Abstract
Integration-deficient lentiviral vectors (IDLVs) have been shown to transduce a wide spectrum of target cells and organs in vitro and in vivo and to maintain long-term transgene expression in nondividing cells. However, epigenetic silencing of episomal vector genomes reduces IDLV transgene expression levels and renders these safe vectors less efficient. In this article, we describe for the first time a complete correction of factor IX (FIX) deficiency in hemophilia B mice by IDLVs carrying a novel, highly potent human FIX cDNA. A 50-fold increase in human FIX cDNA potency was achieved by combining two mechanistically independent yet synergistic strategies: (i) optimization of the human FIX cDNA codon usage to increase human FIX protein production per vector genome and (ii) generation of a highly catalytic mutant human FIX protein in which the arginine residue at position 338 was substituted with leucine. The enhanced human FIX activity was not associated with liver damage or with the formation of human FIX-directed inhibitory antibodies and rendered IDLV-treated FIX-knockout mice resistant to a challenging tail-clipping assay. A novel S1 nuclease-based B1-quantitative polymerase chain reaction assay showed low levels of IDLV integration in mouse liver. Overall, this study demonstrates that IDLVs carrying an improved human FIX cDNA safely and efficiently cure hemophilia B in a mouse model.
Introduction
In a recent human clinical trial for the treatment of β-thalassemia, lentiviral vectors integrated into the third intron of the protooncogene HMGA2 led to overexpression of truncated HMGA2 mRNA and to benign clonal expansion.1 Molecular analysis of the mechanisms involved in the dysregulation of proliferation following retro- and lentiviral vector integration implicated vector-contained regulatory elements in transcriptional and posttranscriptional alterations of host oncogene expression.1,2,3 These included enhancer elements, polyadenylation signals, and splice sites (either major or cryptic). The range of mechanisms by which integrating vectors can alter host gene expression renders the task of developing mutation-free lentiviral vectors more challenging and has been the impetus for the development of various integration-deficient lentiviral vector (IDLV) systems.
To date, most IDLVs have been generated by packaging genomic vector RNAs into vector particles containing class I integrase mutants, which render the vector integration defective and yet support all other vector functions required for efficient gene transfer.4,5 IDLVs maintain long-term expression in slowly dividing or nondividing cells in vitro and in vivo.6,7,8,9,10,11,12,13,14,15 However, recent studies have demonstrated that epigenetic silencing renders IDLV transgene expression inefficient.6,16,17 Given the transcriptional silencing of episomal human immunodeficiency virus (HIV)-1-based vectors, achieving effective gene replacement therapy on systemic administration of IDLVs remains a major challenge. Indeed, in a recent preclinical study by Mátrai et al., intravenous administration of IDLVs carrying a canine factor IX cDNA resulted in a merely transient detectable increase (1–2%) in canine factor IX (cFIX) activity in hemophilia B mice.18 The relatively high dose of IDLVs (total p24gag 260 µg) used by Mátrai et al. suggests that a further increase in IDLV dosage would not achieve long-term therapeutic levels of FIX activity in hemophilia B mice.
In this article, we report on the development of novel lentiviral vectors carrying a highly potent human FIX cDNA as a means for curing hemophilia B mice through systemic administration of IDLVs. Two strategies for improving human FIX cDNA function were combined to synergistically enhance overall human FIX potency. Specifically, a fivefold increase in human FIX protein production per lentiviral vector genome was obtained by optimizing the codon usage of the human FIX cDNA. We further improved the effectiveness of vector-delivered human FIX by using more catalytically active human FIX mutants. This strategy was premised on earlier studies showing that the FIX variants R338A (the arginine 338 residue replaced by alanine) and R338L were three- to sevenfold more catalytically active than the wild-type (WT) human FIX protein.19,20 In line with these studies, an eightfold increase in human FIX-specific activity was demonstrated in vitro by lentiviral vectors carrying the R338L human FIX. To maximize the effectiveness of vector-delivered human FIX, the two aforementioned strategies of improving human FIX cDNA were combined to generate novel codon-optimized human FIX cDNAs encoding a highly catalytically active R338L human FIX mutant. Indeed, the increased human FIX production and the enhanced catalytic activity of the R338L mutant synergistically increased overall human FIX activity per vector genome by >50-fold.21,22 Of note, systemic administration of IDLVs carrying the novel human FIX cDNA achieved complete long-term cure (with maximal human FIX activity >500%) of hemophilia B in mice. Vector-treated mice survived a challenging tail-clipping assay. Furthermore, vector administration did not induce liver damage or the development of human FIX-directed humoral immune response.
Results
Lentiviral vectors carrying highly potent human FIX cDNAs exhibit superior therapeutic potential
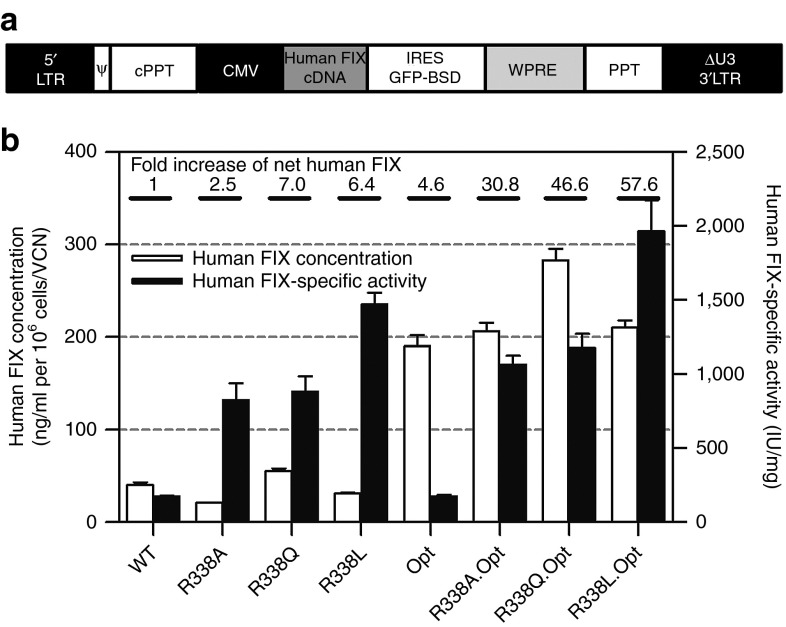
To facilitate effective IDLV-mediated gene replacement therapy for hemophilia B, we sought to enhance the therapeutic potency of IDLV-delivered human FIX cDNAs. With that aim, a series of WT and codon-optimized (Opt.) human FIX cDNA variants, with the arginine 338 residue replaced by alanine (R338A), glutamine (R338Q), or leucine (R338L), were cloned into a bicistronic lentiviral vector expressing the green fluorescent protein and blasticidin fusion protein (Figure 1a). Premised on earlier studies,19,20 we expected that the novel human FIX cDNAs would be more potent than their WT counterpart. To test this hypothesis, HepG2 cells were transduced with the aforementioned lentiviral vectors and selected for blasticidin resistance. The efficiency of human FIX production was evaluated by normalizing the concentration of secreted human FIX to the vector copy number (VCN). Specific clotting activities of the variants and WT human FIX proteins secreted into the culture medium were determined by a standard activated partial thromboplastin time assay, normalized to human FIX protein concentration. The overall effect of the various human FIX cDNA modifications on human FIX clotting activity was calculated as the multiplication product of the fold increase in human FIX production per vector genome and the fold increase in human FIX-specific activity. As shown in Figure 1b, lentiviral vectors carrying codon-optimized human FIX cDNAs expressed nearly fivefold more human FIX protein per vector genome than vectors carrying the WT human FIX cDNA. All three human FIX variant proteins carrying the 338-residue mutations exhibited increased clotting activity. Of the three codon-optimized mutations, the R338L variant exhibited the highest specific activity (1964.4 IU/mg), which is 11-fold higher than the parental WT human FIX clotting activity (Figure 1b). Furthermore, the overall clotting activity per vector genome obtained from the HepG2 cell line (expressing the codon-optimized R338L human FIX variant) was >50-fold higher than the clotting activity yielded by WT human FIX-expressing cells. Encouraged by this result, we sought to evaluate the therapeutic potential of IDLVs carrying the codon-optimized R338L human FIX in vivo.
Figure 1.
Codon optimization and R338 mutations synergistically enhance the efficacy of human FIX cDNA delivered by lentiviral vector in vitro. (a) Depiction of the lentiviral vector backbone used in this study. All vectors contain 5′ and large U3-deleted 3′ long terminal repeats (5′LTR and ΔU3 3′LTR), packaging signal (Ψ), central polypurine tract (cPPT), cytomegalovirus promoter, human FIX transgene, internal ribosome entry site (IRES), a fusion protein encoded by the green fluorescent protein (GFP) reporter and the blasticidin resistance genes, woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), and polypurine tract (PPT). (b) Concentration and clotting activity of wild-type (WT) and modified human FIX proteins expressed in HepG2 cells transduced with integrase-competent lentiviral vectors harboring a series of human FIX cDNAs, including WT and three R338 mutants (R338A, R338Q, and R338L) either in the background of the parental human FIX cDNA or following codon optimization. Concentration of human FIX in culture medium (obtained from HepG2 cell lines transduced with the aforementioned human FIX cDNAs) was determined by enzyme-linked immunosorbent assay and normalized to vector copy number (VCN) as determined by quantitative polymerase chain reaction. The human FIX-specific activity was determined by an activated partial thromboplastin time assay normalized to human FIX level. Each sample was analyzed in duplicates. Data were normalized to cell number and VCN and are presented as means ± SEM from triplicate samples. The overall effect of the various human FIX cDNA modifications on human FIX clotting activity was calculated as the multiplication product of the fold increase in human FIX production per vector genome and the fold increase in human FIX-specific activity.
IDLVs carrying codon-optimized R338L human FIX cDNA mediate complete long-term correction of FIX deficiency in vivo
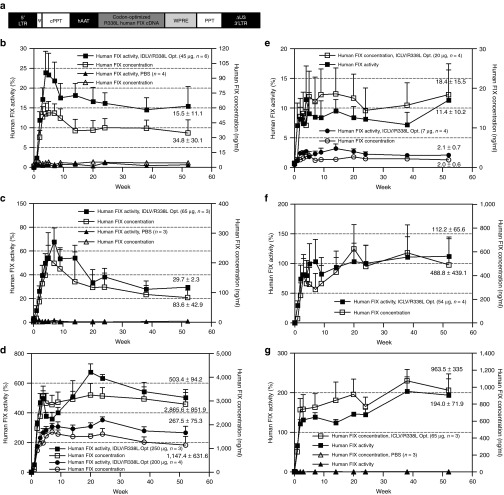
To evaluate IDLVs' ability to correct FIX deficiency in a mouse model of hemophilia B, the codon-optimized R338L human FIX cDNA was incorporated into a lentiviral vector under the control of the liver-specific human α1-antitrypsin promoter (pTK1340; Figure 2a). Vector particles containing either the WT HIV-1 integrase (integrase-competent lentiviral vector (ICLV)) or the D64E integrase mutant (IDLV) were generated by transient transfection.23 Escalating doses of IDLVs (45, 65, 200, and 250 µg p24gag) and ICLVs (7, 20, and 65 µg p24gag) were administered to C57BL/6 hemophilia B mice by a single intraperitoneal injection. Concentration of human FIX protein and its clotting activity in mice plasma were periodically determined by the human FIX enzyme-linked immunosorbent assay and activated partial thromboplastin time assay, respectively. As shown in Figure 2b–g, all IDLV-treated mice exhibited long-term therapeutic concentration and activity levels of human FIX in a dose-dependent manner. Note that at 1 year after vector administration, the average plasma human FIX concentration and activity in mice treated with the lowest dose of IDLV (45 µg p24gag) were 34.8 ng/ml and 15.5%, respectively (Figure 2b), which is sufficient to significantly improve the clinical status of patients with hemophilia B.24 As expected, ICLVs were found to be more efficacious than IDLVs at obtaining therapeutic human FIX levels in hemophilia B mice. At 1 year postinjection, human FIX activity in mice treated with 65 µg p24gag of ICLV or IDLV was 194% and 29.7%, respectively. However, at 20 weeks postinjection, the average human FIX concentration and activity in mice administered with the highest dose of IDLV (250 µg p24gag) was 3,250 ng/ml and 675%, respectively.
Figure 2.
Long-term expression of therapeutic levels of R338L human FIX in hemophilia B mice. (a) Depiction of the lentiviral vector (pTK1340) from which the codon-optimized R338L human FIX is expressed under the control of the human α1-antitrypsin promoter (hAAT). (b–g) Levels of human FIX concentration (ng/ml) and clotting activity (percentage normal human FIX clotting activity as measured in vitro) in hemophilia B mice treated with escalating doses of integration-deficient lentiviral vectors (IDLVs): (b) 45 µg p24gag, (c) 65 µg p24gag, and (d) 200 and 250 µg p24gag , compared with mice treated with (e) integrase-competent lentiviral vectors (ICLVs): (e) 7 and 20 µg p24gag, (f) 54 µg p24gag, and (g) 65 µg p24gag. All samples were measured in duplicates. Data are presented as mean ± SEM. cPPT, central polypurine tract; LTR, long terminal repeat; PBS, phosphate-buffered saline; PPT, polypurine tract; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
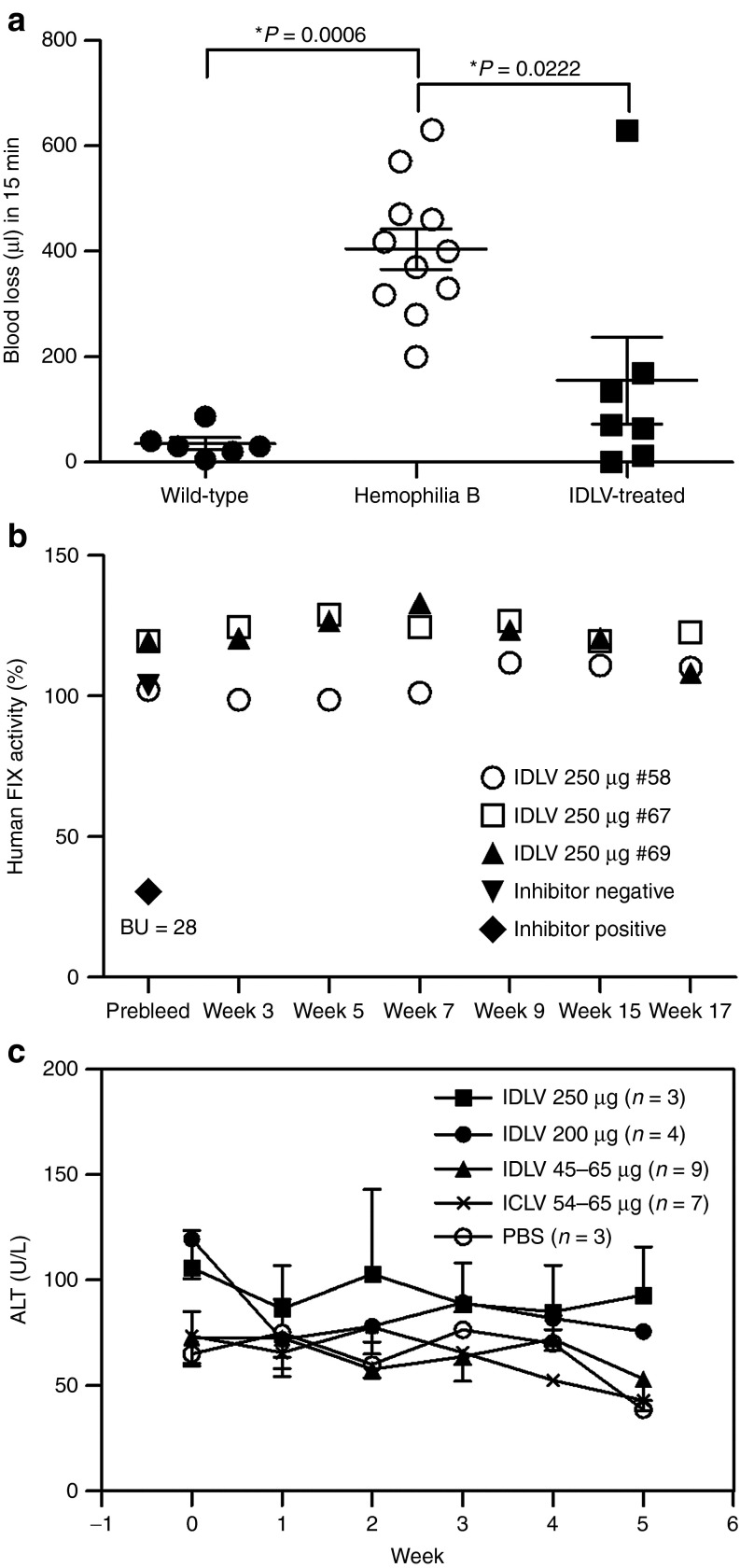
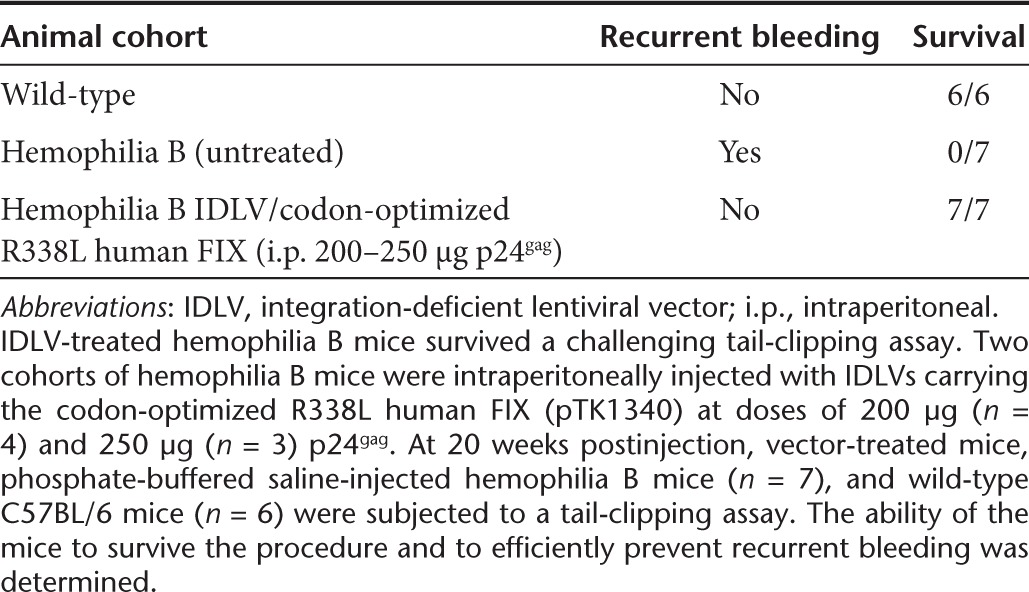
We further directly evaluated the ability of IDLVs carrying the codon-optimized R338L human FIX cDNA to correct FIX deficiency in vivo. Hemophilia B mice treated with total doses of either 200 or 250 µg p24gag IDLVs were subjected to a challenging tail-clipping assay. As shown for the first time (Figure 3a and Table 1), all seven IDLV-treated mice survived the bleeding challenge and demonstrated significantly reduced blood loss. All seven untreated hemophilia B mice exhibited massive blood loss, which necessitated euthanasia (Table 1). Furthermore, in contrast with earlier reports,18,25,26 these data suggest that therapeutically effective doses of IDLVs and ICLVs do not elicit significant humoral or cellular immune responses against vector-delivered human FIX or vector-transduced cells, respectively. Of note, necropsy of two mice revealed tumor development. However, the low VCN in the tumors' DNA suggested that tumor development was not associated with insertional mutagenesis (Supplementary Figure S1 and Supplementary Table S1).
Figure 3.
Integration-deficient lentiviral vector (IDLV) delivery of the codon-optimized R338L human FIX rendered hemophilia B mice resistant to tail-clipping injury without inducing immunologic adverse effects. (a) Functional clotting assay in vivo. Hemophilia B mice were intraperitoneally injected with IDLVs carrying the codon-optimized R338L human FIX cDNA under the control of the human α1-antitrypsin promoter (pTK1340), with doses of either 200 µg (n = 4) or 250 µg (n = 3) p24gag. At 20 weeks postvector administration, treated mice were challenged with a tail-clipping assay. Total volume of blood lost in 15 min was measured in seven vector-treated mice and compared with blood loss of tail-clipped wild-type (n = 6) and phosphate-buffered saline (PBS)-injected hemophilia B mice (n = 11). P value was assessed by a paired Student's t-test. (b) Lack of neutralizing antibodies directed to human FIX protein in IDLV-treated mice. A modified Bethesda assay was used to detect the emergence of inhibitory human FIX antibodies. Plasma samples were collected from the aforementioned IDLV-treated hemophilia B mice (250 µg p24gag, n = 3) before (prebleed) and at various time points postvector administration and were analyzed for the presence of inhibitory antibodies. Plasma samples obtained from a naive (untreated) mouse and from the human FIX inhibitor-producing mouse (a hemophilia B mouse injected with purified human FIX protein and exhibiting 28 Bethesda units (BU)/ml), were used as negative and positive control, respectively. (c) Evaluation of vector-mediated hepatotoxicity. To detect liver damage induced by either uptake of vector particles or due to human FIX production, levels of alanine aminotransferase (ALT) were determined in mouse plasma obtained before and periodically after vector administration. Samples from PBS-treated mice were used as negative controls. ICLV, integrase-competent lentiviral vectors.
Table 1. Tail-clipping assay as a means to functionally evaluate clotting activity in vivo in IDLV-treated hemophilia B mice.
Lack of immune response to human FIX and vector-transduced cells
To identify potential vector-induced immunotoxicity, we sought to determine whether either humoral or cellular immune response to human FIX antigen or to vector-transduced hepatocytes, respectively, had evolved following vector administration and human FIX expression. Thus, mouse plasma samples obtained before and at various time points after vector administration were tested for the presence of inhibitory antibodies to human FIX. In addition, plasma levels of liver enzymes were determined as a means to detect liver damage mediated by cellular cytotoxicity to vector-transduced hepatocytes. As shown in Figure 3b, human FIX activity was not affected by undiluted mouse plasma, thus ruling out the possibility that inhibitory antibodies to human FIX had developed following vector administration. In addition, normal levels of liver enzymes found in all tested mouse plasma samples indicated that liver transduction with lentiviral vectors, carrying human FIX expression under the control of the human α1-antitrypsin promoter, did not induce hepatocyte-directed immune response (Figure 3c). Of note, in contrast with earlier studies,18,25,26 the aforementioned data demonstrated that maintaining therapeutic levels of human FIX expression from conventional lentiviral vectors does not necessitate miRNA-mediated knockdown of human FIX expression in antigen-presenting cells.
Biodistribution and integration of ICLVs and IDLVs in hemophilia B mice
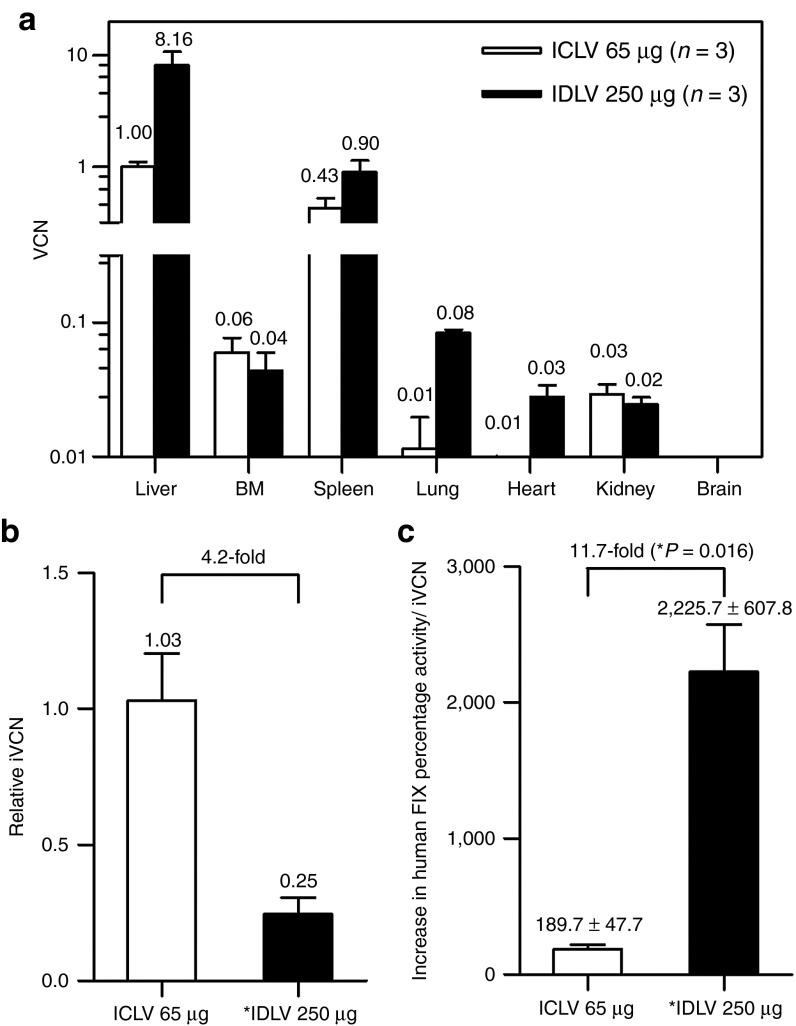
To characterize the ICLV and IDLV biodistribution following a single intraperitoneal administration of codon-optimized R338L human FIX, multiplex quantitative polymerase chain reaction (qPCR) analysis was used on genomic DNA samples from different mouse tissues obtained at 1 year postvector administration (duration of experiments). As shown in Figure 4a, the highest VCN per host genome of both ICLVs (65 µg p24gag) and IDLVs (250 µg p24gag) was found in liver tissues, with 1.0 and 8.2 VCN, respectively. In spleen tissues, the VCN of ICLVs and IDLVs was 0.4 and 0.9, respectively. As expected, neither IDLV nor ICLV vector genomes were detected in brain tissues. Interestingly, up to 0.06 VCN of ICLVs was found in bone marrow samples. This relatively efficient in vivo bone marrow transduction is in line with the data presented in an earlier study by Pan et al.27 Due to the loss of episomal vector genomes, a VCN of merely 0.04 was found in actively dividing bone marrow cells at 1 year post high-dose (250 µg p24gag) IDLV administration. However, these findings raised the possibility that some of the IDLV genomes might have illegitimately integrated into the host chromatin by integrase-independent mechanisms. Recent studies16,28 characterizing the rates of residual IDLV integration in vitro demonstrated that up to 1 in 200 IDLV genomes are likely to integrate into a host cell genome. To accurately compare the number of integrated viral genomes in liver tissues at 1 year postintraperitoneal administration of either ICLVs or IDLVs, we established and used a modified B1-qPCR assay (Supplementary Data), which was described earlier by Tervo et al.29 As shown in Figure 4b, the levels of integrated vector genomes following administration of ICLV (65 µg p24gag) in liver tissues were found to be more than fourfold higher than the IDLV levels (250 µg p24gag). Note that the average total IDLV vector genomes in liver tissues were eightfold higher than the total ICLV genomes, indicating that the level of IDLV integration is nearly 35-fold lower than the level of ICLV integration. To further characterize the mechanism of IDLV integration, unique exactly mapped integration sites (ISs) of IDLVs (257 ISs) and ICLVs (1,697 ISs) were isolated by non-restrictive linear amplification-mediated (LAM)-PCR and LAM-PCR and were sequenced by 454 pyrosequencing.30,31 In line with an earlier study,18 18.6% of IDLV ISs showed long terminal repeat deletions >2 nucleotides, whereas only 3.1% of ISs retrieved from ICLV-administered samples showed such deletions. These findings support the notion that IDLVs carrying the codon-optimized R338L human FIX cDNA can impart a long-term cure of hemophilia B with minimal genotoxic risks. However, to directly evaluate the added biosafety value of IDLVs, we calculated the therapeutic/genotoxic index (TGI) of the lentiviral vectors used in this study as the overall increase in human FIX percentage activity per integrated vector genome in vivo. As shown in Figure 4c, at 1 year postsystemic administration of lentiviral vectors (250 µg p24gag), the calculated TGI of IDLVs was significantly (nearly 12-fold) higher than the TGI of its integrated counterparts. The findings of this study provide the first in vivo experimental evidence that systemic administration of IDLVs is a safe and effective gene replacement methodology exhibiting reduced genotoxicity. Moreover, therapeutic efficacy was not associated with an immune response to either the IDLV-delivered transgene or the vector-transduced hepatocytes.
Figure 4.
Analysis of total and integrated vector copy number (VCN) of integration-deficient lentiviral vectors (IDLVs) and integrase-competent lentiviral vectors (ICLVs) in mouse tissues demonstrated low illegitimate integration and a high therapeutic/genotoxic index of IDLVs. VCN in tissues was assessed at 1 year postintraperitoneal vector administration of ICLVs (65 µg p24gag) and IDLVs (250 µg p24gag). (a) A multiplex quantitative polymerase chain reaction analysis of total VCN per host genome of ICLV and IDLV in mouse tissues. (b) The relative integration level of IDLVs (n = 3) normalized to the level of ICLV integration in mouse livers was analyzed by the S1/B1-quantitative polymerase chain reaction assay. Integrated VCN (iVCN) in a single mouse liver treated with ICLVs (65 µg p24gag) served as a baseline (valued as 1.00) for the analysis of two additional ICLV-treated mice and to establish the standard curve in the analysis of three IDLV (250 µg p24gag)-treated mice. The relative therapeutic/genotoxic index (TGI) of ICLV and IDLV vectors was calculated as the overall increase in human FIX percentage activity per iVCN. (c) Graph demonstrating TGI values of IDLVs and ICLVs following intraperitoneal administration to hemophilia B mice. P value was assessed by a paired Student's t-test. BM, bone marrow.
Discussion
The IDLV system offers an efficient approach to ferry large genetic cargoes to dividing and nondividing cells in vitro and in vivo, without posing the risk of insertional mutagenesis that is associated with ICLVs. However, epigenetic silencing of IDLVs renders this promising gene delivery system inefficient for in vivo applications that require systemic vector administration.6,18 In this study, we sought to use the codon-optimized R338L human FIX cDNA as a means to achieve long-term therapeutic levels of human FIX in hemophilia B while reducing the risk of adverse effects associated with integrating vectors.
In earlier publications,21,22 we demonstrated that combining the aforementioned modifications in a single human FIX cDNA synergistically enhanced the total human FIX activity per vector genome in vitro by more than 50-fold. However, we reasoned that the efficacy and safety of using a highly active human FIX cDNA should be evaluated in a long-term in vivo study. A recent study demonstrated the ability of IDLV carrying codon-optimized R338L human FIX to achieve canine FIX activity of up to 45% of normal level in hemophilia B mice,32 but the ability of IDLVs carrying human FIX to support a long-term cure for hemophilia B mice—without inducing cellular or humoral immune response—and their long-term biodistribution and number of illegitimately integrated vector genomes per host genome have not been documented. In this study, we demonstrate for the first time the ability of IDLVs carrying the codon-optimized R338L human FIX to achieve a long-term (1-year) cure of hemophilia B in a mouse model. Levels of IDLV-delivered human FIX antigen and activity were dose dependent. Furthermore, survival of the IDLV-treated mice after a challenging tail-clipping assay provided the strongest attestation to the therapeutic potential of the IDLV system.
Although the C57BL/6 hemophilia B mice used in this study were devoid of all murine FIX coding sequences, long-term expression of human FIX following IDLV and ICLV administration was not associated with either an increase in liver enzymes or with the development of inhibitory antibodies to human FIX. These findings are in line with an earlier report demonstrating long-term hepatic expression of firefly luciferase following gene delivery by IDLV and ICLV.6 The lack of immune response to transgene-expressing hepatocytes and to circulating human FIX can be attributed in part to liver-mediated immune tolerance.33 Of note, in contrast with earlier studies,18,25,26 we report that long-term expression of human FIX did not require the incorporation of mir142 complementary sequences as a means to avoid human FIX-directed immune response by reducing its expression in hematopoietic cells.
Although novel polypurine tract-deleted vectors, generating primarily one-long terminal repeat circles, exhibited significantly lower levels of illegitimate IDLV integration,28 the ability to quantitatively characterize the level of illegitimate integration of IDLVs in vivo is required to accurately evaluate their biosafety value. In this article, we describe the development of the S1/B1-qPCR methodology (Supplementary Figure S2). Tervo et al. described the two-round, nested B1-qPCR methodology earlier as a means to evaluate HIV-1 integration in mouse cell lines in vitro.29 However, this study demonstrated significant background readouts, which were attributed to linear amplification of HIV-1 sequences independent of viral integration (Supplementary Figure S3). This phenomenon significantly reduces the sensitivity of the B1-qPCR system and renders it unsuitable for quantifying low levels of IDLV integration in vivo. Thus, to accurately evaluate IDLV integration in mouse liver, three modifications were incorporated into the conventional B1-qPCR methodology.29 (i) To minimize the synthesis of single-stranded DNA templates, the viral anchor primer was directed to sequences upstream to the viral polypurine tract (rather than to sequences in the viral long terminal repeats; Supplementary Figure S2). (ii) To further reduce the levels of single-stranded DNAs, amplification products of the first round of PCR were subjected to S1-nuclease degradation before the second nested-PCR round (Supplementary Figure S4). (iii) Earlier studies34,35 demonstrated tissue-specific patterns of HIV-1 vector integration. Because our studies focused on hepatic human FIX delivery, we sought to characterize the rate of illegitimate IDLV integration relative to the levels of ICLV integration in mouse liver. Therefore, the standard curve used in this study was based on liver genomic DNA of an ICLV-treated mouse.
The novel S1/B1-qPCR assay, for the first time, allowed accurate detection of low levels of integration and thus facilitated the comparative evaluation of the IDLV and ICLV levels of integration, genotoxicity, and biosafety in vivo. Premised on the S1/B1-qPCR methodology, we calculated the vector TGI as the overall increase in human FIX percentage activity per integrated vector genome in vivo. The aforementioned results indicate that the TGI of IDLVs was nearly 12-fold higher than the calculated index obtained for their integrated counterparts. Incorporating the highly efficacious humanized R338L into the IDLVs provides the ability to cure hemophilia B with reduced vector load. This constitutes a major leap forward in vector biosafety and opens a new avenue in the gene therapy of hemophilia B using IDLVs, in addition to other viral and nonviral-based vectors including adeno-associated virus-based vectors and naked DNA, respectively.36
Overall, in this study, we demonstrated for the first time that IDLVs carrying the codon-optimized R338L human FIX variant supported a long-term (1-year) complete cure of hemophilia B in a mouse model of FIX deficiency. IDLV-treated hemophilic mice survived a challenging tail-clipping assay and did not develop a detectable immune response to either vector-delivered human FIX or vector-transduced cells. In addition, we report the development of a novel S1 nuclease-based B1-qPCR methodology to quantify low levels of vector integration in vivo for evaluating the biosafety of IDLV gene delivery. The findings of this study indicate that the IDLV is a safe and efficacious gene delivery system suitable for gene replacement applications in nonterminal genetic diseases.
Materials and Methods
Lentiviral vector constructs. A series of human FIX cDNA fragments, including WT cDNA (pTK1201), codon-optimized cDNA (pTK1206), three R338 mutants (R338A, pTK912; R338Q, pTK1414; and R338L, pTK1367), and each 338-residue mutant on the background of codon-optimized human FIX cDNA (codon-optimized R338A, pTK1207; codon-optimized R338Q, pTK1336; and codon-optimized R338L, pTK1335), were cloned into the Hpa I site of a lentiviral vector (pTK642). All the above human FIX constructs were expressed under the control of the cytomegalovirus promoter and contained an internal ribosome entry site conjugated with the green fluorescent protein reporter gene and the blasticidin resistance gene. The codon-optimized R338L human FIX cDNA (pTK1340) used in in vivo studies was cloned under a liver-specific human α1-antitrypsin promoter.
Lentiviral particle production, concentration, and titration. All vector-derived lentiviral particles were produced in 293T cells using three-plasmid transient transfection, as previously described.37 Lentiviral vectors harboring human FIX cDNA used in the in vitro studies were packaged with integrase-competent packaging vector (NRF).37 Codon-optimized R338L human FIX (pTK1340) lentiviral vectors used in in vivo studies were packaged by either NRF or integrase-defective packaging vector (pTK939), which expresses D64E-mutant integrase. Vector titer was determined by measuring the p24 capsid concentration using enzyme-linked immunosorbent assay, as previously described.17 All viral vector preparations were confirmed for the absence of replication-competent retrovirus, as previously documented.38
Generation of HepG2 cell lines stably expressing WT and modified human FIX cDNAs. HepG2 cells were transduced with lentiviral vectors harboring relevant human FIX constructs at equivalent amounts of p24gag. Blasticidin-resistant cells were selected and their VCNs were determined using a multiplex qPCR.
Determination of protein concentration and clotting activity of human FIX secreted from HepG2 cells. To compare the yield and clotting function of human FIX proteins generated from each HepG2 cell line, cells were plated at 0.5 × 106 cells per well on a six-well plate for 24 h. Subsequently, the culture medium was replaced with fresh medium containing vitamin K (Hospira, Lake Forest, IL). After an additional 24-h incubation, the medium containing human FIX clotting proteins was collected and analyzed for protein concentration and clotting activity using enzyme-linked immunosorbent assay and activated partial thromboplastin time, respectively.
Determination of human FIX protein concentration and coagulation activity. Human FIX protein concentration was measured using a sandwich enzyme-linked immunosorbent assay, modified based on the original method described previously.39 Materials and procedures are detailed in the Supplementary Materials and Methods. Clotting activity was measured by activated partial thromboplastin time as previously described,40 with a minor change in the standard curve, which was generated by performing clotting assay on serial dilutions of a plasma-derived, highly purified human FIX protein (CSL Behring, King of Prussia, PA).
Animal studies. All procedures involving animal study were performed in accordance with the Guide for the Care and Use of Laboratory Animals; the in vivo study protocol was approved by the University of North Carolina Institutional Animal Care and Usage Committee. The FIX-deficient mouse model (murine FIX-knockout C57BL/6 strain) used in this study was described previously.41
In vivo coagulation assay. At 20 weeks postviral treatment, mice injected with total doses of either 200 or 250 µg p24gag IDLVs were subjected to a tail-clipping assay performed under anesthesia, following the procedure described previously.42 Blood was collected into citrated tubes over a 15-min period. Following tail clipping, mice were monitored hourly for 8 hours to observe recurrent bleeding; mice experiencing recurrent bleeding episodes >8 hours after the tail-clipping procedure required euthanasia.
Evaluation of viral vector toxicity and human FIX-directed inhibitory antibodies. As a test of liver function, mouse plasma samples were collected weekly during the first 5 weeks postviral injection and submitted to the Animal Clinical Chemistry and Gene Expression Laboratories for alanine aminotransferase analysis using an automatic chemical analyzer (Johnson & Johnson's VT350, New Brunswick, NJ). To detect human FIX inhibitory antibodies, a modified Bethesda assay was performed according to the protocol previously described.42
Preparation of mouse tissue DNA for quantification of VCN. At 1 year postviral administration, mouse tissues, including liver, brain, heart, lung, spleen, kidney, and bone marrow, were harvested. Genomic DNAs were isolated from tissues using a Blood & Tissue DNeasy kit (Qiagen, Valencia, CA), according to the manufacturer's instructions; RNAs were removed using RNase A (Fermentas, Pittsburgh, PA). All samples were treated with DpnI (New England Biolabs, Ipswich, MA). Total VCN and integrated vector genomes were quantified using multiplex qPCR and S1/B1-qPCR, respectively.
Multiplex qPCR. Multiplex qPCR is a probe-based PCR optimized in this study for use in quantification of HIV-1 VCNs. Amplifications of both vector and reference gene are carried out simultaneously in a single PCR reaction using two different readout probes. The multiplex qPCR approach is uniquely designed to quantify vector genomes in both human and mouse systems (using an identical set of primers/probe for vector amplification, and a relevant reference gene; β-glucuronidase for humans and glyceraldehyde-3-phophate dehydrogenase for mouse). Materials and procedures used in the multiplex qPCR are detailed in the Supplementary Materials and Methods.
S1/B1-qPCR. The novel S1/B1-qPCR was optimized based on the conventional B1-qPCR29 to enable accurate quantification of integrated vector genomes in sample DNAs containing episomal viral vector genomes. The protocol, including sequences of primers and probes, is provided in the Supplementary Materials and Methods.
Integration site analysis. The pattern of vector integration mediated by ICLVs and IDLVs was assessed at 1 year postviral treatment. DNA samples extracted from liver tissues of hemophilia B mice, injected with either IDLV or ICLV at 65 µg p24gag (n = 3 per group), were mapped to identify vector ISs using non-restrictive LAM-PCR and LAM-PCR, followed by 454 pyrosequencing, as previously described.30,31
SUPPLEMENTARY MATERIAL Figure S1. Hematoxylin/eosin staining of tumor-containing liver and spleen tissues. Figure S2. Optimization of the S1/B1 qPCR methodology. Figure S3. Significant background amplification of nonintegrating vector genomes by conventional nested B1-qPCR in the absence of the B1-directed primers. Figure S4. S1-nuclease treatment reduces background B1-qPCR amplification of nonintegrating HIV-1 vectors. Table S1. Vector copy number in tumor and in normal liver and spleen tissues. Data. Materials and Methods.
Acknowledgments
This study was supported by the National Institutes of Health (NIH) grant R01-DK058702-10 to T.K. and T.S. and by the University of North Carolina (UNC) Center for AIDS Research. It was also supported by the NIH grants RC3 HL103396-01 and P01 HL112761 and by a Research Award from the Hemostasis and Thrombosis Research Society to P.E.M. We are grateful to Mckeon and Chiune Sugihara. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: pD64E from Vinay K. Pathak and HIV-1 p24 monoclonal antibody (183-H12-5c) from Bruce Cheseboro and Kathy Wehrly. T.K. receives royalties for the polypurine tract-deleted vector technology licensed by the UNC to Immune Design. The University of North Carolina receives funding to support P.E.M.'s research laboratory from Asklepios Biopharmaceuticals (Chapel Hill, NC), Baxter Bioscience (Thousand Oaks, CA), Novo Nordisk (Plainsboro, NJ), and Prolor Biotech (Nes Ziona, Israel). P.E.M. has served on advisory committees for Asklepios, Baxter, Bayer (Leverkusen, Germany), Novo Nordisk, and Pfizer (New York, NY). Asklepios Biopharmaceuticals is the sponsor of an open gene therapy clinical trial for hemophilia B using an AAV vector. This manuscript is dedicated to the Gold Star families and to the US Marine Corps–Semper Fi.
Supplementary Material
References
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human ß-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Moiani A, Paleari Y, Sartori D, Mezzadra R, Miccio A, Cattoglio C, et al. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J Clin Invest. 2012;122:1653–1666. doi: 10.1172/JCI61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasik MB, McCray PB., Jr Integrase-defective lentiviral vectors: progress and applications. Gene Ther. 2010;17:150–157. doi: 10.1038/gt.2009.135. [DOI] [PubMed] [Google Scholar]
- Wanisch K, Yáñez-Muñoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M, Kantor B, Cockrell A, Ma H, Zeithaml B, Li X, et al. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol Ther. 2008;16:1968–1976. doi: 10.1038/mt.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick HE, Nowrouzi A, Fronza R, McDonald RA, Kane NM, Alba R, et al. Integrase-deficient lentiviral vectors mediate efficient gene transfer to human vascular smooth muscle cells with minimal genotoxic risk. Hum Gene Ther. 2012;23:1247–1257. doi: 10.1089/hum.2012.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutant F, Frenkiel MP, Despres P, Charneau P. Protective antiviral immunity conferred by a nonintegrative lentiviral vector-based vaccine. PLoS ONE. 2008;3:e3973. doi: 10.1371/journal.pone.0003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nunzio F, Félix T, Arhel NJ, Nisole S, Charneau P, Beignon AS. HIV-derived vectors for therapy and vaccination against HIV. Vaccine. 2012;30:2499–2509. doi: 10.1016/j.vaccine.2012.01.089. [DOI] [PubMed] [Google Scholar]
- Hu B, Dai B, Wang P. Vaccines delivered by integration-deficient lentiviral vectors targeting dendritic cells induces strong antigen-specific immunity. Vaccine. 2010;28:6675–6683. doi: 10.1016/j.vaccine.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacz K, Mukherjee S, Apolonia L, Blundell MP, Bouma G, Escors D, et al. Nonintegrating lentivector vaccines stimulate prolonged T-cell and antibody responses and are effective in tumor therapy. J Virol. 2009;83:3094–3103. doi: 10.1128/JVI.02519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri DR, Michelini Z, Baroncelli S, Spada M, Vendetti S, Bona R, et al. Nonintegrating Lentiviral Vector-Based Vaccine Efficiently Induces Functional and Persistent CD8+ T Cell Responses in Mice. J Biomed Biotechnol. 2010;2010:534501. doi: 10.1155/2010/534501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri DR, Michelini Z, Baroncelli S, Spada M, Vendetti S, Buffa V, et al. Successful immunization with a single injection of non-integrating lentiviral vector. Mol Ther. 2007;15:1716–1723. doi: 10.1038/sj.mt.6300241. [DOI] [PubMed] [Google Scholar]
- Philippe S, Sarkis C, Barkats M, Mammeri H, Ladroue C, Petit C, et al. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc Natl Acad Sci USA. 2006;103:17684–17689. doi: 10.1073/pnas.0606197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Cornu TI, Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- Kantor B, Ma H, Webster-Cyriaque J, Monahan PE, Kafri T. Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc Natl Acad Sci USA. 2009;106:18786–18791. doi: 10.1073/pnas.0905859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátrai J, Cantore A, Bartholomae CC, Annoni A, Wang W, Acosta-Sanchez A, et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Jin J, Lollar P, Bode W, Brandstetter H, Hamaguchi N, et al. Changing residue 338 in human factor IX from arginine to alanine causes an increase in catalytic activity. J Biol Chem. 1998;273:12089–12094. doi: 10.1074/jbc.273.20.12089. [DOI] [PubMed] [Google Scholar]
- Simioni P, Tormene D, Tognin G, Gavasso S, Bulato C, Iacobelli NP, et al. X-linked thrombophilia with a mutant factor IX (factor IX Padua) N Engl J Med. 2009;361:1671–1675. doi: 10.1056/NEJMoa0904377. [DOI] [PubMed] [Google Scholar]
- Suwanmanee T, Hannah W, Gui T, Ma H, Cockrell AS, Monahan P, et al. A combination of codon optimization and amino acid substitution in the human factor IX gene yields a high level of superactive factor IX. ASGCT Annual Meeting Abstracts. 2010;18:S206. [Google Scholar]
- Suwanmanee T, Hu G, Ma H, Gui T, Monahan P, Kafri T. Complete long-term correction of hemophilic B mice using integration-defective lentiviral vectors expressing codon-optimized R338L human factor IX. ASGCT Annual Meeting Abstracts. 2012;20:S89. [Google Scholar]
- Cockrell AS, Ma H, Fu K, McCown TJ, Kafri T. A trans-lentiviral packaging cell line for high-titer conditional self-inactivating HIV-1 vectors. Mol Ther. 2006;14:276–284. doi: 10.1016/j.ymthe.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Walsh CE, Batt KM. Hemophilia clinical gene therapy: brief review. Transl Res. 2013;161:307–312. doi: 10.1016/j.trsl.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- Brown BD, Cantore A, Annoni A, Sergi LS, Lombardo A, Della Valle P, et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144–4152. doi: 10.1182/blood-2007-03-078493. [DOI] [PubMed] [Google Scholar]
- Pan D, Gunther R, Duan W, Wendell S, Kaemmerer W, Kafri T, et al. Biodistribution and toxicity studies of VSVG-pseudotyped lentiviral vector after intravenous administration in mice with the observation of in vivo transduction of bone marrow. Mol Ther. 2002;6:19–29. doi: 10.1006/mthe.2002.0630. [DOI] [PubMed] [Google Scholar]
- Kantor B, Bayer M, Ma H, Samulski J, Li C, McCown T, et al. Notable reduction in illegitimate integration mediated by a PPT-deleted, nonintegrating lentiviral vector. Mol Ther. 2011;19:547–556. doi: 10.1038/mt.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo HM, Goffinet C, Keppler OT. Mouse T-cells restrict replication of human immunodeficiency virus at the level of integration. Retrovirology. 2008;5:58. doi: 10.1186/1742-4690-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- Paruzynski A, Arens A, Gabriel R, Bartholomae CC, Scholz S, Wang W, et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- Cantore A, Nair N, Della Valle P, Di Matteo M, Màtrai J, Sanvito F, et al. Hyperfunctional coagulation factor IX improves the efficacy of gene therapy in hemophilic mice. Blood. 2012;120:4517–4520. doi: 10.1182/blood-2012-05-432591. [DOI] [PubMed] [Google Scholar]
- Herzog RW. Hepatic AAV gene transfer and the immune system: friends or foes. Mol Ther. 2010;18:1063–1066. doi: 10.1038/mt.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomae CC, Arens A, Balaggan KS, Yáñez-Muñoz RJ, Montini E, Howe SJ, et al. Lentiviral vector integration profiles differ in rodent postmitotic tissues. Mol Ther. 2011;19:703–710. doi: 10.1038/mt.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan PE, Sun J, Gui T, Wichlan DG, McPhee SW, Samulski R. Employing factor IX variants to avoid limitations imposed by immune recognition of AAV vector in hemophilia B gene therapy. ASH Annual Meeting Abstracts. 2011;118:801. [Google Scholar]
- Xu K, Ma H, McCown TJ, Verma IM, Kafri T. Generation of a stable cell line producing high-titer self-inactivating lentiviral vectors. Mol Ther. 2001;3:97–104. doi: 10.1006/mthe.2000.0238. [DOI] [PubMed] [Google Scholar]
- Kafri T, van Praag H, Ouyang L, Gage FH, Verma IM. A packaging cell line for lentivirus vectors. J Virol. 1999;73:576–584. doi: 10.1128/jvi.73.1.576-584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, You Q, Hagstrom JN, Sands M, High KA. Successful expression of human factor IX following repeat administration of adenoviral vector in mice. Proc Natl Acad Sci USA. 1996;93:3056–3061. doi: 10.1073/pnas.93.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DY, Zhang TP, Gui T, Stafford DW, Monahan PE. Creation of a mouse expressing defective human factor IX. Blood. 2004;104:1733–1739. doi: 10.1182/blood-2004-01-0138. [DOI] [PubMed] [Google Scholar]
- Lin HF, Maeda N, Smithies O, Straight DL, Stafford DW. A coagulation factor IX-deficient mouse model for human hemophilia B. Blood. 1997;90:3962–3966. [PubMed] [Google Scholar]
- Kung SH, Hagstrom JN, Cass D, Tai SJ, Lin HF, Stafford DW, et al. Human factor IX corrects the bleeding diathesis of mice with hemophilia B. Blood. 1998;91:784–790. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.