experiments (**P<0.01, ***P<0.001). (c) MTT cell viability assay results showing percentage of surviving HCT116 cells transfected for 24 h with siFGFR1, siFGFR2, siFGFR3 or siFGFR4 before treatment with solvent control or IC30 (48 h) doses of 5-FU or oxaliplatin. (d) Immunoprecipitation using a phospho-tyrosine antibody was performed on HCT116 cells pre-treated with an IC50 (48 h) dose of BGJ398 for 24 h followed by a further IC50 (48 h) dose of BGJ398±100 ng/ml of FGF19 before immunoblotting for FGFR4
FGFR4 is differentially expressed in human colon adenocarcinoma versus normal colonic mucosal tissue
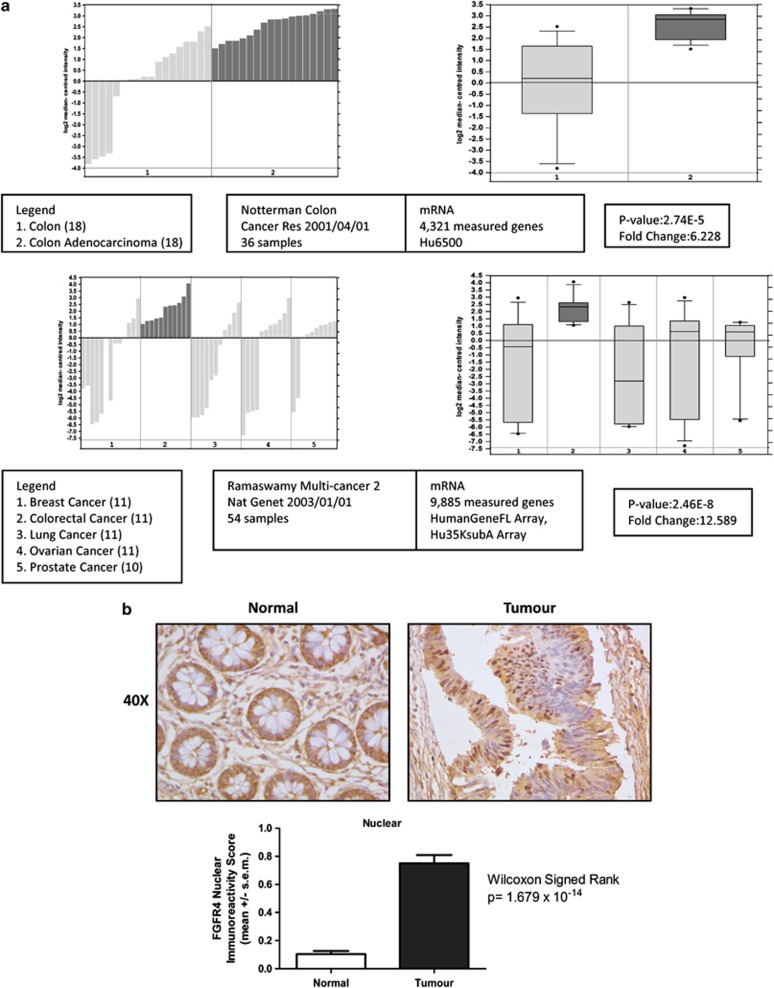
Analysis of the Oncomine database of publically available microarray expression data revealed upregulation of FGFR4 mRNA in CRC compared with normal colon tissues and in relation to other cancers (Figure 2a). FGFR2 mRNA was found to be upregulated in CRC compared with normal colon in a single data set, but no significant differences were found for FGFR1 or FGFR3 (data not shown). Having demonstrated an increase in FGFR4 expression at the mRNA level, we sought to examine FGFR4 expression in tumour tissue. Using a tissue microarray (TMA) compiled from 149 early stage CRC patients (Supplementary Table S2), we investigated the expression of FGFR4 in matched tumour and adjacent normal tissues using an antibody to the C-terminus portion of the receptor. Analysis of staining in the normal colonic epithelium showed predominant absence or mild staining (Figure 2b, upper panel). In addition, significantly higher nuclear staining was observed in the tumour tissue compared with normal colon (P=1.679e-14) (Figure 2b, lower panel). Considering this data and the previous results from the siRNA-mediated and pharmacological inhibition of FGFR4, we selected FGFR4 for further functional analysis as a novel and potentially druggable target in CRC.2
Figure 2.
FGFR4 is differentially expressed in human colon adenocarcinoma versus normal colonic mucosal tissue. (a) FGFR4 transcriptional profiling in the Oncomine database. The Oncomine database of publicly available microarray data was searched for differential expression of FGFR4 between normal colon and colonic adenocarcinoma tissue. The bar graphs and box plots are shown for each data set as well as the sample breakdown, reference, platform used, fold change and level of significance as determined by the Student's t-test. (b) Representative high-powered ( × 40) images illustrating weak nuclear staining for FGFR4 in normal colon and strong nuclear staining in colonic adenocarcinomas. Bar graph representing the relative immunoreactivity score calculated for FGFR4 nuclear staining in normal and malignant colorectal epithelial cells
FGFR4 silencing synergistically enhances the effects of chemotherapy in a panel of colon cancer cells
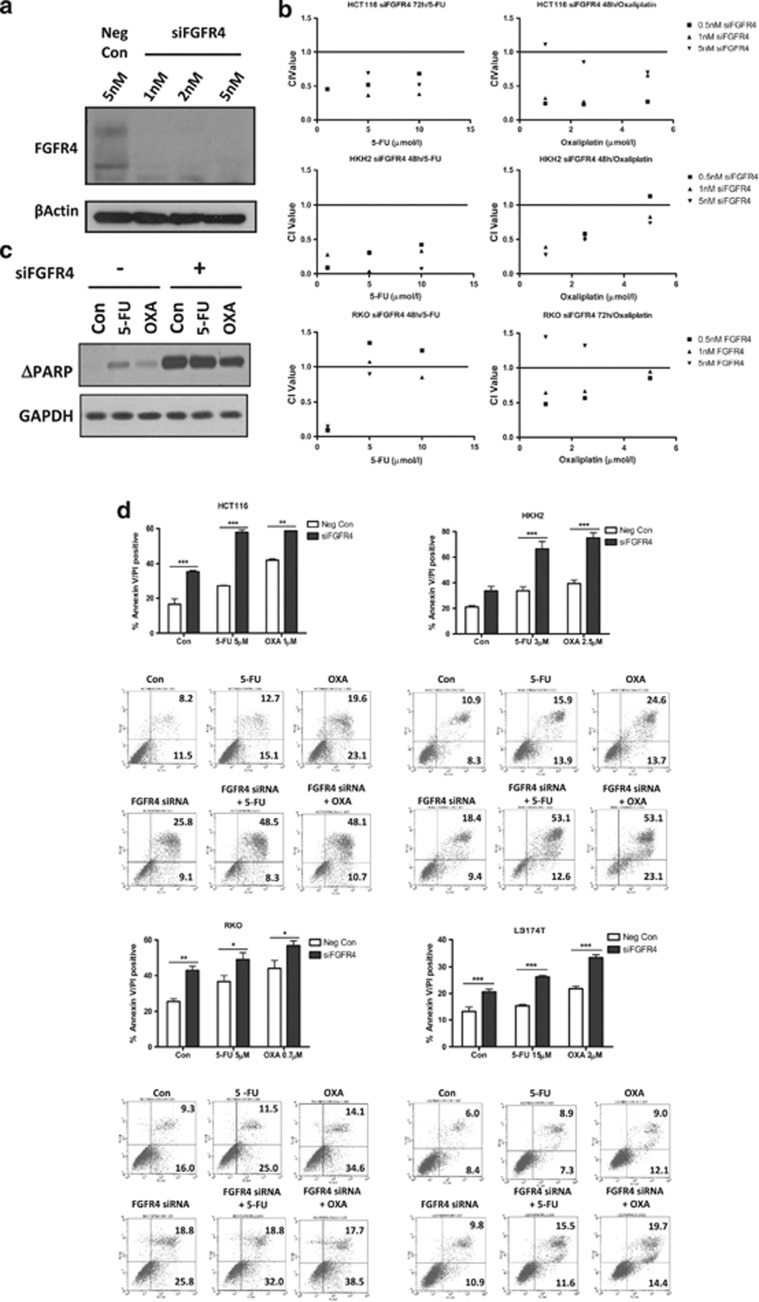
HCT116 cells were transfected for 48 h with 1–5 nM siRNA targeting FGFR4 (siFGFR4), and silencing was confirmed by western blotting (Figure 3a). Quantitative-PCR analysis showed that silencing of FGFR4 did not significantly affect the expression of FGFR1-3 in HCT116 and RKO cells at the 24 or 48 h timepoints (Supplementary Figure S1A). In order to examine the effects of FGFR4 silencing in a range of genetic backgrounds, we utilised a panel of colon cancer cell lines displaying mutations in KRAS (HCT116, LS174T), BRAF (RKO) and PIK3CA (HCT116, HKH2, LS174T, RKO). HCT116, HKH2 and RKO cells were transfected for 24 h with siFGFR4 before a 24/48 h co-treatment with a range of doses of 5-FU/oxaliplatin. Combination Index (CI) values were calculated and indicated that FGFR4 silencing synergised with 5-FU and oxaliplatin treatment in HCT116, HKH2 and RKO cell line models, particularly at low drug concentrations (Figure 3b). Similar results were obtained using two further FGFR4 siRNA sequences (Supplementary Figures S1B and C).
Figure 3.
FGFR4 silencing synergistically enhances the effects of chemotherapy in a panel of colon cancer cells. (a) Transfection of HCT116 cells was carried out using 1–5 nM of siFGFR4, and FGFR4 silencing was confirmed by western blotting. The unmodified receptor is present at 88 kDa while the phosphorylated and glycosylated forms are present at 88–125 kDa. (b) Cell viability assays were conducted in the HCT116, HKH2 and RKO cell lines using a MTT assay following FGFR4 silencing (siFGFR4 1–5 nM) for 24 h before co-treatment with parental IC30 (48 h) and IC50 (48 h) doses of chemotherapy for 24/48 h. The method of Chou and Talalay was used to evaluate the interaction between siFGFR4 and either 5-FU or oxaliplatin at 48 or 72 h. CI values were calculated using Calcusyn v2.0 where values<1,=1 and >1 indicate synergism, additivity and antagonism, respectively. (c) HCT116 cells were transfected with negative control siRNA or siFGFR4 for 24 h before a 48 h co-treatment with IC30 (48 h) doses of either 5-FU or oxaliplatin. Western blotting was performed for PARP cleavage with equal loading assessed by probing for GAPDH. (d) Apoptosis was assessed using Annexin V/PI flow cytometric analysis. HCT116, HKH2 and RKO cells were transfected with 5 nM siFGFR4 and LS174T cells with 10 nM siFGFR4 for 24 h before a 48 h co-treatment with IC30 (48 h) doses of either 5-FU or oxaliplatin. Percentage of cells in early apoptosis (Annexin V positive/PI negative, bottom right quadrant) and late apoptosis (Annexin V positive/PI positive, upper right quadrant) are indicated. Significance and synergism were determined by two-way analysis of variance. Results are presented as mean values±S.E.M. for three replicate experiments (*P<0.05, **P<0.01, ***P<0.001)
To examine the mechanism of synergy between FGFR4 siRNA and chemotherapy, apoptosis was assessed. PARP (poly-ADP ribose polymerase-1) cleavage was induced by FGFR4 silencing alone, and this was further increased in the presence of chemotherapy (Figure 3c). In HCT116 cells, FGFR4 silencing resulted in significantly higher levels of apoptosis compared with cells transfected with the negative control siRNA (P<0.05; Figure 3d). In the presence of chemotherapy treatment, apoptosis in FGFR4-silenced cells was further enhanced for both 5-FU and oxaliplatin (P<0.001 and P<0.01, respectively), with a synergistic interaction between 5-FU and siFGFR4 confirmed using two-way ANOVA (P=0.0275). In the HKH2 cell line, the addition of siFGFR4 to 5-FU or oxaliplatin significantly increased apoptosis for both drugs (P<0.001), and the interaction was synergistic (P=0.0208 and 0.0028 for 5-FU and oxaliplatin, respectively). Similar results were observed in the RKO and LS174T cell lines (Figure 3d) and with the use of two further FGFR4 siRNA sequences (Supplementary Figure S1D). These results demonstrate that FGFR4 silencing reduces cell viability, induces apoptosis and enhances the apoptotic effects of both 5-FU and oxaliplatin. Moreover, these effects are not dependent on the mutational status of the KRAS oncogene as indicated by the results from HCT116 and HKH2 KRAS mutant and wild-type paired cell lines.
FGFR4 silencing induces caspase-dependent apoptosis in colon cancer cell lines
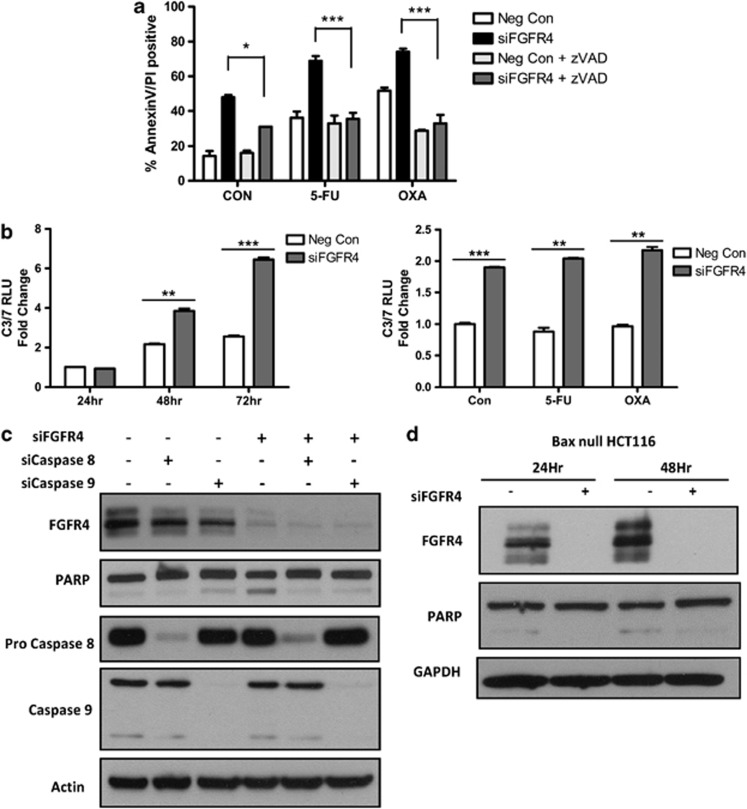
Caspase-dependent apoptosis following FGFR4 silencing was assessed using the pan-caspase inhibitor zVAD, which completely attenuated apoptosis (Figure 4a). Moreover, activity of the executioner caspases 3 and 7 was significantly induced following transfection of HCT116 and HKH2 cells with siFGFR4 (Figure 4b and data not shown). IC30 (48 h) doses of 5-FU or oxaliplatin failed to significantly increase caspase 3/7 activity but did further increase caspase 3/7 activity when FGFR4 was silenced in the HCT116 and HKH2 cell lines (Figure 4b and data not shown). To define the relative importance of the extrinsic and intrinsic apoptotic pathways in mediating siFGFR4-induced apoptosis, we used siRNAs targeting caspase 8 (extrinsic pathway) or caspase 9 (intrinsic pathway). Notably, silencing of either caspase prevented apoptosis induced by FGFR silencing (Figure 4c). In addition, transfection of Bax null HCT116 cells with siFGFR4 failed to induce PARP cleavage, confirming the role of the intrinsic apoptotic pathway in siFGFR4-induced apoptosis (Figure 4d). Together, these results suggest that siFGFR4-induced apoptosis proceeds via caspase 8-mediated activation of the Bax-regulated, caspase 9-dependent intrinsic apoptotic pathway, consistent with HCT116 cells being type II with respect to the extrinsic pathway.29
Figure 4.
FGFR4 silencing induces caspase-dependent apoptosis in colon cancer cell lines. (a) Annexin V/PI flow cytometric analysis was used to measure levels of apoptosis following transfection with siFGFR4 for 24 h before co-treatment with an IC30 (48 h) doses of either 5-FU or oxaliplatin±10 μM zVAD. (b) Caspase 3/7 activity levels were measured in HCT116 cells following 24–72 h FGFR4 silencing alone (5 nM siFGFR4) or co-treatment for 24 h with IC30 (48 h) doses of 5-FU or oxaliplatin following 24 h FGFR4 silencing. (c) Western blotting was used to measure the expression of FGFR4, PARP, Pro-Caspase 8 and Caspase 9 following transfection with either 20 nM siCaspase 8 or 10 nM siCaspase 9 for 24 h followed by transfection with 5 nM siFGFR4 or 5 nM siRNA negative control and harvested after a further 24 h. (d) Western blotting showing the effects of FGFR4 silencing at 24 and 48 h in Bax null HCT116 cells by probing for expression of FGFR4 and PARP. Equal loading was assessed by probing for GAPDH or actin. Significance and synergism were determined by two-way analysis of variance (a) or Student's t-test (b). Results are presented as mean values±S.E.M. for three replicate experiments (*P<0.05, **P<0.01, ***P<0.001)
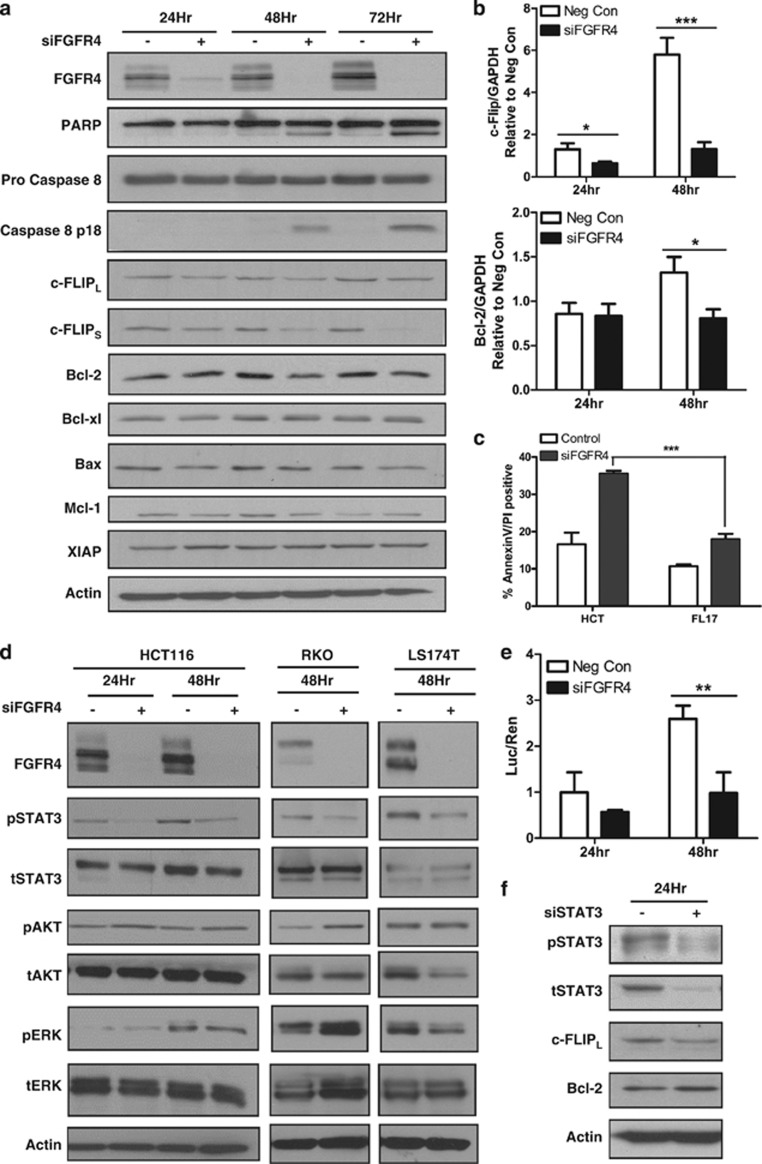
FGFR4 silencing downregulates the anti-apoptotic proteins c-FLIP and Bcl-2 and reduces STAT3 activity in colon cancer cells
To further study the mechanism of apoptosis in HCT116 cells, the expression levels of pro- and anti-apoptotic proteins were assessed following transfection with siFGFR4 for 24–72 h. In keeping with our previous data, FGFR4 silencing induced PARP cleavage, processing of procaspase 8 and downregulation of the anti-apoptotic proteins Bcl-2 and both the long and short splice forms of c-FLIP compared with time-matched controls (Figure 5a). No significant changes were noted in the expression of other apoptosis-regulating proteins. Analysis of mRNA expression confirmed reduction of c-FLIP mRNA expression following transfection with siFGFR4, and a reduction in Bcl-2 expression was also observed at 48 h (Figure 5b). A similar decrease in c-FLIP and Bcl-2 expression was observed in the RKO cell line (Supplementary Figure S2A). Given the role of caspase 8 in siFGFR4-induced apoptosis and the downregulation of the endogenous inhibitor of caspase 8, c-FLIP, we next examined the effect of FGFR4 silencing in the c-FLIPL overexpressing cell line FL17.30 Flow cytometric analysis demonstrated that the FL17 cell line was significantly more resistant to siFGFR4-induced apoptosis compared with the parental HCT116 cell line (P<0.001, Figure 5c).
Figure 5.
FGFR4 silencing downregulates the anti-apoptotic proteins c-FLIP and Bcl-2 and reduces pSTAT3 activity in colon cancer cells. (a) Western blotting was used to measure expression of FGFR4, PARP, Pro-Caspase 8, Caspase 8 p18, FLIPL, FLIPS, Bcl-2, BCLXL, Bax, MCL1 and XIAP at 24–72 h following transfection with 5 nM siFGFR4. (b) Q-PCR of c-FLIP and Bcl-2 expression following transfection of HCT116 cells with siFGFR4 for 24–48 h. (c) Annexin V/PI flow cytometric analysis was used to measure apoptosis in HCT116 parental cells and the overexpressing cell line, FL17, following 72 h FGFR4 silencing. (d) Western blotting was used to measure expression of FGFR4, pSTAT3, tSTAT3, pAKT, tAKT, pERK and tERK following transfection of 5 nM siFGFR4 for 24–48 h in HCT116, 48 h in RKO and 10 nM siFGFR4 for 48 h in LS174T cells. (e) A luciferase construct STAT3 reporter assay was used to measure STAT3 activity in HCT116 at 24 and 48 h following transfection with 5 nM siFGFR4. (f) Western blotting was used to measure the expression of pSTAT3, tSTAT3, c-FLIPL and Bcl-2. Equal loading was assessed by probing for GAPDH or actin. Significance was assessed by two-way analysis of variance (c) or Student's t-test (b and d). Results are presented as mean values±S.E.M. for three replicate experiments (*P<0.05, **P<0.01, ***P<0.001)
To assess changes in pro-survival signal transduction pathways induced by FGFR4 silencing that may be affecting c-FLIP and Bcl-2 expression, HCT116, RKO and LS174T cells were transfected with siFGFR4 for 24–48 h. Most notably, STAT3 activation as assessed by Y705 phosphorylation was reduced after 24 and 48 h following transfection in each of these cell lines (Figure 5d). These results were confirmed using other FGFR-targeting siRNAs (Supplementary Figure S2B) and BGJ398 (Supplementary Figure S2C). A STAT3-driven luciferase reporter gene confirmed a significant reduction in STAT3 activity 48 h after transfection with siFGFR4 (P<0.01) (Figure 5e). Furthermore, silencing of STAT3 in HCT116 cells resulted in downregulation of c-FLIP protein expression although expression of Bcl-2 was unaffected (Figure 5f), suggesting that FGFR4 may regulate FLIP, but not Bcl-2, expression via STAT3.
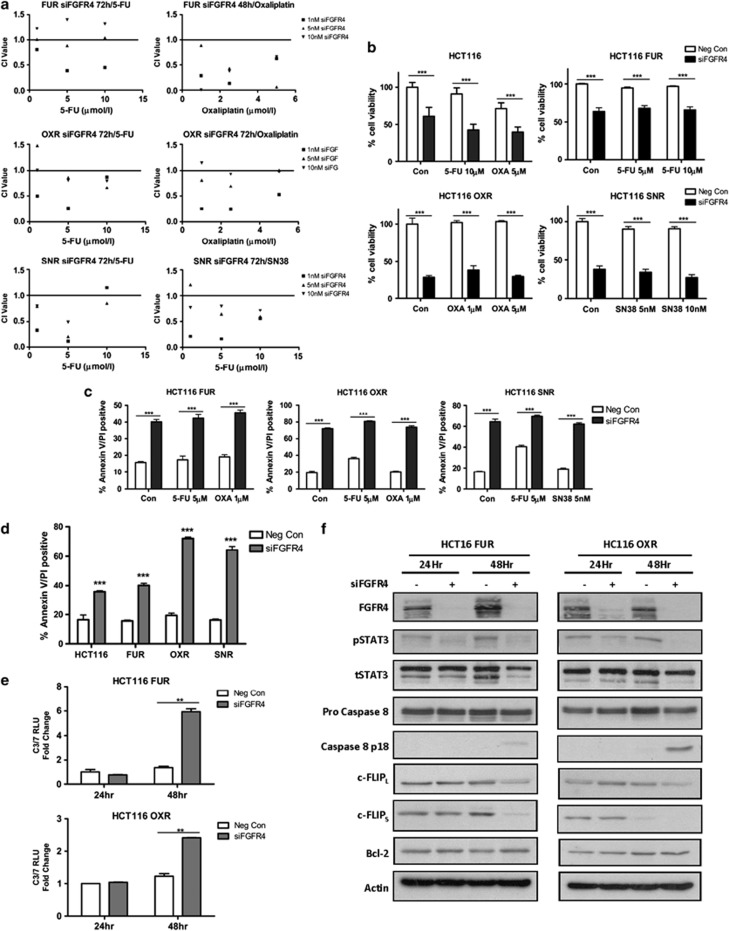
FGFR4 silencing induces apoptosis in drug-resistant colon cancer cell lines
To assess the effects of FGFR4 silencing in the context of acquired drug resistance, previously described drug-resistant HCT116 models were used.31 CI values were calculated following transfection with 1–10 nM siFGFR4 before 24/48 h co-treatment with a range of doses of 5-FU, oxaliplatin or SN38 (the active metabolite of irinotecan) (Figure 6a). Silencing of FGFR4 interacted synergistically with the chemotherapeutic agent to which the cells were resistant and a second agent. Basal expression of FGFR4 in these resistant cell lines was similar to the parental cell line (Supplementary Figure S3). A significant reduction in cell viability was observed in the HCT116 5-FU- (HCT116 FUR), oxaliplatin- (HCT116 OXR) and SN38-resistant (HCT116 SNR) cells following FGFR4 silencing alone or in combination with IC30 (48 h) chemotherapy (Figure 6b). Markedly elevated levels of apoptosis were observed following silencing of FGFR4 in all three drug-resistant models (Figure 6c compared to Figure 3d). The oxaliplatin- and SN38-resistant models were more sensitive to loss of FGFR4 than the parental cells, as assessed by flow cytometric measurement of apoptosis (Figure 6d), indicating addiction of these drug-resistant cells to FGFR4 signalling. Similar to the parental cells, activation of caspase 3/7 was observed following transfection with siFGFR4 in both the HCT116 FUR and OXR cell lines (Figure 6e); this was accompanied by downregulation of c-FLIPL, c-FLIPS and pSTAT3 (Y705) (Figure 6f). In addition, processing of caspase 8 to its active p18-form was observed after 48 h following transfection of the HCT116 FUR and OXR cell lines with FGFR4 siRNA; this was particularly apparent in the OXR cell line (Figure 6f). However, no changes in Bcl-2 were observed following FGFR4 silencing in these drug-resistant cell lines.
Figure 6.
FGFR4 silencing induces apoptosis in drug-resistant colon cancer cell lines. (a) Cell viability assays were conducted in the HCT116 parental, 5-FU-, oxaliplatin- and SN38-resistant cell lines using a MTT assay following FGFR4 silencing (siFGFR4 1–10 nM) for 24 h before co-treatment with parental IC30 (48 h) and IC50 (48 h) doses of chemotherapy for 24/48 h. The method of Chou and Talalay was used to evaluate the interaction between siFGFR4 and either 5-FU oxaliplatin or SN38 at 48 or 72 h. CI values were calculated using Calcusyn v2.0 where values<1,=1 and>1 indicate synergism, additivity and antagonism, respectively. (b) Cell viability of HCT116 parental, 5-FU-, oxaliplatin- and SN38-resistant cell lines (HCT116 FUR, OXR and SNR, respectively) was assessed using a MTT assay following FGFR4 silencing (siFGFR4 10 nM for resistant lines) for 24 h before co-treatment with parental IC30 (48 h) and IC50 (48 h) doses of chemotherapy. (c) Apoptosis was assessed using Annexin V/PI flow cytometric analysis. HCT116 FUR, OXR and SNR cells were transfected with 10 nM siFGFR4 for 24 h before a 48-h co-treatment with IC30 (48 h) parental cell line doses of either 5-FU, oxaliplatin or SN38. Percentage of cells in early apoptosis (Annexin V positive/PI negative, bottom right quadrant) and late apoptosis (Annexin V positive/PI positive, upper right quadrant) are indicated. Significance and synergism were determined by two-way analysis of variance. Results are presented as mean values±S.E.M. for three replicate experiments (*P<0.05, **P<0.01, ***P<0.001). (d) Apoptosis was assessed using Annexin V/PI flow cytometric analysis following transfection of HCT116 parental and drug-resistant cell lines with siFGFR4 according to the conditions previously described. (e) Caspase 3/7 activity levels were measured in HCT116 FUR and OXR cells following 24–72 h FGFR4 silencing (10 nM siFGFR4). (f) Western blotting was used to measure expression of FGFR4, pSTAT3, tSTAT3, Pro-Caspase 8, Caspase 8 p18, FLIPL, FLIPS and BCL2 at 24 and 48 h following transfection with 10 nM siFGFR4 in the HCT116 FUR and OXR cell lines. Equal loading was assessed by probing for actin
Discussion
Despite significant developments in conventional chemotherapy and targeted agents over the past two decades, overall survival for patients with metastatic CRC remains poor.32 The effectiveness of current chemotherapeutic agents is limited by drug resistance, thus the discovery of the mechanisms governing the cellular response to chemotherapy is a priority.33 This study provides evidence of FGFR4 as a novel determinant of chemo-resistance and its potential as a drug target in CRC.
Recently, a role for FGFR4 has been described in the resistance of breast cancer cells to the DNA-damaging agents doxorubicin and cyclophosphamide.27 Upregulation of FGFR4 was found in a doxorubicin-resistant MDA-MB-453 breast cancer cells, and silencing of FGFR4 significantly increased apoptosis. FGFR4 was highly expressed in our tumour CRC TMA, and we demonstrate for the first time that FGFR4 has an important role in resistance to oxaliplatin and 5-FU treatment in a range of CRC cell line models. Furthermore, silencing of FGFR4 in combination with chemotherapy was not only capable of inducing a significant increase in apoptosis in parental cell lines but also in 5-FU-, oxaliplatin- and SN38-resistant cell lines. In fact, the oxaliplatin-resistant cell line and a cell line resistant to SN38 (the active metabolite of irinotecan) were more sensitive to FGFR4 depletion than the parental cell line, suggesting increased dependence on FGFR4 in the setting of acquired drug resistance.
FGFR4 acts through a number of pathways via intracellular molecules, such as FGFR substrate 2α (FRS2α), phospholipase C (PLCγ) and STAT3.34, 35, 36, 37 Inhibition of FGFR4 in breast and RMS cell lines has been shown to downregulate ERK activity, indicating a role for MAPK pathway inhibition in the induction of apoptosis following FGFR4 blockade in these models.27, 38 However, we did not observe consistent changes in ERK or Akt activation in response to FGFR4 silencing, whereas changes in STAT3 activity were consistently observed. FGFR4, along with FGFR1 and FGFR3, has been shown to modulate the activity of STAT3, and in pituitary tumours the polymorphic FGFR4 G388R variant mediates pituitary oncogenesis through activation of STAT3, but not the MAPK pathway.37, 39 Further downstream of FGFR4, we found that expression of the anti-apoptotic proteins Bcl-2, c-FLIPL and c-FLIPS were downregulated in response to FGFR downregulation. Moreover, c-FLIP expression was both FGFR4- and STAT3-dependent, suggesting that FGFR4 regulates c-FLIP expression via STAT3. In contrast, Bcl-2 expression was not STAT3-dependent, indicating that FGFR4 regulates its expression via an alternative mechanism. In drug-resistant cell lines, FGFR4 silencing also downregulated c-FLIP expression, although Bcl-2 expression was unaltered. Notably, apoptosis induced by FGFR4 depletion was found to be FLIP-, caspase 8-, caspase 9- and Bax-dependent, suggesting that FLIP downregulation following FGFR4 silencing leads to caspase 8 activation, which then cross-talks with the mitochondria to promote Bax-mediated release of cytochrome c and activation of caspase 9 followed by activation of the executioner caspases 3 and 7.29
Recent data regarding FGFR1 in breast cancer has shown that, following activation, the receptor is cleaved by the protease Granzyme B and the C-terminus trafficks to the nucleus, where it regulates cell migration.40 Interestingly, high nuclear FGFR4 staining was found in our tumour CRC tissue, and our phenotypic effects following FGFR4 inhibition could be explained by a nuclear function of FGFR4 on STAT3 and expression of c-FLIP. In addition, nuclear localisation of other receptor tyrosine kinases, such as epidermal growth factor receptor, has been correlated with a migratory phenotype and poor prognosis.41, 42, 43, 44
In summary, we have described a novel role for FGFR4 in resistance to 5-FU and oxaliplatin chemotherapy in CRC. Silencing of FGFR4 downregulates c-FLIP expression and, in combination with chemotherapy, induces apoptosis in parental and drug-resistant cell line models. The development of selective small-molecule inhibitors will allow inhibition of the FGFR4 to be explored as a potential therapeutic strategy in CRC.
Materials and Methods
Materials
5-FU, oxaliplatin and SN38 were purchased from Sigma Chemicals (St. Louis, MO, USA), Sanofi-Synthelabo (Malvern, PA, USA) and Abatra Technology (Xi'an, China), respectively. zVAD (OMe)-FMK was purchased from Calbiochem (Darmstadt, Germany). BGJ398 was kindly provided by Novartis, and all siRNAs were purchased from Qiagen (Crawley, UK).
Cell lines
Authentication and culture of CRC cell lines HCT116, LS174T and RKO has been described previously.45 The HCT116 parental and HCT116 Bax−/− isogenic human colon cancer cell lines were kindly provided by Professor Bert Vogelstein (Johns Hopkins University, Baltimore, MD, USA) and maintained in Dulbecco's Modified Eagle's Medium (DMEM). The 5-FU-, oxaliplatin-, SN38-resistant and c-FLIPL-overexpressing HCT116 sub-lines were generated as previously described.30, 31 The RKO and LS174T human CRC cell lines were provided by the National Cancer Institute and the American Type Culture Collection (ATCC), respectively, and maintained in DMEM.
CI values
siRNA transfection conditions have been previously described.45 CI values were calculated as previously described.46
Western blotting analysis
Western blots were performed as previously described.47 PARP (eBioscience, San Diego, CA, USA), tSTAT3 (Cell Signalling Technology Inc, Beverly, MA, USA), pERK (Santa Cruz Biotechnology, Santa Cruz, CA, USA), FLIP (NF-6) and Caspase 8 (Abexis Biochemical, Birmingham, UK) mouse monoclonal antibodies were used in conjunction with a horseradish peroxidase-conjugated sheep anti-mouse secondary antibody (Amersham, Chalfont St Giles, UK). FGFR4 XP, pAKT, tAKT, Caspase 9, Bcl-2, Bcl-xl, Bax, XIAP (Cell Signalling Technology Inc), FGFR4 C16, tERK, Mcl-1 (myeloid cell leukemia sequence 1; Santa Cruz Biotechnology) and pSTAT3 (Abcam, Cambridge, UK) were used in conjunction with anti-rabbit secondary antibody (Amersham). Equal loading was assessed using GAPDH (glyceraldehyde-3-phosphate dehydrogenase; AbD Serotec, Oxford, UK).
Annexin V/propidium iodide (PI) analysis
Cells were harvested, stained for Annexin V–fluoroscein isothiocyanate and PI uptake and analysed using a BD FACSCalibur instrument according to the manufacturer's instructions (BD Biosciences, Oxford, UK). Levels of apoptosis were calculated as the sum of the Annexin V positive/PI negative (early apoptosis) and Annexin V positive/PI positive (late apoptosis) cell populations.
Caspase-activation assays
Caspase activity in cell protein isolates was measured using Caspase Glo 3/7 and 8 assays (Promega, Madison, WI, USA).
Cell viability assays
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) (Sigma Chemicals) assay was used to assess cell viability as previously described.30
STAT3 reporter assay
The STAT3 reporter, negative and positive control constructs were obtained from Qiagen (Crawley, UK) and transfected with FGFR4 or negative control siRNA using GeneJuice (Novagen, Darmstadt, Germany). The Biotech Synergy 4 Multi-mode Luminometer and the Gen5 software programme measured luminescence after 50 μl of luciferase assay reagent was injected into each well. Readings were taken after 1-s incubation (100–200 sensitivity) and were recorded as relative luminescence units (RLU).
Immunohistochemistry
A human CRC TMA was obtained from the Northern Ireland Biobank. The TMA was constructed using samples from patients entered into a phase III randomised controlled trial of treatment with 5-FU/FA chemotherapy versus observation alone following surgical resection of stage II and III tumours.48 The trial was conducted in compliance with the Declaration of Helsinki and ethical approval for this study was obtained from the local ethics committee. The TMA consists of 1836 cores representing colon or rectal adenocarcinoma with matched normal mucosa from 149 individuals (Supplementary Table S2).
Staining for FGFR4 (FGFR4 C16, Santa Cruz Biotechnology) was conducted with the assistance of the Northern Ireland Molecular Pathology laboratory. The TMA was scored by two independent observers, and for each core, two scores of FGFR4 positivity were determined on the basis of plasma membrane/cytoplasmic staining and nuclear staining. A semiquantitative scale of FGFR4 intensity was assigned as an integer (0=no staining, 1=mild, 2=moderate, 3=strong staining). In cases where there were replicate cores from a single tumour, an average score was assigned. Nuclear staining was defined as negative for scores ≤1 and positive if >1. Fisher's exact test was used to compare nuclear staining scores between normal and tumour samples.
Acknowledgments
We thank Cathy Fenning and Gail Stewart for their technical assistance, Cheryl McFarlane for help with the manuscript and Novartis for kindly supplying the inhibitor BGJ398. We also thank the patients who took part in the Northern Ireland adjuvant chemotherapy trial, the staff who collected the tissues and clinical data and the trial funders Wyeth Ltd (now Pfizer Inc) and the Friends of Montgomery House (now Friends of the Cancer Centre). Grant support: This study was supported by Cancer Research UK and the Bobby Moore fund. Financial support: This work was supported by Cancer Research UK (C212/A7402) (to PGJ); a Cancer Research UK Bobby Moore Fellowship and the Research and Development Office Northern Ireland, Department of Health, Social Services and Public Safety (RRG/3261/05, RRG 6.42).
Glossary
- 5-FU
5-fluorouracil
- Bcl-2
B-cell lymphoma-2
- CI
Combination Index
- c-FLIP
cellular FLICE-inhibitory protein
- CRC
colorectal cancer
- FGFR4
fibroblast growth factor receptor 4
- MAPK
mitogen-activated protein kinase
- PARP
poly-ADP ribose polymerase-1
- PI
propidium iodide
- siRNA
small interfering RNA
- STAT3
signal transducer and activator of transcription 3
- TMA
tissue microarray
PGJ is employed by Almac Diagnostics and has an ownership interest in both Almac Diagnostics and Fusion Antibodies. He is a consultant/advisor for, and has received honoraria from, Chugai pharmaceuticals, Sanofi-Aventis and Pfizer. All the other authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by H-U Simon
Supplementary Material
References
- Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- Stevenson L, Allen WL, Turkington R, Jithesh PV, Proutski I, Stewart GE, et al. Identification of galanin and its receptor GalR1 as novel determinants of resistance to chemotherapy and potential biomarkers in colorectal cancer. Clin Cancer Res. 2012;18:5412–5426. doi: 10.1158/1078-0432.CCR-12-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Wu X, Ge H, Lemon B, Weiszmann J, Gupte J, Hawkins N, et al. Selective activation of FGFR4 by an FGF19 variant does not improve glucose metabolism in ob/ob mice. Proc Natl Acad Sci USA. 2009;106:14379–14384. doi: 10.1073/pnas.0907812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ge H, Lemon B, Vonderfecht S, Weiszmann J, Hecht R, et al. FGF19-induced hepatocyte proliferation is mediated through FGFR4 activation. J Biol Chem. 2010;285:5165–5170. doi: 10.1074/jbc.M109.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowardhan B, Douglas DA, Mathers ME, McKie AB, McCracken SR, Robson CN, et al. Evaluation of the fibroblast growth factor system as a potential target for therapy in human prostate cancer. Br J Cancer. 2005;92:320–327. doi: 10.1038/sj.bjc.6602274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penault-Llorca F, Bertucci F, Adelaide J, Parc P, Coulier F, Jacquemier J, et al. Expression of FGF and FGF receptor genes in human breast cancer. Int J Cancer. 1995;61:170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- Shah RN, Ibbitt JC, Alitalo K, Hurst HC. FGFR4 overexpression in pancreatic cancer is mediated by an intronic enhancer activated by HNF1alpha. Oncogene. 2002;21:8251–8261. doi: 10.1038/sj.onc.1206020. [DOI] [PubMed] [Google Scholar]
- Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require Klotho beta. J Biol Chem. 2007;282:27277–27284. doi: 10.1074/jbc.M704244200. [DOI] [PubMed] [Google Scholar]
- Jaakkola S, Salmikangas P, Nylund S, Partanen J, Armstrong E, Pyrhonen S, et al. Amplification of fgfr4 gene in human breast and gynecological cancers. Int J Cancer. 1993;54:378–382. doi: 10.1002/ijc.2910540305. [DOI] [PubMed] [Google Scholar]
- Leung HY, Gullick WJ, Lemoine NR. Expression and functional activity of fibroblast growth factors and their receptors in human pancreatic cancer. Int J Cancer. 1994;59:667–675. doi: 10.1002/ijc.2910590515. [DOI] [PubMed] [Google Scholar]
- Olson DC, Deng C, Hanahan D. Fibroblast growth factor receptor 4, implicated in progression of islet cell carcinogenesis by its expression profile, does not contribute functionally. Cell Growth Differ. 1998;9:557–564. [PubMed] [Google Scholar]
- Sahadevan K, Darby S, Leung HY, Mathers ME, Robson CN, Gnanapragasam VJ. Selective over-expression of fibroblast growth factor receptors 1 and 4 in clinical prostate cancer. J Pathol. 2007;213:82–90. doi: 10.1002/path.2205. [DOI] [PubMed] [Google Scholar]
- Ezzat S, Zheng L, Zhu XF, Wu GE, Asa SL. Targeted expression of a human pituitary tumor-derived isoform of FGF receptor-4 recapitulates pituitary tumorigenesis. J Clin Invest. 2002;109:69–78. doi: 10.1172/JCI14036. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nicholes K, Guillet S, Tomlinson E, Hillan K, Wright B, Frantz GD, et al. A mouse model of hepatocellular carcinoma: ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am J Pathol. 2002;160:2295–2307. doi: 10.1016/S0002-9440(10)61177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French DM, Lin BC, Wang M, Adams C, Shek T, Hotzel K, et al. Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One. 2012;7:e36713. doi: 10.1371/journal.pone.0036713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roidl A, Foo P, Wong W, Mann C, Bechtold S, Berger HJ, et al. The FGFR4 Y367C mutant is a dominant oncogene in MDA-MB453 breast cancer cells. Oncogene. 2010;29:1543–1552. doi: 10.1038/onc.2009.432. [DOI] [PubMed] [Google Scholar]
- Taylor JGt, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–847. [PubMed] [Google Scholar]
- Wang J, Stockton DW, Ittmann M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin Cancer Res. 2004;10 (18 Pt 1:6169–6178. doi: 10.1158/1078-0432.CCR-04-0408. [DOI] [PubMed] [Google Scholar]
- Streit S, Mestel DS, Schmidt M, Ullrich A, Berking C. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients. Br J Cancer. 2006;94:1879–1886. doi: 10.1038/sj.bjc.6603181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Andrade VC, Parise O, Jr, Hors CP, de Melo Martins PC, Silva AP, Garicochea B. The fibroblast growth factor receptor 4 (FGFR4) Arg388 allele correlates with survival in head and neck squamous cell carcinoma. Exp Mol Pathol. 2007;82:53–57. doi: 10.1016/j.yexmp.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Liu R, Li J, Xie K, Zhang T, Lei Y, Chen Y, et al. FGFR4 promotes stroma-induced epithelial-to-mesenchymal transition in colorectal cancer. Cancer Res. 2013;73:5926–5935. doi: 10.1158/0008-5472.CAN-12-4718. [DOI] [PubMed] [Google Scholar]
- Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- Roidl A, Berger HJ, Kumar S, Bange J, Knyazev P, Ullrich A. Resistance to chemotherapy is associated with fibroblast growth factor receptor 4 up-regulation. Clin Cancer Res. 2009;15:2058–2066. doi: 10.1158/1078-0432.CCR-08-0890. [DOI] [PubMed] [Google Scholar]
- Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamin o]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem. 2011;54:7066–7083. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- Wilson TR, McEwan M, McLaughlin K, Le Clorennec C, Allen WL, Fennell DA, et al. Combined inhibition of FLIP and XIAP induces Bax-independent apoptosis in type II colorectal cancer cells. Oncogene. 2009;28:63–72. doi: 10.1038/onc.2008.366. [DOI] [PubMed] [Google Scholar]
- Longley DB, Wilson TR, McEwan M, Allen WL, McDermott U, Galligan L, et al. c-FLIP inhibits chemotherapy-induced colorectal cancer cell death. Oncogene. 2006;25:838–848. doi: 10.1038/sj.onc.1209122. [DOI] [PubMed] [Google Scholar]
- Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, Carey PD, et al. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res. 2004;10:2158–2167. doi: 10.1158/1078-0432.ccr-03-0362. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- Longley DB, Allen WL, Johnston PG. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim Biophys Acta. 2006;1766:184–196. doi: 10.1016/j.bbcan.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Ong SH, Guy GR, Hadari YR, Laks S, Gotoh N, Schlessinger J, et al. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Honegger AM, Rotin D, Fischer R, Bellot F, Li W, et al. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet. 1999;8:35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- Hart KC, Robertson SC, Kanemitsu MY, Meyer AN, Tynan JA, Donoghue DJ. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene. 2000;19:3309–3320. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- Crose LE, Etheridge KT, Chen C, Belyea B, Talbot LJ, Bentley RC, et al. FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clin Cancer Res. 2012;18:3780–3790. doi: 10.1158/1078-0432.CCR-10-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno T, Asa SL, Zheng L, Mayr T, Ullrich A, Ezzat S. The FGFR4-G388R polymorphism promotes mitochondrial STAT3 serine phosphorylation to facilitate pituitary growth hormone cell tumorigenesis. PLoS Genet. 2011;7:e1002400. doi: 10.1371/journal.pgen.1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197:801–817. doi: 10.1083/jcb.201108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam RM, Danciu T, McLellan DL, Borer JG, Lin J, Zurakowski D, et al. A nuclear form of the heparin-binding epidermal growth factor-like growth factor precursor is a feature of aggressive transitional cell carcinoma. Cancer Res. 2003;63:484–490. [PubMed] [Google Scholar]
- Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WL, Stevenson L, Coyle VM, Jithesh PV, Proutski I, Carson G, et al. A systems biology approach identifies SART1 as a novel determinant of both 5-fluorouracil and SN38 drug resistance in colorectal cancer. Mol Cancer Ther. 2012;11:119–131. doi: 10.1158/1535-7163.MCT-11-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Wilson TR, McLaughlin KM, McEwan M, Sakai H, Rogers KM, Redmond KM, et al. c-FLIP: a key regulator of colorectal cancer cell death. Cancer Res. 2007;67:5754–5762. doi: 10.1158/0008-5472.CAN-06-3585. [DOI] [PubMed] [Google Scholar]
- Oladipo O, Conlon S, O'Grady A, Purcell C, Wilson C, Maxwell PJ, et al. The expression and prognostic impact of CXC-chemokines in stage II and III colorectal cancer epithelial and stromal tissue. Br J Cancer. 2011;104:480–487. doi: 10.1038/sj.bjc.6606055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.