Abstract
MYOGENIN is a member of the muscle regulatory factor family that orchestrates an obligatory step in myogenesis, the terminal differentiation of skeletal muscle cells. A paradoxical feature of alveolar rhabdomyosarcoma (ARMS), a prevalent soft tissue sarcoma in children arising from cells with a myogenic phenotype, is the inability of these cells to undergo terminal differentiation despite the expression of MYOGENIN. The chimeric PAX3-FOXO1 fusion protein which results from a chromosomal translocation in ARMS has been implicated in blocking cell cycle arrest, preventing myogenesis from occurring. We report here that PAX3-FOXO1 enhances glycogen synthase kinase 3β (GSK3β) activity which in turn represses MYOGENIN activity. MYOGENIN is a GSK3β substrate in vitro on the basis of in vitro kinase assays and MYOGENIN is phosphorylated in ARMS-derived RH30 cells. Constitutively active GSK3β(S9A) increased the level of a phosphorylated form of MYOGENIN on the basis of western blot analysis and this effect was reversed by neutralization of the single consensus GSK3β phosphoacceptor site by mutation (S160/164A). Congruently, GSK3β inhibited the trans-activation of an E-box reporter gene by wild-type MYOGENIN, but not MYOGENIN with the S160/164A mutations. Functionally, GSK3β repressed muscle creatine kinase (MCK) promoter activity, an effect which was reversed by the S160/164A mutated MYOGENIN. Importantly, GSK3β inhibition or exogenous expression of the S160/164A mutated MYOGENIN in ARMS reduced the anchorage independent growth of RH30 cells in colony-formation assays. Thus, sustained GSK3β activity represses a critical regulatory step in the myogenic cascade, contributing to the undifferentiated, proliferative phenotype in alveolar rhabdomyosarcoma (ARMS).
Keywords: alveolar rhabdomyosarcoma, PAX3-FOXO1, MYOGENIN, GSK3β, cell proliferation, tumorigenicity
Rhabdomyosarcoma (RMS) is the most common pediatric soft tissue sarcoma, accounting for 5% of all childhood cancers and approximately 50% of soft tissue sarcomas.1, 2, 3 There are two main subtypes: embryonal and alveolar RMS and although embryonal RMS is more common, alveolar RMS is considered to carry a worse prognosis. A gene fusion resulting in the t(2;13)(q35;q14) somatic cell chromosomal translocation fuses PAX3 and Foxo1 to create a potent transcription factor (PAX3-FOXO1) which is a predominant causative genetic lesion for the development of alveolar rhabdomyosarcoma (ARMS).1 ARMS is a highly malignant mesenchymal tumor that has properties of immature striated muscle tissue resulting in dense aggregates of poorly differentiated cells that are separated by fibrous membranes resulting in a loss in cellular cohesion.2, 3 PAX3 is a key determinant of somatic myogenesis and, is involved in the migration of progenitor cells to the dermomyotome region of the somite where they grow and divide in the presence of growth factors.4 PAX3 is also required to activate the myogenic determination gene, MYOD.5 MYOD is one of four myogenic regulatory factors (MRFs, which include MYF-5, MRF4 and MYOGENIN) from the basic helix-loop-helix superfamily of transcription factors which interact with myocyte enhancer factor-2 (MEF2) proteins in the hierarchical control of muscle-specific gene expression.6 Two kinases that potently exert effects on this myogenic regulatory cascade are p38 mitogen activated protein kinase (MAPK) and glycogen synthase kinase 3β (GSK3β). p38 MAPK is a key regulator of skeletal myogenesis that critically interacts with and activates MEF2 in the somite myotome during development.7, 8, 9 Conversely, GSK3β activation leads to a repression in skeletal and cardiac muscle differentiation, in part by antagonizing p38 MAPK-mediated activation of MEF2.10, 11 GSK3β usually targets proteins that have already been phosphorylated by another kinase at a ‘priming' serine or threonine residue located four amino acids C-terminal to a consensus (S/T)XXX(S/T)-PO4 motif.12, 13 Regulation of MEF2 and the MRFs leads to morphological changes including epithelial to mesenchymal transition, cell alignment and fusion to form multinucleated myotubes that eventually develop into functional, contractile muscle fibers. In particular, cells that express MYOD and MYOGENIN are typically fusion competent14, 15 with the exception of ARMS cell types. To date, lack of myogenic differentiation of PAX3-FOXO1 expressing ARMS cells has been attributed to their inability to upregulate p57Kip2 activity, hence destabilizing the DNA binding affinity of MYOD transcription complexes.16 Dysfunctional MYOD/E-protein complex association and transcriptional control is a common feature between ARMS and the non-PAX3-FOXO1 expressing embryonal rhabdomyosarcoma (ERMS). Subsequent restoration of the MYOD/E12 complex has been shown to switch ERMS cells from an arrested myofibroblast phase to a more differentiated state.17 Similarly p38 MAPK activity can potentiate myogenic differentiation in ERMS cells by enhancing MYOD trans-activation properties.18 Therefore, it is fairly clear that in both rhabdomyosarcoma subtypes the ability of MYOD to potentiate transcription is compromised. However, the role of MYOGENIN in RMS is more equivocal. For normal myogenesis to occur, both in vitro and in vivo, an absolute requirement for MYOGENIN is evident. Thus, MYOGENIN activity constitutes a pivot point for irreversible commitment to terminal differentiation.19, 20 The combination of data from gene targeting studies of the MRFs21, 22 supports the prevailing consensus that while the other three MRFs can compensate each other's functional roles,23, 24, 25, 26 MYOGENIN is absolutely essential for skeletal muscle fiber formation.20 Despite its expression in RMS, the paradox as to why MYOGENIN cannot mediate competence for differentiation is unknown.
Here, we examined the posttranslational regulation of MYOGENIN in ARMS. On the basis of the in silico prediction of a single consensus phosphorylation site for GSK3β on the MYOGENIN protein and also high levels of GSK3β activity in these cells, we determined that MYOGENIN function is potently repressed by GSK3β activity in ARMS. Moreover, pharmacological inhibition of GSK3β results in a profound decrease in size and, to a certain extent, number of RMS colonies in a colony-formation assay. This effect is mimicked by introduction of MYOGENIN bearing neutralizing mutations in the GSK3β consensus site. In combination, these data reveal MYOGENIN as a key target of GSK3β activity in ARMS, indicating that pharmacologic manipulation of this signaling axis may provide an opportunity for therapeutic intervention.
Results
MYOGENIN is expressed in PAX3-FOXO1 expressing RH30 cells
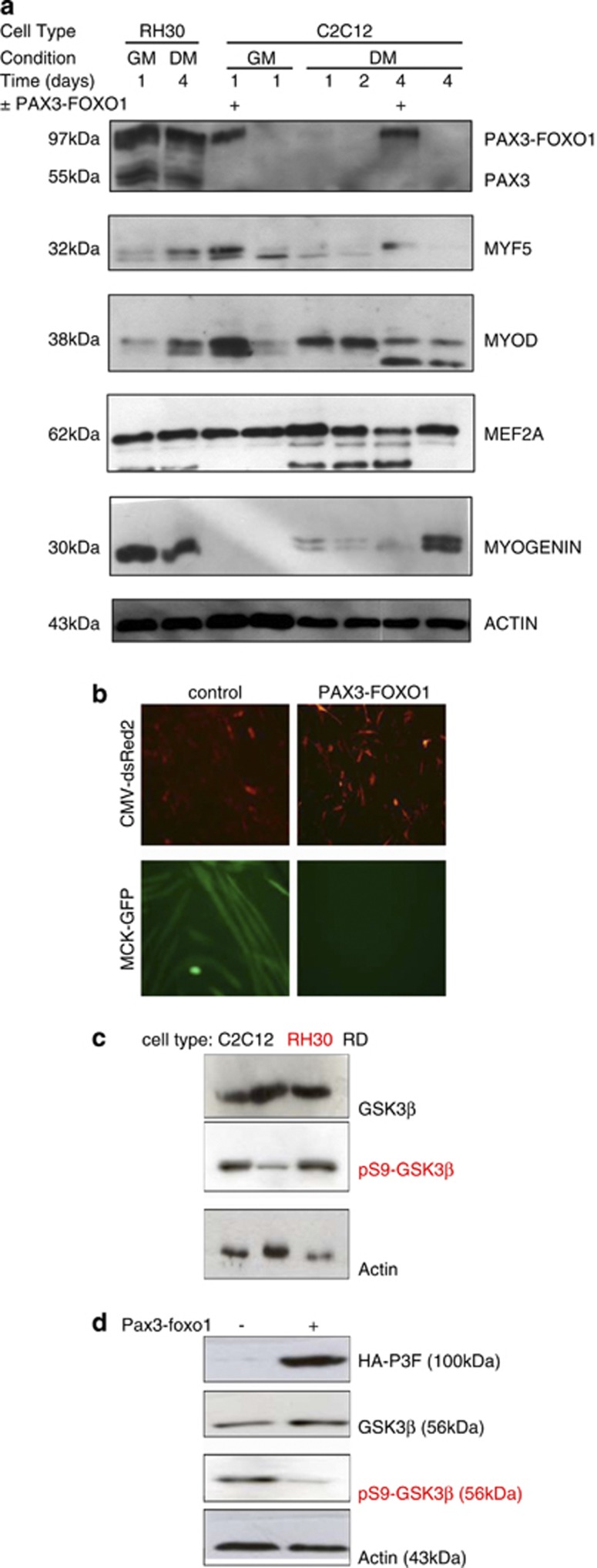
Serum (10% FBS) contains growth factors that repress the transcriptional activity of MRFs and also stimulate cell cycle progression hence rendering C2C12 myoblasts proliferative. In tissue culture, serum withdrawal (2% HS) results in activation of MEF2 and MRFs causing cell alignment and fusion to form multinucleated myotubes. Initially, in order to investigate the effect of PAX3-FOXO1 on this differentiation program, proliferating C2C12 myoblasts were transiently transfected with CMV-dsRed2, MCK-eGFP, and either HA-PAX3-FOXO1 or pcDNA3.1 control vector. Growth media (GM) was replaced with differentiation media (DM) 19 h after transfection and cells were allowed to differentiate for 96 h. SDS-PAGE samples were prepared from populations of myoblasts that either expressed or did not express PAX3-FOXO1, (a) before serum withdrawal (time=0; GM=10% FBS) and (b) at 24 h increments upon serum withdrawal (days 1–4; DM=2% HS). Protein expression levels of these samples were then compared with protein samples from PAX3-FOXO1 expressing RH30 cells in GM and DM, by western blotting. These data indicate that despite the expression of PAX3-FOXO1, MYOGENIN protein expression is maintained in human ARMS-derived RH30 cells (Figure 1a). In addition, PAX3-FOXO1 repressed myotube formation in C2C12 myoblasts (Figures 1a and b). Detection of myogenic differentiation using an MCK promoter driving GFP expression27 revealed GFP expressing, multinucleated myotubes in the controls but not in cells expressing PAX3-FOXO1 (Figure 1b).
Figure 1.
MYOGENIN protein expression and GSK3β activity are both maintained in ARMS: (a) C2C12 myoblasts were transfected with HA-PAX3-FOXO1 or pcDNA3.1 control plasmid for 1 day before extraction or serum withdrawal and then extraction at 1 day increments for up to 4 days as indicated. Protein levels were compared with protein extracts from PAX3-FOXO1 expressing RH30 cells 1 day in growth media (GM) and 4 days in differentiation media (DM). The results show that despite the expression of PAX3-FOXO1, RH30 cells also express MYOGENIN. On the other hand, HA-PAX3-FOXO1 overexpression in C2C12 inhibits MYOGENIN expression and subsequent myogenic differentiation. (b) C2C12 myoblasts were transfected with CMV-dsRed2, MCK-eGFP and, either HA-PAX3-FOXO1 or pcDNA3.1 control plasmid. HA-PAX3-FOXO1 overexpression repressed the formation of multinucleated myotubes. (c) Endogenous GSK3β protein levels and phosphorylation at serine 9 were compared in C2C12 myoblasts, RH30 and ERMS RD cells. Although GSK3β is expressed in all three cell types, it is predominantly phosphorylated and hence inactive in C2C12 myoblasts and RD cells but not PAX3-FOXO1 expressing RH30 cells. (d) C2C12 myoblasts were transfected with HA-PAX3-FOXO1 or pcDNA3.1 control plasmid for 1 day before extraction. Overexpression of HA-PAX3-FOXO1 resulted in decreased phosphorylation of GSK3β at serine 9 indicating its activation
It is well documented that MRFs and MEF2 proteins are highly sensitive to pro-myogenic kinases such as p38 MAPK9, 28, 29, 30 and also kinases such as GSK3β which are repressive to myogenesis.10, 31 Therefore we tested for GSK3β activity under conditions when myogenesis is supressed. As GSK3β is constitutively active until it is repressed by phosphorylation at serine 9 (by PKB), we assessed both total GSK3β protein expression levels and S9 phosphorylation levels using appropriate antibodies as indicated. We document that GSK3β is expressed in proliferative C2C12 myoblasts, PAX3-FOXO1 expressing ARMS cells (RH30) and, non-PAX3-FOXO1 ERMS cells (RD). However only in PAX3-FOXO1 expressing RH30 cells, is GSK3β predominantly in its unphosphorylated form (at serine 9) and, hence fully active state (Figure 1c). In addition, ectopic expression of PAX3-FOXO1 resulted in reduced phosphorylation of GSK3β at serine 9 in C2C12 myoblasts (Figure 1d).
MYOGENIN trans-activation function is repressed by GSK3β
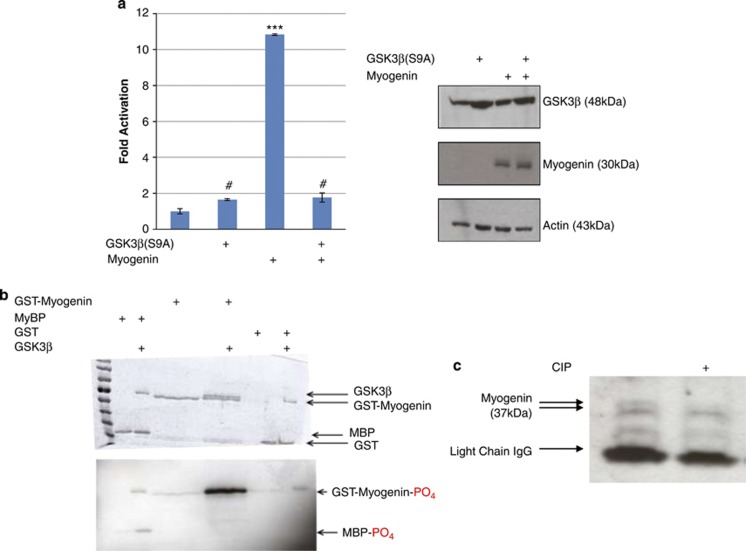
To assess the effect of GSK3β activity on MYOGENIN function, trans-activation of a 4x E-box Luciferase construct was measured in proliferating C2C12 myoblasts that were transfected with different combinations of constitutively active GSK3β(S9A) and MYOGENIN as indicated in Figure 2a. The data indicate that MYOGENIN potentiates the 4x E-box Luc reporter gene and that GSK3β(S9A) abrogates this effect (P<0.001) indicating repression of MYOGENIN by active GSK3β (Figure 2a, left panel) without affecting the MYOGENIN protein expression levels (Figure 2a, right panel).
Figure 2.
Overexpressed, constitutively active GSK3β (S9A) represses MYOGENIN trans-activation of E-box. (a) C2C12 myoblasts were transfected with 4x E-box Luc reporter and different combinations of HA-GSK3β(S9A) and MYOGENIN or pcDNA3.1 control plasmid as indicated. Overexpressed HA-GSK3β(S9A) repressed MYOGENIN transcriptional activity (P<0.001). (b) GSK3β directly phosphorylates MYOGENIN in vitro: Purified GST-MYOGENIN was incubated in vitro with GST-GSK3β and (γ-32P) ATP. GST and MBP proteins were used as negative and positive control respectively as indicated. Bands were resolved using SDS-PAGE and visualized by Coomassie Blue staining. Gels were dried and exposed to X-ray film for 21 h after the assay. (c) Calf-intestinal phosphatase (CIP) treatment of immunoprecipitated MYOGENIN that was obtained from 1000 μg of RH30 protein extract. The data shows that CIP treatment causes a loss of a high-molecular weight, phosphorylated form of MYOGENIN. #ns, ***P<0.001
GSK3β directly phosphorylates MYOGENIN in vitro
In order to determine whether MYOGENIN is a substrate for GSK3β, an in vitro kinase assay was performed using GST-MYOGENIN (1–225), purified GST-GSK3β and γ-32P ATP. Bands were resolved using SDS-PAGE and subsequent autoradiography showed 32P labeled bands for MYOGENIN, autophosphorylated GSK3β and MyBP (positive control, Figure 2b). In addition, Coomassie Blue staining revealed a lower mobility band indicative of phosphorylation (Figure 2b). To further test the idea that the lower mobility band is hyperphosphorylated, we used calf-intestinal phosphatases on RH30 cell lysates and found that the low mobility band was eradicated (Figure 2c). Collectively these data suggest that MYOGENIN is a GSK3β substrate in vitro.
Pharmacologic manipulation of GSK3β activity alters MYOGENIN properties
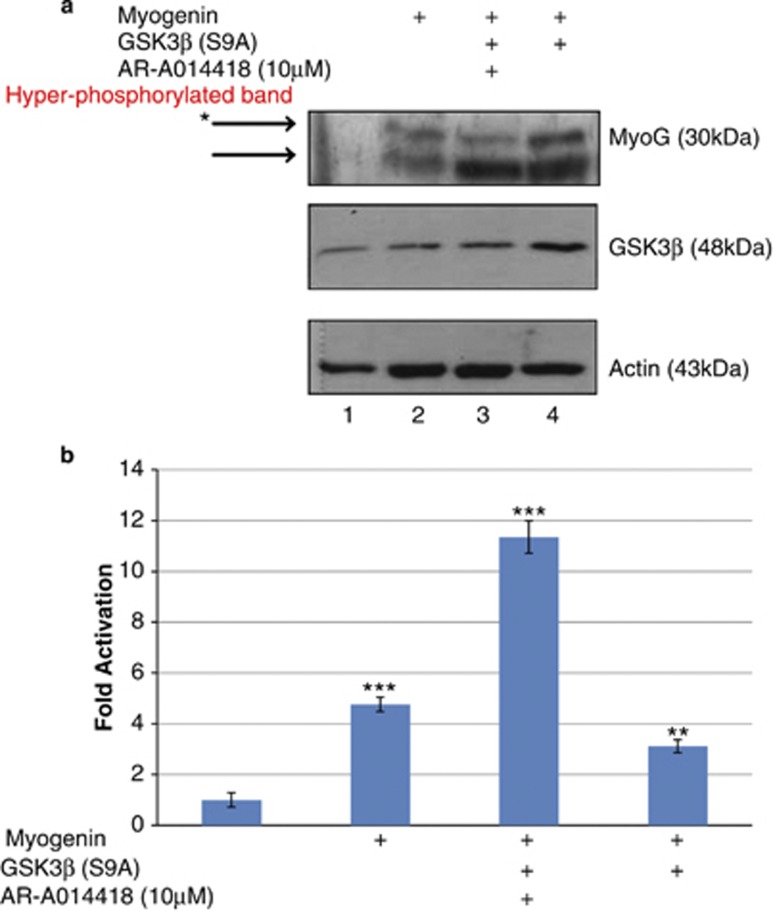
To further investigate the effect of GSK3β on MYOGENIN, COS7 cells were co-transfected with MYOGENIN and GSK3β(S9A) and, then treated with or without 10 μM GSK3β inhibitor, AR-A014418, as indicated in Figures 3a and b. Western blot analysis revealed two predominant forms of MYOGENIN, a low mobility hyperphosphorylated isoform and a high mobility, hypophosphorylated isoform (Figure 3a, lane 2). The lower mobility, hyperphosphorylated band is reduced upon pharmacological treatment with AR-A014418 as indicated (Figure 3a, lane 3). This corresponded with a significant increase in trans-activation of an E-box cis element driven reporter gene (P<0.001, Figure 3b). In contrast, constitutively active GSK3β(S9A) without pharmacological inhibition resulted in an increase in the low mobility, hyperphosphorylated band (Figure 3a, lane 4) which corresponded to a decrease in E-box luciferase activity in reporter gene assays (P<0.05, Figure 3b).
Figure 3.
GSK3β increases MYOGENIN protein, possibly through phosphorylation and this corresponds with decreased transcriptional activity. (a) Cos7 cells were transiently transfected with or without MYOGENIN and/or GSK3β(S9A) and then treated for 19 h with either 10 μM GSK3β inhibitor or DMSO 24 h after transfection as indicated. Protein samples were extracted and western blot analysis revealed an increase in a slower migrating, hyperphosphorylated MYOGENIN band (lane 4) in the presence of overexpressed HA-GSK3β(S9A), which was reduced in the presence of GSK3β inhibitor (lane 3). (b) E-box Luc reporter gene was co-transfected in Cos7 cells using the same conditions that were described above. Overexpressed MYOGENIN significantly enhanced transcriptional activity of the E-box promoter (***P<0.001) and, this effect was further increased in the presence of GSK3β inhibitor despite overexpression of GSK3β(S9A) (P<0.001). Overexpression of GSK3β(S9A) repressed MYOGENIN transcriptional activity (**P<0.05)
Mutation of a consensus GSK3β phosphoacceptor site on MYOGENIN (S160/164A) prevents GSK3β-mediated repression
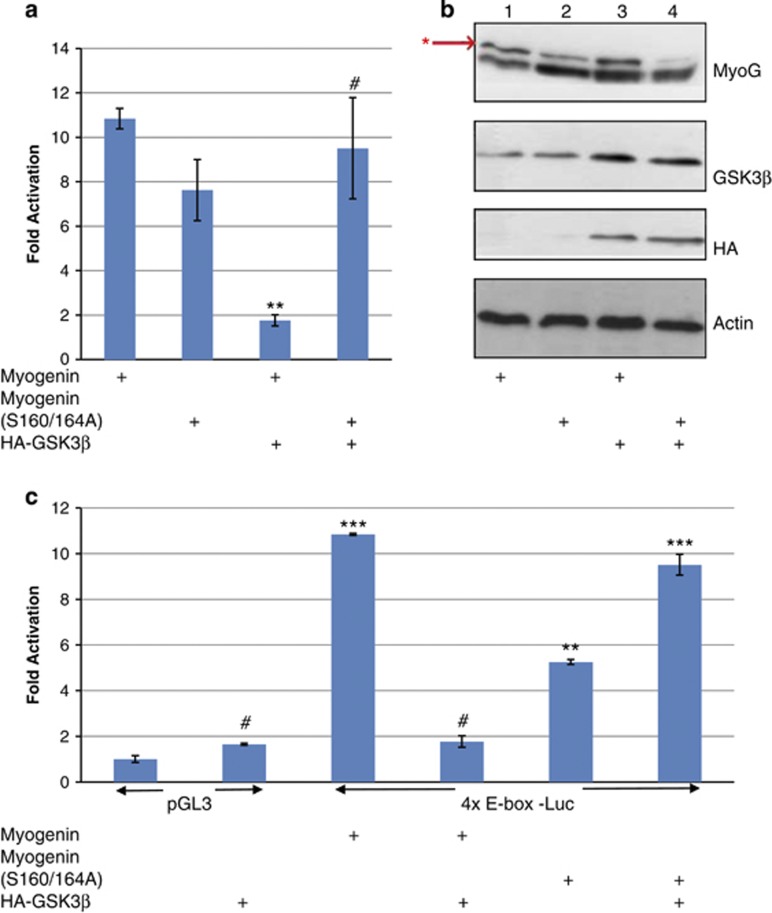
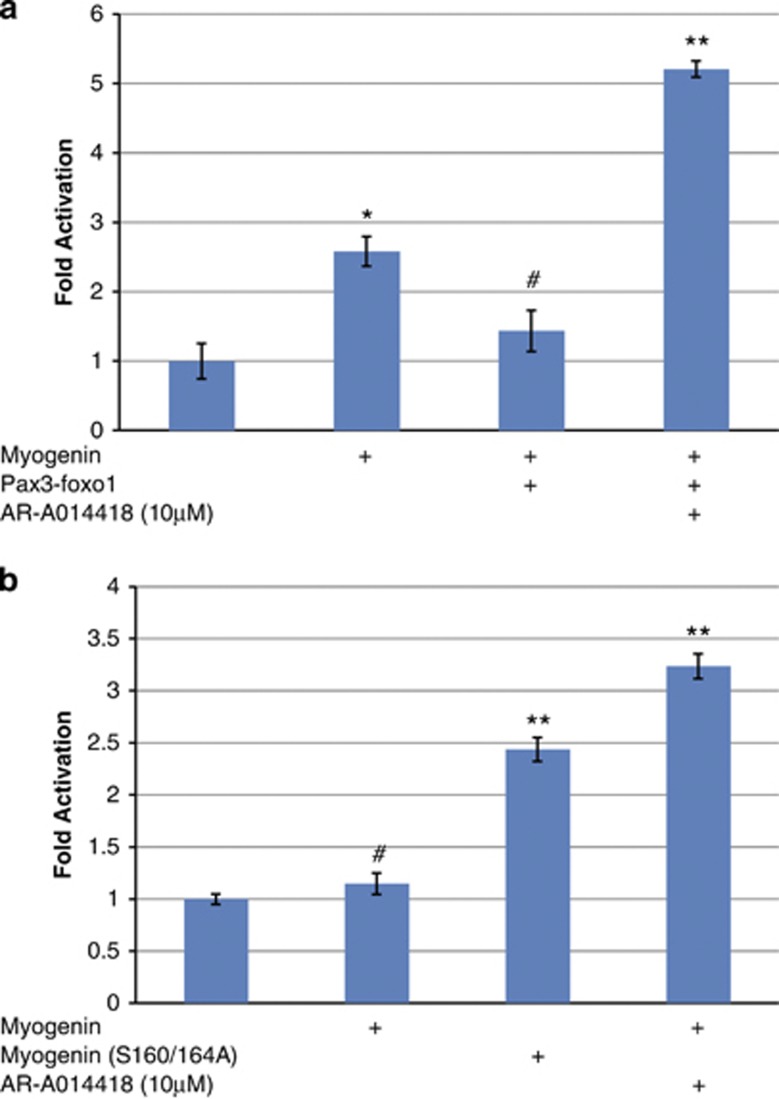
By in silico analysis, MYOGENIN contains a highly conserved putative GSK3β consensus phosphoacceptor site (Table 1), which we targeted by neutralizing site-directed mutagenesis. We observed that although wild-type MYOGENIN is sensitive to the repressive effects of constitutively active GSK3β(S9A), MYOGENIN (S160/164A) was not (Figure 4a). Western blot analysis revealed that MYOGENIN (S160/164A) mutations correspond with a decrease in the low mobility, hyperphosphorylated upper band (Figure 4b, lane 2) and that this effect was not altered by ectopically expressed HA-GSK3β(S9A). Together these data indicate that S160/164A mutations in MYOGENIN render it insensitive to the repressive effect of GSK3β. GSK3β(S9A) expression resulted in an increase in the low mobility, hyperphosphorylated form of wild-type MYOGENIN (Figure 4b, lane 3) and this corresponded with decreased E-box luciferase activity (P<0.001, Figure 4a). Although trans-activation of the skeletal muscle gene E-box cis-element by mutated MYOGENIN (S160/164A) is marginally less potent than wild-type MYOGENIN (P<0.05, Figure 4c); it is resistant to inhibition by activated GSK3β (P<0.001, Figure 4c).
Table 1. GSK3β consensus sequence within Myogenin.
| Myogenin sequence: | Species: |
|---|---|
| 158 VPSECSSHSASCSP 171 | Human |
| 158 VPSECNSHSASCSP 171 | Mouse |
| 158 VPSECNSHSASCSP 171 | Rat |
Figure 4.
MYOGENIN neutralizing phosphomutant (S160/164A) is resistant to GSK3β repression of transcription activity as well as an increased slower migrating, hyperphosphorylated MYOGENIN band. (a) 4x E-box Luc activity was assessed in C2C12 myoblasts that were transfected with either wild-type MYOGENIN or MYOGENIN (S160/164A) and, co-transfected with HA-GSK3β(S9A) or pcDNA3.1 control plasmid as indicated. HA-GSK3β(S9A) repressed MYOGENIN trans-activation of the 4x E-box promoter region (P<0.001) but had no effect on mutated MYOGENIN (S160/164A) transcriptional activity. (b) Western blot analysis of the same samples revealed a decrease in a slower migrating, hyperphosphorylated band for overexpressed MYOGENIN (S160/164A, lane 2) with respect to overexpressed wild-type MYOGENIN (lane 1). Co-transfected HA-GSK3β(S9A) caused an increase in the slow migrating, hyperphosphorylated MYOGENIN band (lane 3) but not with overexpressed mutated MYOGENIN (S160/164A, lane 4). (c) Independent analysis of E-box Luc activity in C2C12 myoblasts with different combinations of overexpressed MYOGENIN, mutated MYOGENIN (S160/164A), HA-GSK3β(S9A) or pcDNA3.1 control plasmid as indicated. ***P<0.001, **P<0.05, #ns
PAX3-FOXO1 activation of GSK3β antagonizes muscle creatine kinase promoter activation
To further examine the functional significance of our findings, we used MCK promoter activity, as a key indicator of the activation of myogenic differentiation, in C2C12 myoblasts that were transfected with or without the PAX3-FOXO1 oncogene (Figure 5a). These data depict that PAX3-FOXO1 represses MCK promoter activation in myoblasts that have been co-transfected with MYOGENIN (P<0.01) and this effect is not only abrogated by pharmacological inhibition of GSK3β, but further activated (P<0.001, Figure 5a). Interestingly, in PAX3-FOXO1 expressing, human ARMS-derived RH30 cells, ectopically expressed MYOGENIN had no effect on MCK promoter activity unless it was coupled with pharmacological inhibition of GSK3β using AR-A014418 (P<0.001, Figure 5b). Conversely, mutated MYOGENIN (S160/164A) was able to potentiate MCK promoter activity regardless of GSK3β inhibition (P<0.05, Figure 5b). Taken together, these data provide evidence that S160/164 on MYOGENIN are likely key targets of GSK3β signaling in alveolar rhabdomyosarcoma resulting in a diminution of the critical E-box dependent gene activation that is necessary and sufficient for differentiation.
Figure 5.
Pharmacological inhibition of GSK3β rescues PAX3-FOXO1 repression of MYOGENIN's transcriptional activation of MCK promoter in both C2C12 myoblasts and RH30 human ARMS cells. (a) MCK-Luc promoter activity was assessed in C2C12 myoblasts that were transfected with different combinations of MYOGENIN, PAX3-FOXO1 and pcDNA3.1 control plasmid as indicated and then treated with either 10 μM AR-A014418 or DMSO solvent. MYOGENIN enhanced MCK-Luc activity as expected (P<0.001) and this effect was repressed by co-expression of PAX3-FOXO1 (P<0.01). Pharmacological inhibition of GSK3β not only reversed the effect of PAX3-FOXO1 but resulted in a super-activation (P<0.001). (b) To assess the importance of these findings in human-derived ARMS, RH30 cells were transfected with either MYOGENIN or mutated MYOGENIN(S160/164A) and MCK-Luc promoter activity was assessed. The data shows that wild-type MYOGENIN could not trans-activate the MCK promoter region unless it was coupled with pharmacological inhibition of GSK3β (P<0.001). This was in contrast to mutated MYOGENIN (S160/164A), which could potentiate MCK promoter activity (P<0.001) regardless of GSK3β inhibition. (c) Summary of our findings: GSK3β activity in ARMS represses the activation of muscle-specific genes by repressing the transcriptional activity of MYOGENIN. #ns, *P<0.01, **P<0.001
Manipulation of GSK3β and MYOGENIN activity reduces tumorigenic properties of ARMS-derived RH30 cells
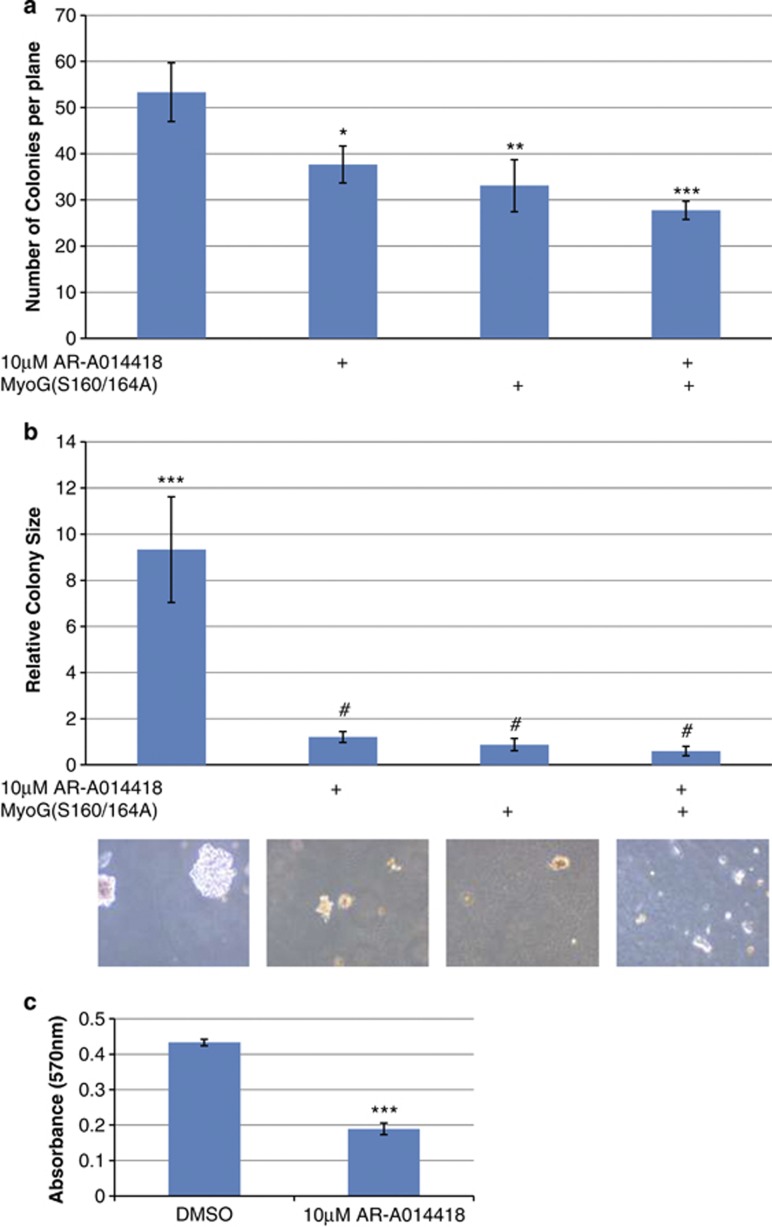
Colony-formation assays were performed as previously described using RH30 cells32 which can grow in an anchorage independent manner. Equal numbers of RH30 cells that have been transiently transfected with or without MYOGENIN containing the S160/164A mutations were seeded in growth media with or without 10 μM AR-A014418 (GSK3β inhibitor) and allowed to form colonies for 21 days (Figure 6). The addition of 10 μM AR-A014418 significantly impaired the ability of RH30 cells to form colonies (P<0.05) and remarkably reduced the size of the colonies (P<0.0001). A similar reduction in colony numbers and size were also evident in RH30 cells that were transfected with MYOGENIN (S160/164A) mutations (Figures 6a and b). In addition, we confirmed that pharmacological inhibition of GSK3β significantly reduced cell proliferation of PAX3-FOXO1 expressing cells (Figure 6c). Collectively these findings strongly indicate that GSK3β activity promotes the tumorigenicity of RH30 cells and that this effect is neutralized by expression of MYOGENIN bearing mutations that render it insensitive to GSK3β.
Figure 6.
Soft agarose colony formation and MTT cell proliferation assays: (a) Equal numbers of RH30 cells were seeded under different conditions as depicted, and allowed to form colonies for 21 days. On the 22nd day the colonies were stained with 0.005% Crystal Violet overnight. Colonies were counted at different planes (n=10) in four independent experiments done in triplicate. The total number of colonies was reduced by (i) 10 μM AR-A014418, P<0.05 (ii) Transient transfection of MYOGENIN containing the S160/164A neutralizing mutations, P<0.01 and (iii) both, P<0.001. Also see Supplementary Figure 1 for visual representation of the data. (b) We searched for the three largest colonies in each of the 12 plates for each condition and calculated the area using ImageJ software. The data revealed that the control colonies could grow up to 9 × bigger than any of the experimental conditions, P<0.001. (c) MTT cell proliferation assays were performed in PAX3/FOXO1A-expressing cells with and without 10 μM AR-A014418 treatment. The experiment revealed that GSK3β inhibition reduces cell proliferation by ∼2-fold, P<0.001. #ns, *P<0.05, **P<0.01, ***P<0.001
Electrical stimulation of ARMS-derived RH30 cells reduces GSK3β activity through Akt (PKB)
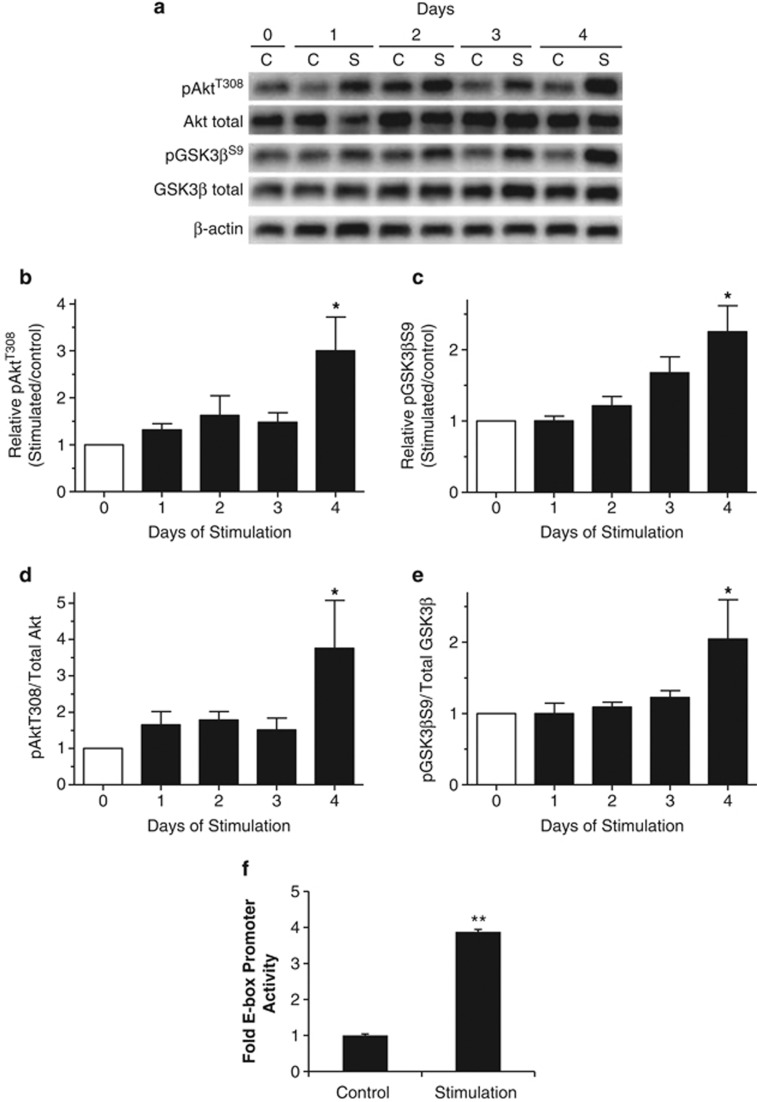
Electrical stimulation of skeletal muscle cells in cell culture has been shown to induce phenotype alterations and differentiation.33 Given that rhabdomyosarcoma shares properties of the skeletal muscle lineage, we electrically stimulated cultured RH30 cells for 4 h/day (5 Hz) for up to 4 days with the idea that it might promote differentiation by affecting the Akt/GSK3β signaling pathway.34 Stimulation of these cells resulted in an increase in pAktT308 to levels that were 3.00±0.72-fold higher than those in non-stimulated cells after 4 days of stimulation (Figures 7a and b). Concomitantly, pGSKβS9 was also increased 2.25±0.37 fold following 4 days of stimulation (Figures 7a and c). These increases in pAktT308 and pGSKβS9 were not a result of increases in total protein (Figure 7a) as indicated by the 3.76±1.32 and 2.05±0.55 increases in relative phosphorylation, respectively (Figures 7d and e). These changes in kinase activity corresponded with increased E-box promoter activity in stimulated cells compared with controls (Figure 7f). Collectively, these data indicate that electrical stimulation suppresses GSK3β activity and correspondingly activates MRF activity supporting our previous findings and also highlighting the possibility of using electrical stimulation as a therapeutic intervention in ARMS patients.
Figure 7.
In vitro electrical stimulation of RH30 cells. (a) Western blot analysis revealed that electrical stimulation increased PKB/Akt activity and that this corresponded with increased phosphorylation of GSK3β at S9. Relative increase of phosphorylation at: (b) Akt at T308 and (c) GSK3β at S9, over time. Graphical representation of phosphorylated to total amounts of: (d) Akt and (e) GSK3β. (f) E-box promoter activity decreased with electrical stimulation and this also corresponded with inhibition of GSK3β at S9. *P<0.01, **P<0.001
Discussion
ARMS, unlike ERMS, has a well-characterized cytogenetic basis in the majority of patients resulting from chromosomal translocations between chromosomes 1 and 13 and also 2 and 13 that result in fusion of the DNA binding domains of either Pax7 or PAX3 with the trans-activation domain of the Forkhead (FKHR) transcription factor family member Foxo1.1, 2, 35 In view of the well-substantiated crucial role of PAX3 and 7 in the development of skeletal muscle4, 5 it is therefore not surprising that the signature of ARMS tumor cells is a muscle-like phenotype and the expression of a variety of structural muscle marker genes such as myosin heavy chain and desmin.36 What is surprising is the sustained expression of MYOD and MYOGENIN in ARMS,37, 38 which are transcription factors that are intimately associated with the terminally differentiated, non- proliferative phenotype of normal myogenic cells, begging the question as to why they cannot exert this effect in ARMS. In particular, the function of MYOGENIN in the myogenic regulatory hierarchy places it at a pivotal and required step in the terminal commitment of myogenic progenitors to the differentiation program.19, 20, 34 Thus, our observations reported here, that MYOGENIN function in ARMS is repressed by inappropriate sustained signaling by the kinase GSK3β, may be of considerable significance for understanding the etiology of this disease. Moreover, as repression of kinase activity is, in many cases, a tractable pharmacologic approach, we now propose targeting GSK3β activity as a tangible therapeutic strategy for ARMS.
In support of the above, a recent study showed that ARMS-associated PAX3/7-Foxo1 fusion proteins inhibit MYOD target genes.39 It was also reported that forced MYOD/E-protein dimer expression could not rescue PAX3/7-Foxo1 repression of myogenic factors.39 Here, we also report that ectopically expressed PAX3-FOXO1 represses the induction of muscle genes, even when MRFs are expressed. We propose that the posttranslational repression of MYOGENIN activity is due to sustained GSK3β activity and, through a cross-talk mechanism, subsequent repression of p38 MAPK (Supplementary Figure 1) as we have previously described.10 p38 MAPK and PKB/Akt are both required for activation of MEF2/MYOD transcriptional control and chromatin remodeling events at crucial myogenic loci for the differentiation program.11, 40
In other systems, GSK3β phosphorylation of its protein substrates results in subsequent targeting for proteasomal degradation.12, 13 However, GSK3β does not appear to affect MYOGENIN protein stability in our experiments as we observe an increase in a slow migrating, hyperphosphorylated form of MYOGENIN in response to GSK3β signaling that is not reduced in terms of its level of expression suggesting that proteasomal degradation of MYOGENIN is not enhanced by GSK3β. Conversely, neutralizing mutations of the GSK3β consensus enhanced MYOGENIN trans-activation of the muscle creatine kinase promoter, and also reduced the tumorigenic properties of ARMS cells (RH30) in a colony-formation assay. These findings suggest that GSK3β-mediated inhibition of MYOGENIN trans-activation properties impairs MYOGENIN's ability to promote terminal differentiation in tumorigenic RH30 cells.
Cell cycle control is an essential component of normal growth control and development which goes awry in tumorigenesis. To date several growth-promoting PAX3-FOXO1 target genes have been implicated in RMS such as the IGF-R and c-Met although, while their contribution to proliferation is likely, the extent of their precise involvement in ARMS is still not clear.41 During normal skeletal myogenesis, upregulation of a cyclin-dependent kinase inhibitor, p21, stalls myoblasts in the G2/M phase of the cell cycle thus priming them for differentiation by promoting cell cycle exit, which is a requirement for subsequent muscle-specific gene expression.42 Consistent with the idea that GSK3β activation may contribute to the oncogenic properties resulting from PAX3-FOXO1 expression in ARMS, we observed that the number of proliferative RH30 cells is approximately halved by pharmacological inhibition of GSK3β. So far, the exact mechanism by which GSK3β regulates cell proliferation in ARMS is unknown. However, GSK3β has recently been shown to activate KLF643 and we recently identified that KLF6 enhances cell proliferation in myogenic cells through a TGFβ/Smad3 dependent pathway.44 We therefore speculate that PAX3-FOXO1/GSK3β enhancement of cell proliferation may involve KLF6 as a downstream effector as it is also highly expressed in various RMS cell types.
In summary, MYOGENIN normally activates genes that regulate cell fusion and terminal differentiation of skeletal muscle. In PAX3-FOXO1 expressing ARMS cells, our data indicate that sustained GSK3β activity represses MYOGENIN function, contributing to the transformed, proliferative phenotype of these cells. On the basis of this evidence, we propose that pharmacologic targeting of GSK3β kinase activity may constitute a tractable therapeutic strategy for ARMS.
Materials and Methods
Plasmids
E-box, MYOGENIN and MCK reporter constructs in pGL3 and expression vectors for MYOGENIN in EMSV were used in reporter gene assays. HA-tagged PAX3-FOXO1 was cloned into pcDNA3.1 and kindly donated by Dr. Malkin at MaRS, Toronto. HA-tagged GSK3β(S9A) was cloned in pcDNA3 ORF 995–2305.
Antibodies
Anti-MYOGENIN and anti-HA mouse monoclonal antibodies as well as anti-MEF2A rabbit polyclonal antibody were produced with the assistance of the York University Animal Care Facility; anti-PAX3 (1 : 250; Cell Signaling, Whitby, ON, Canada) GSK3β, phospho-GSK3β (1 : 1000; Cell Signaling); actin, MYOD, Myf-5, GFP, dsRed2 (1 : 2000; SantaCruz, Santa Cruz, CA, USA) were used for immunoblotting experiments.
Cell culture and transfection
C2C12, Cos7 and RH30 cells were maintained in DMEM supplemented with 10% fetal bovine serum (HyClone, Burlington, ON, Canada), 1% L-glutamine and 1% penicillin-streptomycin. Cells were maintained in a humidified, 37 °C incubator with a 5% CO2 atmosphere. For transfections, cells were seeded 1 day before transfection and transfected according to the standard calcium phosphate method previously described. A mixture of 50 μl 2.5 M CaCl2 per 25 μg DNA with an equal volume of 2x HeBS (2.8 M NaCl, 15 mM Na2HPO4, 50 mM HEPES, pH=7.15) was used and the cells were incubated overnight followed by washing and addition of fresh media. The cells were counted and transferred to pre-gelatin-coated plates.
Protein extractions, immunoblotting and reporter gene assays
Cells were collected using an NP-40 lysis buffer (0.5% NP-40, 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10 mM sodium pyrophosphate, 1 mM EDTA (pH 8.0), 0.1 M NaF) containing 10 μg/ml leupetin and aprotinin, 5 μg/ml pepstatin A, 0.2 mM phenylmethylsulfonyl fluoride and 0.5 mM sodium orthovanadate. Protein concentrations were determined using the Bradford method (Bio-Rad, Mississauga, ON, Canada) with bovine serum albumin (BSA) as a standard. An amount 20 μg of total protein extracts were used for immunoblotting, diluted in sample buffer containing 5% β-mercaptoethanol and boiled.
Transcriptional assays were done using luciferase reporter plasmids. The cells were collected for these assays using 20 mM Tris, (pH 7.4) and 0.1% Triton-X 100 and the values obtained were normalized to β-galactosidase activity expressed from a constitutive SV40 driven expression vector and represented as relative light units (RLU) or in some cases corrected Luciferase values for control, reporter alone transfections were arbitrarily set to 1.0, and fold activation values were calculated. Bars represent the mean (n=3) and error bars represent the standard error of the mean (n=3). Independent two sample t-tests of all quantitative data were conducted using R software. P-values are indicated with respect to controls where appropriate.
In vitro kinase assay
A total of 3 μg of purified recombinant GST-MYOGENIN was mixed with either 0.5 μg purified recombinant GST-GSK3β (1–433; Cell Signaling) and with (γ-32P) ATP and incubated for 30 min at 30 °C. Samples were denatured for 5 min at 95 °C in SDS sample buffer. Protein samples were then separated by 10% SDS-PAGE and exposed on X-ray film (Kodak X-Omat, Toronto, ON, Canada) for 21 h to detect 32P incorporation. The lanes containing GST-MYOGENIN are elongated because two lanes were pooled to fit a higher total reaction volume to accommodate for the low concentration of purified GST-MYOGENIN (0.06 μg/μl). All lanes contain equal total amounts of proteins (3 μg).
Electrical stimulation
Cells were plated onto 0.1% gelatin-coated 6-well plates. The lids of the plates were fitted with two parallel platinum wire electrodes, placed at the opposite ends of each well and extending into the media. The wires from all wells were arranged in parallel and connected to an electrical stimulator (Harvard Apparatus Canada, Saint-Laurent, Quebec, Canada). Cells were stimulated at 5 V and a frequency of 5 Hz for 4 hours/day and allowed a subsequent 20 h recovery period. Cells were collected following the recovery period throughout the 4 days of the protocol.
Soft agarose colony-formation assay
Materials: 0.7% (w/v) DNA grade Agarose, 1% (w/v) DNA grade Agar, 0.005% Crystal Violet (Sigma-Aldrich, Oakville, ON, Canada), 2X Media+20% (v/v) FBS. After 48 h of transfection with MYOGENIN containing the S160/164A mutations or empty vector, RH30 cells were assayed for their capacity to form colonies as previously described.45 A total of 1 × 104 cells were suspended on a layer of 0.35% agarose in DMEM (10% FBS) with or without 10 μM AR-A014418, in 6-well plates. Medium was refreshed every 3–5 days as needed and on the 22nd day, the amount of colonies were counted using a contrast phase microscope. The relative colony sizes were calculated using ImageJ software (Scion Corporation, Frederick, MD, USA). Four independent experiments were carried out in triplicate.
Acknowledgments
We thank Dr. D Malkin from The Research Institute at The Hospital for Sick Children, Toronto, Ontario, Canada, for providing the HA-PAX3-FOXO1 plasmids as well as the RH30 and RD cells. This work was supported by funding provided by Canadian Institutes for Health Research (CIHR) and Natural Sciences and Engineering Research Council of Canada (NSERC).
Glossary
- GFP
green fluorescent protein
- GSK3β
glycogen synthase kinase 3β
- Luc
luciferase
- MAPK
mitogen activated protein kinase
- MCK
muscle creatine kinase
- MEF2
myocyte enhancer factor-2
- PI3K
phosphoinositide 3-kinase
- PKB
protein kinase B
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Galili N, Davis RJ, Fredericks S, Mukhopadhyay FJ, Rauscher BS, III, Rovera EG, et al. Fusion of a forkhead domain gene to PAX3 in the solid tumor alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Barr FG. The role of chimeric paired box domain transcription factors in the pathogenesis of pediatric rhabdomyosarcoma. Cancer Res. 1999;59 (7 suppl:1711s–1715s. [PubMed] [Google Scholar]
- Paulino AC, Okcu MF. Rhabdomyosarcoma. Curr Probl Cancer. 2008;32:7–34. doi: 10.1016/j.currproblcancer.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The role of pax genes in the development of tissues and organs: PAX3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C R Biol. 2007;330:530–533. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Currie PD, Bryson-Richardson RJ. The genetics of vertebrate myogenesis. Nat Rev. 2008;9:632–646. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- Han J, Molkentin J. Regulation of MEF2 by p38 MAPK and its implication in cardiomyocyte biology. Trends Cardiovasc Med. 2000;10:19–22. doi: 10.1016/s1050-1738(00)00039-6. [DOI] [PubMed] [Google Scholar]
- Muñoz JP, Collao A, Chiong M, Maldonado C, Adasme T, Carrasco L, et al. The transcription factor MEF2C mediates cardiomyocyte hypertrophy induced by IGF-1 signaling. Biochem Biophys Res Commun. 2009;388:155–160. doi: 10.1016/j.bbrc.2009.07.147. [DOI] [PubMed] [Google Scholar]
- de Angelis L, Zhao J, Andreucci JJ, Olson EN, Cossu G, McDermott JC. Regulation of vertebrate myotome development by the p38 MAP kinase-MEF2 signaling pathway. Dev Biol. 2005;283:171–179. doi: 10.1016/j.ydbio.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Dionyssiou MG, Nowacki NB, Hashemi S, Zhao J, Kerr A, Tsushima RG, et al. Cross-talk between glycogen synthase kinase 3β (GSK3β) and p38MAPK regulates myocyte enhancer factor 2 (MEF2) activity in skeletal and cardiac muscle. J Mol Cell Cardiol. 2013;54:35–44. doi: 10.1016/j.yjmcc.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Frame S. The renaissance of GSK3. Nat Rev. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- Grossi A, Yadav K, Lawson MA. Mechanical stimulation increases proliferation, differentiation and protein expression in culture: Stimulation effects are substrate dependent. J Biomech. 2007;40:3354–3362. doi: 10.1016/j.jbiomech.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Roeb W, Boyer A, Cavenee WK, Arden KC. PAX3-FOXO1 controls expression of p57Kip2 cell-cycle regulator through degredation of EGR1. Proc Natl Acad Sci USA. 2007;104:18085–18090. doi: 10.1073/pnas.0708910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, MacQuarrie KL, Analau E, Tyler AE, Dilworth FJ, Cao Y, et al. MYOD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev. 2009;23:694–707. doi: 10.1101/gad.1765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Wu Z, Zhang P, Wood LD, Bhakta KS, Han J, et al. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev. 2000;14:574–584. [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the MYOGENIN gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Rawls A, Morris JH, Rudnicki M, Braun T, Arnold HH, Klein WH, et al. MYOGENIN's functions do not overlap with those of MYOD or Myf-5 during mouse embryogenesis. Dev Biol. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- Megeney LA, Rudnicki MA. Determination versus differentiation and the MYOD family of transcription factors. Biochem Cell Biol. 1995;73:723–732. doi: 10.1139/o95-080. [DOI] [PubMed] [Google Scholar]
- Weintraub AJ. The MYOD family and myogenesis: redundancy, networks and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MYOD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, et al. Mrf4 determines skeletal muscle identity in MYF5:MYOD double mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kaul A, Koster M, Neuhaus H, Braun T. Myf-5 revisited: loss of early myotome formation does not lead to rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- Rawls A, Valdez MR, Zhang W, Richardson J, Klein WH, Olson EN. Overlapping functions of the myogenic bHLH genes MRF4 and MYOD revealed in double mutant mice. Development. 1998;125:2349–2358. doi: 10.1242/dev.125.13.2349. [DOI] [PubMed] [Google Scholar]
- Kollias HD, Perry RLS, Miyake T, Aziz A, McDermott JC. Smad7 promotes and enhances skeletal muscle differentiation. Mol Cell Biol. 2006;26:6248–6260. doi: 10.1128/MCB.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornatsky OI, Cox DM, Tangirala P, Andreucci JJ, Quinn ZA, Wrana JL, et al. Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 1999;27:2646–2654. doi: 10.1093/nar/27.13.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis F, Perdiguero E, Nebreda AR, Munoz-Canoves P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 2006;16:36–44. doi: 10.1016/j.tcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Keren A, Bengal E, Frank D. p38 MAP kinase regulates the expression of XMYF5 and affects distinct myogenic programs during Xenopus development. Dev Biol. 2005;288:73–86. doi: 10.1016/j.ydbio.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Du M, Perry RL, Nowacki NB, Gordon JW, Salma J, Zhao J, et al. Protein kinase A represses skeletal myogenesis by targeting myocyte enhancer factor 2D. Mol Cell Biol. 2008;28:2952–2970. doi: 10.1128/MCB.00248-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi L, Ciaarapica R, De Salvo M, Verginelli F, Gueguen M, Martini C, et al. Inhibition of Notch3 signalling induces rhabdomyosarcoma cell differentiation promoting p38 phosphorylation and p21(Cip1) expression and hampers tumour cell growth in vitro and in vivo. Cell Death Differ. 2012;19:871–881. doi: 10.1038/cdd.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor MK, Irrcher I, Hood DA. Contractile activity-induced transcriptional activation of cytochrome c involves Sp1 and Is proportional to mitochondrial ATP synthesis in C2C12 muscle cells. J Biol Chem. 2001;276:15898–15904. doi: 10.1074/jbc.M100272200. [DOI] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- Tonin PN, Scrable H, Shimada H, Cavenee WK. Muscle-specific gene expression in rhabdomyosarcomas and stages of human fetal skeletal muscle development. Cancer Res. 1991;51:5100–5106. [PubMed] [Google Scholar]
- Dias P, Chen B, Dilday B, Palmer H, Hosoi H, Singh S, et al. Strong immunostaining for MYOGENIN in rhabdomyosarcoma is significantly associated with tumors of the alveolar subclass. Am J Pathol. 2000;156:399–408. doi: 10.1016/S0002-9440(10)64743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebire NJ, Malone M. MYOGENIN and MYOD1 expression in paediatric rhabdomyosarcomas. J Clin Pathol. 2003;56:412–416. doi: 10.1136/jcp.56.6.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhabeu F, Hayashi S, Morgan JE, Relaix F, Zammit PS. Alveolar rhabdomyosarcoma-associated proteins PAX3/FOXO1A and PAX7/FOXO1A suppress the transcriptional activity of MYOD-target genes in muscle stem cells. Oncogene. 2013;32:651–662. doi: 10.1038/onc.2012.73. [DOI] [PubMed] [Google Scholar]
- Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Guttridge DC. Mechanisms of impaired differentiation in rhabdomyosarcoma. FEBS J. 2013;17:4323–4334. doi: 10.1111/febs.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung R, Parnaik VK. Cyclin D3 promotes myogenic differentiation and Pax7 transcription. J Cell Biochem. 2012;113:209–219. doi: 10.1002/jcb.23346. [DOI] [PubMed] [Google Scholar]
- Lang UE, Kocabayoglu P, Cheng GZ, Ghiassi-Nejad Z, Munoz U, Vetter D, et al. GSK3β phosphorylation of the KLF6 tumor suppressor promotes its transactivation of p21. Oncogene. 2012;32:4557–4564. doi: 10.1038/onc.2012.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionyssiou MG, Salma J, Bevzyuk M, Wales S, Zakharyan L, McDermott JC. Kruppel-like factor 6 (KLF6) promotes cell proliferation in skeletal muscle cells in response to TGFβ/Smad3 signaling. Skelet Muscle. 2013;3:7. doi: 10.1186/2044-5040-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarapica R, Annibali D, Raimondi L, Savino M, Nasi S, Rota R. Targeting Id protein interactions by an engineered HLH domain induces human neuroblastoma cell differentiation. Oncogene. 2009;28:1881–1891. doi: 10.1038/onc.2009.56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.