Abstract
New evidence indicates the involvement of protein degradation dysfunctions in neurodegeneration, innate immunity response and alcohol hepatotoxicity. We recently demonstrated that ethanol increases brain proinflammatory mediators and causes brain damage by activating Toll-like receptor 4 (TLR4) signaling in glia. However, it is uncertain if the ubiquitin-proteasome and autophagy-lysosome pathways are involved in ethanol-induced brain damage and whether the TLR4 response is implicated in proteolytic processes. Using the cerebral cortex of WT and TLR4-knockout mice with and without chronic ethanol treatment, we demonstrate that ethanol induces poly-ubiquitinated proteins accumulation and promotes immunoproteasome activation by inducing the expression of β2i, β5i and PA28α, although it decreases the 20S constitutive proteasome subunits (α2, β5). Ethanol also upregulates mTOR phosphorylation, leading to a downregulation of the autophagy-lysosome pathway (ATG12, ATG5, cathepsin B, p62, LC3) and alters the volume of autophagic vacuoles. Notably, mice lacking TLR4 receptors are protected against ethanol-induced alterations in protein degradation pathways. In summary, the present results provide the first evidence demonstrating that chronic ethanol treatment causes proteolysis dysfunctions in the mouse cerebral cortex and that these events are TLR4 dependent. These findings could provide insight into the mechanisms underlying ethanol-induced brain damage.
Keywords: ubiquitin-proteasome pathway, autophagy-lysosome pathway, ethanol, TLR4, brain damage
Intracellular proteolysis has a crucial role in maintaining cellular homeostasis and it affects various essential cellular processes such as cell cycle regulation, transcriptional control, signal transduction, antigen presentation and protein turnover.1 Dysfunctions in the proteolytic process have been found in a wide range of pathologies,2, 3 including inflammatory and neurodegenerative disorders.4, 5 Indeed, many neurodegenerative diseases are characterized by protein misfolding and an abnormal accumulation and aggregation of specific proteins, which can result for deficient clearance systems, such as the ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway (ALP). Examples of these disorders are Alzheimer's or Parkinson's disease, which present an accumulation of aberrant proteins that produce a toxic neuronal effect, also called proteotoxicity.6
Digestion through the UPS is a selective process responsible for the vast majority of protein degradation in mammalian cells.7 Conjugation of proteins to ubiquitin is the hallmark of their degradation by the 26S proteasome, which is composed of a central 20S core containing catalytic subunits (β1, β2, β5), as well as two 19S regulatory particles that enable the recognition of proteins. Several proinflammatory cytokines, in particular IFN-γ and TNF-α,8 are able to induce the expression of alternative catalytic subunits (β1i, β2i, β5i), and the union of the 20S core to PA28 regulatory particles, which leads to the formation of the so-called immunoproteasome.9 The function of the immunoproteasome improves the major histocompatibility complex (MHC) class I presentation during immune responses, and also regulates proinflammatory cytokine production, and T-cell differentiation and proliferation.10 Moreover, the immunoproteasome, and its subunit β5i in particular, has been recently involved in autoimmune disorders and other inflammatory diseases.11, 12
The ALP comprises the process in which any cellular component is delivered to the lysosome in order to be degraded by hydrolases such as cathepsins. Autophagy is finely regulated by a complex set of molecules known as ATG (autophagy-related genes) that participate in the formation of autophagic vacuoles or autophagosomes, which deliver the cargo to the lysosome.13 This process is mediated by two different conjugation events: the assembly of the ATG5-ATG12 complex and the formation of the LC3-phospatidyl ethanolamine complex.14 Autophagy is also regulated by two different kinase complexes: a beclin-1 complex involved in membrane nucleation; and the mTOR complex, which is the main regulator of this proteolytic pathway.15 Emerging evidence indicates that autophagy dysregulation participates in several neurodegenerative and inflammatory disorders.16 Treatment of these pathologies with rapamycin, a mTOR inhibitor, reduces inflammation and ameliorates the course of the disease.17
Alcohol is a neurotoxic compound and its abuse induces brain damage and can lead to neurodegeneration.18 Although the neuropathological processes underlying these effects remain poorly understood, we have demonstrated that ethanol, by activating the brain innate immune system and TLR4 receptors in both cultures of glial cells and in brain,19, 20 induces gliosis, production of cytokines and neuroinflammation, brain damage, myelin derangements and neurodegeneration.21, 22 Elimination of TLR4 receptors abolishes the neuroinflammation and brain damage induced by chronic alcohol intake in mice.19, 20, 21 Besides the overactivation of TLRs signaling, dysregulation of proteolytic processes also contributes to the pathology of several neurodegenerative disorders, such as Alzheimer's disease, Parkinson's disease or amyotrophic lateral sclerosis.23, 24, 25 Notably, recent evidence indicates the participation of protein degradation processes in alcoholic liver pathology.26, 27 However, it is uncertain whether alterations in proteolytic processes are involved in ethanol-induced brain damage and neurodegeneration. Therefore, the aim of this study is to evaluate if chronic ethanol treatment impairs the UPS and ALP processes in the brain and whether these proteolytic processes are influenced by the innate immune TLR4 receptor.
Here we report that in vivo chronic ethanol treatment alters both UPS and ALP, leading to an accumulation of ubiquitinated proteins in the mice cerebral cortex. However, alcohol upregulates the immunoproteasome by activating the neuroimmune system. Consistently, we provide evidence that the effects of ethanol on proteolytic processes are mediated by innate immune receptor TLR4 signaling, as minimal changes in protein degradation pathways were observed in the cerebral cortex of ethanol-treated TLR4-knockout (KO) mice. These findings provide new insights into the mechanisms underlying ethanol-induced brain damage.
Results
Role of TLR4 in the ethanol-induced accumulation of ubiquitinated proteins
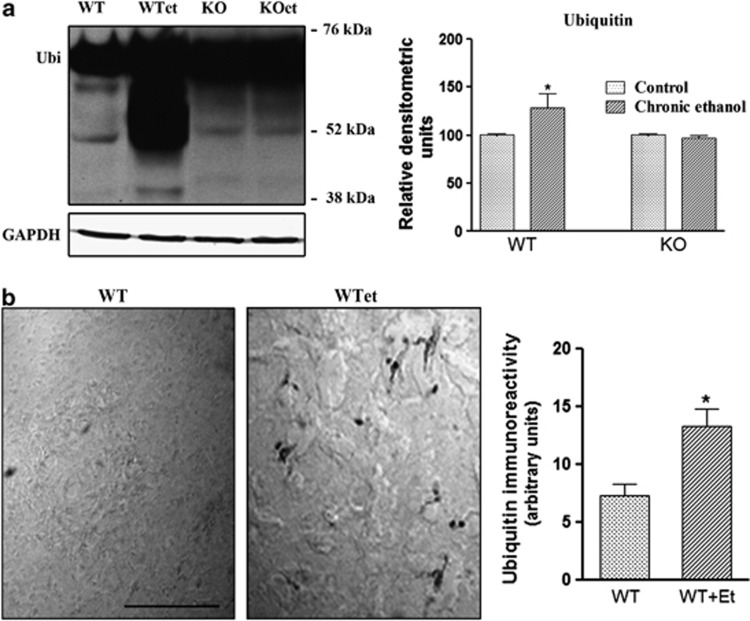
The UPS is the major degradation system in the cell that is involved in the degradation of short-lived, misfolded and defective proteins. Impairment of UPS along with an accumulation of ubiquitinated proteins is the major contributor to the pathogenesis of many neurodegenerative disorders.16, 28 We therefore first explored if chronic ethanol intake affects the ability to remove ubiquitinated proteins and whether TLR4 signaling participates in these effects. To answer this question, we performed western blot analyses and we used an ubiquitin antibody capable of detecting all ubiquitinated proteins. Figure 1a shows that in vivo chronic ethanol treatment increased the ubiquitinated proteins in the WT cerebral cortex, inducing a smear of proteins of about approximately 45–76 kDa. Conversely, the same ethanol treatment did not increase the amount of ubiquitinated proteins in the cerebral cortex of TLR4-KO mice, as no significant differences between ethanol-treated and control TLR4-KO mice were noted. Likewise, an immunohistochemical analysis of ubiquitinated proteins (Figure 1b) further demonstrated that chronic ethanol treatment promotes the accumulation of ubiquitinated proteins in the cerebral cortex and that this event is associated with the TLR4 function.
Figure 1.
Chronic ethanol intake increases poly-ubiquitinated proteins in the cortices of WT mice, but not in TLR4-KO mice. The immunoblot (a) and immunohistochemistry (b) analyses and the quantification of the expression of poly-ubiquitinated proteins in the cortical extracts or brain cortical sections of the WT and TLR4-KO mice treated or not with ethanol for 4 months. Data represent mean±S.E.M., n=7 mice/group. *P<0.05 compared with the ethanol-treated WT or TLR4-KO mice with their respective untreated control groups, according to an unpaired Student's t-test. Blots were stripped and the total quantity of GAPDH was also assessed. A representative immunoblot of each protein is shown. Scale bar: 50 μm
Chronic ethanol treatment impairs the ubiquitin-proteasome pathway
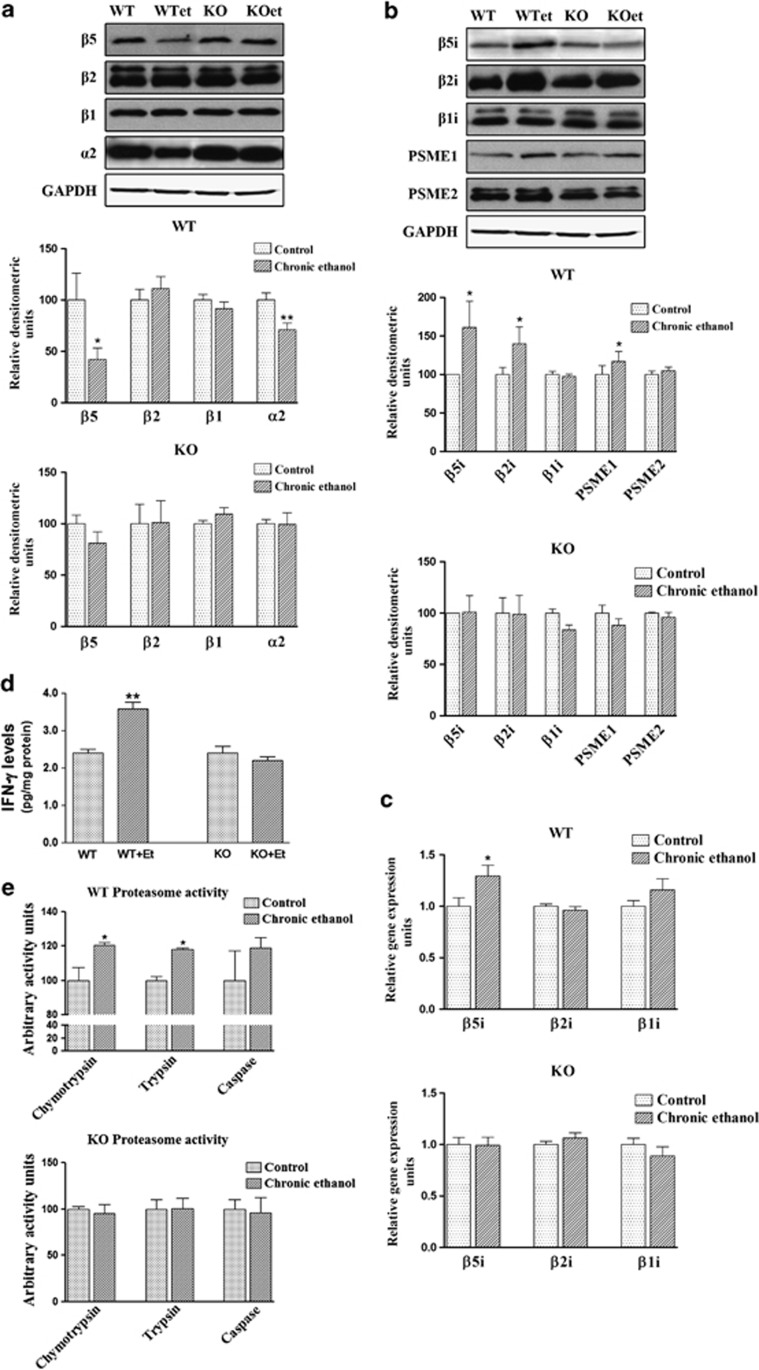
The 20S proteasome constitutes the proteolytic core of a large protein degradation complex and allows the degradation of unneeded or misfolded proteins by activating several proteolytic activities capable of removing ubiquitinated proteins. The 20S proteasome contains several catalytic subunits (β1, β2, β5) and is associated with two 19S regulatory particles that enable protein recognition. Therefore, to search the potential role of ethanol in the UPS pathway, we first measured the levels of the 20S proteasome constitutive subunits. As shown in Figure 2a, chronic in vivo treatment with ethanol decreased the β5 and α2 subunits expression in the cerebral cortices of WT mice, whereas no changes were noted in the β1 and β2 subunits. However, ethanol treatment did not affect the expression levels of any of the proteasome constitutive subunits in the cerebral cortices of TLR4-KO mice.
Figure 2.
Chronic ethanol treatment alters the UPS in the cerebral cortices of WT mice, but causes no changes in the cortices of TLR4-KO mice. (a) The immunoblot analysis and quantification of the β5, β2, β1 and α2 subunits expressions in the brain extracts of the WT and TLR4-KO mice treated or not with ethanol for 4 months. (b) The immunoblot analysis and quantification of the β5i, β2i, β1i, PSME1 and PSME2 subunit expressions in the brain extracts of the WT and TLR4-KO mice treated or not with ethanol for 4 months. (c) The mRNA levels of the β5i, β2i and β1i subunits of the WT and TLR4-KO mice treated or not with ethanol for 4 months. (d) The IFN-γ levels of the WT and TLR4-KO mice treated or not with ethanol for 4 months. (e) Levels of the three proteolytic activities, chymotrypsin-like, trypsin-like and caspase-like in the WT and TLR4-KO mice treated or not with ethanol for 4 months. Data represent mean±S.E.M., n=7 mice/group. *P<0.05, **P<0.01 compared with the ethanol-treated WT or TLR4-KO mice with their respective untreated control groups, according to an unpaired Student's t-test. Blots were stripped, and the total quantity of GAPDH was also assessed. A representative immunoblot of each protein is shown
In many neurodegenerative diseases, the functional decline of the constitutive proteasome is associated with an induction of the catalytic subunits (β1i, β2i and β5i) of the immunoproteasome, and also with the union of the 20S core to PA28 regulatory particles9, 29 by the upregulation of proinflammatory cytokines IFN-γ and TNF-α.8 We therefore determined whether alcohol treatment could induce the immunoproteasome subunits in the cortices of WT mice. Figure 2 shows that the protein expression of β5i and β2i (Figure 2b), and the β5i mRNA levels (Figure 2c) significantly increased in the cortices of ethanol-treated WT mice. Similarly, the levels of the PSME1 (or PA28α) subunit, a proteasome activator implicated in immunoproteasome assembly and antigen processing,30 also increased in ethanol-treated WT animals. Notably, as in most neurodegenerative pathologies, ethanol treatment significantly enhanced the TNF-α21 and IFN-γ levels in the cerebral cortices of WT mice (Figure 2d). Conversely, no significant changes in the immunoproteasome subunits and IFN-γ levels were noted in the cortices of ethanol-treated TLR4-KO when compared with untreated TLR4-KO (Figures 2b–d).
To establish whether the upregulation of some immunoproteasome subunits in the cortices of ethanol-treated animals was associated with changes in proteasome activity, we measured the three proteolytic chymotrypsin-like, trypsin-like and caspase-like activities. Figure 2e shows that the chymotrypsin-like and trypsin-like activities were enhanced in the cerebral cortices of ethanol-treated WT mice, whereas no changes were observed in ethanol-treated TLR4-KO mice.
In summary, the above results suggest that ethanol treatment reduces the 20S constitutive proteasome expression in the cerebral cortex and promotes the accumulation of poly-ubiquitinated proteins, while it also stimulates the production of proinflammatory cytokines, probably through TLR4 signaling, which, in turn, induce the activation of the immunoproteasome.
The ALP is also impaired by ethanol treatment: role of innate immune receptors TLR4 activation
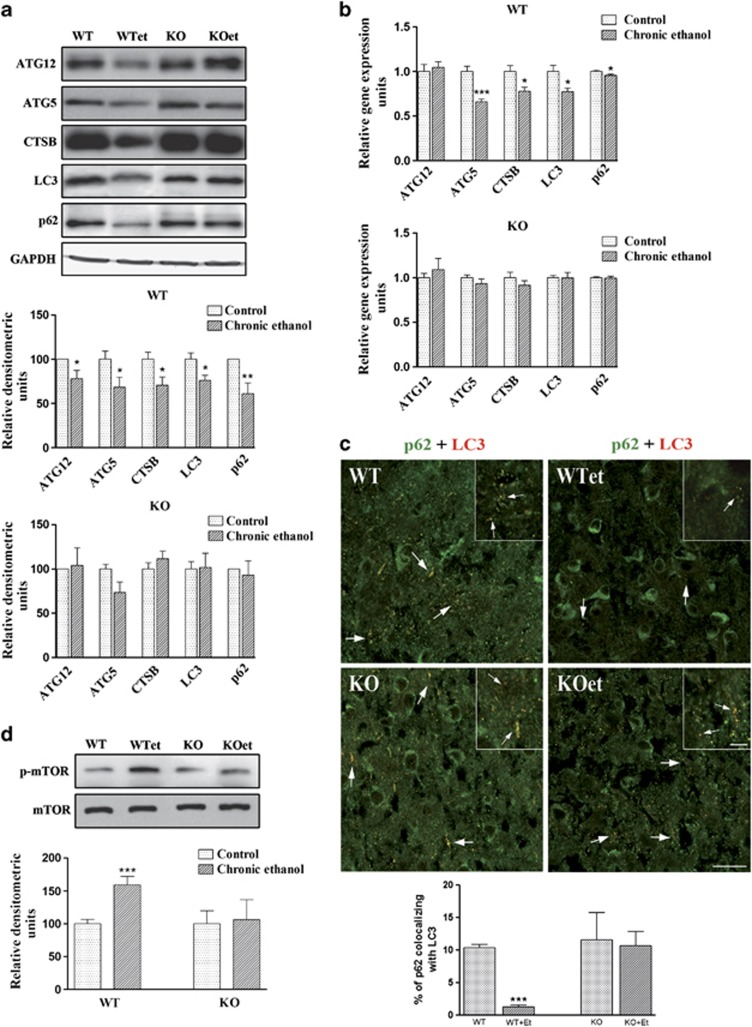
We next evaluated the potential effects of in vivo chronic ethanol treatment on the ALP pathway. For this aim, we first assessed the proteins involved in the formation of autophagosomes, such as LC3, ATG5 and ATG12, as well as the major lysosomal enzyme required for the degradation of contents in the autophagosome, cathepsin B, in the cerebral cortex. Figures 3a and b show that ethanol treatment decreased the mRNA and protein levels of cathepsin B, LC3 and ATG5, and only the protein levels of ATG12 in WT mice. No significant changes were observed between the TLR4-KO mice treated with or without ethanol.
Figure 3.
The ALP is impaired in the cortices of ethanol-treated WT mice. (a) The immunoblot analysis and quantification of the expressions of ATG5, ATG12, LC3, cathepsin B and p62 in the cortical extracts of the WT and TLR4-KO mice treated or not with ethanol for 4 months. (b) The mRNA levels of ATG5, ATG12, LC3, cathepsin B and p62 of the WT and TLR4-KO mice treated or not with ethanol for 4 months. (c) The double-labeling immunofluorescence of LC3 and p62 in the brain cortices of the WT and TLR4-KO mice treated with or without ethanol for 4 months. The scale bar of the insets is 6.25 μm, the scale bar of the low magnification images is 25 μm. (d) The immunoblot analysis and quantification of the expression of p-mTOR in the brain extracts of the WT and TLR4-KO mice treated or not with ethanol for 4 months. Data represent mean±S.E.M., n=7–12 mice/group. *P<0.05, **P<0.01, ***P<0.001 as compared with the ethanol-treated WT or TLR4-KO mice with their respective untreated control groups, according to an unpaired Student's t-test. Blots were stripped, and the total quantities of GAPDH and mTOR were also assessed. A representative immunoblot of each protein is shown
We next evaluated whether the expression of p62, a protein that can bind to LC3 and ubiquitinated proteins,31 has a role in the selective autophagy of ubiquitinated protein aggregates and organelles. Figures 3a and b depict a downregulated p62 in the cortices of ethanol-treated WT mice, but no significant changes were noted in TLR4-KO mice (Figures 3a and b), suggesting that TLR4 signaling is involved in the ethanol-induced impairment of p62 and the subsequent autophagic process. Immunofluorescence studies further established that LC3 and p62 interact, and that this interaction diminished in ethanol-treated WT mice (Figure 3c).
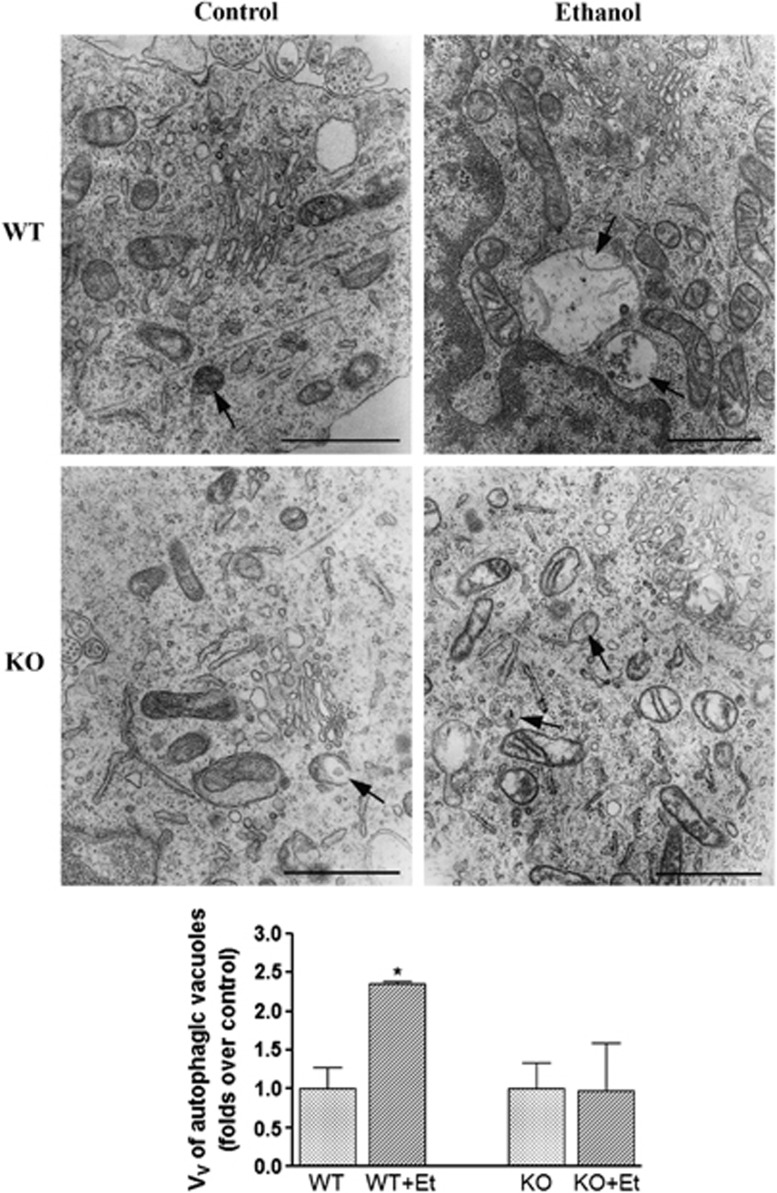
To further confirm that ethanol treatment affects the ALP, electron microscopy was employed to evaluate the number and size of autophagic vacuoles. These studies revealed that ethanol treatment increased the volume density of the autophagic vacuoles in the cytoplasm of WT mice, whereas minor changes were observed in TLR4-KO mice (Figure 4). These results suggest that ethanol either impairs the clearance of autophagosomes or affects authophagic catabolism. Deficient maturation or impaired clearance of autophagosomes might affect the degradation of their cargo, which could lead to cell damage and neurodegeneration.32
Figure 4.
The electron microscopy analysis shows that chronic ethanol consumption increases the volume density of autophagic vacuoles in WT mice. The representative transmission electron micrographs of the cerebral cortices obtained from the ultrathin tissue sections of the WT and TLR4-KO mice treated or not with ethanol for 4 months is shown. Bars represent the volume density (Vv) of autophagic vacuoles (μm3/100 μm3 per cell). Scale bars represent 1 μm. Arrows indicate autophagic vacuoles. Data represent mean±S.E.M., n=3–4 mice/group. *P<0.05 as compared with the ethanol-treated WT or TLR4-KO mice with their respective untreated control groups, according to an unpaired Student's t-test
Finally, in order to evaluate the mechanism by which ethanol induces impairment of autophagy, we measured the mTOR phosphorylation levels. The kinase mTOR is a critical regulator of the autophagic process, as mTOR activation inhibits this pathway.33 As depicted in Figure 3d, ethanol upregulated the phosphorylation of mTOR in WT mice, whereas similar phosphorylation levels were found between control and ethanol TLR4-KO mice. We also demonstrated that ethanol consumption does not alter the levels of beclin-1 in WT and TLR4-KO mice (data not shown), suggesting that ethanol-induced inhibition of autophagy could be mediated by the mTOR pathway.
In summary, the above results provide evidence that ethanol treatment reduces the autophagic process by impairing autophagosomes clearance and/or the degradation of their cargo proteins and organelles, events that might promote neurodegeneration. The obtained results also suggest that innate immune activation participates in the ethanol-induced reduction of the autophagic process.
Effect of an in vivo administration of bortezomib and rapamycin on ethanol-treated mice
Finally, in order to evaluate the in vivo relevance of protein degradation impairment in ethanol-induced neuroinflammation and brain damage, an UPS modulator, bortezomib, which inhibits 26S proteasome catalytic activity,34 was administered to the animals. The effects of ethanol on the ALP pathway were also assessed by administering rapamycin to the animals, a compound which inhibits mTOR signaling (e.g., AMPK and p53 signaling)35 and induces autophagy.36 Both inhibitors were injected to the control and chronic ethanol-treated WT mice.
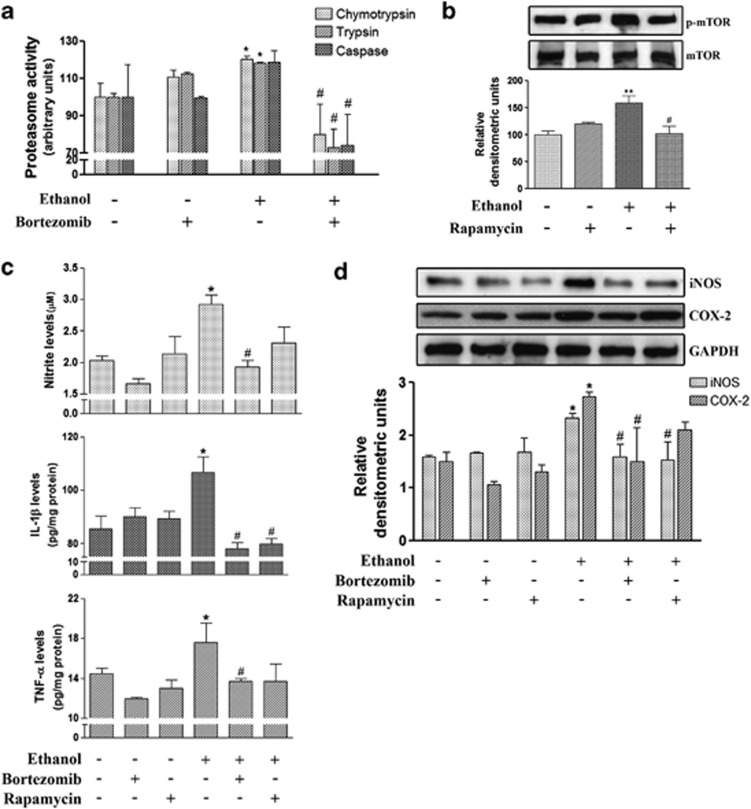
The results in Figure 5a demonstrate that bortezomib administration markedly reduces the ethanol-induced activation of the proteasome activities, chymotrypsin- and trypsin-like, in the cerebral cortex of WT mice. Similarly, administration of rapamycin significantly inhibits the induction of mTOR phosphorylation produced by chronic ethanol intake in the cerebral cortex of WT mice (Figure 5b). These results indicate that under our experimental conditions, bortezomib and rapamycin are able to inhibit ethanol-induced alterations in the UPS and ALP pathways.
Figure 5.
Effect of the in vivo administration of bortezomib and rapamycin to ethanol-treated WT mice. (a) Levels of the three proteolytic activities, chymotrypsin-like, trypsin-like and caspase-like, in the brain cortical extracts of these groups: control; ethanol exposure (4 months); bortezomib (1 mg/kg); ethanol exposure (4 months) plus bortezomib (1 mg/kg). (b) The immunoblot analysis and quantification of the expression of p-mTOR in the brain cortical extracts of these groups: control; ethanol exposure (4 months); rapamycin (3 mg/kg); ethanol exposure (4 months) plus rapamycin (3 mg/kg). The levels of TNF-α, IL-1β and NO (c) and the immunoblot analysis and quantification of COX-2 and iNOS (d) were measured in the extracts of the brain cortices of these groups: control; ethanol exposure (4 months); rapamycin (3 mg/kg); bortezomib (1 mg/kg); ethanol exposure (4 months) plus rapamycin (3 mg/kg); ethanol exposure (4 months) plus bortezomib (1 mg/kg). Rapamycin or bortezomib was administered once weekly intraperitoneally or intravenously, respectively. Data represent mean±S.E.M., n=4–6 mice/group. *P<0.05, **P<0.01 as compared with the control group, #P<0.05 as compared with the ethanol group, according to the one-way ANOVA followed by the Bonferroni's post hoc test. Blots were stripped, and the total quantity of GAPDH was also assessed. A representative immunoblot of each protein is shown
We next determined whether ethanol-induced neuroinflammation was associated with the protein degradation impairment observed in the cerebral cortex of chronic ethanol-treated mice. Figure 5c illustrates that whereas bortezomib completely suppresses the upregulation in the levels of TNF-α, IL-1β and NO in cortex of ethanol-treated WT mice, rapamycin only restores the IL-1β expression in the cortices of these animals. The analysis of the inflammatory mediators, iNOS and COX-2, showed that whereas bortezomib abolishes the upregulation of the iNOS and COX-2 expressions in the cortex of ethanol-treated WT mice, rapamycin only restores the levels of iNOS in the ethanol-treated animals (Figure 5d). There were no significant changes in the expression of proinflammatory cytokines and mediators when either inhibitor was administered alone.
Discussion
Emerging evidence indicates the participation of UPS and ALP in both the innate immune response and the pathology of inflammatory and neurodegenerative disorders.37, 38 We demonstrated that ethanol, by activating the innate immune receptors TLR4 in glial cells, increases the production of cytokines and inflammatory mediators, causing neuroinflammation and brain damage.19, 20, 21 However, whether ethanol-induced neuroimmune activation can impair the neural proteolytic processes, which contribute to brain damage is presently unknown. Here we show for the first time that chronic ethanol intake impairs both the UPS and ALP pathways by causing an accumulation of ubiquitinated proteins in the brain, events that might participate in ethanol-induced brain damage and neurodegeneration. We also provide evidence that the elimination of TLR4 abolishes the ethanol-induced dysregulation of UPS and ALP, as well as the accumulation of ubiquitinated proteins, suggesting that TLR4 receptors have a key role in these pathological processes.
The intracellular accumulation and aggregation of proteins are a common feature across many neurodegenerative diseases, including Alzheimer's disease, Huntington's disease, spinocerebellar ataxia, Parkinson's disease, amyotrophic lateral sclerosis and prion disease.25 Many of these neurodegenerative disorders are caused by mutations, which result in the production of misfolded proteins that are specific for each disease. Therefore, the ubiquitin-dependent degradation of misfolded proteins by proteasomes constitutes a critical part of the cytoprotective quality control machinery, as defects in this system can lead to the accumulation of aberrant proteins. However, in later disease stages, mutant proteins tend to form aggregates as their continuous production might exceed the capacity of an UPS, which might already be dysfunctional in several pathologies. We hereby show that chronic ethanol consumption is able to lead to an accumulation of ubiquitinated proteins in WT mice, effects that may be associated with the impairment of UPS and/or immunoproteasome functioning. Consistently with our results, the microarray studies of the frontal cortex of alcoholics revealed a downregulation in the expression of the genes related with the UPS along with changes in the gene expression associated with lysosomes, apoptosis, myelination and the immune/stress response.39, 40
The constitutive 20S proteasome is replaced with the immunoproteasome in the presence of an inflammatory environment, principally through the expression of cytokines IFN-γ and TNF-α.8 Activation of the immunoproteasome occurs in different neurodegenerative diseases associated with neuroinflammation,41, 42 and also in brain injury.43 Nevertheless, although the immunoproteasome can have a protective role in acute neuroinflammation,44 its overactivation has deleterious effects on these neurodegenerative diseases. We demonstrated that ethanol consumption induces neuroinflammation by increasing the levels of several cytokines (IL-1β, TNF-α, IL-6) and other inflammatory mediators, leading to brain damage.21 The present findings further reveal that ethanol treatment upregulates IFN-γ, an event that can mediate the induction of the β2i and β5i immunoproteasome subunits and the regulatory particle PA28α subunit; nevertheless, it reduces some constitutive subunits (e.g., β5 and α2) of the 20 proteasome. This composition shift toward the immunoproteasome correlates with an increase in the chymotrypsin- and trypsin-like proteasomal activities, as demonstrated in the present study and which coincides with others.45, 46 Yet despite the increase in proteasome activities, ethanol was also seen to cause a massive buildup of ubiquitinated aggregates. This accumulation of aggregates might be attributable to either an excess of ubiquitinated proteins, which overwhelm the degradation capacity of the proteasome, or the inability of ubiquitinated proteins to enter the proteasome cavity.47 Likewise, the immunoproteasome is specialized in cleaving small peptides for antigen presentation, but the change in its activity does not affect the rate of ubiquitinated proteins' complete breakdown.45 Notably, activation of β5i chymotrypsin-like activity has been recently reported to be involved in the generation and sustainment of an inflammatory environment,11, 12, 48 whereas the use of inhibitors of this activity, such as bortezomib, has been described in many cancer therapies,49, 50 neurodegenerative disorders51 and other chronic inflammatory conditions.52 We also observed that bortezomib completely restores the levels of the cytokines and inflammatory mediators induced by ethanol in WT brain cortices, thus corroborating the role of ethanol in UPS impairment.
Our previous studies revealed the involvement of TLR4 activation in the ethanol-induced release of cytokines and inflammatory mediators, and also in brain damage.19, 20, 21 The present findings also support the role of the TLR4 response and innate immune activation in ethanol-induced dysfunctions of proteolytic pathways in the brain. Similarly, activation of the TLR4 response promotes the upregulation of immunoproteasome subunits and proteasome activity in LPS-stimulated microglial cells.42 Therefore, elimination of TLR4 might reduce the ethanol-induced inflammatory environment, which, in turn, can reduce or abolish the immunoproteasome activation. According to this hypothesis, here we demonstrate for the first time that ethanol does not produce proteasome or immunoproteasome alterations when the TLR4 response is lacking in TLR4-KO mice. Lack of the effects of ethanol in TLR4-KO mice might be associated with the absence of a brain inflammatory cellular environment. Indeed, TLR4 deficiency prevents the accumulation of ubiquitinated proteins induced by alcohol abuse in the brain cortex.
Besides the UPS, the other major route for intracellular protein degradation is the ALP. They both work together during the clearance of ubiquitinated protein aggregates53, 54 through, for example, protein p62, which links these structures to the autophagic machinery by a direct interaction with LC3.31 We herein demonstrate that ethanol treatment downregulates p62, as well as its colocalization with LC3, and other autophagic proteins, which suggests that the accumulation of ubiquitinated proteins may also be correlated with an impairment of the autophagy machinery. Our electron microscopy results further support that ethanol impairs the autophagic process, as it induces the formation of autophagic vacuoles with greater volume density. Based on these data, we postulate that the accumulation of autophagic vacuoles can aggravate the neurophathological process given the difficulty of the clearing process of defective vacuoles and the generation of new ones. Hence, ethanol can impair the autophagic flux and causes effects in autophagic catabolism, and these events may contribute to neurodegeneration.32
One important target of the autophagy pathway is mTOR, one of the main kinase complexes that negatively regulates the autophagy pathway.55 The pharmacological modulation of mTOR with inhibitors, such as rapamycin, has been reported to promote the autophagic degradation of aggregation-prone proteins in vitro and to diminish the severity of neurodegeneration in several in vivo models.56 We also observed that rapamycin administration partially reduces neuroinflammation in ethanol-treated WT mice. Moreover, as mTOR is activated by the TLR4/PI3K-I/Akt or TLR4/MAPK/Erk1/2 cascade,57, 58 the upstream signals of both cascades (e.g., TLR4 activation) may modulate the autophagy. In line with this, we observed that ethanol-treated TLR4-KO mice show no changes in mTOR phosphorylation, and hence no affection of the autophagy pathway.
Taken together, we conclude for the first time that these results support the role of TLR4 in ethanol-induced proteasome and autophagy impairment in the cerebral cortex, which leads to the accumulation and aggregation of proteins. Furthermore, blocking this receptor can ameliorate neuroinflammation, and possibly neurodegeneration, induced by ethanol through the modulation of UPS and ALP. Our results are consistent with the changes noted in the expression of those genes involved in ubiquitination and proteolysis in cortices of alcohol abusers, and provide novel mechanisms to help understand brain damage caused by alcohol abuse.
Materials and Methods
Animals and treatments
Male C57BL/6 WT (Harlan Ibérica, Barcelona, Spain) and TLR4-knockout (KO) mice (C57BL/6 background, kindly provided by Dr. S Akira, Osaka University, Suita, Japan) aged 6–7 weeks were used. All the animals were kept under controlled light and dark (12/12 h), temperature (23 °C) and humidity (60%) conditions. All the experimental procedures were carried out in accordance with the guidelines approved by the European Communities Council Directive (86/609/ECC) and by Spanish Royal Decree 1201/2005. The animal experiments were also approved by the Ethical Committee of Animal Experimentation of the Príncipe Felipe Research Center (Valencia, Spain).
For the chronic ethanol treatment, male 7-week-old C57BL/6 (WT, TLR4+/+) and TLR4 knockout (KO, TLR4−/−) mice weighing 18–20 g were housed (4 animals/cage) and maintained with water (WT and KO control) or water containing 10% (v/v) ethanol, and solid diet ad libitum for 4 months. During this period, daily food and liquid intake was similar for WT and KO mice, and also for the ethanol-treated and untreated groups.21 Thus, mice consumed about 12.6±2.0 g ethanol/kg of body weight. The blood ethanol levels reached in ethanol-treated WT and KO mice were 122±18 mg/dl. Body weight gain at the end of the 4 months period was similar in male WT (C57BL/6) and in TLR4-KO mice treated with or without ethanol.21
For the in vivo UPS and ALP modulation, we used bortezomib and rapamycin, respectively. We used control and ethanol-treated WT mice for 4 months, and the treatment with both inhibitors was carried out over the last 8 weeks of this period. Bortezomib (1 mg/kg; LC Laboratories, Woburn, MA, USA), dissolved in DMSO and physiological saline, was administered intravenously once weekly in anesthetized mice with 2–4% isoflurane inhalation. Rapamycin (3 mg/kg; LC Laboratories), dissolved in DMSO and physiological saline, was administered intraperitoneally once weekly.
Mice were killed by cervical dislocation; brains were removed, dissected and brain cortices were immediately snap-frozen in liquid nitrogen until their analysis. Some animals were anesthetized, perfused with paraformaldehyde (PF) and used for the immunohistochemistry analyses.
Western blot analysis
Brain tissue from the cerebral cortex was homogenized in lysis buffer (1% Nonidet P-40, 20 mM Tris-HCl, pH 8, 4 mM sodium chloride, 40 mM sodium fluoride and protease inhibitors) for 30 min on ice. An equal amount of cell lysate of each sample (40 μg of protein/lane) was loaded onto SDS-polyacrylamide gels, and was then blotted onto polyvinylidene fluoride membranes. Membranes were blocked with 5% non-fat dried milk in TBS containing 0.1% Tween-20, and were then incubated overnight with the following primary antibodies: anti-Ubiquitin, anti-Proteasome 20S α2, anti-Proteasome 20S LMP7 or β5i, anti Proteasome 20S LMP2 or β1i, anti-PSME1, anti-PSME2, anti-APG5L/ATG5, anti-LC3A/B and anti-SQSTM1/p62 (Abcam, Cambridge, UK); anti-ATG12, anti-Cathepsin B, anti-20S Proteasome β5, anti-20S Proteasome β2, anti-20S Proteasome β1, anti-MECL-1 or β2i, anti-p-mTOR and anti-mTOR (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-iNOS (BD Transduction Laboratories, San Jose, CA, USA) and anti-COX-2 (Cayman Chemical, Ann Arbor, MI, USA). After washing with TBS containing 0.1% Tween-20, blots were incubated with HRP-conjugated antibodies. Blots were developed using the ECL system (ECL Plus; Thermo Scientific, Rockford, IL, USA). All the membranes were stripped for 30 min in SDS solution (0.4% SDS and 200 mM glycine, pH 2.5), and were washed and incubated with anti-GAPDH mAb (Chemicon, Temecula, CA, USA) for 2 h as a loading control. The intensity of the bands was quantified with the image analysis software ImageJ 1.44p (National Institutes of Health, Bethesda, MD, USA), and the densitometry analysis is shown in arbitrary units normalized to the GAPDH loading control.
RNA isolation and quantitative RT-PCR
Total RNA from brain cortices was isolated using the Tri Reagent Method (Sigma-Aldrich, St. Louis, MO, USA), as described in the manufacturer's protocol. Reverse transcription of 1 μg of total RNA was performed using the Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics, Basel, Switzerland). Amplification of the target and housekeeping (β-glucuronidase) genes was performed using the Taqman Gene Expression Master Mix (Applied Biosystems, Foster City, CA, USA) in a LightCycler 480 System (Roche Diagnostics) following the manufacturer's instructions. The assay codes of the primers are shown in Table 1. Data were analyzed using the LightCycler 480 relative quantification software, and were normalized to the amplification product of β-glucuronidase.
Table 1. mRNA primer assay codes.
| Gene | Assay code |
|---|---|
| β5i | Mm01278979_m1 |
| β2i | Mm00479052_g1 |
| β1i | Mm00479004_m1 |
| Atg12 | Mm00503201_m1 |
| Atg5 | Mm00504340_m1 |
| Cathepsin B | Mm01310506_m1 |
| LC3 | Mm00782868_sH |
| p62 | Mm00448091_m1 |
| GUSB | Mm00446953_m1 |
Proteasomal activity assay
Cerebral cortices were placed on ice and homogenized in extraction buffer (20 mM Tris-HCl, pH 7.8, 1 mM dithiothreitol, 1 mM ATP, 10% glycerol, 0.5 mM EDTA, 0.5% Igepal and 5 mM MgCl2). Lysates were centrifuged at 14 000 × g at 4 °C for 15 min. The three activities of the 20S proteasomes (chymotrypsin-like, trypsin-like and peptidylglutamyl-peptide hydrolase or caspase-like activities) were analyzed using fluorogenic substrates: Suc-LLVY-MCA, Boc-LSTR-MCA and Z-LLE-β-naphthylamide (Sigma-Aldrich), respectively. The brain lysates (50 μg of protein) and the fluorogenic substrate (final concentration, 50–100 μM) were incubated together in a buffer containing 20 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 1 mM DTT and 1 mM ATP. All the assays were done in triplicate. Incubation took place at 37 °C for 60 min in a 96-well plate. Fluorescence was determined at 355 nm excitation/460 nm emission in a microplate spectrofluorometer (Perkin-Elmer, Waltham, MA, USA). Proteasomal activity was expressed in arbitrary units.
Cytokine analysis
The levels of TNF-α, IL-1β, NO and IFN-γ were determined in cerebral cortex lysates. The TNF-α, IL-1β and IFN-γ levels were measured using an ELISA kit (eBioscience, Vienna, Austria) following the manufacturer's protocols, and NO production was assessed by measuring the nitrite levels by the Griess reaction as described previously.59
Brain tissue preparation and immunohistochemistry
Mice were anesthetized by an intraperitoneal injection of sodium penthobarbital (60 mg/kg) and fentanyl (0.05 mg/kg) for analgesia. Animals were then perfused transcardially with 0.9% cold saline containing heparin, followed immediately by 4% PF in 0.1 m phosphate buffer, pH 7.4, for tissue fixation. PF-fixed brains were removed, postfixed overnight at 4 °C with the same fixative solution (4% PF) and stored at 4 °C in PBS containing 30% sucrose for cryoprotection. Coronal brain sections (40 μm) were obtained with a cryostat (Microm HM 505E) and collected on polysine glass slides (Menzel-Gläser, Thermo Scientific). Brain sections were treated with citrate buffer, pH 6.0, for 15 min for epitope retrieval and were incubated with a peroxide quenching solution and 3% methanol-hydroperoxide for 10 min, followed by a blocking solution (5% serum in 0.1% PBS/Tween 20) for 1 h. Then, they were incubated overnight at 4 °C with mouse monoclonal anti-Ubiquitin (1 : 1000; Abcam). Sections were then incubated for 1 h with the secondary biotinylated antibody (1 : 200; Vector Laboratories, Burlingame, CA, USA). The avidin-biotin-peroxidase method (ABC kit; Vector Laboratories) was performed using 3,3′-diaminobenzidine (0.7 mg/ml) as a substrate (SIGMA FAST 3,3′-Diaminobenzidine; Sigma-Aldrich) according to manufacturer's protocol. Sections were mounted onto glass slides with distyrene-plasticizer-xylene-mounting medium (Sigma-Aldrich). Immunostainings were visualized and digitally recorded with a camera (DFC-480; Leica Microsystems, Wetzlar, Germany) mounted onto a microscope (DM-6000B; Leica Microsystems). The quantitative image analysis of the immunoreactivity of ubiquitin in the cerebral cortex was assessed with the MetaMorph software analysis (version 7.0; Molecular Devices, Sunnyvale, CA, USA). Approximately 18–20 fields of the cerebral cortex from five to eight coronal brain sections of at least three animals per group were analyzed.
For double immunofluorescence labeling, coronal brain sections were treated with citrate buffer, pH 6.0, for 15 min for epitope retrieval and were then incubated for 30 min with 0.3% sudan black solution to quench autofluorescence. After blocking with 5% normal serum for 1 h, sections were subsequently incubated overnight at 4 °C with anti-LC3A/B (1 : 50) and anti-SQSTM1/p62 (1 : 250; Abcam). Brain sections were then incubated with the respective secondary Alexa Fluor-conjugated antibodies (1 : 500; Invitrogen, Carlsbad, CA, USA). Hoechst 33342 dye (0.5 mg/ml, Molecular Probes, Eugene, OR, USA) was used as a nuclei marker. Negative controls were performed by replacing the respective primary antibodies with isotype-matched irrelevant IgG. Sections were mounted onto glass slides with fluorescent mounting medium (Merck KGaA, Darmstadt, Germany). Images were acquired using a TCS-SP2-AOBA confocal laser-scanning microscope (Leica Microsystems) and a × 63 HCX PL APO lbd.BL63xN.A.1.4 oil objective. All the confocal images were acquired with the same settings. The fluorescence distribution was analyzed with the MetaMorph software analysis (version 7.0), and the results were expressed as the % of p62 colocalizing with LC3. The quantitative analysis was performed in 18–20 fields of the cerebral cortex from five to eight coronal brain sections of at least three animals per group.
Electron microscopy
Mice were perfused via the left ventricle under deep anesthesia with 1% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4). Small blocks of the cerebral cortex were processed for electron microscopy and were embedded in Epon resin, as previously described in detail.60 The ultrathin tissue sections obtained were contrasted with uranyl acetate and lead citrate and were examined under a Philips CM100 transmission electron microscope (Philips, Amsterdam, Netherlands). From each sample 20–25 micrographs were randomly taken (primary magnification, × 10 000). Next the volume of autophagic vesicles was estimated by morphometry with the Visilog program (Noesis Vision, Inc., Mérignac, France). The results are expressed as the volume density (Vv) of autophagic vacuoles (μm3/100 μm3 per cell).
Acknowledgments
We thank M Gómez and MJ Morillo for their technical assistance. We also thank the Confocal Microscopy Service at the Centro de Investigación Príncipe Felipe. This work has been supported by grants from the Spanish Ministry of Economics and Competitiveness (SAF2012-33747), the Spanish Ministry of Health: The Institute Carlos III and FEDER funds (RTA-Network RD12-0028-007), and PNSD (Ex. 20101037), Generalitat Valenciana-Consellería de Educación: PROMETEO/2009/072.
Glossary
- UPS
ubiquitin-proteasome system
- ALP
autophagy-lysosome pathway
- MHC
major histocompatibility complex
- ATG
autophagy-related genes
- KO
knockout
The authors declare no conflict of interest.
Footnotes
Edited by A Verkhratsky
References
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. Bioessays. 2008;30:1172–1184. doi: 10.1002/bies.20852. [DOI] [PubMed] [Google Scholar]
- Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Rami A. Review: autophagy in neurodegeneration: firefighter and/or incendiarist. Neuropathol Appl Neurobiol. 2009;35:449–461. doi: 10.1111/j.1365-2990.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Basler M, Kirk CJ, Groettrup M. The immunoproteasome in antigen processing and other immunological functions. Curr Opin Immunol. 2012;25:74–80. doi: 10.1016/j.coi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Nijholt DA, de Graaf TR, van Haastert ES, Oliveira AO, Berkers CR, Zwart R, et al. Endoplasmic reticulum stress activates autophagy but not the proteasome in neuronal cells: implications for Alzheimer's disease. Cell Death Differ. 2011;18:1071–1081. doi: 10.1038/cdd.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles A, Fung G, Luo H. Immune and non-immune functions of the immunoproteasome. Front Biosci. 2012;17:1904–1916. doi: 10.2741/4027. [DOI] [PubMed] [Google Scholar]
- Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- Basler M, Dajee M, Moll C, Groettrup M, Kirk CJ. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol. 2010;185:634–641. doi: 10.4049/jimmunol.0903182. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Wang-Rosenke Y, Scholl V, Binder E, Loof T, Khadzhynov D, et al. Low-dose mTOR inhibition by rapamycin attenuates progression in anti-thy1-induced chronic glomerulosclerosis of the rat. Am J Physiol Renal Physiol. 2008;294:F440–F449. doi: 10.1152/ajprenal.00379.2007. [DOI] [PubMed] [Google Scholar]
- Hayes DM, Deeny MA, Shaner CA, Nixon K. Determining the threshold for alcohol-induced brain damage: new evidence with gliosis markers. Alcohol Clin Exp Res. 2013;37:425–434. doi: 10.1111/j.1530-0277.2012.01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco AM, Valles SL, Pascual M, Guerri C. Involvement of TLR4/type I IL-1 receptor signaling in the induction of inflammatory mediators and cell death induced by ethanol in cultured astrocytes. J Immunol. 2005;175:6893–6899. doi: 10.4049/jimmunol.175.10.6893. [DOI] [PubMed] [Google Scholar]
- Fernandez-Lizarbe S, Pascual M, Guerri C. Critical role of TLR4 response in the activation of microglia induced by ethanol. J Immunol. 2009;183:4733–4744. doi: 10.4049/jimmunol.0803590. [DOI] [PubMed] [Google Scholar]
- Alfonso-Loeches S, Pascual-Lucas M, Blanco AM, Sanchez-Vera I, Guerri C. Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. J Neurosci. 2010;30:8285–8295. doi: 10.1523/JNEUROSCI.0976-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso-Loeches S, Pascual M, Gomez-Pinedo U, Pascual-Lucas M, Renau-Piqueras J, Guerri C. Toll-like receptor 4 participates in the myelin disruptions associated with chronic alcohol abuse. Glia. 2012;60:948–964. doi: 10.1002/glia.22327. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda E, Isidoro C, Carresi C, Mollace V. Defective autophagy in Parkinson's disease: role of oxidative stress. Mol Neurobiol. 2012;46:639–661. doi: 10.1007/s12035-012-8318-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Lin A, Li J, French BA, French SW. Proteasome inhibitor up regulates liver antioxidative enzymes in rat model of alcoholic liver disease. Exp Mol Pathol. 2011;90:123–130. doi: 10.1016/j.yexmp.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osna NA, Thomes PG, Jr TM. Involvement of autophagy in alcoholic liver injury and hepatitis C pathogenesis. World J Gastroenterol. 2011;17:2507–2514. doi: 10.3748/wjg.v17.i20.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 1998;21:516–520. doi: 10.1016/s0166-2236(98)01276-4. [DOI] [PubMed] [Google Scholar]
- Bendotti C, Marino M, Cheroni C, Fontana E, Crippa V, Poletti A, et al. Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: implication for protein aggregation and immune response. Prog Neurobiol. 2012;97:101–126. doi: 10.1016/j.pneurobio.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Kloetzel PM, Soza A, Stohwasser R. The role of the proteasome system and the proteasome activator PA28 complex in the cellular immune response. Biol Chem. 1999;380:293–297. doi: 10.1515/BC.1999.040. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Huber R, Moroder L. The persisting challenge of selective and specific proteasome inhibition. J Pept Sci. 2009;15:58–66. doi: 10.1002/psc.1107. [DOI] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziotto JJ, Cao K, Collins FS, Krainc D. Rapamycin activates autophagy in Hutchinson-Gilford progeria syndrome: implications for normal aging and age-dependent neurodegenerative disorders. Autophagy. 2012;8:147–151. doi: 10.4161/auto.8.1.18331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ZH, Ip NY. Autophagy deregulation in neurodegenerative diseases—recent advances and future perspectives. J Neurochem. 2011;118:317–325. doi: 10.1111/j.1471-4159.2011.07314.x. [DOI] [PubMed] [Google Scholar]
- Koga H, Cuervo AM. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol Dis. 2011;43:29–37. doi: 10.1016/j.nbd.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72:756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Diaz-Hernandez M, Hernandez F, Martin-Aparicio E, Gomez-Ramos P, Moran MA, Castano JG, et al. Neuronal induction of the immunoproteasome in Huntington's disease. J Neurosci. 2003;23:11653–11661. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre M, Kamphuis W, Dooves S, Kooijman L, Chan ET, Kirk CJ, et al. Reactive glia show increased immunoproteasome activity in Alzheimer's disease. Brain. 2013;136:1415–1431. doi: 10.1093/brain/awt083. [DOI] [PubMed] [Google Scholar]
- Yao X, Liu J, McCabe JT. Alterations of cerebral cortex and hippocampal proteasome subunit expression and function in a traumatic brain injury rat model. J Neurochem. 2008;104:353–363. doi: 10.1111/j.1471-4159.2007.04970.x. [DOI] [PubMed] [Google Scholar]
- Pintado C, Gavilan MP, Gavilan E, Garcia-Cuervo L, Gutierrez A, Vitorica J, et al. Lipopolysaccharide-induced neuroinflammation leads to the accumulation of ubiquitinated proteins and increases susceptibility to neurodegeneration induced by proteasome inhibition in rat hippocampus. J Neuroinflammation. 2012;9:87. doi: 10.1186/1742-2094-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaczynska M, Rock KL, Goldberg AL. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- Gaczynska M, Rock KL, Spies T, Goldberg AL. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP7. Proc Natl Acad Sci USA. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canu N, Barbato C, Ciotti MT, Serafino A, Dus L, Calissano P. Proteasome involvement and accumulation of ubiquitinated proteins in cerebellar granule neurons undergoing apoptosis. J Neurosci. 2000;20:589–599. doi: 10.1523/JNEUROSCI.20-02-00589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi N, Morrison DC, Reis J. Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim Biophys Acta. 2012;1823:2087–2093. doi: 10.1016/j.bbamcr.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MD, Liu JC, Barthel TK, Hussain S, Lovria E, Cheng D, et al. A proteasome inhibitor, bortezomib, inhibits breast cancer growth and reduces osteolysis by downregulating metastatic genes. Clin Cancer Res. 2010;16:4978–4989. doi: 10.1158/1078-0432.CCR-09-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Ramakrishnan V, Rajkumar SV. Bortezomib combination therapy in multiple myeloma. Semin Hematol. 2012;49:228–242. doi: 10.1053/j.seminhematol.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah IM, Di NM. The ubiquitin-proteasome system and proteasome inhibitors in central nervous system diseases. Cardiovasc Hematol Disord Drug Targets. 2007;7:250–273. doi: 10.2174/187152907782793572. [DOI] [PubMed] [Google Scholar]
- Chen FT, Liu YC, Yang CM, Yang CH. Anti-inflammatory effect of the proteasome inhibitor bortezomib on endotoxin-induced uveitis in rats. Invest Ophthalmol Vis Sci. 2012;53:3682–3694. doi: 10.1167/iovs.12-9505. [DOI] [PubMed] [Google Scholar]
- Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol. 2013;4:1–26. [PMC free article] [PubMed] [Google Scholar]
- Cecarini V, Bonfili L, Cuccioloni M, Mozzicafreddo M, Rossi G, Buizza L, et al. Crosstalk between the ubiquitin-proteasome system and autophagy in a human cellular model of Alzheimer's disease. Biochim Biophys Acta. 2012;1822:1741–1751. doi: 10.1016/j.bbadis.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cao R, Cai T, Aschner M, Zhao F, Yao T, et al. The role of autophagy dysregulation in manganese-induced dopaminergic neurodegeneration. Neurotox Res. 2013;24:478–490. doi: 10.1007/s12640-013-9392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Rubinsztein DC. Small molecule enhancers of autophagy for neurodegenerative diseases. Mol Biosyst. 2008;4:895–901. doi: 10.1039/b804606a. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lizarbe S, Pascual M, Gascon MS, Blanco A, Guerri C. Lipid rafts regulate ethanol-induced activation of TLR4 signaling in murine macrophages. Mol Immunol. 2008;45:2007–2016. doi: 10.1016/j.molimm.2007.10.025. [DOI] [PubMed] [Google Scholar]
- Romero AM, Esteban-Pretel G, Marin MP, Ponsoda X, Ballestin R, Canales JJ, et al. Chronic ethanol exposure alters the levels, assembly, and cellular organization of the actin cytoskeleton and microtubules in hippocampal neurons in primary culture. Toxicol Sci. 2010;118:602–612. doi: 10.1093/toxsci/kfq260. [DOI] [PubMed] [Google Scholar]