Abstract
Abasic lesions are a family of DNA modifications that lack Watson-Crick bases. The parent member of this family, the apurinic/apyrimidinic lesion (AP), occurs as an intermediate during DNA repair, following nucleobase alkylation, and from random hydrolysis of native nucleotides. In a given day, each cell produces between 10,000 and 50,000 AP lesions. A variety of oxidants including γ-radiolysis produce oxidized abasic sites, such as C4-AP, from the deoxyribose backbone. A number of potent, cytotoxic antitumor agents, such as bleomycin and the enediynes (e.g. calicheamicin, esperamicin, and neocarzinostatin) also lead to oxidized abasic sites in DNA.
Keywords: DNA damage, reaction mechanism, DNA repair
The absence of Watson-Crick bases prevents DNA polymerases from properly determining which nucleotide to incorporate opposite abasic lesions. Consequently, several studies have revealed that (oxidized) abasic sites are highly mutagenic. Abasic lesions are also chemically unstable, prone to strand scission, and possess electrophilic carbonyl groups. However, researchers have only uncovered the consequences of the inherent reactivity of these electrophiles within the past decade. The development of solid phase synthesis methods for oligonucleotides that both place abasic sites in defined positions and circumvent their inherent alkaline lability has facilitated this research.
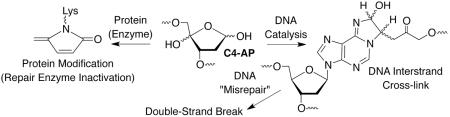
Chemically synthesized oligonucleotides containing abasic lesions provide substrates that have allowed researchers to discover a range of interesting chemical properties of potential biological importance. For instance, abasic lesions form DNA-DNA interstrand cross-links, a particularly important family of DNA damage because they block replication and transcription absolutely. In addition, bacterial repair enzymes can convert an interstrand crosslink derived from C4-AP into a double-strand break, the most deleterious form of DNA damage. Oxidized abasic lesions can also inhibit DNA repair enzymes that remove damaged nucleotides. DNA polymerase β, an enzyme that is irreversibly inactivated, is vitally important in base excision repair and is overproduced in some tumor cells. Nucleosome core particles, the monomeric components that make up chromatin, accentuate the chemical instability of abasic lesions. In experiments using synthetic nucleosome core particles containing abasic sites, the histone proteins catalyze strand cleavage at the sites that incorporate these lesions. Furthermore, in the presence of the C4-AP lesion, strand scission is accompanied by modification of the histone protein.
The reactivity of (oxidized) abasic lesions illustrates how seemingly simple nucleic acid modifications can have significant biochemical effects and may provide a chemical basis for the cytotoxicity of the chemotherapeutic agents that produce them.
1. Introduction
A DNA abasic site (AP) results from hydrolysis of a nucleotide's glycosidic bond, which occurs 10,000–50,000 times per day per cell under typical aerobic conditions.1 Oxidized abasic sites arise following hydrogen atom abstraction from the 2'-deoxyribose ring of a nucleotide (Scheme 1) and are produced by some of the most potent chemotherapeutic agents that damage DNA.2 Abasic sites are highly mutagenic, but this important, well-documented topic will not be discussed.3–7 In addition, although there is still a lot to learn about the mechanisms for oxidized abasic site formation, this topic was recently reviewed and also is not covered.2 The chemical reactivity of various abasic sites in free DNA and as part of nucleosome core particles (NCPs), as well as their inactivation of repair enzymes is the focus of this account.
Scheme 1.
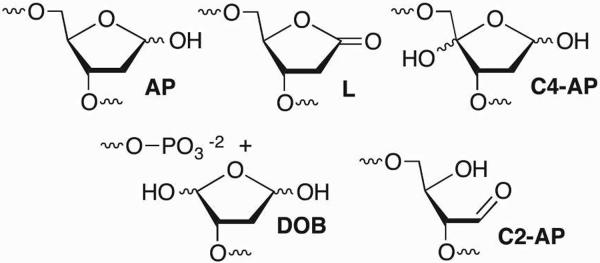
Examples of DNA abasic sites.
It is well established that abasic sites destabilize duplex DNA due to the loss of hydrogen bonding and other noncovalent interactions. Abasic sites themselves are electrophilic and chemically unstable. For instance, an AP site has a half-life on the order of weeks at pH 7.4 but is completely cleaved upon mild alkaline treatment.8,9 With the exception of C2-AP, DNA containing an abasic site is readily cleaved under mild alkaline conditions. Strand scission via β-elimination introduces another electrophilic site that can react with DNA and proteins. Nonetheless, abasic sites are often considered less biologically significant than other more rare families of lesions, such as interstrand cross-links (ICLs) and double-strand breaks (DSBs). This is primarily due to the efficiency by which AP is repaired by enzymes of the base excision repair (BER) pathway. However, recent discoveries indicate that abasic site electrophilicity plays an important role in biologically relevant reactions, resulting in DNA interstrand cross-links, irreversible inhibition of DNA repair enzymes, and histone modification.
2. Synthesis of Oligonucleotides Containing Abasic Lesions at Defined Sites
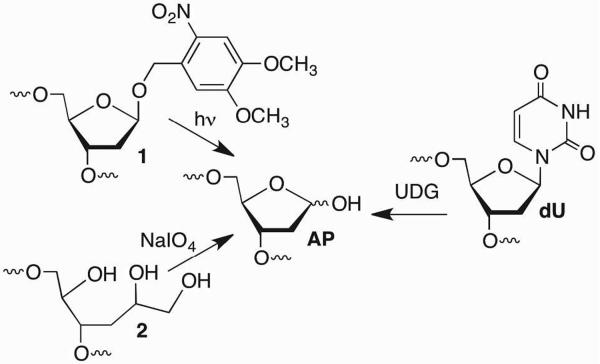
Although AP and oxidized abasic sites are produced by a variety of DNA damaging agents, it is impractical to use this approach for preparing oligonucleotides containing a single lesion at a defined position. Studies on abasic lesions are greatly facilitated by their incorporation in oligonucleotides using solid phase synthesis methods. In order to maximize compatibility with standard solid phase synthesis protocols, the alkaline lability of these molecules requires that they be unmasked following oligonucleotide deprotection and cleavage from the solid support. Oligonucleotides containing AP are most frequently prepared by reacting an incorporated dU nucleotide with glycosylase, uracil DNA glycosylase (UDG) (Scheme 2). Alternatively, AP sites are introduced into chemically synthesized oligonucleotides as the photolabile o-nitrobenzyl acetal (1), which protects the lesion from alkaline deprotection/cleavage conditions.10–12 The AP site is revealed post-synthetically by photolyzing at 350 nm. The syntheses of the requisite phosphoramidites are straight forward from 2-deoxyribose. AP sites are also introduced via periodate oxidation of a vicinal diol (2). The aldehyde produced during this reaction spontaneously cyclizes to yield the desired product.13 A similar approach has also been used to synthesize oligonucleotides containing C2-AP.14
Scheme 2.
Synthetic methods for preparing oligonucleotides containing AP sites.
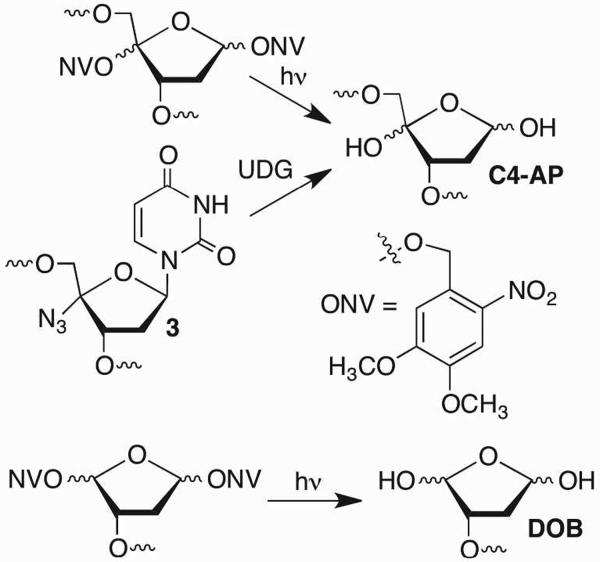
The o-nitrobenzyl photoredox reaction was also used to produce DNA containing the DOB, C4-AP, and L lesions (Schemes 3, 4).15–18 A clever combination of the o-nitrobenzyl photoredox reaction and a C1-cyano substituent (4) provided oligonucleotides containing L upon photolysis and spontaneous loss of HCN.19 Photoexcitation of the 7-nitroindole (5), which abstracts the C1'-hydrogen atom has also proven to be a very reliable method for generating the lactone lesion.20 In addition, a derivative of 2'-deoxyadenosine that utilizes this photochemistry enables enzymatic incorporation of L at defined sites.21 Finally, C4-AP can be generated chemoenzymatically by subjecting oligonucleotides containing 4'-azido-2'-deoxyuridine (3) to UDG.22 The scalability of this method was demonstrated by producing sufficient material to characterize duplex DNA containing C4-AP by NMR.23
Scheme 3.
Synthetic methods for preparing oligonucleotides containing C4-AP and DOB lesions.
Scheme 4.
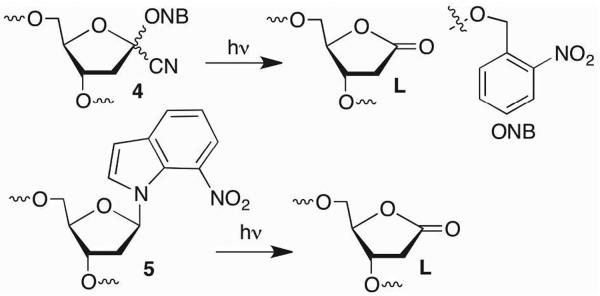
Synthetic methods for preparing oligonucleotides containing L lesions.
3. Irreversible Inhibition of Base Excision Repair Enzymes by Oxidized Abasic Lesions
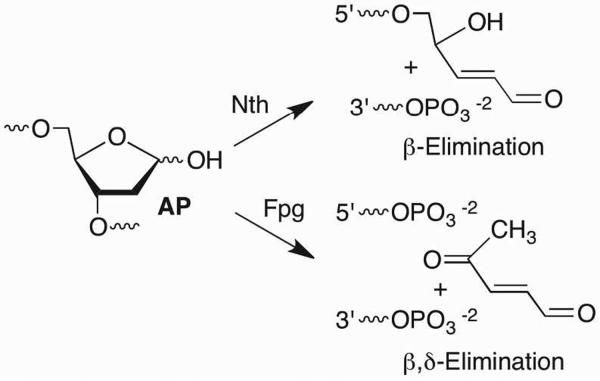
The genome is protected against AP sites by the BER pathway (Scheme 5). In mammalian cells the primary repair pathway begins with 5'-incision of the AP lesion by apurinic endonuclease 1 (Ape1).24 The remnants of the AP site (dRP, Scheme 6) are removed in a β-elimination reaction that is catalyzed by the lyase domain (dRPase) of DNA polymerase β (Pol β).25 The resulting single nucleotide gap is filled in by Pol β and rejoined by DNA ligase. A variety of endonucleases in other species possess the same activity as Ape1. Some BER glycosylases (e.g. Nth) also possess the ability to induce β-elimination creating cleaved DNA in which the 5'-fragment contains a 3'-pent-2-en-1-al terminus, which is subsequently removed by a nuclease such as Ape1 (Scheme 7).26 One BER protein in E. coli, formamidopyrimidine N-glycosylase (Fpg) activates AP by using a N-terminal proline residue to form an iminium ion, which is followed by the β,δ-elimination reaction to fully remove the lesion.27
Scheme 5.
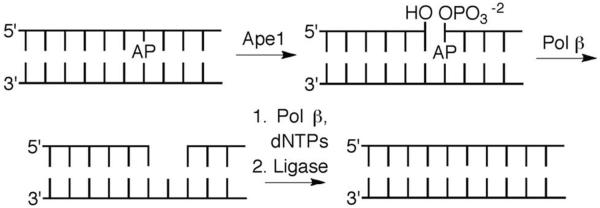
Overview of base excision repair (BER) of AP sites.
Scheme 6.
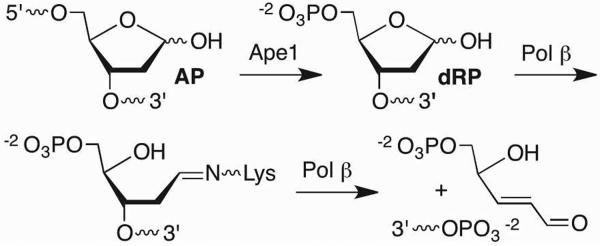
Removal of AP lesion by BER.
Scheme 7.
Reaction of bifunctional BER glycosylases with AP.
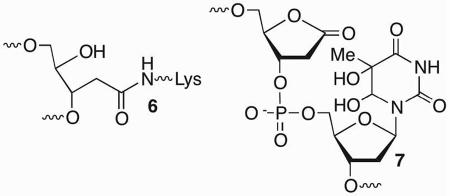
C2-AP is a substrate for phosphodiesterases, but not for the lyase activities of glycosylase enzymes or Pol β because it lacks a β-phosphate group relative to the aldehyde.28,29 This obstacle to repair is overcome by Pol β and FEN1, which carry out long patch BER via strand displacement synthesis (Scheme 8). The displaced DNA containing the lesion is hydrolyzed by FEN1. Although L contains the requisite leaving group in the β-position relative to its carbonyl, it inactivates E. coli endonuclease III (Nth), which possesses a relatively strong lyase activity. L forms DNA-protein cross-links (DPCs) with Nth and inhibits the enzyme's activity on AP.30–32 DPC formation was rationalized based upon analogy to Schiff base formation with AP, which involves Lys120. However, attack by this nucleophile on the lactone carbonyl forms a stable amide (6). Cross-linking is eliminated when the Lys120Ala Nth variant is incubated with duplex DNA containing L.30 Nth inhibition by L even extends to tandem lesions (e.g. 7) containing the oxidized abasic site where the enzyme cannot excise a typical substrate such as thymine glycol.33 Consequently, long patch BER is required to remove 7 from DNA. Inactivation of Nth by L was more the exception than the rule, as several other glycosylases, including Fpg were not cross-linked. However, Fpg and Neil1 formed DNA-protein cross links with the β-elimination product from L. Preincubation of the butenolide with thiol prevented cross-linking to Fpg and Neil1 suggesting that the products resulted from 1,4-conjugate addition. Inactivation of BER glycosylases by L and its β-elimination product were the first examples of irreversible inhibition of DNA repair enzymes.34 However, the processes were inefficient and it is uncertain how relevant they are biologically.
Scheme 8.
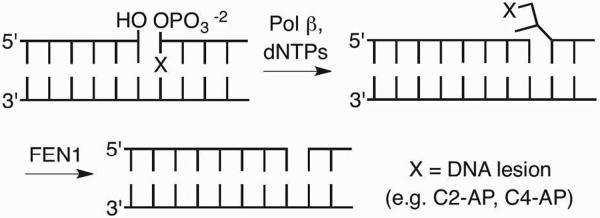
Long patch base excision repair.
Oxidized abasic lesions L, C2-AP, and C4-AP are substrates for the first step of BER (Schemes 5 and 6), incision by 5'-phosphodiesterases, albeit poorer ones than AP.28,35–37 Oxidized abasic sites affect the subsequent BER step more significantly. In mammalian cells, dRP (Scheme 6) is removed by the lyase (dRPase) activity of Pol β.25 Based upon its reactivity with Nth, it was not surprising that Ape1 incised L formed DNA-protein cross-links with Pol β.38 Furthermore, DPC formation with incised L has a similar effect on Pol β activity as the lactone does when it is part of a tandem lesion with thymine glycol.33 However, L cross-linking with Pol β is inefficient, possibly due to the lactone's inherently lower electrophilicity compared to the aldehyde in AP and so one must again question the biological significance.38
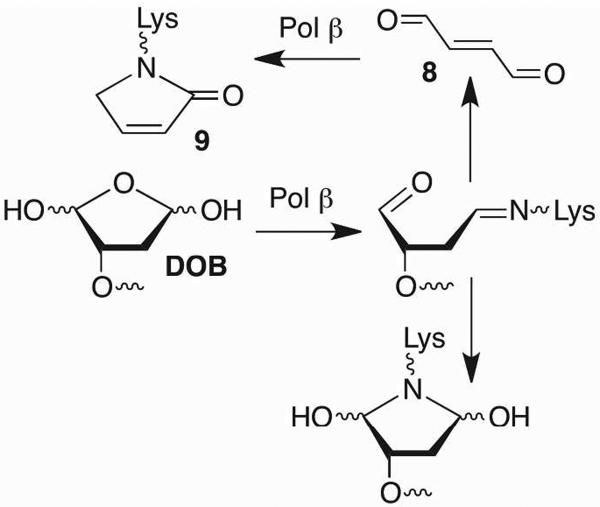
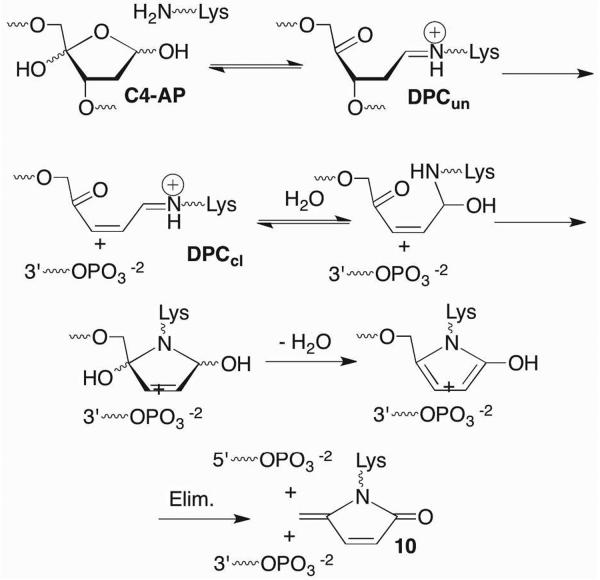
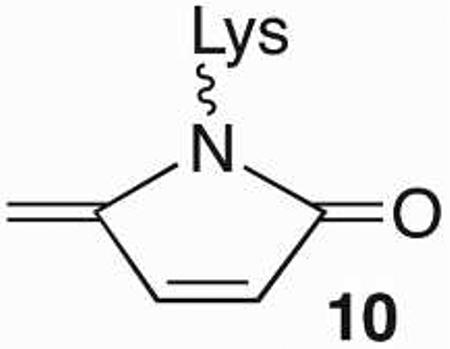
The C4-AP and DOB lesions more closely resemble an AP site more so than does L because they also contain an aldehyde at what was the C1-position. However, the presence of a 1,4-dicarbonyl functional group in C4-AP and DOB, which reacts rapidly with primary amines dramatically alter the outcomes of interactions with lyase enzymes (Scheme 9).18,39,40 Pol β excises on average ~ 4 DOB molecules before the enzyme is inactivated.41,42 Quantitative analysis revealed that DNA containing DOB behaved as an irreversible inhibitor (KI ~ 13 nM, kInact ~ 4 × 10−4 s−1). Radioactive isotopic labeling experiments indicated that the major inhibition pathway involved DPC formation, whereas released 8 accounts for ~10% of the inactivation events. Subsequent MS analysis of digested Pol β provided direct evidence for covalent modification of Lys84 (9) and indirect evidence for reaction at Lys72. Both lysines have been implicated in Schiff base formation during 5'-dRP excision, although Lys72 is believed to be the nucleophile in the majority of reactions.43−46 Qualitatively similar observations were made when the 5'-phosphorylated C4-AP (pC4-AP) produced by the action of Ape1 was incubated with Pol β.37 Inactivation by pC4-AP required on average between 6 and 7 enzyme turnovers, but ultimately resulted in covalent modification of the same Lys residues (e.g. 10) that were modified following reaction with DOB.
Scheme 9.
Inactivation of Pol β by DOB.
DNA polymerase λ (Pol λ), which also possesses dRPase activity has been proposed to play a back-up role for Pol β in DNA repair.47,48 DOB and pC4-AP affect Pol λ similarly as they do Pol β. Approximately one out of every 4 interactions between DOB and Pol λ result in the enzyme's inactivation.42 Furthermore, mass spectral analysis indicates that DOB and pC4-AP modify homologous Lys residues in the Pol λ lyase domain (Lys312, Lys324). The efficient, irreversible inhibition of Pol β, a vital enzyme involved in BER, and its putative back-up (Pol λ) by these lesions sheds light on the mechanisms of action of the potent anti-tumor agents that produce them.2,49 Ultimately, these processes may contribute to the chemical bases for their cytotoxicity.
4. DNA interstrand cross-link formation by abasic sites
Interstrand cross-links (ICLs) are a biologically important family of DNA lesions, as they prohibit the dehybridization necessary for replication and transcription to proceed.50,51 ICLs are produced a by a number of antitumor agents and also result indirectly from lipid oxidation.52 Although it had previously been proposed, Gates was the first to unambiguously characterize a DNA ICL that did not involve an exogenous molecule.53 AP forms ICLs selectively with the dG in 5'-C-AP (11) sequences under mild reducing conditions (NaBH3CN) (Scheme 10). In some sequences ICL yields approach 20% in ~12 h.54 Mass spectrometric analysis, varying the local sequence, and methoxyamine competition experiments all supported cross-linking between the AP aldehyde and 2-amino group of dG. This connectivity pattern also explains why ICL formation occurs preferentially in the 5'-C-AP sequence. The guanine 2-amino group at this position is in closer proximity (~3.7 Å) than when it is either opposite AP (~5.3 Å) or present in the 5'-AP-C (~8.6 Å) sequence.
Scheme 10.
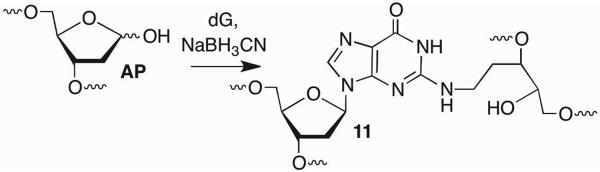
DNA interstrand cross-link formation by AP.
In 2007 C4-AP was also proposed to form ICLs with dC in cellular DNA.55 Ravanat detected a diastereomeric mixture of 12 by LC/MS following enzyme digestion of cellular DNA. The yield of 12 detected in human lymphocyte DNA showed a dependence on bleomycin or ionizing radiation dose, both of which are known to produce C4-AP. This was the first evidence for the formation of ICLs from an abasic site in cells.
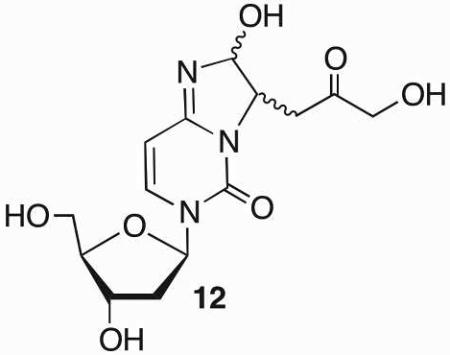
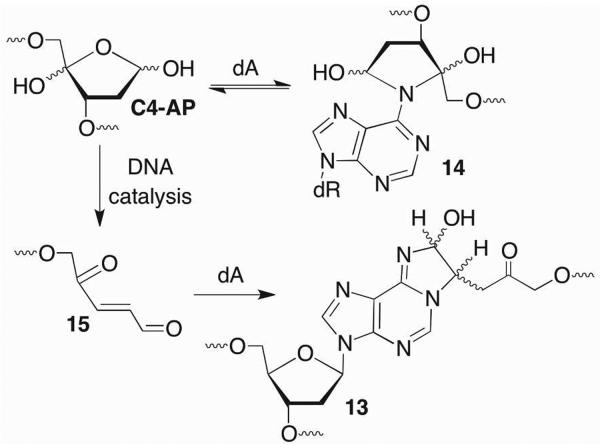
C4-AP reactivity was investigated further using synthetic duplexes, which provided complete control over the local sequence.56 These experiments confirmed the formation of 12 and revealed that the yield of cross-link with dC were highly sequence dependent. The preferred reactive positions did not strictly depend upon the distance between the C1-aldehyde of the lesion and cytosine-N4. Reaction occurred preferentially with dC that was either opposite C4-AP or a 5'-or 3'-flanking dG. In contrast to AP, no cross-linking was detected between C4-AP and dG, although iso-dG paired with iso-dC did yield ICLs.57 C4-AP also cross-linked with dA in a variety of sequence contexts (Scheme 11).17,56 2'-Deoxyadenosine formed two types of cross-links with C4-AP. One was structurally analogous to 12 (13). The other consisted of an intact duplex in which C4-AP remained uncleaved (14). The latter cross-link only formed with a dA opposite thymidine in duplexes containing 5'-C4-AP-T sequences, whereas the location of 13 showed variability similar to 12 with respect to changes in local sequence. In addition to its more exacting structural requirements, 14 formed almost 10-times more rapidly than 13, but proved to be unstable and reverted (t1/2 ~ 3.2 h) to C4-AP with a rate constant that is twice as fast as that at which it is formed.
Scheme 11.
DNA interstrand cross-link formation by C4-AP.
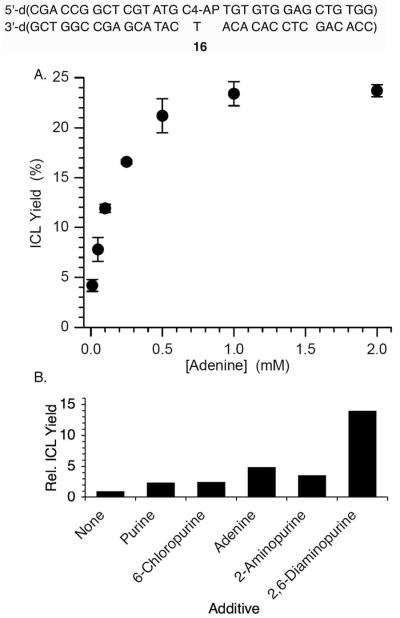
Although 13 formed more slowly than 14, like 12, it was chemically stable. Interestingly, even though no cross-link formed with the dA opposite C4-AP, this nucleotide catalyzed ICL formation at a dA opposite the 3'-adjacent thymidine.56 For instance, DNA containing dA opposite C4-AP was 5-fold more reactive than when thymidine was opposite the abasic site. In addition, cross-linking was rescued upon adding adenine to 16 (Figure 1A). The efficacy of other purines at rescuing cross-linking correlated with their electron density (Figure 1B). Finally, mechanistic studies indicated that the β-elimination from C4-AP (15) was the rate-determining step.56,57
Figure 1.
Effects on ICL formation when C4-AP is opposite thymidine (16). A. ICL rescue as a function of [adenine]. B. Effects of various purines on ICL yield.
Catalysis of ICL formation by the local sequence is an unusual instance in which DNA promotes a process to its own detriment.58 The outcome of nucleotide excision repair (NER) of the cross-link makes the process all the more intriguing. DNA damage is removed as part of an oligonucleotide during NER. ICL repair by NER is a multi-step process, which may occur by incising or “unhooking” one strand, followed by a DNA polymerase filling in the gap produced (Scheme 12).59 It is not understood how the NER system chooses which strand to incise first and for most ICLs the order is unimportant. Because one strand is already nicked in DNA containing 12 or 13, incision of the uncleaved strand first creates a double-strand break (DSB), the most deleterious form of DNA damage (Scheme 13). Indeed DSBs are produced ~15% of the time when ICL containing 12 is subjected to NER.60 This is the first example in which an ICL is misrepaired and converted into a DSB, but since this report two other instances of NER misrepair have been reported.61,62 NER misrepair is the culmination of a series of chemical events that illustrate how an apparently simple abasic lesion (C4-AP) can be transformed into ones that have greater biological impact.
Scheme 12.
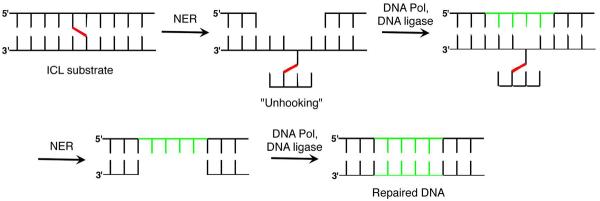
NER of an ICL.
Scheme 13.
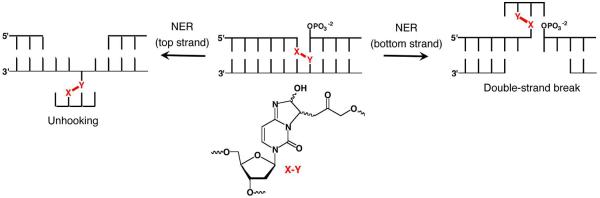
DSB formation of NER misrepair.
5. Abasic site reactivity in nucleosome core particles
In the nucleus, DNA is condensed into chromatin, which is composed of monomeric nucleosome units. In each nucleosome DNA (~145 bp) wraps around an octameric core of 4 different highly positively charged histone proteins. The histones also contain lysine-rich termini (“tails”) that protrude through the nucleosome core particle (NCP). Post-translational modification of the lysines in the conformationally mobile tails plays an important role in gene regulation. Abasic sites in nucleosomal DNA are in close proximity to the lysine-rich tails and other nucleophiles within the octameric core. In studies on the reaction of bleomycin and neocarzinostatin with chromatin, Povirk was the first to observe that the histone proteins induce DNA strand scission at C4-AP and L lesions.63
Elucidating the reactivity between abasic lesions and histone proteins in chromatin was greatly facilitated using nucleosome core particles (NCPs) prepared by combining the synthesis of 145 bp DNA containing appropriate precursors at defined sites with histone protein expression in E. coli. The requisite DNA molecules were prepared by enzymatically ligating chemically synthesized oligonucleotides. Histone expression and assembly of the octameric core enabled exploration of the roles of specific amino acids by preparing variants of the wild type proteins. As anticipated by Povirk's report, strand scission at AP, L, and C4-AP are all accelerated in NCPs. However, the reaction mechanisms and possible consequences are different for each.9,11,64–66
Strand scission at AP sites is accelerated as much as 100-fold in nucleosome core particles.9,11,65 Mechanistic studies, including trapping by NaBH3CN, and deuterium KIEs revealed that reversibly formed DNA-protein cross-links (DPCs) are intermediates en route to strand breaks and that β-elimination is the rate-determining step (Scheme 14). DPC persistence varied depending upon the location of AP within the NCP. For instance, DPCs persisted for more than 24 h at the site 1.5 helical turns from the center of the DNA sequence (SHL 1.5), which is a hot spot for DNA damaging agents.67,68 An important unresolved question is how these persistent DPCs are repaired.
Scheme 14.
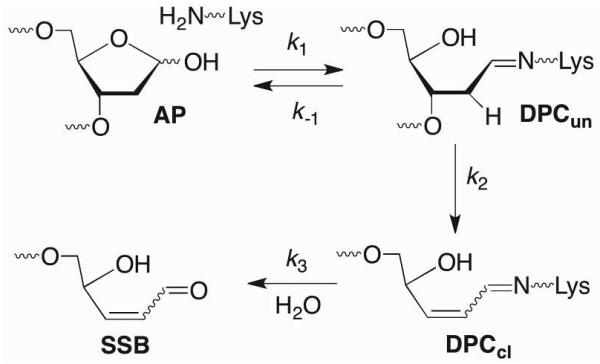
AP cleavage in a nucleosome core particle.
The identities of proteins cross linked to the AP sites were determined using an assay that relied on 32P transfer from the 5'-adjacent phosphate to the protein.11 DPC formation correlated with the proximity of the AP site with the lysine-rich amino terminal tail. For instance, histone H4 accounted for more than 95% of the DPCs with the AP site at SHL 1.5. Mutagenesis experiments confirmed that the histone tails played a vital role in catalyzing strand scission at AP sites.9,65 Approximately 95% of the accelerated cleavage of an AP site at SHL 1.5 is accounted for by mutating the five lysines in the H4 tail to arginine, and substituting alanine for histidine 18. In addition, substituting alanines for lysines in the H4 tail affects the ratio of cross linked products (Table 1), suggesting that the proteins also play a role in the β-elimination step (Scheme 14). Overall, the histone proteins within the NCP behave like lyase enzymes.25 Recently, there have been a number other reports of proteins exhibiting unexpected lyase activity.69,70 For instance, Ku70 was previously thought to play a role as a structural protein in double strand break repair by nonhomologous end joining, possesses dRPase activity (Scheme 6).70
Table 1.
Effect of mutations in histone H4 on the ratio of DNA-protein cross-links at AP89 in a NCP.9
| Histone H4 | DPCun: DPCcl |
|---|---|
| Wild type | 0.15 |
| Lys5,8,12Ala | 0.12 |
| Lys16Ala | 0.26 |
| Lys20Ala | 0.64 |
| Lys16,20Ala | 1.22 |
| 1–20 Deletion | 10.3 |
Another interesting effect was observed when two AP sites were introduced on opposite strands 3 nucleotides apart at SHL 1.5.11 Such clustered lesions are produced by ionizing radiation and are an important family of DNA damage because they are recalcitrant to repair.71,72 Based upon the reactivity of isolated AP lesions at the respective positions, an induction period was expected for double strand break (DSB) formation, but little or none was observed. Kinetic analysis indicated that following reaction at one AP site, the reactivity of the remaining lesion in the cluster was accelerated 10–15-fold compared to when it was the sole damage site incorporated in the NCP. Initial experiments suggested that the formation of a DPC with an AP on one strand resulted in a further acceleration of AP cleavage on the opposing strand by increasing the effective molarity of nucleophilic lysines.11 However, independent synthesis of an appropriate NCP revealed that merely the presence of a proximal strand break in the complementary strand was sufficient to induce the additional acceleration in strand scission at an AP site observed during double strand break formation.9 Although the physical reason behind this acceleration is unknown at this point, given the cytotoxicity of DSBs, their NCP catalyzed formation from proximal AP lesions suggests a chemical basis for the biological significance of clustered lesions.
The acceleration of cleavage at L in the NCP is more modest than that of an AP site.64 Strand scission increases between 11- and ~43 fold compared to free DNA and deprotonation at C2 is involved in the rate determining step. In contrast to AP cleavage, small amounts of DNA-protein cross-links are formed and they result from trapping of the cleaved product rather than as an intermediate en route to DNA cleavage. 2-Deoxyribonolactone is often formed as part of a bistranded lesion in which the other component is a strand break.49 Consequently, accelerated L cleavage rate in NCPs indicates that these bistranded lesions are de facto DSBs in chromatin.
C4-AP is the most reactive abasic lesion reported on as yet in NCPs, where its lifetime (as short as 14 min) is reduced as much as 550-fold compared to in naked DNA.66 As is the case for AP, DPC formation is necessary for strand scission and incubation of the NCP containing C4-AP results in stable cross-links at the expense of strand breaks (Scheme 15). Compared to AP, C4-AP DPCs form more transiently and in lower yields. Furthermore, only those containing uncleaved DNA (DPCun, Scheme 15) are detected. Both DNA cleavage fragments contain phosphate termini indicating that C4-AP is eliminated in its entirety. Fluorescent tagging experiments and MS analysis of digested proteins reveal that C4-AP is transferred to the lysine-rich amino terminal tail of the histone protein. The lactam modification of the histone (10) is analogous to the product formed when C4-AP inactivates Pol β, and represents an unnatural post translational histone modification, akin to formylation by formyl phosphate DNA.73 Its biochemical effects are unknown, nor has it been detected in cellular DNA as yet.
Scheme 15.
C4-AP cleavage in a nucleosome core particle.
Finally, ICLs involving C4-AP or AP were not observed in NCPs. This is not inconsistent with reports on cross-linking summarized above. The rates for AP and C4-AP ICL formation are considerably slower than the lesions' reactivity in NCPs where the limit of detection is ~1%. Lower yields are more readily detected by MS, which was used to detect ICLs from C4-AP in cellular DNA.55
6. Conclusions
AP and related oxidized abasic sites are some of the most commonly observed DNA lesions. Until recently, they were thought of as mutagenic lesions that were readily repairable. The above examples illustrate that the reactivity and biochemical effects are broad, extending to the formation of the most deleterious forms of DNA damage, interstrand cross links and double strand breaks. Abasic site reactivity provides chemical insight into the mechanisms of action of the chemotherapeutics that produce them and presents new questions and opportunities for research at the chemistry-biology interface.
Acknowledgment
I thank my coworkers who contributed to this research and whose names appear in the references. I am particularly grateful to Dr. Jonathan Sczepanski and Dr. Chuanzheng Zhou for carefully reading the manuscript and their suggestions. We appreciate financial support from the National Institute of General Medical Sciences (GM-063028).
Biography
Marc Greenberg received his B.S. (Chemistry) from New York University and B.E. from The Cooper Union School of Engineering in 1982. He carried out undergraduate research with Professor David Schuster. After receiving his Ph.D. from Yale University in 1988 under the guidance of Professor Jerome A. Berson, he carried out postdoctoral research as an American Cancer Society fellow with Professor Peter Dervan at the California Institute of Technology. He began his independent career in 1990 at Colorado State University and moved to Johns Hopkins University in 2002 where he is Professor of Chemistry. His research interests encompass fundamental and applied studies on nucleic acid chemistry and biochemistry.
References
- (1).Lindahl T. Instability and Decay of the Primary Structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- (2).Pitié M, Pratviel G. Activation of DNA Carbon Hydrogen Bonds by Metal Complexes. Chem. Rev. 2010;110:1018–1059. doi: 10.1021/cr900247m. [DOI] [PubMed] [Google Scholar]
- (3).Loeb LA, Preston B,D. Mutagenesis by Apurinic/Apyrimidinic Sites. Ann. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- (4).Kroeger KM, Goodman MF, Greenberg MM. A Comprehensive Comparison of DNA Replication Past 2-Deoxyribose and Its Tetrahydrofuran Analog in Escherichia Coli. Nucleic Acids Res. 2004;32:5480–5485. doi: 10.1093/nar/gkh873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kroeger KM, Jiang YL, Kow YW, Goodman MF, Greenberg MM. Mutagenic Effects of 2-Deoxyribonolactone in Escherichia Coli. An Abasic Lesion That Disobeys the A Rule. Biochemistry. 2004;43:6723–6733. doi: 10.1021/bi049813g. [DOI] [PubMed] [Google Scholar]
- (6).Kroeger KM, Kim J, Goodman MF, Greenberg MM. Effects of the C4'-Oxidized Abasic Site on Replication in Escherichia Coli. An Unusually Large Deletion Is Induced by a Small Lesion. Biochemistry. 2004;43:13621–13627. doi: 10.1021/bi048337r. [DOI] [PubMed] [Google Scholar]
- (7).Kroeger KM, Kim J, Goodman MF, Greenberg MM. Replication of an Oxidized Abasic Site in Escherichia Coli by a dNTP-Stabilized Misalignment Mechanism That Reads Upstream and Downstream Nucleotides. Biochemistry. 2006;45:5048–5056. doi: 10.1021/bi052276v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sugiyama H, Fujiwara T, Ura A, Tashiro T, Yamamoto K, Kawanishi S, Saito I. Chemistry of Thermal Degradation of Abasic Sites in DNA. Mechanistic Investigation on Thermal DNA Strand Cleavage of Alkylated DNA. Chem. Res. Toxicol. 1994;7:673–683. doi: 10.1021/tx00041a013. [DOI] [PubMed] [Google Scholar]
- (9).Zhou C, Sczepanski JT, Greenberg MM. Mechanistic Studies on Histone Catalyzed Cleavage of Apyrimidinic/Apurinic Sites in Nucleosome Core Particles. J. Am. Chem. Soc. 2012;134:16734–16741. doi: 10.1021/ja306858m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Horn T, Downing K, Gee Y, Urdea MS. Controlled Chemical Cleavage of Synthetic DNA at Specific Sites. Nucleosides Nucleotides. 1991;10:299–302. [Google Scholar]
- (11).Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-Protein Cross-Linking and Strand Scission by an Abasic Site in a Nucleosome Core Particle. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Peoc'h D, Meyer A, Imbach JL, Rayner B. Efficient Chemical Synthesis of Oligodeoxynucleotides Containing a True Abasic Site. Tetrahedron Lett. 1991;32:207–210. [Google Scholar]
- (13).Shishkina IG, Johnson F. A New Method for the Postsynthetic Generation of Abasic Sites in Oligomeric DNA. Chem. Res. Toxicol. 2000;13:907–912. doi: 10.1021/tx000108s. [DOI] [PubMed] [Google Scholar]
- (14).Kim J, Weledji YN, Greenberg MM. Independent Generation and Characterization of a C2'-Oxidized Abasic Site in Chemically Synthesized Oligonucleotides. J. Org. Chem. 2004;69:6100–6104. doi: 10.1021/jo049033d. [DOI] [PubMed] [Google Scholar]
- (15).Kim J, Gil JM, Greenberg MM. Synthesis and Characterization of Oligonucleotides Containing the C4'-Oxidized Abasic Site Produced by Bleomycin and Other DNA Damaging Agents. Angew. Chem. Int. Ed. 2003;42:5882–5885. doi: 10.1002/anie.200352102. [DOI] [PubMed] [Google Scholar]
- (16).Kodama T, Greenberg MM. Preparation and Analysis of Oligonucleotides Containing Lesions Resulting from C5'-Oxidation. J. Org. Chem. 2005;70:9916–9924. doi: 10.1021/jo051666k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sczepanski JT, Jacobs AC, Greenberg MM. Self Promoted DNA Interstrand Cross Link Formation by an Abasic Site. J. Am. Chem. Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- (18).Usui K, Aso M, Fukuda M, Suemune H. Photochemical Generation of Oligodeoxynucleotide Containing a C4′-Oxidized Abasic Site and Its Efficient Amine Modification: Dependence on Structure and Microenvironment. J. Org. Chem. 2008;73:241–248. doi: 10.1021/jo702080r. [DOI] [PubMed] [Google Scholar]
- (19).Lenox HJ, McCoy CP, Sheppard TL. Site Specific Generation of Deoxyribonolactone Lesions in DNA Oligonucleotides. Org. Lett. 2001;3:2415–2418. doi: 10.1021/ol016255e. [DOI] [PubMed] [Google Scholar]
- (20).Kotera M, Roupioz Y, Defrancq E, Bourdat A-G, Garcia J, Coulombeau C, Lhomme J. The 7-Nitroindole Nucleoside as a Photochemical Precursor of 2'-Deoxyribonolactone: Access to DNA Fragments Containing This Oxidative Abasic Lesion. Chem. Eur. J. 2000;6:4163–4169. doi: 10.1002/1521-3765(20001117)6:22<4163::aid-chem4163>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- (21).Berthet N, Crey-Desbiolles C, Kotera M, Dumy P. Chemical Synthesis, DNA Incorporation and Biological Study of a New Photocleavable 2'-Deoxyadenosine Mimic. Nucleic Acids Res. 2009;37:5237–5245. doi: 10.1093/nar/gkp562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chen J, Stubbe J. Synthesis and Characterization of Oligonucleotides Containing a 4'-Keto Abasic Site. Biochemistry. 2004;43:5278–5286. doi: 10.1021/bi0495376. [DOI] [PubMed] [Google Scholar]
- (23).Chen J, Dupradeau F-Y, Case DA, Turner CJ, Stubbe J. Nuclear Magnetic Resonance Structural Studies and Molecular Modeling of Duplex DNA Containing Normal and 4'-Oxidized Abasic Sites. Biochemistry. 2007;46:3096–3107. doi: 10.1021/bi6024269. [DOI] [PubMed] [Google Scholar]
- (24).Wilson DMI, Barsky D. The Major Human Abasic Endonuclease: Formation, Consequences and Repair of Abasic Lesions in DNA. Mutat. Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- (25).Matsumoto Y, Kim K. Excision of Deoxyribose Phosphate Residues by DNA Polymerase B During DNA Repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- (26).Chou K-M, Cheng Y-C. An Exonucleolytic Activity of Human Apurinic/Apyrimidinic Endonuclease on 3' Mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- (27).Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. Substrate Specificity of Fpg Protein. Recognition and Cleavage of Oxidatively Damaged DNA. J. Biol. Chem. 1994;269:15318–24. [PubMed] [Google Scholar]
- (28).Greenberg MM, Weledji YN, Kroeger KM, Kim J. In Vitro Replication and Repair of DNA Containing a C2'-Oxidized Abasic Site. Biochemistry. 2004;43:15217–15222. doi: 10.1021/bi048360c. [DOI] [PubMed] [Google Scholar]
- (29).Wong RS, Sczepanski JT, Greenberg MM. Excision of a Lyase-Resistant Oxidized Abasic Lesion from DNA. Chem. Res. Toxicol. 2010;23:766–770. doi: 10.1021/tx9003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hashimoto M, Greenberg MM, Kow YW, Hwang J-T, Cunningham RP. The 2-Deoxyribonolactone Lesion Produced in DNA by Neocarzinostatin and Other DNA Damaging Agents Forms Cross-Links with the Base-Excision Repair Enzyme Endonuclease III. J. Am. Chem. Soc. 2001;123:3161–3162. doi: 10.1021/ja003354z. [DOI] [PubMed] [Google Scholar]
- (31).Kroeger KM, Hashimoto M, Kow YW, Greenberg MM. Cross-Linking of 2-Deoxyribonolactone and Its β-Elimination Product by Base Excision Repair Enzymes. Biochemistry. 2003;42:2449–2455. doi: 10.1021/bi027168c. [DOI] [PubMed] [Google Scholar]
- (32).Kow YW, Wallace SS. Mechanism of Action of Escherichia Coli Endonuclease III. Biochemistry. 1987;26:8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- (33).Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA Tandem Lesion Repair by Strand Displacement Synthesis and Nucleotide Excision Repair. Biochemistry. 2008;47:4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nakano T, Terato H, Asagoshi K, Masaoka A, Mukuta M, Ohyama Y, Suzuki T, Makino K, Ide H. DNA-Protein Cross-Link Formation Mediated by Oxanine. A Novel Genotoxic Mechanism of Nitric Oxide-Induced DNA Damage. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- (35).Xu Y.-j., DeMottt MS, Hwang JT, Greenberg MM, Demple B. Action of Human Apurinic Endonuclease (Ape1) on C1'-Oxidized Deoxyribose Damage in DNA. DNA Repair. 2003;2:175–185. doi: 10.1016/s1568-7864(02)00194-5. [DOI] [PubMed] [Google Scholar]
- (36).Greenberg MM, Weledji YN, Kim J, Bales BC. Repair of Oxidized Abasic Sites by Exonuclease III, Endonuclease IV, and Endonuclease III. Biochemistry. 2004;43:8178–8183. doi: 10.1021/bi0496236. [DOI] [PubMed] [Google Scholar]
- (37).Jacobs AC, Kreller CR, Greenberg MM. Long Patch Base Excision Repair Compensates for DNA Polymerase β Inactivation by the C4'-Oxidized Abasic Site. Biochemistry. 2011;50:136–143. doi: 10.1021/bi1017667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).DeMott MS, Beyret E, Wong D, Bales BC, Hwang J-T, Greenberg MM, Demple B. Covalent Trapping of Human DNA Polymerase β by the Oxidative DNA Lesion 2-Deoxyribonolactone. J. Biol. Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- (39).Aso M, Usui K, Fukuda M, Kakihara Y, Goromaru T, Suemune H. Photochemical Generation of C4'-Oxidized Abasic Site Containing Oligodeoxynucleotide and Its Efficient Amine Modification. Org. Lett. 2006;8:3183–3186. doi: 10.1021/ol060987v. [DOI] [PubMed] [Google Scholar]
- (40).Aso M, Kondo M, Suemune H, Hecht SM. Chemistry of the Bleomycin-Induced Alkali-Labile DNA Lesion. J. Am. Chem. Soc. 1999;121:9023–9033. [Google Scholar]
- (41).Guan L, Greenberg MM. Irreversible Inhibition of DNA Polymerase β by an Oxidized Abasic Lesion. J. Am. Chem. Soc. 2010;132:5004–5005. doi: 10.1021/ja101372c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Guan L, Bebenek K, Kunkel TA, Greenberg MM. Inhibition of Short Patch and Long Patch Base Excision Repair by an Oxidized Abasic Site. Biochemistry. 2010;49:9904–9910. doi: 10.1021/bi101533a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA Polymerase B with the Mutagenic DNA Lesion 8-Oxodeoxyguanine Reveals Structural Insights into Its Coding Potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- (44).Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. Functional Analysis of the Amino-Terminal 8-kDa Domain of DNA Polymerase β as Revealed by Site-Directed Mutagenesis. J. Biol. Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- (45).Matsumoto Y, Kim K, Katz DS, Feng J-A. Catalytic Center of DNA Polymerase β for Excision of Deoxyribose Phosphate Groups. Biochemistry. 1998;37:6456–6464. doi: 10.1021/bi9727545. [DOI] [PubMed] [Google Scholar]
- (46).Feng J-A, Crasto CJ, Matsumoto Y. Deoxyribose Phosphate Excision by the N-Terminal Domain of the Polymerase β: The Mechanism Revisited. Biochemistry. 1998;37:9605–9611. doi: 10.1021/bi9808619. [DOI] [PubMed] [Google Scholar]
- (47).Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an Intrinsic 5'-Deoxyribose-5-Phosphate Lyase Activity in Human DNA Polymerase Λ. J. Biol. Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- (48).Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA Polymerase Lambda Mediates a Back-up Base Excision Repair Activity in Extracts of Mouse Embryonic Fibroblasts. J. Biol. Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- (49).Xi Z, Goldberg IH. In: Comprehensive Natural Products Chemistry. Kool ET, editor. Vol. 7. Elsevier; Amsterdam: 1999. pp. 553–592. [Google Scholar]
- (50).Guainazzi A, Scharer OD. Using Synthetic DNA Interstrand Crosslinks to Elucidate Repair Pathways and Identify New Therapeutic Targets for Cancer Chemotherapy. Cell. Mol. Life Sci. 2010;67:3683–3697. doi: 10.1007/s00018-010-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Noll DM, Mason TM, Miller PS. Formation and Repair of Interstrand Cross-Links in DNA. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Dutta S, Chowdhury G, Gates KS. Interstrand Cross-Links Generated by Abasic Sites in Duplex DNA. J. Am. Chem. Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Johnson KM, Price NE, Wang J, Fekry MI, Dutta S, Seiner DR, Wang Y, Gates KS. On the Formation and Properties of Interstrand DNA,Äìdna Cross-Links Forged by Reaction of an Abasic Site with the Opposing Guanine Residue of 5'-Cap Sequences in Duplex DNA. J. Am. Chem. Soc. 2013;135:1015–1025. doi: 10.1021/ja308119q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Regulus P, Duroux B, Bayle P-A, Favier A, Cadet J, Ravanat J-L. Oxidation of the Sugar Moiety of DNA by Ionizing Radiation or Bleomycin Could Induce the Formation of a Cluster DNA Lesion. Proc. Nat. Acad. Sci. USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. Scope and Mechanism of Interstrand Cross-Link Formation by the C4'-Oxidized Abasic Site. J. Am. Chem. Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Sczepanski JT, Hiemstra CN, Greenberg MM. Probing DNA Interstrand Cross-Link Formation by an Oxidized Abasic Site Using Nonnative Nucleotides. Bioorg. Med. Chem. 2011;19:5788–5793. doi: 10.1016/j.bmc.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Riggins JN, Pratt DA, Voehler M, Daniels JS, Marnett LJ. Kinetics and Mechanism of the General-Acid-Catalyzed Ring-Closure of the Malondialdehyde-DNA Adduct, N2-(3-Oxo-1-Propenyl)Deoxyguanosine (N2-OpdG), to 3'-(2'-Deoxy-β-D-Erythro-Pentofuranosyl)Pyrimido [1,2−2]Purin-10(3h)-one (M1dG) J. Am. Chem. Soc. 2004;126:10571–10581. doi: 10.1021/ja040010q. [DOI] [PubMed] [Google Scholar]
- (59).Friedberg EC, Walker G,C, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed ASM Press; Washington, D.C.: 2006. [Google Scholar]
- (60).Sczepanski JT, Jacobs AC, Van Houten B, Greenberg MM. Double Strand Break Formation During Nucleotide Excision Repair of a DNA Interstrand Cross-Link. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Peng X, Ghosh AK, Van Houten B, Greenberg MM. Nucleotide Excision Repair of a DNA Interstrand Cross-Link Produces Single- and Double-Strand Breaks. Biochemistry. 2010;49:11–19. doi: 10.1021/bi901603h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Weng M.-w., Zheng Y, Jasti VP, Champeil E, Tomasz M, Wang Y, Basu AK, Tang M-S. Repair of Mitomycin C Mono- and Interstrand Cross-Linked DNA Adducts by Uvrabc: A New Model. Nucleic Acids Res. 2010;38:6976–6984. doi: 10.1093/nar/gkq576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Bennett RAO, Swerdlow PS, Povirk LF. Spontaneous Cleavage of Bleomycin-Induced Abasic Sites in Chromatin and Their Mutagenicity in Mammalian Shuttle Vectors. Biochemistry. 1993;32:3188–3195. doi: 10.1021/bi00063a034. [DOI] [PubMed] [Google Scholar]
- (64).Zhou C, Greenberg MM. Histone-Catalyzed Cleavage of Nucleosomal DNA Containing 2-Deoxyribonolactone. J. Am. Chem. Soc. 2012;134:8090–8093. doi: 10.1021/ja302993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Sczepanski JT, Zhou C, Greenberg MM. Nucleosome Core Particle-Catalyzed Strand Scission at Abasic Sites. Biochemistry. 2013;52:2157–2164. doi: 10.1021/bi3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Zhou C, Sczepanski JT, Greenberg MM. Histone Modification Via Rapid Cleavage of C4'-Oxidized Abasic Sites in Nucleosome Core Particles. J. Am. Chem. Soc. 2013;135:5274–5277. doi: 10.1021/ja400915w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kuduvalli PN, Townsend CA, Tullius T,D. Cleavage by Calicheamicin G1 of DNA in a Nucleosome Formed on the 5S Rna Gene of Xenopus Borealis. Biochemistry. 1995;34:3899–3906. doi: 10.1021/bi00012a005. [DOI] [PubMed] [Google Scholar]
- (68).Davey G, Wu B, Dong Y, Surana U, Davey CA. DNA Stretching in the Nucleosome Facilitates Alkylation by an Intercalating Antitumor Agent. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku Is a 5'-dRp/AP Lyase That Excises Nucleotide Damage near Broken Ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Muller TA, Andrzejak MM, Hausinger RP. A Covalent Protein, DNA 5'-Product Adduct Is Generated Following Ap Lyase Activity of Human AlkBH1 (AlkB Homologue 1) Biochemical Journal. 2013;452:509–518. doi: 10.1042/BJ20121908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Sage E, Harrison L. Clustered DNA Lesion Repair in Eukaryotes: Relevance to Mutagenesis and Cell Survival. Mutat. Res. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).O'Neill P, Wardman P. Radiation Chemistry Comes before Radiation Biology. Int. J. Radiat. Biol. 2009;85:9–25. doi: 10.1080/09553000802640401. [DOI] [PubMed] [Google Scholar]
- (73).Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-Formylation of Lysine in Histone Proteins as a Secondary Modification Arising from Oxidative DNA Damage. Proc. Nat. Acad. Sci. USA. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]