Abstract
Rectal swabs from 155 sheep and 252 goats from Grenada were evaluated to determine the prevalence of Campylobacter spp., antibiotic resistance, and multilocus sequence types. Fifteen Campylobacter isolates were obtained (14 C. jejuni and 1 C. coli). The prevalence (3.7%) did not differ significantly between sheep (4.5%) and goats (3.2%). Among the seven antimicrobials tested, resistance was only detected for tetracycline (30.8%) and metronidazole (38.5%). Campylobacter isolates showed no significant difference between sheep and goats for type of antimicrobial resistance or percent of resistant isolates. Twelve of the isolates were successfully genotyped consisting of four recognized clonal complexes and three novel sequence types. Importantly, one isolate from one goat was identified as the C. jejuni sequence type-8, a zoonotic and tetracycline-resistant clone reported to be a highly virulent clone associated with ovine abortion in the USA. Although most samples were from comingled sheep and goat production units, there were no shared sequence types between these two host species. None of the sequence types identified in this study have previously been reported in poultry in Grenada, suggesting sheep- and goat-specific Campylobacter clones in Grenada. This is the first report of genotyping of Campylobacter isolates from sheep and goats in the Eastern Caribbean.
1. Introduction
Since its initial recognition in the 1970s, Campylobacter has become one of the most common causative agents of bacterial foodborne gastroenteritis in humans [1, 2]. The incidence of human Campylobacter infections is increasing worldwide along with the proportion of isolates resistant to antibiotics [3]. Animals, including sheep and goats, are the natural reservoir hosts for Campylobacter spp. Although most infections in small ruminants are asymptomatic, there is recent recognition of a zoonotic clone, sequence type- (ST)-8, which is associated with ovine abortions in a number of states within the United States [4, 5]. Recent studies report a wide range (5% to 49%) of Campylobacter prevalence in healthy sheep and goats from different geographic areas [6–9]. Several studies conducted in high sheep and goat producing areas indicate that these small ruminants may be important sources of human exposure [3, 6–15].
Grenada is a small, developing island state in the Caribbean where many residents are involved in small-scale sheep and goat production for meat, milk, and milk products. It is likely that, in a predominantly rural country like Grenada, sheep and goats serve as important sources for human exposure by direct contact or via environmental or food exposure. The goal of this study was to determine whether sheep and goats are a reservoir host for pathogenic and antibiotic-resistant Campylobacter spp. Another goal was to characterize Campylobacter population structure among these animals by multilocus sequence type (MLST) genotyping of the isolates.
2. Materials and Methods
2.1. Study Population and Sample Collection
Rectal swabs from 155 sheep and 252 goats from the islands of Grenada and Carriacou, islands in the tri-island state of Grenada, were obtained between May and July, 2011. The much smaller island of Carriacou is similar in land mass and population to the individual parishes in Grenada [16] and thus, for the purposes of this study, the island of Carriacou was considered as an island consisting of one parish. Based on the Grenada Ministry of Agriculture, Forestry and Fisheries report on the number of sheep and goats inspected in Grenada [17], we estimated a population size of 3000 sheep and 2500 goats on these 2 islands at the time of our sampling. Given these population estimates we sampled greater than 5% of the sheep and greater than 10% of the goats on these two islands. At the 95% confidence level our sample sizes were sufficient to detect a 5% fecal shedding rate with confidence intervals of ±3.34% and ±2.55% in sheep and goats, respectively (http://www.surveysystem.com/sscalc.htm).
Groups of sheep and/or goats owned by one person or family were defined as a production unit. Twenty-one production units were selected from the 6 parishes of Grenada and 8 production units were selected from the sister island of Carriacou. Production units ranged in size from 10 to 120 animals. Approximately 25% of the sheep and goats within these production units were sampled. Samples were from clinically healthy animals that were at least 6 months old. Health status was determined by physical exam by a veterinarian. Culture swabs containing Cary-Blair transport medium (BBL, Becton, Dickinson and Company, Sparks, MD, USA) were used for sample collection. Samples were transported on ice to the St George's University School of Veterinary Medicine Diagnostic and Research Laboratory for processing the same day.
2.2. Bacterial Culture and Identification
Rectal swabs were plated onto Campylobacter blood-free selective agar with cefoperazone and amphotericin B supplements (Modified CCDA-Preston plates, Oxoid Ltd., Basingstoke, UK). The plates were incubated under microaerophilic conditions at 42°C for 48 h using a microair atmosphere generating system (GENbox microaer system, 96126 BioMerieux UK Ltd, Basingstoke UK). Presumptive Campylobacter colonies were identified by their typical grey-white, mucoid flat appearance. Campylobacter morphology was confirmed by Gram staining. Growths of pure cultures were transferred into 2% sterile skim milk in cryovials and stored at −85°C. To speciate the isolates, the hippurate test was used as previously described [18]. A multiplex PCR that differentiates between C. jejuni, C. coli, C. lari, and C. upsaliensis on the basis of amplicon size of the IpxA gene was also used [19].
2.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility/resistance of Campylobacter isolates (n = 15) was determined for the following seven antibiotics: gentamicin, chloramphenicol, ciprofloxacin, ampicillin, tetracycline, erythromycin, and metronidazole. Susceptibility and resistance to metronidazole has been proposed as an epidemiological marker for C. jejuni of avian origin [20] and may have relevance to ruminant-associated C. jejuni as well. The other six antibiotics are all used to treat human cases of campylobacteriosis. The Epsilometer test (E-test, AB Biodisk, Solna, Sweden) for minimum inhibitory concentration (MIC) was conducted according to the manufacturer's instructions on Mueller Hinton agar (Remel, Lennexa, KS, USA) as previously described [18]. A C. jejuni strain of poultry origin susceptible to all seven drugs and giving reproducible MICs was used as a control. The MIC of a drug was read directly from the scale printed on the E-test strip at the point of intersection between the zone of bacterial growth and the rest of the strip. Break points used by Guévremont et al. [21] and those established by the Clinical and Laboratory Standards Institute [22] for the microbroth dilution test on aerobic bacteria were used to interpret MICs. The break point for resistance to metronidazole was set at ≥16 μg mL−1, as recommended by Lorian [23]. MIC values used to classify a strain as resistant were ampicillin ≥32 μg mL−1, chloramphenicol ≥32 μg mL−1, ciprofloxacin ≥4 μg mL−1, erythromycin ≥8 μg mL−1, gentamicin ≥16 μg mL−1, tetracycline ≥16 μg mL−1, and metronidazole ≥16 μg mL−1.
2.4. Multilocus Sequence Typing (MLST)
Stock cultures of the 7 isolates from sheep and 8 isolates from goats were grown on blood agar plates for 24 h at 42°C under microaerophilic conditions. Total DNA was extracted using DNA blood and tissue purification kits (DNeasy Blood & Tissue Kits, Qiagen Inc., Valencia, CA, USA) according to manufacturer's instructions and samples were shipped to the College of Veterinary Medicine, University of Minnesota, St. Paul, MN, for MLST determination. PCR amplification of seven genes was carried out as described previously [24–26] using guidelines and PCR primer sequences obtained from the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/mlst-info/Cjejuni/primers.html). Briefly, PCR was performed using commercial ready-to-use master mixes in a master cycler (Mastercycler (Eppendorf AG), HotStarTaq Master Mix kit, Qiagen Inc., Valencia, CA, USA). The reaction mixture contained 10 μM of each primer, 25 μL of PCR master mix, and 2 μL of template DNA and water to make total volume of 50 μL. The cycling program consisted of an initial denaturation at 95°C for 15 min, followed by 35 cycles of 94°C for 30 sec, respective annealing temperature for each primer for 1 min, and extension at 72°C for 3 min with a final extension at 72°C for 10 min. To confirm the presence of amplicon of the expected size, the PCR amplification products were run on 1.2% agarose gel. Purified PCR products were quantified and sequenced (ACGT, Inc., Wheeling, IL, USA) using the same primers used for the PCR amplification. The obtained sequences were analyzed using sequence analysis software (Sequencherver 10.1, Gene Codes Corporation, MI, USA). Sequence data were submitted to the Campylobacter PubMLST database for allele assignments.
2.5. Data Analysis
A two-tailed Fischer's Exact test was used to compare the percent of sheep and goats positive for fecal Campylobacter, prevalence rates among parishes, and the percent of isolates resistant to the various antibiotics. A value of P ≤ 0.05 was considered significant. Sequence analysis and determination of sequence types (ST) and clonal complex (CC) were performed with the integrated network MLST application for Campylobacter (SmartGene Inc, Raleigh, North Carolina, USA) which uses the PubMLST database http://pubmlst.org/campylobacter/ for ST and CC designation. A neighbor-joining tree based on allelic sequences was constructed using a suite of analysis programs (PHYLIP analysis programs) available on the PubMLST website http://pubmlst.org/analysis/. Tree drawing was performed using PubMLST Phylodendron software also available at the same PubMLST website. Tree branches are labeled by animal source (sheep or goat) and the corresponding ST assignment. Isolates not yet assigned to an ST are labeled as “Novel.”
3. Results
3.1. Production Unit Information
Of the 21 production units sampled in Grenada, 15 consisted of both sheep and goats and 6 consisted only of goats. None of the production units consisted only of sheep. Managers of sheep and goat production units on both islands reported no antibiotic use in their flocks.
3.2. Campylobacter Species
Fifteen Campylobacter spp. isolates were obtained from the 407 animals sampled (Table 1). All isolates were phenotypically typed by the hippurate test as either C. jejuni or C. coli, with 14 of the 15 isolates (93.3%) identified as C. jejuni (6 from sheep and 8 from goats) and 1 (6.7%) of the isolates identified as C. coli (from a sheep on the island of Carriacou). Results of speciation were further confirmed by multiplex PCR. The overall prevalence of Campylobacter fecal shedding was 3.7% and did not differ significantly between sheep (4.5%) and goats (3.2%) (P = 0.59). The sheep/goat prevalence ranged from 0% in the parish of St. George to 20% in the parish of St. Patrick. These were the only two parishes that differed significantly from each other for percent positive animals (P = 0.0019).
Table 1.
The prevalence of Campylobacter in sheep and goats from the six parishes of Grenada and from Carriacou.
| Parish | Number of sheep/goat sampled | Number (%) of Campylobacter in sheep | Number (%) of Campylobacter in goats | Total number (%) of Campylobacter isolates | ||
|---|---|---|---|---|---|---|
| C. jejuni | C. coli | C. jejuni | C. coli | |||
| St. George | 4/66 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| St. David | 31/60 | 0 (0.0) | 0 (0.0) | 2 (3.3) | 0 (0.0) | 2 (2.2) |
| St. Andrew | 23/47 | 0 (0.0) | 0 (0.0) | 2 (3.3) | 0 (0.0) | 2 (2.9) |
| St. Mark | 6/14 | 1 (16.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (5.0) |
| St. Patrick | 19/1 | 4 (21.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (20.0) |
| St. John | 18/29 | 0 (0.0) | 0 (0.0) | 1 (3.4) | 0 (0.0) | 1 (2.1) |
| Carriacou | 54/35 | 0 (0.0) | 2 (3.7) | 3 (8.6) | 0 (0.0) | 5 (5.6) |
| Total | 155/252 | 5(3.2) | 2 (1.3) | 8 (3.1) | 0 (0.0) | 15 (3.7) |
3.3. MLST Profiles and Antibiotic Resistance
MLST genotyping was completed on 13 of the 15 isolates (Table 2). For the other 2 isolates neither CC nor ST determinations were possible because amplification products were not detected for one or more loci. Although PCR conditions were optimized and sequencing was repeated, additional primers were not used to fill the missing gaps for ST designation due to limitation of sample. Five CCs identified in this study have previously been described (ST-21, ST-52, ST-353, ST-61, and ST-677). In addition, three novel clones not yet assigned to an ST were detected, one from a sheep and two from goats. Two CCs, ST-21 and ST-52, were identified in both sheep and goats. Within the CC ST-21, clones ST-50 and ST-454 were isolated from sheep and clones ST-8 and ST-5340 were isolated from goats. The two isolates from ST-52 were both novel. The only ST clone isolated from more than one animal was ST-50 which was identified in samples from 3 sheep. Among the C. jejuni isolates, five CCs were identified. The most frequent C. jejuni CC was ST-21 containing 7 of the 14 C. jejuni isolates.
Table 2.
Clonal complexes and allelic profiles from 15 Campylobacter jejuni/coli strains isolated from sheep and goats in Grenada and Carriacou.
| Host | CCa | STb | Allele number | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ASP | GLN | GLT | GLY | PGM | TKT | UNC | |||
| Sheep | ST-21 | 50 (3)c | 2 | 1 | 12 | 3 | 2 | 1 | 5 |
| 454 (1) | 2 | 1 | 1 | 5 | 22 | 1 | 8 | ||
| ST-52 | #1 Novel (1)d | 9 | 25 | 2 | 10 | 104 | 3 | 6 | |
| ST-353 | 5908 (1) | 7 | 2 | 5 | 2 | 104 | 3 | 6 | |
| Untypable sheep | ? (1) | Close 28 | 4 | 12 | Close 28 | Close 1 | ? | 5 | |
|
| |||||||||
| Goats | ST-21 | 8 (1) | 2 | 1 | 1 | 3 | 2 | 1 | 6 |
| 5340 (1) | 2 | 1 | 12 | 3 | 104 | 1 | 5 | ||
| ST-52 | 52 (1) | 9 | 25 | 2 | 10 | 22 | 3 | 6 | |
| ST-61 | 61 (1) | 1 | 4 | 2 | 2 | 6 | 3 | 17 | |
| ST-677 | 5020 (1) | 10 | 2 | 50 | 62 | 120 | 76 | 52 | |
| ST-21 | #1 Novel (1)d | 2 | 39 | 12 | 3 | 2 | 1 | 5 | |
| ST-677 | #2 Novel (1)d | 33 | 39 | 50 | Close 47e | 120 | 76 | 52 | |
| Untypable goat | ? (1) | Close 28 | ? | Close 2 | Close 28 | Close 1 | ? | 6 | |
aClonal complex.
bSequence types.
cNumbers in parentheses after each ST denote the number of isolates.
dNo exact matches yet in the PubMLST data base for ST determination.
eAllele 47 for GLY was used for CC determination.
Eight of the 15 Campylobacter isolates were resistant to either metronidazole or tetracycline or both (Table 3). There was no significant difference between the percent of isolates resistant to tetracycline (30.8%) and the percent resistant to metronidazole (38.5%) (P = 1.0). All of the metronidazole-resistant isolates were from Carriacou. This included 3 of the goat C. jejuni isolates and both of the C. coli isolates from sheep. Tetracycline-resistant isolates were from Carriacou, St. Patrick, and St. David and included the C. coli metronidazole-resistant Carriacou sheep isolate. All four of the tetracycline resistant isolates belonged to the ST-21 clonal complex and encompassed 3 of the 4 ST-21 clones.
Table 3.
Antibiotic resistance of the 15 Campylobacter jejuni/coli isolates from sheep and goats in Grenada and Carriacou.
| Antibiotica | Sheep (7 isolates) Number of resistant isolates/% |
Goats (8 isolates) Number of resistant isolates/% |
Total (15 isolates) Number of resistant isolates/% |
|---|---|---|---|
| Tetracycline | 3b/42.9% | 1/12.5% | 4/30.8% |
| Metronidazole | 2b/28.5% | 3/37.5% | 5/38.5% |
|
| |||
| Total | 4 b /57.1% | 4/50% | 8 b /53.3% |
aAntibiotics tested but no resistance found: Ampicillin, Chloramphenicol, Ciprofloxacin, Erythromycin, Gentamicin.
bOne sample was resistant to both tetracycline and metronidazole.
A cladogram constructed from the 13 typable Campylobacter MLST allelic profiles illustrates the clustering of the STs identified in this study and demonstrates the genetic relatedness of sheep and goat isolates and the lack of any shared STs between sheep and goats (Figure 1).
Figure 1.
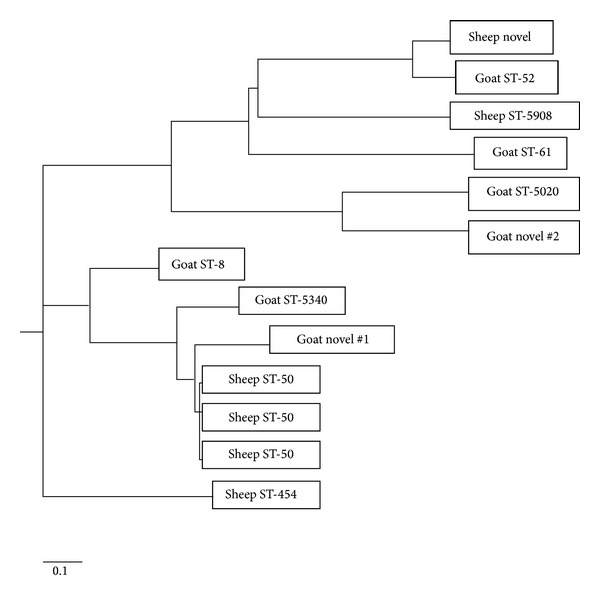
Neighbor-joining tree constructed from 8 ST clones and 3 novel clones identified among 13 Campylobacter jejuni isolates from Grenadian sheep and goat fecal samples. Tree branches are labeled with the corresponding ST assignments.
4. Discussion
The aims of this study were to determine the prevalence, genetic diversity, and antibiotic resistance of Campylobacter isolates from sheep and goats in Grenada. The Campylobacter prevalence in sheep (4.5%) and goats (3.2%) in this study is generally lower than that reported from other geographic areas [6, 7, 9, 12, 15]. However, similar to these reports and the PubMLST data base, the majority of our isolates were C. jejuni. C. coli was isolated from only one sheep from the island of Carriacou. Because this isolate was not MLST typable, one must consider the possibility that the isolate was not a Campylobacter. The low prevalence results from our study may reflect sample size, sampling biases, or the lack of Campylobacter MLSTs in Grenada that predominantly colonize sheep/goats. The lower prevalence noted in this study may also be due to the fact that the animals in Grenada are kept in a free range system and there is no intensive farming on the island. Enrichment methods were not used because, compared to direct plating, they were shown to be inefficient for isolation of Campylobacters from fecal samples from animals [27–29]. Musgrove et al. [30] noted that large numbers of non-Campylobacter species that inhabit the intestinal tract may outcompete Campylobacters during enrichment, confounding detection. However, information on enrichment techniques with regard to fecal sample of sheep and goats is lacking, and further research may elucidate this aspect. Most of the ST clones identified in our study are STs that are globally associated with poultry as a host species (Campylobacter PubMLST Database, 2012). Regardless, C. jejuni and possibly C. coli are present in small ruminants in Grenada.
In this study we identified eleven ST clones including eight previously reported STs within five recognized CCs and three novel clones not yet assigned. Of major interest is the detection of the previously reported clone ST-8 in CC ST-21 in a single goat in our study. It was recently reported that the vast majority of C. jejuni isolates associated with sheep abortion in several US states belonged to this single ST-8 clone, referred to as clone SA for sheep abortion. Further, as with the isolate from the goat in this study, all were resistant to tetracycline, the only antibiotic class currently approved in the USA for treatment of Campylobacter abortion in sheep [4]. More recently clone SA was reported in association with human disease [5]. Although the PubMLST database only shows one report of ST-8 in a goat, other goat ST-8 isolates have been reported in association with abortions [5]. Because not all ST-8 isolates may be the SA clone, additional testing of our ST-8 isolate would be required to determine if it is indeed identical to the SA clone. Because sheep or goat abortion cases in Grenada are rarely seen by a veterinarian and never diagnostically evaluated, there is no information on Campylobacter-associated ovine or caprine abortions on the island. Unlike in the USA, antibiotics are not used in small ruminant production in Grenada and thus the detection of the tetracycline resistant ST-8 clone on the island cannot be explained by tetracycline use in these animals. It is possible that importation of sheep and/or goats into Grenada accounts for clone ST-8 being present on the island. Of interest is that documented importation of sheep into Grenada during the last 20 years has only occurred from the island of Barbados and prior to 2004. However, there is one documented shipment of goats into Grenada in 2007 from the United States (B. Louison, Grenada Ministry of Agriculture, Forestry and Fisheries, personal communication, 2011). Undocumented importations may also have occurred.
The major lineages identified in this study correspond to CCs that are primarily associated with sporadic human disease and poultry and are geographically widely distributed (Table 4). However, none of the sheep/goat isolates from this study correspond to any of the STs previously identified in poultry in Grenada [18]. Because of the type of agriculture practiced in Grenada where biosecurity between various animal production units is minimal and comingling of species is common, these observations support host species-associated Campylobacter STs due to factors other than host species containment. This observation is similar to results from a study in Scotland of sheep and cattle exposed to the same farming environment, but that consistently showed significant differences in their carriage of Campylobacter species, STs, and CCs [31].
Table 4.
Campylobacter Clonal complexes from sheep and goats in Grenada for source, country, and human epidemiology as reported in pubMLST (accessed November 21, 2012).
| CCa | ST-21 | ST-21 | ST-21 | ST-21 | ST-61 | ST-52 | ST-677 |
|---|---|---|---|---|---|---|---|
| STb | 50 | 454 | 8 | 5340 | 61 | 52 | 5020 |
| (number of isolates) | (570) | (2) | (40) | (1) | (282) | (95) | (1) |
| (%C.j/%C.c) | (100/0) | (100/0) | (100/0) | (100/0) | (98.58/1.42) | (98.95/1.05) | (100/0) |
| Source | |||||||
| Human | 77.03% | 100% | 50% | 100% | 62.41% | 83.16% | 100% |
| Poultry | 16.92% | 17.50% | 3.19% | 10.53% | |||
| Cattle | 2.59% | 12.5% | 24.12% | — | |||
| Sheep | o.69% | 12.5% | 6.74% | 5.26% | |||
| Goat | 0.17% | 2.5% | — | — | |||
| Environment | 0.52% | 5% | 0.35% | — | |||
| Country | |||||||
| UK | 48.42% | 7.5% | 100% | 63.12% | 67.37% | 100% | |
| Netherlands | 17.37% | 0 | 7.45% | 9.47% | |||
| Germany | 8.42% | 0 | 2.84% | — | |||
| Canada | 7.72% | 37.5% | 12.41% | 12.63% | |||
| Thailand | 2.81% | 0 | — | — | |||
| Japan | 2.28% | 7.5% | 2.84% | 1.05% | |||
| USA | 0 | 100% | 47.5% | 4.61% | — | ||
| Curacao | 2.11% | 0 | — | 1.05% | |||
| Human epidemiology | |||||||
| Sporadic | 43.33% | 100% | 22.0% | No value | 32.98% | 49.47% | No value |
| Outbreaks | 0.18% | 27.5% | 1.06% | No value | No value |
aClonal complex.
bSequence types.
The Campylobacter isolates (n = 15) genotyped into 11 different STs, indicating considerable population diversity in both sheep and goats. Four of the 8 previously reported STs identified in this study (ST-21 sequence type 454, ST-21 sequence type 5340, ST-677 sequence type 5020, and ST-353 sequent type 5908) have never been reported from sheep before. Five of the 8 previously reported STs identified in this study have never been reported in goats before, with only ST-21 sequence type 50 and sequence type 8 being previously reported. Most of our isolates represent STs reported from the UK, The Netherlands, Germany, and Canada (Table 4). Two of the STs from our study have previously been reported from the Caribbean island of Curacao, all 12 of which were from human cases of gastroenteritis (Campylobacter PubMLST Database, 2012).
CC ST-21 was the dominant CC for both sheep and goat isolates (Table 2) and is represented globally and from multiple host species including sheep and goats (Table 4). Within this complex, clone ST-50 accounted for 3 of the 4 sheep isolates. CC ST-21 was also the only shared complex between sheep and goats and the only complex with multiple isolates from both host species. At the ST level, however, there were no shared clones between sheep and goats. Because the majority of our samples were from sheep and goat comingled production units, these results further support the existence of different sheep-associated and goat-associated Campylobacter clones. Currently the PubMLST database has only 9 Campylobacter submissions from goats and 162 from sheep (Campylobacter PubMLST Database, 2012). Thus, it is not yet possible to know whether different sheep- and goat-associated Campylobacter STs exist globally. Determining the clonal population structure for Campylobacter among small ruminants is of interest in terms of potential links to antibiotic resistance and transmission to humans.
The Campylobacter isolates showed no significant difference between sheep and goats for either the type of antibiotic resistance or the percent of antibiotic resistant isolates (Table 3). Resistance was only detected for tetracycline and metronidazole. The level of Campylobacter tetracycline resistance in this study (57% of ovine isolates and 50% of caprine isolates) is in close agreement with those from Canada [32], Spain [33], and Iran [8, 12]. The E-test results are known to correlate well with agar dilution and broth dilution methods for erythromycin and ciprofloxacin. However, it has been noted that the overall agreement between E-test and broth microdilution method is only 90%, and even less with agar dilution methods [34, 35].
Results from this study contribute to the understanding of Campylobacter population structure within and among host species in Grenada. Sheep and goat rearing in this island state consists of small production units which are often in close association with back yard or small-scale poultry and swine production. This type of animal agriculture is common in many parts of the world and findings from Grenada may shed light on Campylobacter population structure, spread, and antimicrobial resistance relevant to many geographic areas.
5. Conclusions
This report of genetic typing of Campylobacter isolates from sheep and goats is the first in the Eastern Caribbean. Results from this study document that both sheep and goats are reservoirs for antibiotic resistant Campylobacter jejuni/coli. Results also document that the previously identified, highly pathogenic Campylobacter jejuni genetic clone for ovine abortion in the USA, ST-8, is also present in Grenada. Identical to the US isolates, the ST-8 isolate from a goat in Grenada is also tetracycline-resistant. Campylobacter isolates in this study are primarily from comingled sheep and goat production units and yet no shared genetic clones were identified between these two host species. None of the sequence types identified in this study match those previously reported in poultry in Grenada, suggesting the presence of sheep- and goat-specific Campylobacter clones in Grenada.
Acknowledgments
This project was supported by a grant from the St. George's University Small Research Grant award and was approved by the St. George's University's Institutional Animal Care and Use Committee. The authors would like to acknowledge Ms. Vanessa Matthew for her technical assistance, and Angela Pashain and Kayla Pashain, students from Michigan State University, East Lansing, USA, for assistance with sample collection and processing during their research externship in Grenada.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.CDC. 2012, http://www.cdc.gov/nczved/divisions/dfbmd/diseases.
- 2.Smith K. Epidemiology of Campylobacter in humans, food and drug administration, US department of health & human services. 2009, http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/ucm083597.htm.
- 3.Moore JE, Barton MD, Blair IS, et al. The epidemiology of antibiotic resistance in Campylobacter . Microbes and Infection. 2006;8(7):1955–1966. doi: 10.1016/j.micinf.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 4.Sahin O, Plummer PJ, Jordan DM, et al. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. Journal of Clinical Microbiology. 2008;46(5):1663–1671. doi: 10.1128/JCM.00031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin O, Fitzgerald C, Stroika S, et al. Molecular evidence for zoonotic transmission of an emergent, highly pathogenic Campylobacter jejuni clone in the United States. Journal of Clinical Microbiology. 2012;50(3):680–687. doi: 10.1128/JCM.06167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adesiyun AA, Kaminjolo JS, Loregnard R, Kitson-Piggott W. Campylobacter infections in calves, piglets, lambs and kids in Trinidad. British Veterinary Journal. 1992;148(6):547–556. doi: 10.1016/0007-1935(92)90011-O. [DOI] [PubMed] [Google Scholar]
- 7.Garcia AB, Steele WB, Taylor DJ. Prevalence and carcass contamination with Campylobacter in sheep sent for slaughter in Scotland. Journal of Food Safety. 2010;30(1):237–250. doi: 10.1016/j.prevetmed.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Rahimi E, Kazemeini HR, Safaei S, et al. Detection and identification of Campylobacter spp. from retail raw chicken, turkey, sheep and goat meat in Ahvaz, Iran. African Journal of Microbiology Research. 2010;4(15):1620–1623. [Google Scholar]
- 9.Salihu MD, Junaidu AU, Oboegbulem SI, Egwu GO. Prevalence and biotypes of Campylobacter species isolated from sheep in Sokoto State, Nigeria. International Journal of Animal and Veterinary Advances. 2009;1:6–9. [Google Scholar]
- 10.Harris NV, Kimball TJ, Bennett, et al. Campylobacter jejuni enteritis associated with raw goat’s milk. American Journal of Epidemiology. 1987;126(2):179–186. doi: 10.1093/aje/126.2.179. [DOI] [PubMed] [Google Scholar]
- 11.Lévesque S, Frost E, Arbeit RD, Michaud S. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. Journal of Clinical Microbiology. 2008;46(10):3404–3411. doi: 10.1128/JCM.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahimi E, Ameri M, Kazemeini HR. Prevalence and antimicrobial resistance of Campylobacter species isolated from raw camel, beef, lamb, and goat meat in Iran. Foodborne Pathogens and Disease. 2010;7(4):443–447. doi: 10.1089/fpd.2009.0421. [DOI] [PubMed] [Google Scholar]
- 13.Raji MA, Adekeye JO, Kwaga JKP, Bale JOO. Bioserogroups of Campylobacter species isolated from sheep in Kaduna State, Nigeria. Small Ruminant Research. 2000;37(3):215–221. doi: 10.1016/s0921-4488(00)00125-5. [DOI] [PubMed] [Google Scholar]
- 14.Sheppard SK, Dallas JF, MacRae M, et al. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland. International Journal of Food Microbiology. 2009;134(1-2):96–103. doi: 10.1016/j.ijfoodmicro.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woldemariam T, Asrat D, Zewde G. Prevalence of thermophilic Campylobacter species in carcasses from sheep and goats in an abattoir in Debre Zeit area, Ethiopia. The Ethiopian Journal of Health Development. 2009;23:229–233. [Google Scholar]
- 16.Government of Grenada Official Web Site. 2012, http://www.gov.gd.
- 17.Grenada Ministry of Agriculture. Annual Agriculture Review publication, 2009.
- 18.Hariharan H, Sharma S, Chikweto A, Matthew V, DeAllie C. Antimicrobial drug resistance as determined by the E-test in Campylobacter jejuni, C. coli, and C. lari isolates from the ceca of broiler and layer chickens in Grenada. Comparative Immunology, Microbiology and Infectious Diseases. 2009;32(1):21–28. doi: 10.1016/j.cimid.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Werno AM, Klena JD, Shaw GM, Murdoch DR. Fatal case of Campylobacter lari prosthetic joint infection and bacteremia in an immunocompetent patient. Journal of Clinical Microbiology. 2002;40(3):1053–1055. doi: 10.1128/JCM.40.3.1053-1055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobbin G, Hariharan H, Daoust P-Y, et al. Bacterial flora of free-living double-crested cormorant (Phalacrocorax auritus) chicks on Prince Edward Island, Canada, with reference to enteric bacteria and antibiotic resistance. Comparative Immunology, Microbiology and Infectious Diseases. 2005;28(1):71–82. doi: 10.1016/j.cimid.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Guévremont E, Nadeau É, Sirois M, Quessy S. Antimicrobial susceptibilities of thermophilic Campylobacter from humans, swine, and chicken broilers. Canadian Journal of Veterinary Research. 2006;70(2):81–86. [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 19th informational Supplement M100-S19. Wayne, Pa, USA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 23.Lorian V. Antibiotics in Laboratory Medicine. 3rd edition. Baltimore, Md, USA: Williams & Wilkins; 1991. [Google Scholar]
- 24.Dingle KE, Colles FM, Wareing DRA, et al. Multilocus sequence typing system for Campylobacter jejuni . Journal of Clinical Microbiology. 2001;39(1):14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dingle KE, Colles FM, Ure R, et al. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerging Infectious Diseases. 2002;8(9):949–955. doi: 10.3201/eid0809.02-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schouls LM, Reulen S, Duim B, et al. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. Journal of Clinical Microbiology. 2003;41(1):15–26. doi: 10.1128/JCM.41.1.15-26.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin E, Lee Y. Comparison of three different methods for Campylobacter isolation from porcine intestines. Journal of Microbiology and Biotechnology. 2009;19(7):647–650. [PubMed] [Google Scholar]
- 28.Kiess AS, Parker HM, McDaniel CD. Evaluation of different selective media and culturing techniques for the quantification of Campylobacter ssp. from broiler litter. Poultry Science. 2010;89(8):1755–1762. doi: 10.3382/ps.2009-00587. [DOI] [PubMed] [Google Scholar]
- 29.Williams LK, Sait LC, Cogan TA, Jorgensen F, Grogono-Thomas R, Humphrey TJ. Enrichment culture can bias the isolation of Campylobacter subtypes. Epidemiology and Infection. 2012;140:1227–1235. doi: 10.1017/S0950268811001877. [DOI] [PubMed] [Google Scholar]
- 30.Musgrove MT, Berrang ME, Byrd JA, Stern NJ, Cox NA. Detection of Campylobacter spp. in Ceca and crops with and without enrichment. Poultry Science. 2001;80(6):825–828. doi: 10.1093/ps/80.6.825. [DOI] [PubMed] [Google Scholar]
- 31.Sproston EL, Ogden ID, MacRae M, et al. Temporal variation and host association in the Campylobacter population in a longitudinal ruminant farm study. Applied and Environmental Microbiology. 2011;77(18):6579–6586. doi: 10.1128/AEM.00428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott L, Menzies P, Reid-Smith RJ, et al. Antimicrobial Resistance in Campylobacter spp. isolated from Ontario sheep flocks and associations between antimicrobial use and antimicrobial resistance. Zoonoses and Public Health. 2012;59(4):294–301. doi: 10.1111/j.1863-2378.2011.01450.x. [DOI] [PubMed] [Google Scholar]
- 33.Oporto B, Juste RA, Hurtado A. Phenotypic and genotypic antimicrobial resistance profiles of Campylobacter jejuni isolated from cattle, sheep, and free-range poultry faeces. International Journal of Microbiology. 2009;2009:8 pages. doi: 10.1155/2009/456573.456573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luber P, Bartelt E, Genschow E, Wagner J, Hahn H. Comparison of broth microdilution, E test, and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni and Campylobacter coli . Journal of Clinical Microbiology. 2003;41(3):1062–1068. doi: 10.1128/JCM.41.3.1062-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge B, Wand F, Sjolund-Karlsson M, McDermott PF. Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. Journal of Microbiological Methods. 2013;95(1):57–67. doi: 10.1016/j.mimet.2013.06.021. [DOI] [PubMed] [Google Scholar]


