Abstract
Pancreatic ductal adenocarcinoma, an aggressively invasive, treatment-resistant malignancy and the fourth leading cause of cancer deaths in the United States, is usually detectable only when already inevitably fatal. Despite advances in genetic screening, mapping and molecular characterization, its pathology remains largely elusive. Renewed research interest in longstanding doctrines of tumor metabolism has led to the emergence of aberrant signaling pathways as critical factors modulating central metabolic networks that fuel pancreatic tumors. Such pathways, including those of Ras signaling, glutamine-regulatory enzymes, lipid metabolism and autophagy, are directly affected by genetic mutations and extreme tumor microenvironments that typify pancreatic tumor cells. Elucidation of these metabolic networks can be expected to yield more potent therapies against this deadly disease.
Keywords: pancreatic cancer, oncogenic Ras, metabolism, glutamine metabolism, glycolysis, pentose phosphate pathway
Facts
Pancreatic cancer is the 10th most prevalent cancer in men and the 9th in women, and each year leads for approximately 3% of new cases of cancer in the United States.
Despite comprehensive endeavor and major progress made toward improving its prognosis and increasing survival rates (mostly via surgical intervention coupled with chemotherapy and radiation), survival rate for pancreatic cancer has not improved markedly and above 80% of patients develop a local metastatic tumor at progressive stage at prognosis time.
Ample scientific evidence suggest that oncogenic constitutively active K-Ras and its overactivated downstream signaling factors, such as B-Raf, phosphatidylinositol-3-kinase (PI3K) and Akt are strong promoters of pancreatic cancer tumorigenicity.
Oncogenic Ras-driven signals propel abnormal chain of metabolic alterations – including enhanced glycolysis, diverted glutamine consumption, anomalous pentose phosphate pathway (PPP) and autophagy – leading to phenomenon defined as ‘metabolic addiction' of pancreatic tumor cells, which ultimately contribute extensively to development, survival and invasiveness of pancreatic tumor cells.
Open Questions
What are the main metabolic pathways, molecular components and their relation to oncogenic Ras pathway, which are taking part in metabolic addiction of pancreatic cancer cells?
Does Salirasib, a well-established anti-Ras inhibitor, holds promise for the treatment of pancreatic tumor cells?
Could a combinatorial drug–therapy that design to hit critical addictive metabolic pathways be the clue for eradicating pancreatic tumors, while protecting normal cells?
Pancreatic cancer is the 9th most common cancer in women and the 10th in men, and accounts for an estimated 3% of new cancer cases in the United States each year. Despite its relatively low epidemiological ranking, it is one of the most lethal of all types of cancer, and with a 5-year survival rate of only 4% it is rated as the fourth leading cause of cancer deaths for both men and women in the United States. Thus, each year nearly 37 000 patients in the United States and >213 000 people worldwide die from this form of cancer.1, 2 Despite extensive efforts and the considerable progress made toward improving its detection3, 4, 5, 6 and survival rates over the past few decades,7, 8, 9 the 5-year survival rate for pancreatic cancer has not changed significantly, increasing – according to data from surveillance, epidemiology and end results2 – from 3% in 1975 to only 5.4% in 2005 (see other reviews Conroy et al.,10 Muller et al.11 and Poruk et al.12).
The most common type of pancreatic cancer (accounting for 95% of cases) is adenocarcinoma, which arises in the exocrine part of the pancreas and is classified as pancreatic ductal adenocarcinoma (PDAC). A minority (∼5%) of pancreatic tumors originates from islet cells and is classified as neuroendocrine tumors, and specifically as pancreatic endocrine tumors. The symptoms that lead to diagnosis of the disease depend on tumor location, size and tissue of origin, and may include distressing symptoms of biliary drainage obstruction, infection, abdominal pain, lower back pain, and – where the tumor compresses the bile duct – jaundice.
Although the main beneficial treatment for pancreatic cancer is surgical resection followed by radiation and/or chemotherapy, >80% of patients with pancreatic cancer suffer from a progressive local metastatic tumor that is unresectable by the time of diagnosis.13, 14 The posterior location of the pancreas, adjacent to the common bile duct, duodenum, celiac plexus, superior mesenteric artery and portal vein, makes diagnosis extremely difficult. As a result, clinical symptoms of pancreatic cancer, often referred to as the ‘silent killer', are usually unremarkable until the neoplasm has progressed to an advanced stage typified by metastatic spread to regional lymph nodes, liver, peritoneal cavity, and – rarely – to the lungs, bone or brain.1, 2 Therefore, early-stage detection and diagnosis are crucial for improving the death rates of this disease.15 Importantly, familial pancreatic cancer kindreds may become the major beneficiaries of newly discovered diagnostic means and risk factors, as they are known to be at high risk of developing the disease.16, 17
The National Familial Pancreas Tumor Registry, first established in 1994 at Johns Hopkins Hospital in Baltimore, MD, USA, paved the way for the screening and gene discovery studies that demonstrated the familial clustering of pancreatic cancer.16, 17 These seminal studies quantified the risk of hereditary pancreatic cancer and promoted knowledge of the etiology of the disease by identifying several germline mutations that predispose their carriers to pancreatic cancer, including those of BRCA2, PALB2, ATM, p16, PRSS1, STK11, hMLH1 and FANCG genes.18, 19, 20, 21, 22 These mutated genes can provide targets for personalized therapy, and carriers can be counseled regarding their increased risk and the available therapeutic choices among treatments such as preventive surgery, screening and chemopreventive interventions.16 Although a number of novel pancreatic tumor markers have recently been discovered through the use of global analyses of gene expression,23, 24 given the rarity of pancreatic cancer in the general population (approximately 9 cases per 100 000 people in the United States) it is questionable whether the application of screening tests to selected groups at high risk of developing this disease can be justified. Hence, the prognosis of patients with pancreatic tumors remains dismal. The malignancy is profoundly active and responds poorly to any form of therapy.25 Therefore, a substantial research effort is currently focused on identifying novel therapeutic targets for this deadly disease. An intriguing concept that marks a promising future in cancer therapy is the linkage between oncogene addiction and metabolic pathways. Renewed scientific interest in this association has yielded a variety of novel disease targets that should be considered for the development of new therapeutic combinations aimed at eradicating pancreatic tumors and possibly other forms of cancer.
The concept of ‘oncogene addiction', a term coined by Bernard Weinstein,26 refers to the common phenomenon that tumor cells, despite the striking number of their genetic aberrations, develop marked dependency on a particular oncogenic pathway for their sustained survival and proliferation. As a result, tumorigenic cells paradoxically often lose the function of activating alternative cascades that normally act in parallel. Thus, although inactivation of the normal counterpart cascade in wild-type cells can typically be tolerated without conspicuous ramifications, cancer cells (which are inherently less adaptable) often respond markedly to inactivation of their addictive pathway. The potent implication of this abnormal characteristic of cancer cells is that blocking of the crucial cascade upon which they rely should have deleterious effects on the mutated cells while sparing the normal, nonaddicted cells. This type of effective discriminative activity is the desired attribute of any efficient anticancer agent. A growing body of data on aberrant metabolic pathways implicated in cancer etiology now suggests that progressively accumulating mutations and epigenetic aberrations in genes encoding the main enzymes of specific metabolic pathways are prime contributors to oncogenic addiction in many cancers.27, 28 In this review, we discuss aberrations in signal transduction pathways found to be associated with metabolic reprogramming of transformed pancreatic neoplasms (Figures 1 and 2). Understanding of these critical circuits, which may be the Achilles' heel of pancreatic tumors, could bring us closer to the discovery of novel targets that will promote the development of effective therapies against pancreatic cancer.
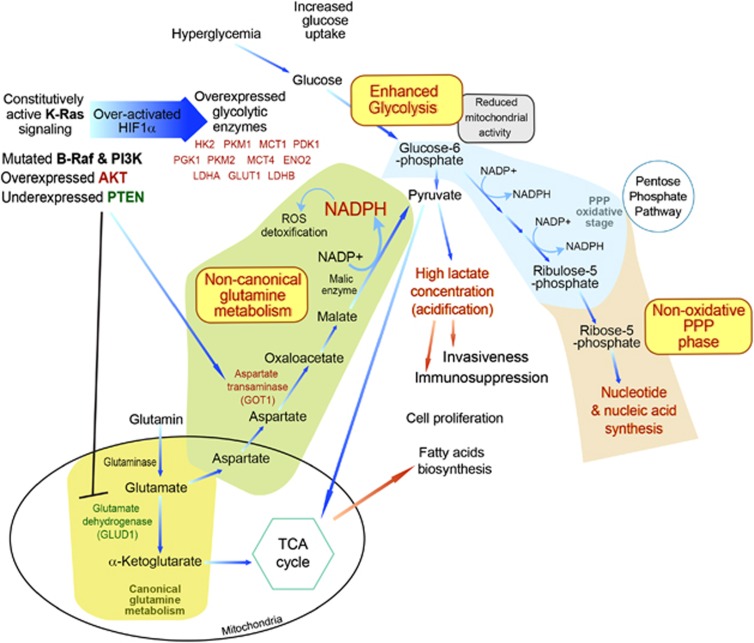
Figure 1.
Schematic overview of key Ras-regulated metabolic pathways that become aberrantly active in pancreatic tumor cells
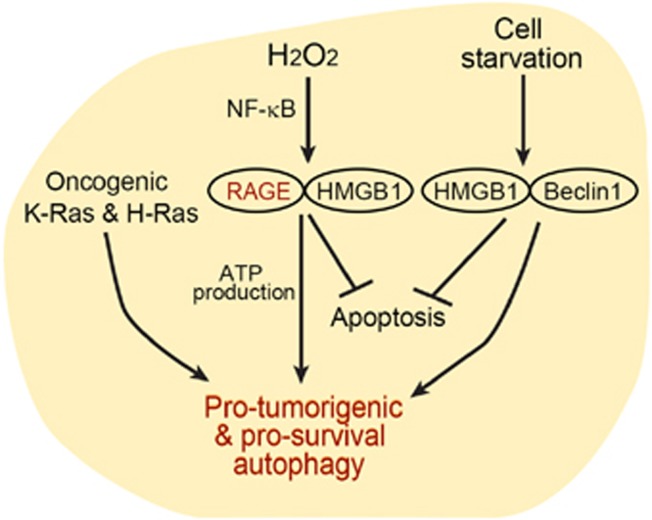
Figure 2.
Illustrative of RAGE/HMGB1 interaction and regulation of autophagic metabolism and promotion of cell survival in pancreatic tumor cells
The Warburg Phenomenon
One of the earliest enigmas raised in cancer etiology was the observation that most cancer cells, even under nonhypoxic conditions, predominantly utilize cytosolic aerobic glycolysis and lactate fermentation rather than mitochondrial oxidative phosphorylation of pyruvate for their energy production. This metabolic anomaly, known as the ‘Warburg effect',29, 30 was first described in the 1920s by Otto Warburg as a shift in cancer cells to the primitive metabolism exhibited by yeast.31 Recent studies have implicated a multitude of oncogenic aberrations that mechanistically stimulate activation of glucose uptake in triggering this metabolic shift.32 Constitutively active components of the Ras pathway stimulate cellular glucose uptake and metabolic rate, thereby overcoming the capacity of the cell to utilize mainly glucose for its bioenergetic requirements. As a result, tumorigenic cells secrete excess metabolites of the glycolytic pathway in the form of lactic acid. Recent studies have strongly implicated aerobic glycolysis in the malignant etiology of pancreatic tumor cells. Thus, for example, the expression of key enzymes that participate in glycolytic metabolism was strongly increased in pancreatic tumor cells compared with normal pancreatic cells, leading to a significant induction of aerobic glycolytic metabolism. These overexpressed enzymes included hexokinase 2 (HK2), phosphoglycerokinase 1, pyruvate dehydrogenase kinase isozyme 1 (PDK1), lactate dehydrogenase A and B (LDHA, LDHB), enolase 2 (ENO2) and pyruvate kinase muscle (PKM1 and PKM2), in addition to the glucose and lactate transporters, PDK1 and LDHA, LDHB, glucose transporter 1 (GLUT1) and monocarboxylate transporters 1 and 4 (MCT1 and MCT4).33, 34, 35, 36, 37, 38, 39 The shift to enhanced glucose metabolism in hypoxic pancreatic cancer cells is clearly manifested by the substantial accumulation of the glycolysis end-product lactic acid in the tumor microenvironment.33 Interestingly, pancreatic cancer patients frequently also suffer from diabetes and hyperglycemia, conditions typified by high blood sugar.40 A recent investigation of the link between hyperglycemia and stimulation of pancreatic cancer cell growth showed that incubation of hypoxic MiaPaCa-2 pancreatic cancer cells with excess glucose results in increased expression of hypoxia-inducible factor (HIF)-1α, accompanied by an increase in cellular ATP and a decrease in mitochondrial activity. Glucose metabolism could also be stimulated by extracellular glucose and hypoxia independently of HIF-1α, as shown by the metabolic response of a MiaPaCa-2 subline devoid of HIF-1α. Nevertheless, hypoxic pancreatic cells harboring HIF-1α showed an increased capacity for migration relative to their HIF-1α-null cellular counterparts, indicating that glucose stimulates pancreatic cancer cell migration by both HIF-1α-dependent and HIF-1α-independent mechanisms.41
Although glycolysis is much less efficient than mitochondrial respiration (producing only two molecules of ATP per glucose molecule compared with the 38 ATP molecules produced by aerobic respiration), there are several reasons why the increased glucose uptake for glycolytic ATP production is advantageous for the tumorigenic properties of pancreatic cancer cells. First, by using aerobic glycolysis under aerobic conditions, pancreatic tumor cells can still thrive independently of an inconstant oxygen diffusion that would otherwise be deleterious for normal cells that are largely dependent on oxidative phosphorylation for their ATP production.42
Second, lactic and bicarbonic acids, as metabolic end-products of aerobic glycolysis, are generated robustly by pancreatic cancer cells and form a major acidity buffer43 that preferentially promotes the invasiveness of these cells.33, 37, 44 Intriguingly, in the course of exploration of the positive correlation between tumor burden and high lactate levels, a close linkage was discovered between the intracellular accumulation of lactic acid and the antitumorigenic immune response. As activated cytotoxic tumor-infiltrating T lymphocytes rely on glycolysis (which is tightly dependent on efficient secretion of lactic acid), increased concentrations of lactate in the tumor environment lead to a decline in the intracellular/extracellular lactate gradient, thereby blocking lactate secretion. Thus, the glycolysis rate (and consequently the entire metabolism) in T cells is markedly reduced. As a result, the degree of immunosuppression in the pancreatic tumor niche is strongly increased, as manifested by inhibition of T-cell proliferation and suppression of their cytokine production.45 In this regard, it is interesting that an increase in ENO1-specific regulatory T cells (Tregs) was detected in pancreatic cancer tumor cells exhibiting high levels of the key glycolytic enzyme ENO1. These Tregs were found to efficiently impose immunosuppression in vitro by suppressing the proliferative response of ENO1-specific effector T cells, implying a possible role for Tregs in stimulating pancreatic cancer progression.46
Third, tumor cells utilize glycolytic intermediates to fuel anabolic processes necessary for cell growth. Thus, for example, alanine aminotransferase synthesizes alanine and malate from pyruvate, glucose 6-phosphate is converted by phosphoglucomutase and glycogen phosphorylase to glycogen, and dihydroxyacetone phosphate is converted to triacylglyceride and phospholipid.47 Several studies have shown that in proliferating cancer cells pyruvate can enter the tricarboxylic acid (TCA) cycle, leading to the export of acetyl coenzyme A (acetylCoA) from the mitochondrial matrix to the cytosol and thus making acetylCoA available for synthesis of fatty acids, cholesterol and isoprenoids.32 Expression of fatty acid synthase, known to catalyze the synthesis of long-chain fatty acids from nicotinamide adenine dinucleotide phosphate (NADPH), acetylCoA and malonyl coenzyme A, was found to correlate with an advanced stage of pancreatic cancer.48, 49
Numerous studies have pointed to the pragmatic advantage of developing therapies against the addictive glycolytic pathway with the aim of eradicating pancreatic cancer cells. As glycolysis can be separated into two phases: (i) the preparatory phase (or ‘energy investment' phase), in which two ATP molecules are consumed and one glucose molecule is split, and (ii) the harvesting phase (or ‘payoff' phase), in which energy is extracted in the form of four ATP molecules – inhibitors of specific glycolytic enzymes can be divided accordingly. Thus, for example, inhibitors of HK, the first glycolytic enzyme that phosphorylates glucose to produce glucose-6-phosphate, interfere with the preparatory phase, while inhibitors of the last glycolytic enzyme LDHA, which catalyzes the reversible conversion reaction of pyruvate to lactate coupled with the recycling of NAD+, abolish the glycolytic harvesting step. Studies related to the preparatory glycolytic phase demonstrated the potent anti-glycolytic effect of everolimus, a rapamycin analog, on pancreatic Panc-1 human cancer cells. The suggested mechanism by which the drug acts is through a gradual increase in expression of miR-143, which consequently targets and reduces expression of the preparatory enzyme HK2.50 Other studies of the harvesting glycolytic stage have demonstrated the effect of enzymatic inhibition of LDHA. As LDHA utilizes NADH as a co-factor for catalyzing its reaction, the ratio of NADH to NAD+ is increased as result of LDHA inhibition. NAD+ is a vital electron acceptor that sustains the glycolytic pathway through its reaction with GAPDH;51 therefore, inhibition of LDHA has a critical effect on glycolysis. Recent studies have shown that LDHA performs a critical function as a generator of glycolysis in cancer cells. Inhibition of its activity in a mouse pancreatic cancer model either by FX11, a small-molecule inhibitor of LDHA or by siRNA knockdown, imposes glycolysis shutdown, ATP reduction and significant induction of oxidative stress.52 Importantly, glycolytic blockade culminates in tumor growth inhibition of pancreatic cells.52
In our own studies, in which the potent Ras inhibitor Salirasib was implemented on a range of glioblastoma cell lines, we discovered that inhibition of oncogenic Ras has robust anti-glycolytic effects53, 54, 55 (see below).
The Pentose Phosphate Pathway
In addition to the above-mentioned glycolysis-related biochemical mechanisms, another vital biogenesis channel, namely the PPP, is attracting renewed research attention in relation to pancreatic cancer. The PPP (also called the phosphogluconate pathway) is a biphasic cytosolic process, which, like glycolysis, utilizes glucose-6-phosphate as its initial metabolite, and therefore represents a biochemical alternative to glycolysis.56 Unlike the glycolysis pathway, in which the main end-product is ATP, the PPP is primarily an anabolic process that in the first (‘oxidative') phase produces NADPH, which is utilized by cells as a reducing agent in biosynthetic reactions, and in the second (‘nonoxidative') phase produces 5-carbon sugars. Tumor cells, by using the oxidative phase, are capable of utilizing glucose for generating NADPH.56 Besides fueling the biogenesis reactions that rebuild macromolecules, NADPH also serves as an antioxidative metabolite in the detoxification of reactive oxygen species (ROS) through the regeneration of reduced glutathione that scavenges ROS and maintains the normal reduced state of the cell, thereby providing protection against hostile microenvironments during accelerated cell proliferation or in the course of chemotherapy.57 Notably, glutathione reductase, which together with NADPH-generating pathways and glutathione provides this cellular defense system against oxidants, was reported to be highly expressed in pancreatic islet cells.58
The pentose sugars generated in the nonoxidative PPP phase serve as primary intermediates in the synthesis of nucleotides and nucleic acids. At times of increased cellular demand for the pentose monosaccharide, such as during cell growth and repair, these metabolic reactions enable excess pentoses to be recycled as intermediates of the glycolytic pathway and essential precursors for nucleic acid biosynthesis.59 Recent work by Ying et al.60 highlighted the importance of the reversible nonoxidative PPP phase in pancreatic tumor cells. Maintenance of pancreatic tumors was found to be strongly driven by mutated K-Ras, which stimulates glucose uptake and conveyance of its biosynthetic intermediates into the nonoxidative branch of the PPP, generating ribose-5-phosphate, which is then utilized for the synthesis of nucleic acids. Nevertheless, that study revealed, remarkably, that in contrast to the canonical paradigm in which generation of nonoxidative mediators is coupled with the synthesis of oxidative ones, the K-Ras-driven accelerated glycolytic flux in pancreatic tumor cells is reprogrammed to bypass the oxidative NADPH biosynthesis phase of the PPP and instead robustly enhances the nonoxidative branch60 (Figure 1). Thus, this study disclosed a novel connection between oncogenic K-Ras and the nonoxidative PPP phase, linking K-Ras directly to DNA biosynthesis. The finding that the generation of pentoses was decoupled from NADPH generation led to the suggestion that a substitute metabolic pathway may be responsible for maintaining cellular redox homeostasis PDAC. An alternative glutamine-consuming pathway generating NADPH was indeed discovered recently, as discussed below in the chapter on glutamine addiction.
Ras Signaling, Glycolysis Addiction and Pancreatic Cancer
In mammals, three genes (H-Ras, K-Ras and N-Ras) are responsible for encoding four highly homologous small GTPase Ras proteins (H-Ras, K-Ras4A, K-Ras4B and N-Ras).61, 62 These proteins belong to a larger Ras-related GTPase protein superfamily whose members function as molecular switches that alternate between inactive (GDP-bound) and active (GTP-bound) conformations. Extracellular signals that activate plasma membrane-bound receptor tyrosine kinases induce activation of Ras guanine nucleotide exchange factors (GEFs), which prompt Ras-mediated exchange of GDP for GTP. In its GTP-bound state the affinity of Ras for a multitude of intracellular factors is increased, with consequent triggering of a diversity of signal transduction cascades including PI3K-Akt, Raf-Mek1/2-Erk1/2, RalGEF and Rho/Rac.62, 63 Activated Ras signals coming from the plasma membrane are carried over the intracellular compartments by a variety of molecular mediators before reaching their target in the nucleus, where they ultimately control the activity of transcription factors that effectively govern an extensive array of cellular responses. Among the chief transcription factors regulated by Ras are Myc, NF-κB, E2F, HIF-1α, AP-1 and c-Jun.63 Thus, by virtue of its many arms, and depending on cellular context and both extracellular and intracellular conditions, Ras is capable of orchestrating numerous cellular processes such as cell survival, migration, cell cycle, metabolism and differentiation.
The Ras signaling pathway is arguably the most intensively studied pathway participating in the metabolic reprogramming that actively contributes to tumorigenicity. The finding that the great majority (>90%) of patients with pancreatic cancer harbor a gain-of-function K-Ras mutation on chromosome 12p (at codons 12, 13 and 61)62, 63 strongly supports the concept that constitutively active Ras pathways have an active role in maintaining the carcinogenic phenotype and in altering the metabolic pathways that promote rapid progression of pancreatic tumors. Mutation in K-Ras genes are detectable not only in tumor tissues but also in the plasma DNA of patients with pancreatic cancer, although this diagnostic finding is often correlated with more advanced stage disease, limiting its beneficial use as a prognostic marker.64, 65 K-Ras mutations are often detected in pancreatic intraepithelial neoplasia (PanIN) and PDAC, indicative of the essential role of Ras in pancreatic carcinogenesis. Nevertheless, mutations in K-Ras alone seem to be insufficient for transformation of the pancreatic tissue to PDAC. Endogenous expression of active K-Ras(G12D) directed to progenitor cells that reside in the mouse pancreas was found to result in frequent development of highly proliferative PanIN, but with only infrequent progression of these precursor lesions to more invasive metastatic adenocarcinomas.66 In addition, transfection of K-Ras(G12V) into human pancreatic duct epithelial cells derived from normal human pancreas resulted in only moderately aberrant phenotypes,67 suggesting that in addition to the constitutive activation of K-Ras genetic and/or epigenetic events are required for promoting development of PDAC.
Although activating mutations in Akt and PTEN were not detected in pancreatic tumors,68, 69, 70 other Ras-signaling factors – B-Raf and PI3K – have been reported to harbor activated mutations in pancreatic cancer cells. B-Raf, a serine/threonine protein kinase located second to Ras in the signaling cascade, manifests a common mutational pattern in a few primary cancers, including 10% of colorectal carcinomas and 66% of melanomas.71, 72, 73 Mutations in K-Ras and in B-Raf are nearly always mutually exclusive, and mutations in B-Raf appear in pancreatic cancers with wild-type Ras at a rate of one in every three cases.74 Another prominent molecular mediator of Ras signaling that becomes mutated in pancreatic cancer is PI3K. Genomic evaluation of the role of PIK3Ca, the gene encoding PI3K during tumorigenesis of pancreatic cells, indicates that approximately 10% of pancreatic cancer precursors harbor activating mutations in this gene.75 Nevertheless, even when mutations in PI3K and Akt genes are undetectable, this signaling pathway is found to be constitutively active in most pancreatic cancers, apparently owing to the deregulated expression of the antagonist signaling factor PTEN, which in pancreatic cancers is lost or significantly reduced.76 In addition, 60% of pancreatic cancers exhibit amplification or overactivation of Akt2 kinase, the direct signaling target of PI3K.77, 78, 79, 80, 81
Activated Ras becomes engaged with metabolic pathways through multiple effector pathways, including those of PI3K/Akt and MAPK.82, 83 The PI3K/Akt signaling pathway is strongly associated with regulatory alterations of several prominent metabolic factors. These include elevated expression levels and translocation to the cellular membrane of the GLUT1,82 stimulation of phosphofructokinase enzymatic activity,83 mitochondrial localization of the glycolytic enzymes HK1 and HK284, 85 and stabilization of HIF-1α protein, whereas MAPK/ERK signaling was shown to enhance HIF-1α transcriptional activity.86 Furthermore, constitutively active K-Ras(G12V) in human embryonic kidney cells (HEK293) was found to lead to mitochondrial dysfunction, respiration suppression and increased glycolytic activity.87 Several studies have documented the strong suppressive effects that can be imposed by downregulation of Ras signals on the glycolytic pathway in various tumor cells. For example, a PI3K inhibitor abolished the expression of HIF-1α in Ras-transformed NIH3T3 cells88 and in prostate cancer cells.89 In addition, suppression of ERK in colon cancer cells90 and in hepatic cancer cells91 resulted in suppression of HIF-1α expression and activation.
Over the last two decades, our group has developed conceptually new Ras inhibitors that specifically target the active form of Ras (Ras-GTP). We demonstrated the potent antiproliferative effects of treatment with the Ras inhibitor trans-farnesylthiosalicylic acid (also known as Salirasib) on a variety of cancer cells, including those of melanoma,92 Merkel cell carcinoma,93 LNCaP, CWR-R1,94 and Panc-1 and MIA PaCa-2 pancreatic cancers.95, 96, 97 Our first genome-wide studies of the effects of Ras inhibition on metabolic pathways in tumorigenic cells indicated that Ras inhibition leads to profound anti-oncogenic effects in glioblastoma multiforme cells.52, 53, 54, 98, 99 These strong effects were caused by degradation of HIF-1α, which blocked glycolysis and triggered severe energetic crises.53, 100 Those promising findings encouraged us to adopt a novel strategy in which Salirasib could be combined with other drugs that can shut down glycolysis through mechanisms that do not involve HIF-1α blockage, thereby repressing metabolic pathways independently of the inhibitory activity of Salirasib. We therefore examined the effects of a combination of Salirasib and the synthetic glucose analog, 2-deoxy-D-glucose (2-DG). The latter had been reported to act as a potent metabolic inhibitor that is selectively directed to tumor cells that consume glucose at high rates under hypoxic conditions.98, 101 When introduced into tumorigenic cells, 2-DG competes with endogenous glucose for key glycolytic enzymes, thereby reducing metabolism rate.98, 101 Recent investigations of the antitumorigenic effects of 2-DG administered in combination with the anti-glycolytic agent 3-bromopyruvate (3-BrPA) in MiaPaCa2 and Panc-1 pancreatic cancer cells indeed demonstrated anticancer effects, manifested by energy depletion and increased cell necrosis.99 Additional studies showed that combinatorial treatment of 2-DG and the antidiabetic drug metformin in LNCaP, P69, PC-3 and DU145 prostate cancer cells leads to almost complete cell-cycle blockage and apoptotic cell death, caused by inhibition of mitochondrial respiration and glycolysis.102 Our recent studies demonstrated that combined treatment with Salirasib and 2-DG in Panc-1 pancreatic carcinoma cells leads to inhibition of additive cell growth, as well as to synergistic apoptosis and a complete contraction of Panc-1 tumor in nude mice.103 Taken together, our results and those of others establish proof-of-principle evidence for the therapeutic effects that Ras inhibition, through glycolysis shutdown, can impose in pancreatic tumors. Thus, glycolysis emerges as one of the central metabolic modules to which pancreatic tumor cells become addicted (see below), and targeting of this pathway using combinatorial drug administration to exploit more than one mechanistic approach has proved to be a highly promising strategy.
Glutamine Addiction
In normal mammalian cells, signaling by growth factors directs nutrient uptake from the extracellular environment to support cell survival, ATP production, cellular biosynthesis (of proteins, RNA, DNA and lipids), cell growth and proliferation. In contrast, a well-established feature of cancerous cells is their strong reliance on cell-autonomous nutrient uptake and metabolism. Glutamine – the most abundant amino acid in the cytoplasm – and glucose both serve as primary sources of carbon for ATP production and biosynthesis in tumorigenic cells.104, 105, 106 Oxidation of acetylCoA to CO2 by the TCA cycle is the main process maximizing ATP production in nonproliferative cells.107 In proliferating cells, however, in addition to providing ATP the TCA cycle functions in biosynthetic pathways in which intermediate metabolites exit the cycle and become converted primarily to fatty acids and nonessential amino acids (NEAAs).104 Glutamine is one of the major anaplerotic precursors that fuel the TCA cycle with a carbon source. In addition, it is an important provider of nitrogen for the vital set of biosynthetic reactions underlying the generation of nucleotide, NEAAs and hexosamine. For reloading the TCA cycle, glutamine needs to be converted to α-ketoglutarate, a central metabolite of glutamine metabolism.106 The enzymatic conversion of glutamine can be mediated through one of two alternative enzymatic reactions – either through a canonical anabolic glutamine metabolism mediated by an oxidative deamination reaction catalyzed by the mitochondrial matrix enzyme glutamate dehydrogenase (GLUD1),105 or via a noncanonical metabolic pathway conducted by transaminases,106 which catalyze transamination (transfer of an amino group from glutamate to the corresponding α-ketoglutarate). Accordingly, many cancer cell types exhibit increased glutamine consumption, a phenomenon dubbed ‘glutamine addiction' because the survival of these cancerous cells is evidently dependent on their glutamine content106 (reviewed in Lunt et al.,47 Wise et al.,106 Young et al.108 and Erickson et al.109). Indeed, various cancer cells utilize glutamine deamination mediated by the canonical pathway in which GLUD1 drives glutamine into the TCA cycle, which consequently promotes tumorigenic cell anabolism.110 Apart from its central role as a feeder of the TCA cycle, glutamine serves as a nitrogen donor that is crucial for the production of nucleotides, certain amino acids and nicotinamide. The synthesis of nitrogen-based nucleotides and NEAAs is a fundamental metabolic step in cancer cell growth. Through donation of its amide group and its conversion to glutamic acid, glutamine serves as an essential nitrogen donor in three autonomous enzymatic reactions responsible for purine synthesis and in two responsible for pyrimidine synthesis.109, 111 Thus, the switch of glutamine from serving as a NEAA in normal cells to an essential amino acid in cancerous cells accounts for its role as a major substrate feeding the anabolic growth processes of tumorigenic cells.
A new study by Son et al.111 has recently shown that distinctly from the general biochemical scenario, which is adopted by many cancer cell types, in PDAC cells glutamine seems to support tumorigenic growth by utilizing the noncanonical metabolic pathway. Silencing of glutaminase, the metabolic enzyme that generates glutamate from glutamine, was shown to significantly reduce PDAC cell growth, while addition of glutamate to the media restored cell growth in glutamine-deprived conditions. On the other hand, α-ketoglutarate, the product of canonical oxidative deamination of glutamate, failed to restore pancreatic cancer cell growth. Nevertheless, supplementation of the end-products of the noncanonical transaminase-mediated glutamine pathway – a combination of α-ketoglutarate and NEAA mixture – significantly restored proliferation in multiple PDAC lines, suggesting that PDAC cells can metabolize glutamine in a noncanonical pathway and that transaminases have an important role in metabolizing glutamine in PDAC111. Furthermore, PDAC cells that were incubated with the GLUD1 inhibitor epigallocatechin gallate or with GLUD1 shRNA were not affected by the treatment, while on the contrary growth inhibition was marked when PDAC cells were treated with the transaminase pan-inhibitor aminooxyacetate or when aspartate transaminase – glutamic-oxaloacetic transaminase 1 (GOT1) – was depleted (Figure 1).111 Silencing of K-Ras in these pancreatic tumorigenic cells significantly increased GLUD1 expression, concomitantly with a marked reduction in GOT1. In accord with those findings, five independent tumors derived from inducible K-Ras PDAC model mice exhibited elevated GOT1 expression and diminished levels of GLUD1.60 Taken together, the above findings point to a novel noncanonical metabolic model in which pancreatic tumor cells convey glutamine-derived aspartate into the cytoplasm, where it is further converted by GOT1 into oxaloacetate, then into malate and finally into pyruvate. As NADPH is generated via the final reaction mediated by malic enzyme, the net outcome of this tumorigenic pathway is a reduction in the NADP+/NADPH ratio, which therefore enables PDAC cells to maintain their redox homeostasis and to support cell proliferation.111 Along these lines, these innovative observations are now being considered as a potential ground for testing new candidate drugs for prophylactic studies in humans. Future studies are underway to advance our understanding of the interplay between this tumorigenic metabolic pathway and other cascades, as well as to determine whether these discoveries may apply in other cancer types as well. Extensive effort is also being devoted to developing small-molecule inhibitors against key enzymes that mediate this signaling pathway and testing their potency in models of pancreatic cancer.112
Fatty Acid and Lipid Regulation
Although the metabolism of fatty acids and lipids is not yet well understood, its prominent role in regulating the progression of various types of cancers in well known. An early study showed that lipids can stimulate cell growth of human pancreatic cancer cell lines, but did not enhance the growth of nonpancreatic cell lines.113 The proliferative effect of lipids on pancreatic cancer cells may act on several levels. First, normal pancreatic cells have been shown to utilize lipids as an energy source (as determined by oxidation of palmitic acid) at higher rates than other cell types (e.g., cells of adipose or liver tissue).114 Thus, the proliferative effect of lipids added as a supplement to the growth medium of pancreatic cancer cells may imply that lipids provide an energy source for pancreatic cell lines. Second, enhancement of cell proliferation by hormones and growth factors is known to be coupled with generation of lipids and lipid-derived messengers that participate in the activation of different growth-regulatory pathways.115 Tumorigenic pancreatic cells produce growth factors implicated in cell growth regulation through paracrine or autocrine mechanisms.116, 117, 118 Therefore, exogenous lipids can stimulate cell growth by increasing the levels of the intracellular signals linked to activation of the growth factor receptors.119 Third, lipids may also accelerate cellular growth through the promotion of membrane backbones. The type of dietary fat was shown to significantly influence the composition of fatty acids of pancreatic membranes.120, 121
In addition, a high-fat diet inducing inflammation in K-Ras (G12D) mouse models, which recapitulate human PDAC, was shown to enhance tumor promotion.122 The enhanced pancreatic tumorigenesis was prompted by a marked elevation in metabolic rates and energy uptake through an elevation in expression of genes encoding regulators of fatty acid uptake and oxidation.122 A high-fat diet and obesity are indeed strongly associated with the incidence of human pancreatic cancer.120, 123, 124 A few reports have indicated that consumption of omega-3 fatty acids has a suppressive effect on the progression of breast, prostate and colon cancers.121, 125, 126, 127 Blockage of cell-cycle progression and induction of programmed cell death were recently shown to be the outcomes of an omega-3-based fat diet in (EL)-K-Ras transgenic mice that develop pancreatic neoplasia.128 On the other hand, increased consumption of omega-6 fatty acids is highly correlated with progression of breast, prostate, colon and pancreatic tumors.129, 130, 131, 132, 133
Autophagy as an Essential Tumor Growth Mechanism in Pancreatic Tumors
Autophagy (also known as macroautophagy) is a regulated catabolic process of cytoplasmic organelles and macromolecules.129, 130, 134 The degradation process starts with invagination of a single-membrane vesicle,130 which sequesters cytoplasmic components into a double-membrane vesicle that forms the autophagosome.135 The autophagosome is then fused to the lysosomes, resulting in lysosomal degradation of its cargo.135 The autophagy process is initiated by the function of ULK1 and ULK2 proteins that were inactivated by mTOR activity,131, 136 and is followed by the activity of the ULK1/2-Atg13-FIP200-Atg101 complex, which mediates delocalization of class III PI3K (PI3KC3) from microtubules to the endoplasmic reticulum for initiation of vesicle nucleation.131, 132, 133, 136 The PI3KC3 complex, which contains the autophagy proteins Beclin1, p150 and Ambra 1, generates phosphoinositide 3-phosphate in the nucleation membrane, which stimulates recruitment of other autophagy (Atg) proteins to the autophagosome.137 Next, Atg12 is conjugated to Atg5 and Atg16L1 through an ubiquitination-like process to form the Atg16 complex.138 The Atg16 complex then mediates expansion and progression of autophagy at the autophagosomal membrane.138 In an additional ubiquitination-like process, regarded as the most specific step of autophagy, Atg4 cleaves LC3 protein, exposing its C-terminal glycine, which then binds to phosphatidylethanolamine (PE),139 promoting recruitment of LC3-PE to the autophagosomal membrane.139
The basic role of this metabolic mechanism, which becomes active under cellular starvation, is to facilitate recycling of cellular material for energy production. In cancer cells its function is rather complicated as it has conflicting implications depending on the context and the source of the tumorigenic tissue. In several cancer cell types, including breast cancer,140 Mantle cell lymphoma,141 chronic myeloid leukemia,142 non-small cell lung carcinoma143 and cervical cancer,144 autophagy induction was found to correlate with tumorigenesis suppression. In contrast, recent data indicate that in pancreatic tumor cells autophagy fulfills a pro-oncogenic role in transformation induced by oncogenes and in subsequent tumor maintenance. Evidence for the presence of autophagy in pancreatic tumors was provided at the end of the 1990s, when autophagic characteristics were observed in atypical acinar cell nodules of rats with pancreatic adenocarcinoma.145 A more recent study showed that basal autophagy was elevated in all examined human-derived PDAC cell lines, in 81% of primary PDAC tumor samples, and in all high-grade pancreatic intraepithelial neoplasms. However, only minimal autophagic signaling was observed in low-grade PanIN or normal pancreatic ductal epithelium.146 Retrospective examination of autophagy components in untreated pancreatic tumor tissues from human patients revealed the prominent presence of LC3 and pointed to a positive correlation between heightened autophagy and increased tumorigenic progression.147
Inhibition of autophagy by chloroquine treatment or genetic intervention led to ROS induction, augmentation of DNA damage and alteration of metabolic outcome as result of impaired mitochondrial oxidative phosphorylation. These events led to a marked growth suppression of pancreatic cancer cells, tumor regression and prolonged survival in a K-Ras-driven genetic mouse model of PDAC, and in cancer xenografts, suggesting that autophagy is necessary for tumorigenic growth of pancreatic cancer, and that pharmacologic intervention in this metabolic process may be of clinical benefit in the treatment of pancreatic neoplasms. As chloroquine and its derivatives are efficacious autophagy inhibitors and for some decades have been safely administered within therapeutic protocols for various purposes, these data can be directly translated into treatment of pancreatic cancer patients, providing a rational strategy toward curing this disease.146, 148
Given their dense stroma and poor vascularization, PDAC cells typically exhibit low nutrient intake and weak growth factor flux. The inadequate vascularization leads to hypoxic conditions, which induce autophagy. Autophagy is triggered primarily by the adaptive response action of the chief transcriptional regulator HIF-1α,149 which in addition to activating a large cassette of target genes activates the transcription of Bnip3 and Bnip3L, both essential for hypoxia-induced autophagy.150 Owing to the BH3 domain possessed by Beclin1, both Bnip3L and Bnip3 compete with Beclin 1 for binding to Bcl2, which upon their increased expression releases Beclin 1, causing induction of autophagy.149, 151 Nevertheless, it was suggested that the reaction of Bnip3 to hypoxic conditions and its induction of autophagy as a cellular response to starvation are part of its role during the early stages, but not during the more advanced stages of PDAC development. Recent studies indicate that the expression of Bnip3 is negatively correlated with the progression of pancreatic cancer, as unlike other HIF-1α target genes, Bnip3 was silenced by gene methylation in PDAC tumor cells.152 It was therefore proposed that Bnip3 becomes downregulated as the disease progresses, and that alternative mechanisms account for the continuous induction of autophagy at the more advanced stages of disease.
Recently, the receptor for advanced glycation end products (RAGEs)153 and ROS were shown to be prominent factors in autophagy in PDAC (Figure 2). RAGE, a multiligand receptor of the immunoglobulin superfamily,154 was shown to participate in the intracellular generation of ROS155, 156 and in tumor-promoting inflammation.157, 158 In PDAC tumor cells, RAGE is overexpressed and its expression is correlated with tumor cell survival, migration and invasiveness.159, 160 In both human and murine pancreatic tumor cell lines, depletion of RAGE significantly increased sensitivity to hypoxia, to UV radiation and to cytotoxic chemotherapy, concomitantly with significantly increased induction of cleaved caspase-3.161 RAGE overexpression, on the other hand, enhanced cell survival by reducing apoptosis and promoting autophagy.161 RAGE is a pattern-recognition receptor capable of binding several ligands, among them the nuclear chromatin remodeling protein known as high-mobility group box 1 (HMGB1).162 This protein was shown to be extracted from necrotic and inflammatory cells and, when released in the extracellular environment, to serve as a cytokine that may contribute to inflammation and tumor progression.163, 164 A decrease in intracellular HMGB1 through targeted knockdown was found to reduce autophagy and increase the sensitivity of PDAC-derived cells to apoptosis induced by the chemotherapeutic drug melphalan. It was therefore proposed that interaction of HMGB1with RAGE leads to enhanced cell resistance to programmed cell death in PDAC.161 In addition, exposure of pancreatic tumor cells to H2O2 was shown to increase RAGE expression through activation of the NF-κB signaling pathway, which was suppressed by administration of inhibitors of the NF-κB pathway.165 Further studies conducted in pancreatic tumor cells indicated that the inflammatory pathway mediated by HMGB1 and RAGE is essential for optimal mitochondrial production of ATP. Absence or reduction of either RAGE or HMGB1 significantly reduced ATP production and slowed tumor growth, clearly indicating a link between the HMGB1–RAGE pathway and alterations in bioenergetics.166
Besides activating the RAGE receptor in neighboring cells, HMGB1 was also shown to mediate autophagy in human Panc2.03 and mouse Panc02 pancreatic carcinoma cell lines through an additional mechanism, regulated by its interactions with Beclin 1. Upon cellular starvation, the oxidated HMGB1 translocates from the nucleus to the cytoplasm, where it disrupts interactions between Bcl-2 and Beclin 1 by competitively binding to the latter.167
Several recent studies have confirmed the correlation between autophagy induction and oncogenic constitutively active Ras in several human cancer cell lines, as well as the need for autophagy in Ras-induced malignant cell transformations.146, 148, 168, 169 It was recently suggested that in cancers mediated by Ras oncogenes, cancer cells develop autophagy addiction as a survival mechanism.170 Oncogenic K-Ras and H-Ras were shown to promote autophagy, which supports transformation and cell survival mainly through the maintenance of mitochondrial operation and ATP levels. In addition, in the scope of a recent comprehensive study, a few human cancer cell lines harboring endogenous oncogenic Ras mutations, including bladder, lung, colorectal and pancreatic cancer cell lines, exhibited significantly high levels of basal autophagy. These studies suggested that oncogenic Ras increases depletion of energy sources, rendering cells dependent on autophagy to preserve their mitochondrial function for energy production and possibly by providing catabolically derived metabolic substrates.148 Thus, the accumulated data point to a pro-survival action of autophagy in cancer cell lines transformed by oncogenic Ras, suggesting that Ras-driven pancreatic tumors may be particularly sensitive to inhibition of the autophagy metabolic pathway to which they have become addicted.
Conclusions
Pancreatic tumor cells harbor a unique combination of complicated genetic aberrations, such as mutations in K-Ras that lead to its overactivation and produce uncontrolled cell proliferation. Some of these distinctive aberrations have now been identified and are being successfully exploited in clinical laboratories as diagnostic markers. Nevertheless, pancreatic cancer remains one of the most lethal malignancies, and intense investigation of the pathology of its molecular circuits should lead to further progress in our ability to cure this deadly disease. The large body of accumulated evidence strongly supports the notion of ‘oncogenic addiction', in which these malignant pancreatic cells become highly dependent on their genomic aberrations. Thus, the same mutated signaling pathways that promote their unregulated growth are also responsible for their survival. For this reason, intensive efforts are underway to understand the mechanisms underlying oncogenic addiction of cancer cells, as breaking the chain of oncogenic addiction seems to hold hope for a cure by killing the cancer cells. Over the past two decades, this scientific undertaking has led to the discovery of metabolic pathways shown to be significant factors in the etiology of pancreatic cancer. The experimental evidence presented in this review reflects some of the central metabolic pathways currently known to fuel cellular metabolism in the tumorigenic microenvironment of pancreatic tissue. As described in each of the sections and summarized in Table 1, breakthroughs in the understanding of each of the denoted metabolic pathways can potentially lead to the discovery of novel soft spots, which could herald the advent of new therapies that can effectively target pancreatic cancer.
Table 1. Examples of the effects of various chemical compounds and treatments on metabolic pathways that involved in pancreatic tumorigenicity.
| Drug/compound/treatment | Treatment outcome(s) | Suggested mechanism | Reference |
|---|---|---|---|
| Everolimus (rapamycin analog) | Inhibition of cell proliferation and glycolysis, and induction of apoptotic cell death of Panc-1 human pancreatic cancer cells | Upregulation of levels of miR-143 transcripts, concomitant with decrease in HK2 transcripts levels | 50 |
| FX11 (inhibitor of LDHA) | Cell growth inhibition of P198 human pancreatic cancer cells (increased sensitivity under hypoxia). Inhibition of pancreatic tumor xenograft progression | Reduction of ATP levels and induction of marked oxidative stress and cell death. Decreased NAD+ recycling (increased NADH/NAD+ ratio) | 52 |
| K-Ras(G12D) doxy withdrawal (transgenic mice) | Doxy induction provokes acinar-to-ductal metaplasia and PanIN lesions within 2 weeks. Doxy withdrawal leads to rapid tumor regression with morphological deterioration of tumor cells and rapid degeneration of stromal elements, decreased tumor cell proliferation and increased apoptosis | Significant reduction in expression levels of several glycolytic enzymes, not accompanied by significant alterations in levels of TCA cycle intermediates. Marked reduction in nonoxidative PPP-specific metabolites S7P and SBP, accompanied by reduction in the flux of glucose into the nonoxidative arm of the PPP | 60 |
| Salirasib (Ras inhibitor) | Significant antiproliferative effects in pancreatic cancerous cells (Panc-1 and MIA PaCa-2) and on a variety of tumorigenic cell lines (melanoma, Merkel cell carcinoma, LNCaP, CWR-R1) | Reduction of Ras-mediated downstream signaling pathway (including Akt and Erk) | 92, 93, 94, 95, 96, 97 |
| Salirasib | Significant antiproliferative effects in glioblastoma cells, accompanied by apoptotic cell death | HIF-1α degradation, leading to glycolysis shutdown and severe energy crisis | 53, 100 |
| Salirasib in combination with 2-DG (glucose analog) | Additive inhibition of cell proliferation, synergistic induction of apoptosis and complete shrinkage of Panc-1 pancreatic carcinoma cells | Glycolysis inhibition | 103 |
| 2-DG in combination with 3-BrPA (anti-glycolytic agent) | MiaPaCa2 and Panc-1 pancreatic cancer cells manifest energy depletion and increased cell necrosis | Glycolysis inhibition | 99 |
| RNA interference targeting glutaminase | Significant reduction in PDAC cell growth | Targeting of transaminase, which abolishes the noncanonical pathway in which PDAC cells metabolize glutamine | 111 |
| Docosahexaenoic acid (DHA) diet (omega-3 fatty acid) | Recurrence and proliferative index of pancreatic precancer in EL-K-Ras mice was decreased in mice maintained on DHA diet. DHA treatment in tissue culture resulted in a dose-dependent reduction in cell cycle progression through both G1/G0 blockage and induction of programmed cell death | 128 | |
| RAGE-silencing by shRNA | Diminished autophagy, increased apoptotic rate and decreased tumor cell survival in human panc2.03 and mouse panc02 cancer cell lines | 161 | |
| HMGB1-knockdown by shRNA | Reduction in autophagy and increase in sensitivity of PDAC-derived cells to apoptosis induced by the chemotherapeutic drug melphalan | 161 | |
| Ethyl pyruvate (pharmacological inhibitor of nuclear HMGB1) | In vivo, tumor cell growth was significantly reduced. In vitro, increased apoptotic signal (PARP) and decreased signals of inflammation (p-p65), and autophagy (LC3-II), and reduced ATP production | 166 | |
| Chloroquine (autophagy blocker) | Significant growth suppression of pancreatic cancer cells, tumor regression and prolonged survival | Increased total and mitochondrial ROS levels, along with DNA damage. Severe decrease in oxidative phosphorylation. Significant elevation in uptake of glucose and lactate production | 146, 148 |
Acknowledgments
We thank Ms. Shirley Smith for editorial assistance. This work was supported by an Israel Science Foundation Grant 06049200403 (YK), and by the Prajs-Drimmer Institute for the Development of Anti-Degenerative Disease Drugs (YK). YK is the incumbent of the Jack H Skirball Chair in Applied Neurobiology.
Glossary
- PDAC
pancreatic ductal adenocarcinoma
- HK
hexokinase
- PDK
pyruvate dehydrogenase kinase isozyme
- LDHA
lactate dehydrogenase A
- ENO1
enolase 1
- PKM
pyruvate kinase muscle
- GLUT
glucose transporter
- MCT
monocarboxylate transporter
- (HIF)-1α
hypoxia-inducible factor 1α
- Treg
regulatory T cell
- TCA
tricarboxylic acid
- acetylCoA
acetyl coenzyme A
- NADPH
nicotinamide adenine dinucleotide phosphate
- PPP
pentose phosphate pathway
- ROS
reactive oxygen species
- GEF
guanine nucleotide exchange factor
- PI3K
phosphatidylinositol-3-kinase
- PanIN
pancreatic intraepithelial neoplasia
- 2-DG
2-deoxy-D-glucose
- 3-BrPA
3-bromopyruvate
- NEAA
nonessential amino acid
- GLUD1
glutamate dehydrogenase
- GOT1
glutamic-oxaloacetic transaminase 1
- PE
phosphatidylethanolamine
- RAGE
receptor for advanced glycation end product
- HMGB1
high-mobility group box 1
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37 (Suppl 8:S4–66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Pan S, Chen R, Brand RE, Hawley S, Tamura Y, Gafken PR, et al. Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J Proteome Res. 2012;11:1937–1948. doi: 10.1021/pr201117w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocker JR, Mohammed A, Aston CE, Brewer M, Lightfoot SA, Rao CV, et al. Mass profiling of serum to distinguish mice with pancreatic cancer induced by a transgenic kras mutation. Int J Cancer. 2013;133:2662–2671. doi: 10.1002/ijc.28285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Yu J, Kim H, Wolfgang CL, Canto M, Hruban RH, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600–3610. doi: 10.1158/1078-0432.CCR-12-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Chen H, Han C, Jin G, Wang D, Tang D. Detection and molecular analysis of circulating tumor cells for early diagnosis of pancreatic cancer. Med Hypotheses. 2013;80:833–836. doi: 10.1016/j.mehy.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- Kulke MH, Blaszkowsky LS, Ryan DP, Clark JW, Meyerhardt JA, Zhu AX, et al. Capecitabine plus erlotinib in gemcitabine-refractory advanced pancreatic cancer. J Clin Oncol. 2007;25:4787–4792. doi: 10.1200/JCO.2007.11.8521. [DOI] [PubMed] [Google Scholar]
- Quispe-Tintaya W, Chandra D, Jahangir A, Harris M, Casadevall A, Dadachova E, et al. Nontoxic radioactive Listeriaat is a highly effective therapy against metastatic pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:8668–8673. doi: 10.1073/pnas.1211287110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy T, Gavoille C, Adenis A. Metastatic pancreatic cancer: old drugs, new paradigms. Curr Opin Oncol. 2011;23:390–395. doi: 10.1097/CCO.0b013e3283473610. [DOI] [PubMed] [Google Scholar]
- Muller SA, Tarantino I, Martin DJ, Schmied BM. Pancreatic surgery: beyond the traditional limits. Recent Results Cancer Res. 2012;196:53–64. doi: 10.1007/978-3-642-31629-6_4. [DOI] [PubMed] [Google Scholar]
- Poruk KE, Firpo MA, Adler DG, Mulvihill SJ. Screening for pancreatic cancer: why, how, and who. Ann Surg. 2013;257:17–26. doi: 10.1097/SLA.0b013e31825ffbfb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray CJ, Ahmad SA, Matthews JB, Lowy AM. Surgery for pancreatic cancer: recent controversies and current practice. Gastroenterology. 2005;128:1626–1641. doi: 10.1053/j.gastro.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Reddy S, Wolfgang CL. The role of surgery in the management of isolated metastases to the pancreas. Lancet Oncol. 2009;10:287–293. doi: 10.1016/S1470-2045(09)70065-8. [DOI] [PubMed] [Google Scholar]
- Cleary SP, Gryfe R, Guindi M, Greig P, Smith L, Mackenzie R, et al. Prognostic factors in resected pancreatic adenocarcinoma: analysis of actual 5-year survivors. J Am Coll Surg. 2004;198:722–731. doi: 10.1016/j.jamcollsurg.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- Brune KA, Lau B, Palmisano E, Canto M, Goggins MG, Hruban RH, et al. Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst. 2010;102:119–126. doi: 10.1093/jnci/djp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer J. 2001;7:266–273. [PubMed] [Google Scholar]
- van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–2588. [PubMed] [Google Scholar]
- Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, Hruban RH, et al. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–826. [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis. 2000;21:857–864. doi: 10.1093/carcin/21.5.857. [DOI] [PubMed] [Google Scholar]
- Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe A.Oncogene addiction Cancer Res 2008683077–3080.discussion 3080. [DOI] [PubMed] [Google Scholar]
- Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia signalling controls metabolic demand. Curr Opin Cell Biol. 2007;19:223–229. doi: 10.1016/j.ceb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezene P, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y, Wu W, Ni X, Kuang T, Jin D, Wang D, et al. Lactate dehydrogenase A is overexpressed in pancreatic cancer and promotes the growth of pancreatic cancer cells. Tumour Biol. 2013;34:1523–1530. doi: 10.1007/s13277-013-0679-1. [DOI] [PubMed] [Google Scholar]
- Zhou W, Capello M, Fredolini C, Racanicchi L, Piemonti L, Liotta LA, et al. Proteomic analysis reveals Warburg effect and anomalous metabolism of glutamine in pancreatic cancer cells. J Proteome Res. 2012;11:554–563. doi: 10.1021/pr2009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuriya K, Kuramitsu Y, Ryozawa S, Fujimoto M, Mori S, Oka M, et al. Expression of glycolytic enzymes is increased in pancreatic cancerous tissues as evidenced by proteomic profiling by two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. Int J Oncol. 2007;30:849–855. [PubMed] [Google Scholar]
- Ishihara H, Wang H, Drewes LR, Wollheim CB. Overexpression of monocarboxylate transporter and lactate dehydrogenase alters insulin secretory responses to pyruvate and lactate in beta cells. J Clin Invest. 1999;104:1621–1629. doi: 10.1172/JCI7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Capello M, Fredolini C, Piemonti L, Liotta LA, Novelli F, et al. Proteomic analysis of pancreatic ductal adenocarcinoma cells reveals metabolic alterations. J Proteome Res. 2011;10:1944–1952. doi: 10.1021/pr101179t. [DOI] [PubMed] [Google Scholar]
- Akakura N, Kobayashi M, Horiuchi I, Suzuki A, Wang J, Chen J, et al. Constitutive expression of hypoxia-inducible factor-1alpha renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res. 2001;61:6548–6554. [PubMed] [Google Scholar]
- Permert J, Ihse I, Jorfeldt L, von Schenck H, Arnqvist HJ, Larsson J. Pancreatic cancer is associated with impaired glucose metabolism. Eur J Surg. 1993;159:101–107. [PubMed] [Google Scholar]
- Liu Z, Jia X, Duan Y, Xiao H, Sundqvist KG, Permert J, et al. Excess glucose induces hypoxia-inducible factor-1alpha in pancreatic cancer cells and stimulates glucose metabolism and cell migration. Cancer Biol Ther. 2013;14:428–435. doi: 10.4161/cbt.23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res. 2006;66:632–637. doi: 10.1158/0008-5472.CAN-05-3260. [DOI] [PubMed] [Google Scholar]
- Ota S, Geschwind JF, Buijs M, Wijlemans JW, Kwak BK, Ganapathy-Kanniappan S. Ultrasound-guided direct delivery of 3-bromopyruvate blocks tumor progression in an orthotopic mouse model of human pancreatic cancer. Target Oncol. 2013;8:145–151. doi: 10.1007/s11523-013-0273-x. [DOI] [PubMed] [Google Scholar]
- Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- Amedei A, Niccolai E, Benagiano M, Della Bella C, Cianchi F, Bechi P, et al. Ex vivo analysis of pancreatic cancer-infiltrating T lymphocytes reveals that ENO-specific Tregs accumulate in tumor tissue and inhibit Th1/Th17 effector cell functions. Cancer Immunol Immunother. 2013;62:1249–1260. doi: 10.1007/s00262-013-1429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Witkiewicz AK, Nguyen KH, Dasgupta A, Kennedy EP, Yeo CJ, Lisanti MP, et al. Co-expression of fatty acid synthase and caveolin-1 in pancreatic ductal adenocarcinoma: implications for tumor progression and clinical outcome. Cell Cycle. 2008;7:3021–3025. doi: 10.4161/cc.7.19.6719. [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu H, Li Z, Zhao Z, Yip-Schneider M, Fan Q, et al. Role of fatty acid synthase in gemcitabine and radiation resistance of pancreatic cancers. Int J Biochem Mol Biol. 2011;2:89–98. [PMC free article] [PubMed] [Google Scholar]
- Liu L, Gong L, Zhang Y, Li N. Glycolysis in Panc-1 human pancreatic cancer cells is inhibited by everolimus. Exp Ther Med. 2013;5:338–342. doi: 10.3892/etm.2012.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler NW. Compartmentation of GAPDH. Adv Exp Med Biol. 2013;985:61–101. doi: 10.1007/978-94-007-4716-6_3. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Jacob-Hirsch J, Amariglio N, Rechavi G, Kloog Y. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1alpha, causing glycolysis shutdown and cell death. Cancer Res. 2005;65:999–1006. [PubMed] [Google Scholar]
- Blum R, Elkon R, Yaari S, Zundelevich A, Jacob-Hirsch J, Rechavi G, et al. Gene expression signature of human cancer cell lines treated with the ras inhibitor salirasib (S-farnesylthiosalicylic acid) Cancer Res. 2007;67:3320–3328. doi: 10.1158/0008-5472.CAN-06-4287. [DOI] [PubMed] [Google Scholar]
- Blum R, Jacob-Hirsch J, Rechavi G, Kloog Y. Suppression of survivin expression in glioblastoma cells by the Ras inhibitor farnesylthiosalicylic acid promotes caspase-dependent apoptosis. Mol Cancer Ther. 2006;5:2337–2347. doi: 10.1158/1535-7163.MCT-06-0193. [DOI] [PubMed] [Google Scholar]
- Riganti C, Gazzano E, Polimeni M, Aldieri E, Ghigo D. The pentose phosphate pathway: an antioxidant defense and a crossroad in tumor cell fate. Free Radical Biol Med. 2012;53:421–436. doi: 10.1016/j.freeradbiomed.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Nagaoka Y, Iuchi Y, Ikeda Y, Fujii J. Glutathione reductase is expressed at high levels in pancreatic islet cells. Redox Rep. 2004;9:321–324. doi: 10.1179/135100004225006812. [DOI] [PubMed] [Google Scholar]
- Dashty M. A quick look at biochemistry: carbohydrate metabolism. Clin Biochem. 2013;46:1339–1352. doi: 10.1016/j.clinbiochem.2013.04.027. [DOI] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi DA. Ras by any other name. Mol Cell Biol. 2001;21:1441–1443. doi: 10.1128/MCB.21.5.1441-1443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Kloog Y. Tailoring Ras-pathway—inhibitor combinations for cancer therapy. Drug Resistance Updates. 2005;8:369–380. doi: 10.1016/j.drup.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Blum R, Cox AD, Kloog Y. Inhibitors of chronically active ras: potential for treatment of human malignancies. Recent Patents Anti-Cancer Drug Discovery. 2008;3:31–47. doi: 10.2174/157489208783478702. [DOI] [PubMed] [Google Scholar]
- Tada M, Omata M, Kawai S, Saisho H, Ohto M, Saiki RK, et al. Detection of ras gene mutations in pancreatic juice and peripheral blood of patients with pancreatic adenocarcinoma. Cancer Res. 1993;53:2472–2474. [PubMed] [Google Scholar]
- Yamada T, Nakamori S, Ohzato H, Oshima S, Aoki T, Higaki N, et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin Cancer Res. 1998;4:1527–1532. [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Qian J, Niu J, Li M, Chiao PJ, Tsao MS. In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K-ras oncogenic activation in pancreatic carcinogenesis. Cancer Res. 2005;65:5045–5053. doi: 10.1158/0008-5472.CAN-04-3208. [DOI] [PubMed] [Google Scholar]
- Mohamedali A, Lea NC, Feakins RM, Raj K, Mufti GJ, Kocher HM. AKT1 (E17K) mutation in pancreatic cancer. Tech Cancer Res Treatment. 2008;7:407–408. doi: 10.1177/153303460800700509. [DOI] [PubMed] [Google Scholar]
- Okami K, Wu L, Riggins G, Cairns P, Goggins M, Evron E, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58:509–511. [PubMed] [Google Scholar]
- Sakurada A, Suzuki A, Sato M, Yamakawa H, Orikasa K, Uyeno S, et al. Infrequent genetic alterations of the PTEN/MMAC1 gene in Japanese patients with primary cancers of the breast, lung, pancreas, kidney, and ovary. Jpn J Cancer Res. 1997;88:1025–1028. doi: 10.1111/j.1349-7006.1997.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Berger DH, Jardines LA, Chang H, Ruggeri B. Activation of Raf-1 in human pancreatic adenocarcinoma. J Surg Res. 1997;69:199–204. doi: 10.1006/jsre.1997.5022. [DOI] [PubMed] [Google Scholar]
- Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, Valentine V, et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003;163:1255–1260. doi: 10.1016/S0002-9440(10)63485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonleben F, Qiu W, Ciau NT, Ho DJ, Li X, Allendorf JD, et al. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12:3851–3855. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, Reddy SA. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23:8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- Altomare DA, Tanno S, De Rienzo A, Klein-Szanto AJ, Skele KL, Hoffman JP, et al. Frequent activation of AKT2 kinase in human pancreatic carcinomas. J Cell Biochem. 2002;87:470–476. doi: 10.1002/jcb.10287. [DOI] [PubMed] [Google Scholar]
- Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri BA, Huang L, Wood M, Cheng JQ, Testa JR. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol Carcinog. 1998;21:81–86. [PubMed] [Google Scholar]
- Schlieman MG, Fahy BN, Ramsamooj R, Beckett L, Bold RJ. Incidence, mechanism and prognostic value of activated AKT in pancreas cancer. Br J Cancer. 2003;89:2110–2115. doi: 10.1038/sj.bjc.6601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Tomita Y, Hoshida Y, Morooka T, Nagano H, Dono K, et al. Prognostic significance of activated Akt expression in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2004;10:2846–2850. doi: 10.1158/1078-0432.ccr-02-1441. [DOI] [PubMed] [Google Scholar]
- Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP, Jr, et al. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem. 1999;274:20281–20286. doi: 10.1074/jbc.274.29.20281. [DOI] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi A, Montaner S, Miyazaki H, Gutkind JS. MAPK and Akt act cooperatively but independently on hypoxia inducible factor-1alpha in rasV12 upregulation of VEGF. Biochem Biophys Res Commun. 2001;287:292–300. doi: 10.1006/bbrc.2001.5532. [DOI] [PubMed] [Google Scholar]
- Hu Y, Lu W, Chen G, Wang P, Chen Z, Zhou Y, et al. K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2012;22:399–412. doi: 10.1038/cr.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazure NM, Chen EY, Laderoute KR, Giaccia AJ. Induction of vascular endothelial growth factor by hypoxia is modulated by a phosphatidylinositol 3-kinase/Akt signaling pathway in Ha-ras-transformed cells through a hypoxia inducible factor-1 transcriptional element. Blood. 1997;90:3322–3331. [PubMed] [Google Scholar]
- Gao N, Ding M, Zheng JZ, Zhang Z, Leonard SS, Liu KJ, et al. Vanadate-induced expression of hypoxia-inducible factor 1 alpha and vascular endothelial growth factor through phosphatidylinositol 3-kinase/Akt pathway and reactive oxygen species. J Biol Chem. 2002;277:31963–31971. doi: 10.1074/jbc.M200082200. [DOI] [PubMed] [Google Scholar]
- Fukuda R, Hirota K, Fan F, Jung YD, Ellis LM, Semenza GL. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J Biol Chem. 2002;277:38205–38211. doi: 10.1074/jbc.M203781200. [DOI] [PubMed] [Google Scholar]
- Mottet D, Michel G, Renard P, Ninane N, Raes M, Michiels C. Role of ERK and calcium in the hypoxia-induced activation of HIF-1. J Cell Physiol. 2003;194:30–44. doi: 10.1002/jcp.10176. [DOI] [PubMed] [Google Scholar]
- Jansen B, Schlagbauer-Wadl H, Kahr H, Heere-Ress E, Mayer BX, Eichler H, et al. Novel Ras antagonist blocks human melanoma growth. Proc Natl Acad Sci USA. 1999;96:14019–14024. doi: 10.1073/pnas.96.24.14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen B, Heere-Ress E, Schlagbauer-Wadl H, Halaschek-Wiener J, Waltering S, Moll I, et al. Farnesylthiosalicylic acid inhibits the growth of human Merkel cell carcinoma in SCID mice. J Mol Med (Berl) 1999;77:792–797. doi: 10.1007/s001099900052. [DOI] [PubMed] [Google Scholar]
- Erlich S, Tal-Or P, Liebling R, Blum R, Karunagaran D, Kloog Y, et al. Ras inhibition results in growth arrest and death of androgen-dependent and androgen-independent prostate cancer cells. Biochem Pharmacol. 2006;72:427–436. doi: 10.1016/j.bcp.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Laheru D, Shah P, Rajeshkumar NV, McAllister F, Taylor G, Goldsweig H, et al. Integrated preclinical and clinical development of S-trans, trans-Farnesylthiosalicylic acid (FTS, Salirasib) in pancreatic cancer. Invest New Drugs. 2012;30:2391–2399. doi: 10.1007/s10637-012-9818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haklai R, Elad-Sfadia G, Egozi Y, Kloog Y. Orally administered FTS (salirasib) inhibits human pancreatic tumor growth in nude mice. Cancer Chemother Pharmacol. 2008;61:89–96. doi: 10.1007/s00280-007-0451-6. [DOI] [PubMed] [Google Scholar]
- Weisz B, Giehl K, Gana-Weisz M, Egozi Y, Ben-Baruch G, Marciano D, et al. A new functional Ras antagonist inhibits human pancreatic tumor growth in nude mice. Oncogene. 1999;18:2579–2588. doi: 10.1038/sj.onc.1202602. [DOI] [PubMed] [Google Scholar]
- Maher JC, Krishan A, Lampidis TJ. Greater cell cycle inhibition and cytotoxicity induced by 2-deoxy-D-glucose in tumor cells treated under hypoxic versus aerobic conditions. Cancer Chemother Pharmacol. 2004;53:116–122. doi: 10.1007/s00280-003-0724-7. [DOI] [PubMed] [Google Scholar]
- Xiao H, Li S, Zhang D, Liu T, Yu M, Wang F. Separate and concurrent use of 2-deoxy-D-glucose and 3-bromopyruvate in pancreatic cancer cells. Oncol Rep. 2013;29:329–334. doi: 10.3892/or.2012.2085. [DOI] [PubMed] [Google Scholar]
- Goldberg L, Ocherashvilli A, Daniels D, Last D, Cohen ZR, Tamar G, et al. Salirasib (farnesyl thiosalicylic acid) for brain tumor treatment: a convection-enhanced drug delivery study in rats. Mol Cancer Ther. 2008;7:3609–3616. doi: 10.1158/1535-7163.MCT-08-0488. [DOI] [PubMed] [Google Scholar]
- Dearling JL, Flynn AA, Sutcliffe-Goulden J, Petrie IA, Boden R, Green AJ, et al. Analysis of the regional uptake of radiolabeled deoxyglucose analogs in human tumor xenografts. J Nucl Med. 2004;45:101–107. [PubMed] [Google Scholar]
- Ben Sahra I, Laurent K, Giuliano S, Larbret F, Ponzio G, Gounon P, et al. Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res. 2010;70:2465–2475. doi: 10.1158/0008-5472.CAN-09-2782. [DOI] [PubMed] [Google Scholar]
- Goldberg L, Israeli R, Kloog Y. FTS and 2-DG induce pancreatic cancer cell death and tumor shrinkage in mice. Cell Death Dis. 2012;3:e284. doi: 10.1038/cddis.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- Young VR, Ajami AM.Glutamine: the emperor or his clothes J Nutr 2001131(9 Suppl2449S–2459S.discussion 2486S-2447S. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Cerione RA. Glutaminase: a hot spot for regulation of cancer cell metabolism. Oncotarget. 2010;1:734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanware NP, Mullen AR, DeBerardinis RJ, Abraham RT. Glutamine: pleiotropic roles in tumor growth and stress resistance. J Mol Med (Berl) 2011;89:229–236. doi: 10.1007/s00109-011-0731-9. [DOI] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhill C. Novel pathway identified for glutamine metabolism in PDAC. Nat Rev Gastroenterol Hepatol. 2013;10:260. doi: 10.1038/nrgastro.2013.57. [DOI] [PubMed] [Google Scholar]
- Clerc P, Bensaadi N, Pradel P, Estival A, Clemente F, Vaysse N. Lipid-dependent proliferation of pancreatic cancer cell lines. Cancer Res. 1991;51:3633–3638. [PubMed] [Google Scholar]
- Calderon P, Furnelle J, Christophe J. In vitro lipid metabolism in the rat pancreas. I. Basal lipid metabolism. Biochim Biophys Acta. 1979;574:379–390. doi: 10.1016/0005-2760(79)90234-0. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu Rev Biochem. 1987;56:159–193. doi: 10.1146/annurev.bi.56.070187.001111. [DOI] [PubMed] [Google Scholar]
- Ohmura E, Okada M, Onoda N, Kamiya Y, Murakami H, Tsushima T, et al. Insulin-like growth factor I and transforming growth factor alpha as autocrine growth factors in human pancreatic cancer cell growth. Cancer Res. 1990;50:103–107. [PubMed] [Google Scholar]
- Liehr RM, Melnykovych G, Solomon TE. Growth effects of regulatory peptides on human pancreatic cancer lines PANC-1 and MIA PaCa-2. Gastroenterology. 1990;98:1666–1674. doi: 10.1016/0016-5085(90)91105-f. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Derynck R, Korc M. Production of transforming growth factor alpha in human pancreatic cancer cells: evidence for a superagonist autocrine cycle. Proc Natl Acad Sci USA. 1987;84:7567–7570. doi: 10.1073/pnas.84.21.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa W, Bandyopadhyay GK, Wallace D, Nandi S. Phospholipids containing polyunsaturated fatty acyl groups are mitogenic for normal mouse mammary epithelial cells in serum-free primary cell culture. Proc Natl Acad Sci USA. 1989;86:4122–4126. doi: 10.1073/pnas.86.11.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon EC, Strouch MJ, Barron MR, Ding Y, Melstrom LG, Krantz SB, et al. Alteration of strain background and a high omega-6 fat diet induces earlier onset of pancreatic neoplasia in EL-Kras transgenic mice. Int J Cancer. 2011;128:2783–2792. doi: 10.1002/ijc.25622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau MD, Sohn KH, Rhee SH, Lee SW, Hunt JD, Hwang DH. Suppression of tumor cell growth both in nude mice and in culture by n-3 polyunsaturated fatty acids: mediation through cyclooxygenase-independent pathways. Cancer Res. 2001;61:1386–1391. [PubMed] [Google Scholar]
- Khasawneh J, Schulz MD, Walch A, Rozman J, Hrabe de Angelis M, Klingenspor M, et al. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci USA. 2009;106:3354–3359. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- Michaud DS, Giovannucci E, Willett WC, Colditz GA, Stampfer MJ, Fuchs CS. Physical activity, obesity, height, and the risk of pancreatic cancer. JAMA. 2001;286:921–929. doi: 10.1001/jama.286.8.921. [DOI] [PubMed] [Google Scholar]
- Rose DP. Effects of dietary fatty acids on breast and prostate cancers: evidence from in vitro experiments and animal studies. Am J Clin Nutr. 1997;66 (6 Suppl:1513S–1522S. doi: 10.1093/ajcn/66.6.1513S. [DOI] [PubMed] [Google Scholar]
- Rose DP, Connolly JM. Dietary fat and breast cancer metastasis by human tumor xenografts. Breast Cancer Res Treat. 1997;46:225–237. doi: 10.1023/a:1005971317978. [DOI] [PubMed] [Google Scholar]
- Rose DP, Connolly JM. Omega-3 fatty acids as cancer chemopreventive agents. Pharmacol Ther. 1999;83:217–244. doi: 10.1016/s0163-7258(99)00026-1. [DOI] [PubMed] [Google Scholar]
- Strouch MJ, Ding Y, Salabat MR, Melstrom LG, Adrian K, Quinn C, et al. A high omega-3 fatty acid diet mitigates murine pancreatic precancer development. J Surg Res. 2011;165:75–81. doi: 10.1016/j.jss.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagosomes: biogenesis from scratch. Curr Opin Cell Biol. 2005;17:415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E, Vanden Berghe T, Lippens S, Agostinis P, Vandenabeele P. Autophagy: for better or for worse. Cell Res. 2012;22:43–61. doi: 10.1038/cr.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116 (Pt 9:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117 (Pt 13:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Yazbeck VY, Buglio D, Georgakis GV, Li Y, Iwado E, Romaguera JE, et al. Temsirolimus downregulates p21 without altering cyclin D1 expression and induces autophagy and synergizes with vorinostat in mantle cell lymphoma. Exp Hematol. 2008;36:443–450. doi: 10.1016/j.exphem.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, et al. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–942. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- Gorzalczany Y, Gilad Y, Amihai D, Hammel I, Sagi-Eisenberg R, Merimsky O. Combining an EGFR directed tyrosine kinase inhibitor with autophagy-inducing drugs: a beneficial strategy to combat non-small cell lung cancer. Cancer Lett. 2011;310:207–215. doi: 10.1016/j.canlet.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Hsu KF, Wu CL, Huang SC, Wu CM, Hsiao JR, Yo YT, et al. Cathepsin L mediates resveratrol-induced autophagy and apoptotic cell death in cervical cancer cells. Autophagy. 2009;5:451–460. doi: 10.4161/auto.5.4.7666. [DOI] [PubMed] [Google Scholar]
- Rez G, Toth S, Palfia Z. Cellular autophagic capacity is highly increased in azaserine-induced premalignant atypical acinar nodule cells. Carcinogenesis. 1999;20:1893–1898. doi: 10.1093/carcin/20.10.1893. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Mitsunaga S, Yamazaki M, Hasebe T, Ishii G, Kojima M, et al. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008;99:1813–1819. doi: 10.1111/j.1349-7006.2008.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival. Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64:5338–5346. doi: 10.1158/0008-5472.CAN-04-0089. [DOI] [PubMed] [Google Scholar]
- Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- Bucciarelli LG, Wendt T, Rong L, Lalla E, Hofmann MA, Goova MT, et al. RAGE is a multiligand receptor of the immunoglobulin superfamily: implications for homeostasis and chronic disease. Cell Mol Life Sci. 2002;59:1117–1128. doi: 10.1007/s00018-002-8491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basta G, Lazzerini G, Del Turco S, Ratto GM, Schmidt AM, De Caterina R. At least 2 distinct pathways generating reactive oxygen species mediate vascular cell adhesion molecule-1 induction by advanced glycation end products. Arterioscler Thromb Vasc Biol. 2005;25:1401–1407. doi: 10.1161/01.ATV.0000167522.48370.5e. [DOI] [PubMed] [Google Scholar]
- Cai W, He JC, Zhu L, Lu C, Vlassara H. Advanced glycation end product (AGE) receptor 1 suppresses cell oxidant stress and activation signaling via EGF receptor. Proc Natl Acad Sci USA. 2006;103:13801–13806. doi: 10.1073/pnas.0600362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiou P, Chatzopoulou M, Pegklidou K, Demopoulos VJ. RAGE: a multi-ligand receptor unveiling novel insights in health and disease. Curr Med Chem. 2010;17:2232–2252. doi: 10.2174/092986710791331086. [DOI] [PubMed] [Google Scholar]
- Rojas A, Figueroa H, Morales E. Fueling inflammation at tumor microenvironment: the role of multiligand/RAGE axis. Carcinogenesis. 2010;31:334–341. doi: 10.1093/carcin/bgp322. [DOI] [PubMed] [Google Scholar]
- Logsdon CD, Fuentes MK, Huang EH, Arumugam T. RAGE and RAGE ligands in cancer. Curr Mol Med. 2007;7:777–789. doi: 10.2174/156652407783220697. [DOI] [PubMed] [Google Scholar]
- Abe R, Yamagishi S. AGE-RAGE system and carcinogenesis. Curr Pharm Des. 2008;14:940–945. doi: 10.2174/138161208784139765. [DOI] [PubMed] [Google Scholar]
- Kang R, Tang D, Schapiro NE, Livesey KM, Farkas A, Loughran P, et al. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 2010;17:666–676. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Prasad R, Wilson SH. HMGB1: roles in base excision repair and related function. Biochim Biophys Acta. 2010;1799:119–130. doi: 10.1016/j.bbagrm.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Tang D, Livesey KM, Schapiro NE, Lotze MT, Zeh HJ., 3rd The receptor for advanced glycation end-products (RAGE) protects pancreatic tumor cells against oxidative injury. Antioxid Redox Signal. 2011;15:2175–2184. doi: 10.1089/ars.2010.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Tang D, Schapiro NE, Loux T, Livesey KM, Billiar TR, et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene. 2013;33:567–577. doi: 10.1038/onc.2012.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Woo SJ, Yoon CH, Lee JS, An S, Choi YH, et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem. 2011;286:12924–12932. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Kimmelman AC. Targeting autophagy addiction in cancer. Oncotarget. 2011;2:1302–1306. doi: 10.18632/oncotarget.384. [DOI] [PMC free article] [PubMed] [Google Scholar]