Abstract
Brome mosaic virus (BMV) packages its genomic and subgenomic RNAs into three separate viral particles. BMV purified from barley, wheat and tobacco have distinct relative abundances of the encapsidated RNAs. We seek to identify the basis for the host-dependent differences in viral RNA encapsidation. Sequencing of the viral RNAs revealed recombination events in the 3′ untranslated region of RNA1 of BMV purified from barley and wheat, but not from tobacco. However, the relative amounts of the BMV RNAs that accumulated in barley and wheat are similar and RNA accumulation is not sufficient to account for the difference in RNA encapsidation. Virions purified from barley and wheat were found to differ in their isoelectric points, resistance to proteolysis, and contacts between the capsid residues and the RNA. Mass spectrometric analyses revealed that virions from the three hosts had different post-translational modifications that should impact the physiochemical properties of the virions. Another major source of variation in RNA encapsidation was due to the purification of BMV particles to homogeneity. Highly enriched BMV present in lysates had a surprising range of sizes, buoyant densities, and distinct relative amounts of encapsidated RNAs. These results show that the encapsidated BMV RNAs reflect a combination of host effects on the physiochemical properties of the viral capsids and the enrichment of a subset of virions. The previously unexpected heterogeneity in BMV should influence the timing of the infection and also the host innate immune responses.
Keywords: Brome mosaic virus, coat protein, virion polymorphism, capsid-RNA interaction, RNA encapsidation
INTRODUCTION
RNA viruses can infect multiple host species with very different outcomes in both progeny production and pathologies.1,2 The goal of this work is to better understand the how different hosts can affect virions of Brome mosaic virus (BMV).
BMV is a multipartite positive-strand RNA virus that can infect monocot and dicot plants. BMV infection is locally confined in two Chenopodium species, but can spread systemically in Chenopodium quinoa, the geranium tobacco, N. benthamiana, and barley and wheat.3–5 The outcome of the viral infection is due to the compatibility of the interactions between plant pathogen sensors and the pathogen molecules.6 However, how the host can affect the physiochemical properties of the virions is not well characterized.
The BMV genome consists of three messenger-sense genomic RNAs designated as RNA1, RNA2, and RNA3. Monocistronic RNA1 and RNA2 each encode one protein required for BMV replication.7,8 Dicistronic RNA3 encodes the movement protein required for cell to cell spread and the coat protein (CP) that encapsidates BMV RNAs. The CP is translated from subgenomic RNA4 that is transcribed from the minus-strand RNA3.9,10 The three BMV genomic RNAs are encapsidated in three separate virions. Subgenomic RNA4 is co-encapsidated with RNA3.11 The capsid of the three particles all contain 180 subunits of the CP that are arranged in a T=3 icosahedral symmetry.12
We noticed that the relative abundances of the three subsets of BMV virions differed as a function of the host. The level of RNA synthesis, the sequence of the RNAs, the integrity of the capsids, the contacting residues between the capsid and the RNA were examined in order to elucidate factors that influenced BMV RNA encapsidation. Virions produced in barley and wheat differed in post-translational modification of the capsid and capsid-RNA interactions. In addition, we observed that BMV particles had previously under-appreciated variations in size, density, and encapsidated viral RNAs.
RESULTS
BMV isolated from three different hosts have distinct ratios of viral RNAs
To examine the effects of the host species on virion production, agroinfiltration of N. benthamiana was used to produce the inoculum to infect barley and wheat. Agroinfiltration launches BMV infection and virion production using recombinant cDNAs of the BMV genome that are transcribed by the host polymerase. Therefore, the inoculum should be less heterogeneous than virions that had been passaged in the laboratory for decades. The virions produced by wheat, barley, and N. benthamiana will be denoted as BMVW, BMVB, and BMVN, respectively.
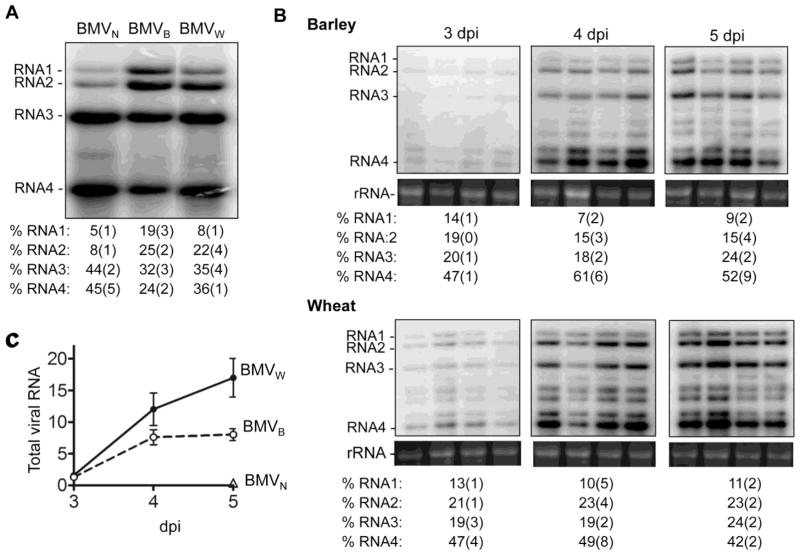
The RNA content of BMV purified by CsCl gradients had distinct relative abundances of the four BMV RNAs as a function of the host (Fig. 1A). Notably, BMVB and BMVw had higher abundances of virions that packaged RNA1 or RNA2 (relative to virions packaging RNA3/4) than did BMVN. The relative abundance of the RNA1 virion from BMVB was also higher when compared to those from BMVW. Interestingly, BMV preparations from barley do not have an equal ratio of RNA3 and RNA4 even though current model states that they are co-packaged in one virion.11 Host-specific differences are reproducible in more than 4 independent virion preparations from each host species. These observations suggest that the host species will impact progeny virus production.
Fig. 1. The encapsidation and replication of BMV RNA in different plant hosts.
A) The relative abundances of the four viral RNAs isolated from the CsCl banded BMVW, BMVB, BMVN. The RNA was detected by a riboprobe that anneals to the common 3′tRNA-like region. The quantification below the image were from four independent experiments. All quantifications show the percentage of the four viral RNAs in individual sample with errors in the brackets. B). Time course of viral RNA accumulation in barley and wheat. 5 μg total RNA isolated from the wheat and barley leaves at the indicated time were separated by gel electrophoresis and blotted for BMV RNA by the riboprobe. rRNA served as a loading control. The relative amounts of the four viral RNA accumulated from the four independent samples are summarized under the Northern blot images. All quantifications show the percentage of the four viral RNAs in individual sample with errors in the brackets. C) Graph summarizing the accumulation of BMV RNAs in barley, wheat and N. benthamiana over time. Quantification used summation of the intensity of all four viral RNAs in quadruplicates at each time point.
BMV RNA replication cannot fully account for the distinct ratios of encapsidated RNAs
We seek to determine whether distinct relative abundances of the encapsidated RNAs are due to the accumulation of the viral RNAs in the plant hosts. Leaves from four independently inoculated pots of barley, wheat, and N. benthamiana inoculated with BMVN were used for the analysis. Throughout the time course, viral RNAs accumulated at a two- to three-fold higher level in wheat than in barley (Fig. 1B, 1C). RNA accumulation was at least ten-fold lower in N. benthamiana and was barely detectable in 5 days after inoculation (Fig. 1C and data not shown). We note that mechanical inoculation leads to reduced RNA accumulation in N. benthamiana when compared to agroinfiltration. In both barley and wheat, RNA4 was the most abundant, followed by RNA3. However, the relative ratios of the four BMV RNAs were fairly similar in barley and wheat (Fig. 1B). These results suggest that the ratios of the subset of BMV virions produced barley and wheat cannot be explained solely by RNA accumulation (Fig. 1A and Table 1).
Table 1.
Summary of the relative amount of the BMV RNAs with different manipulations
| BMVB | BMVW | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| (Plant, 5 dpi) | (Sucrose) | (CsCl) | (Plant, 5 dpi) | (Sucrose) | (CsCl) | ||
|
|
|||||||
| % RNA1: | 9 (2) | 10 (1) | 19 (3) | 11 (2) | 11 (1) | 8 (1) | |
| % RNA2: | 15 (4) | 36 (1) | 25 (2) | 23 (2) | 40 (3) | 22 (4) | |
| % RNA3: | 24 (2) | 30 (1) | 32 (3) | 24 (2) | 32 (1) | 35 (4) | |
| % RNA4: | 52 (9) | 24 (1) | 24 (2) | 42 (2) | 17 (4) | 36 (1) | |
The 3′ untranslated region (UTR) of RNA1 is altered after BMV replicates in barley and wheat
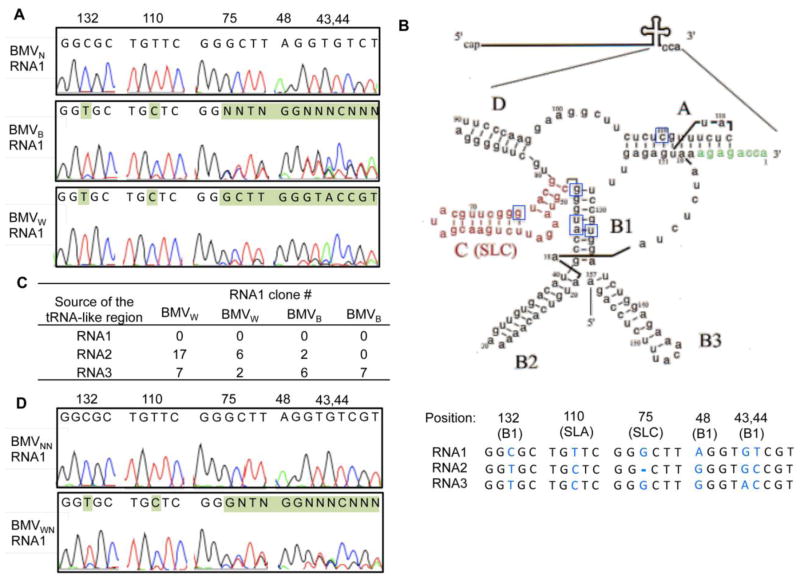
To examine whether the BMV genome had changed from the inoculum, we sequenced cDNAs generated from BMVB, BMVW, and the inoculum BMVN. No obvious changes from the BMV sequences introduced by agroinfiltration were observed in the virion RNAs derived from BMVN. In addition, RNA2 and RNA3 of BMVB and BMVW were largely unchanged from the inoculum. The only changes that are sufficiently abundant to detect in the sequencing output were from the 3′ UTR of RNA1 (Fig. 2A). The changes were found in four independent preparations, two each from BMVB and BMVW. The changes to the 3′ UTR was curious, given that all BMV RNAs share highly similar 3′UTRs.
Fig. 2. RNA recombination occurred within the tRNA-like structure of RNA1 in BMVW and BMVB.
A) Representative sequencing chromatograms of the cDNA population derived from the tRNA-like structure in RNA1 for BMVW, BMVB and BMVN. Residues shaded in green identified mismatches to the sequences present in the Agrobacterium constructs used to start BMV infection. The numbers indicate the position of the nucleotides, with the 3′-most nucleotide being residue 1. B) Schematic of the common tRNA-like structure and the sequences of the diagnostic sites. The schematic adapted from Felden et al. is modeled after the tRNA-like region in RNA3.67 The name of each stem-loop structure is in bold and the core promoter for minus-strand RNA synthesis, SLC, is in red. The nucleotides affected by the recombination in RNA1 are boxed. In the lower panel, the SNPs (nt 43, 44, 132) from the three BMV RNAs are colored blue with their positions shown above. In addition to the three SNPs, substitutions at nt 48 and 110 in RNA1 and a deletion at nt 75 in RNA2 are present in the BMV cDNA in the Agrobacterium. These five positions are used to map the intersegment crossovers in the RNA1 recombinants. C) The number of clones obtained from the RNA1 cDNA library of BMVW or BMVB that contained sequences mapped to the tRNA-like structure in RNA1, RNA2 or RNA3. Two independent preparations of BMVw and BMVB were used to generate the results. D) Representative sequencing chromatograms of the cDNA population derived from the tRNA-like structure in the RNA1 after BMVW and BMVN were passed through N. benthamiana. The progeny virions are denoted as BMVWN and BMVNN respectively.
The 3′ UTR of BMV RNA contains a tRNA-like structure (TLS) that directs minus-strand RNA synthesis (Fig. 2B).13–15 Sequences in the TLS are hotspots for crossovers in intersegment RNA recombination.16 To better characterize the sequence changes, we cloned and sequenced individual cDNAs from two independent preparations of RNA1 of BMVB and BMVW. Three single nucleotide polymorphisms (SNPs) within the TLS diagnostic for each of the three genomic BMV RNAs were used as markers to estimate the crossover sites (Fig. 2B). The TLS of RNA1 all contained the SNPs diagnostic of either RNA2 or RNA3 (Fig. 2C), suggesting that the TLSs of RNA1 from BMVB and BMVW were replaced by those from RNA2 or RNA3. In BMVW, RNA2 served as the preferential donor for the recombinant RNA1. In barley, RNA3 preferentially served as the donor. Notably, BMVN re-inoculated back to N. benthamiana (to generate BMVNN) did not result in detectable RNA recombinants (Fig. 2D). Progeny virions from BMVW passaged to N. benthamiana (to generate BMVWN) retained the heterogeneity in its RNA1 sequence originally present in the BMVW (Fig. 2D). The yield of the BMVWN was not obviously increased compared to BMVN. These results demonstrate that a strong selection exists to change the 3′ UTR of RNA1 when BMV infects barley and wheat. However, the recombinant RNA1 was not selected for or against in infection of N. benthamiana.
Virions produced by wheat and barley have distinct physiochemical properties
While increased recombination in RNA1 is influenced by the host species, similar relative levels of the four BMV RNAs in barley and wheat suggest that RNA accumulation was not primarily responsible for the distinct relative levels of the encapsidated RNAs in these two hosts. To understand the basis for distinct encapsidation of RNAs, we characterized the physiochemical properties of virions produced in barley and wheat.
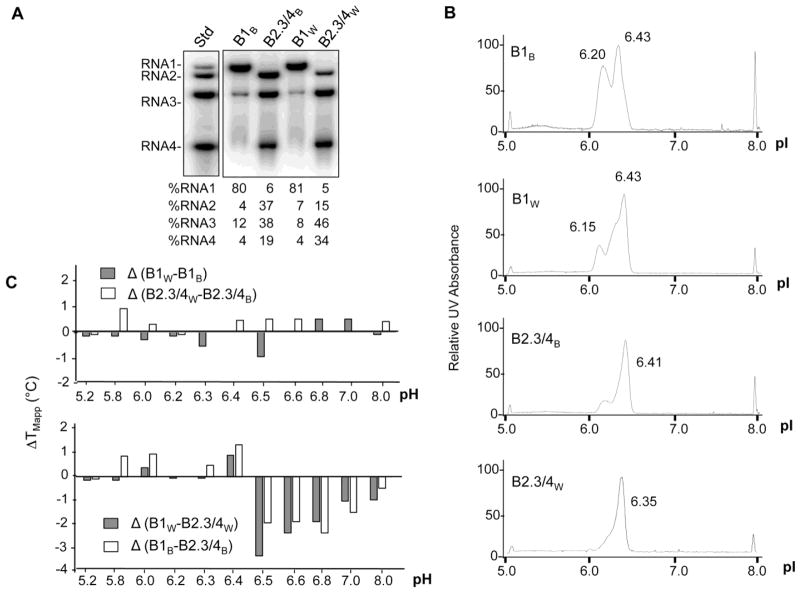
To allow improved resolution of differences in the virions, we separated the three BMV particles into two fractions by modifying the CsCl gradients.17 One fraction was enriched for RNA1 virions and a second enriched for both of the RNA2 and RNA3/4 virions. The fractions will be named B1 or B2.3/4 to designate the enriched RNAs followed by the subscript W or B to denote the host from which the virions were purified. Northern blot analysis showed that over 80% of both the B1B and B1W particles contained RNA1 whereas over 90% of the B2.3/4B or B2.3/4W particles contained RNA2 or RNA3 and RNA4 (Fig. 3A).
Fig. 3. Enrichment of the B1 and B2.3/4 virions from wheat or barley and characterization of their physiochemical properties.
A) Northern blot demonstrating the enrichment of the subsets of the three BMV virions. The proportion of each RNA in the purified virions is below the blot image. B) Isoelectric focusing chromatographs of the subsets of the BMV virions isolated from barley and wheat. The peaks near pI of 5.0 and 8.0 are pI standards added to each sample. C) The difference (ΔTmApp) in apparent melting temperature of the capsid for the subsets of the BMV virions isolated from barley and wheat as a function of pH. The upper panel compares subsets of BMV enriched for the same RNAs but isolated from different hosts. The lower panel compares the subsets of BMV isolated from the same host but enriched for different RNAs.
Isoelectric focusing was used to characterize the subsets of the BMV virions (Fig. 3B). The profile of B1B and B1W both exhibit two peaks of similar abundances with pIs of approximately 6.2 and 6.4. Given that the B1 preparations are at least 80% pure, it is likely that the B1 particles may exist as heterogeneous populations. In addition, the profiles of the peaks differ between B1B and B1W, indicating that there are host-specific effects on the electrostatic charges of the B1 virions. The B2.3/4B and B2.3/4W virions also have distinct isoelectric profiles. These results suggest that heterogeneity exist within the virions and that the host species contribute to the heterogeneity.
Differences in the isoelectric profiles of the virions prompted us to examine the thermostability of virions as a function of pH using differential scanning fluorimetry.18 BMV particles undergo a swelling transition at pH 6.4 that is associated with a ca. 10% change in the virion diameter and a change in the thermostability of the particles. A comparison of the TMapp (the apparent melting point of the capsid) revealed that the B1W particles are consistently less stable than the B1B particles below pH 6.5 (Fig. 3C, top). Another major difference in stability was observed between the B1 and the B2.3/4 virions from both wheat and barley (Fig. 3C, bottom panel). These results suggested that the thermostability of the virions is affected by the host. However, since a large difference was observed between virions that encapsidated RNA1 versus the other RNAs, it is likely that capsid-RNA interactions will substantially impact the thermostability of BMV.
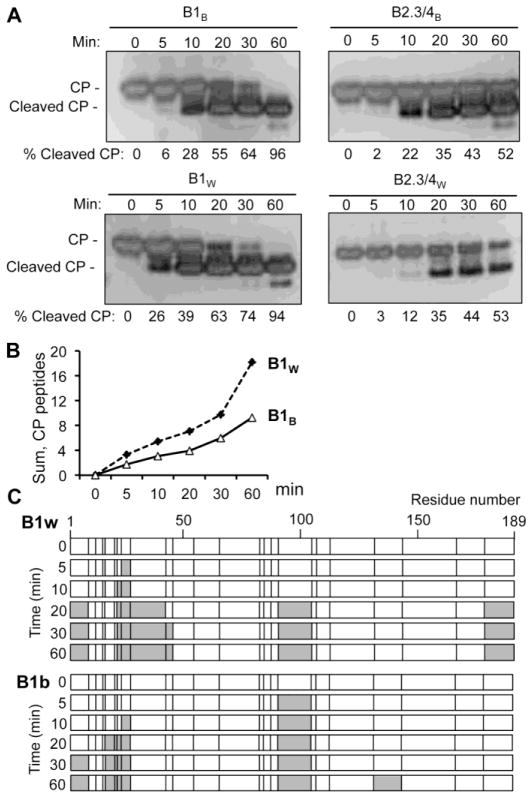
Partial proteolysis was used to confirm differences in the virions from barley and wheat. The state of the digestion was first monitored using denaturing gel electrophoresis and silver staining. The B1W capsid exhibited a proteolyzed CP band sooner than did B1B (Fig. 4A). In contrast, the B2.3/4W particles were more resistant to proteolysis than the B2.3/4B virions at the early points. Notably, the B1W and B1B particles were both digested to completion at a faster rate than the B2.3/4W and B2.3/4B particles (Fig. 4A).
Fig. 4. BMVW and BMVB are differentially sensitive to protease digestion.
A) Partial trypsin digestion of B1 and B2.3/4 particle from BMVW and BMVB. The SDS-PAGE gel was stained with silver. The signal in the cleaved CP and the full length CP were used to calculate the percentage of the cleaved product. B) Quantification of the trypsin cleaved CP peptides by mass-spectrometry. All MALDI-TOF spectra of the CP fragments generated over time in partial trypsin digestion were normalized to the intensity of the bradykinin fragment that was added to the sample prior to the sample processing. The intensities of the CP fragments were summed as a function of digestion time. C) Locations of the CP tryptic peptide fragments released from B1B and B1W. The locations of each arginine or lysine are denoted by a vertical line in the schematics. The proteolytic fragments of the CP detected by mass spectrometry are colored grey.
The identities and quantities of the tryptic peptide fragments released from the virions were identified by mass spectrometry. Quantification was performed by normalization to a trypsin-resistant peptide bradykinin that was spiked into the digestion. Consistent with the results from the gel-based assays, B1W virions released a larger number of peptide fragments than did B1B (Fig. 4B). Additionally, the cleaved CP fragments were different between B1W and B1B. Peptide fragments from B1W CP were mostly assigned to sequences within the N-terminal 46 residues and to the C-terminal tail, whereas the fragments from B1B CP were mostly from the N-terminal 26 residues and the globular domain of the CP (Fig. 4C.). The results show that BMV virions produced from barley and wheat have different sensitivity to proteolysis, even when they encapsidate the same RNA.
BMV virions from different hosts have distinguishable capsid-RNA interaction
Differences in in capsid-RNA interaction likely contributed to the differences in the BMV particles from barley and wheat. To identify these differences, the reversible crosslinking and affinity purification/peptide fingerprinting (RCAP) assay was used to map the capsid residues that contacts RNA. RCAP combines reversible crosslinking, proteolysis, RNA capture, and peptide mass fingerprinting to map the RNA-interacting sites in proteins.19 It was used to analyze protein-RNA interactions in several systems, including wild-type and mutant BMV.18,20 Several differences were observed in the regions within the capsid that contacted RNA (Table 2). First, B1W virions had fewer sites within the capsids contacting RNA1 in comparison to B1B. Capsid-RNA contacts also differed in the B2.3/4 particles in a host species-specific manner (Table 2). The differences can be visualized in models showing a cross-section of the structure of BMV capsid (Fig. 5A). We failed to identify peptides from residues 1–20 of the B2.3/4 capsids, perhaps due to more complete digestion by trypsin resulting in peptides that are difficult to resolve, or reduced crosslinking of these peptides to RNA. These results show that capsid-RNA interactions differ with the host species and also with the identity of the encapsidated RNA.
Table 2.
Capsid peptides mapped by RCAP to interact with the encapsidated RNA.
| Peptide location | B1B | B1W | B2.3/4B | B2.3/4W |
|---|---|---|---|---|
| 1–14 | + | + | ||
| 12–19 | + | |||
| 16–26 | + | |||
| 20–26 | + | + | ||
| 21–26 | + | + | + | + |
| 27–41 | + | + | + | + |
| 27–44 | + | |||
| 42–64 | + | |||
| 45–53 | + | + | ||
| 45–64 | + | + | + | + |
| 65–86 | + | + | ||
| 65–89 | + | + | + | |
| 87–103 | + | + | ||
| 87–105 | + | + | + | + |
| 90–103 | + | + | + | + |
| 90–105 | + | |||
| 112–142 | + | + | + | + |
| 131–142 | + | + | + | + |
| 143–165 | + | |||
| 166–189 | + | + | + |
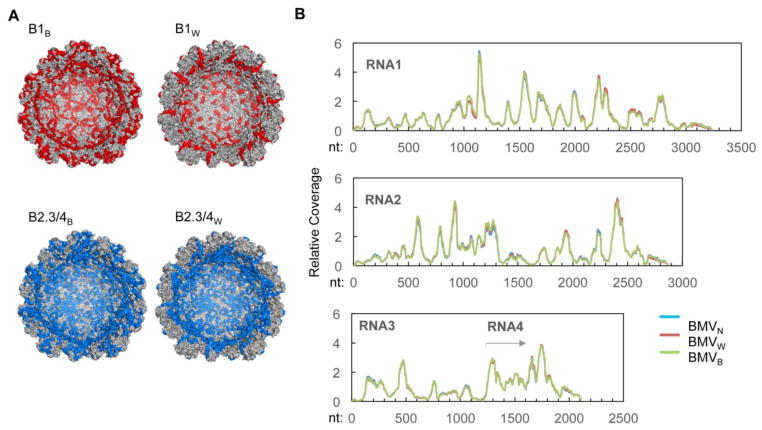
Fig. 5. BMV capsid-RNA interacting residues.
A) Capsid peptide that contact the encapsidated RNA. The peptides associated with RNA1 or RNA2, 3, and 4 in an RCAP assay (summarized in Table 2) are highlighted in red and blue, respectively, for both BMVN and BMVB. A cross-section of the capsid is shown to allow viewing of the CPs from the internal cavity of the BMV virion. B) The RNA sequences that contact the capsid. The normalized coverage of BMV RNAs by the CP-associated RNA fragments in a CLIP-seq assay is shown. The normalized coverage was obtained by dividing the actual coverage at each nucleotide position by the average coverage of the corresponding RNA to account for different depth of sequencing. The traces for BMV isolated from N. benthamiana, wheat or barley were colored blue, red and green, respectively. Note that there was nearly complete overlap of the results.
An amidination interference assay was used to confirm host-specific differences in capsid-RNA interaction.21 Briefly, the amino-modifying agent S-methyl thioacetimidate (SMTA) adds a 41 Da mass tag to the side chains of accessible lysine residues and to unmodified N-terminus of proteins.19 SMTA can freely penetrate the BMV virion and report on the residues both external and internal to the capsid shell.20 Should specific lysines contact RNA or other capsid residues, they will become more protected from amidination. The extent of amidination was quantified by mass spectrometry following proteolysis. Six of the eleven lysine residues in the B1W particles were modified to a higher level when compared to those from B1B, indicating that these residues of the B1W capsid are more exposed than their counterparts from B1B (Table 3). Differential amidination was also observed between B2.3/4W and B2.3/4B. These results are consistent with those from the RCAP assay and show that the BMV virions purified from barley and wheat vary in the capsid conformation and capsid-RNA interaction.
Table 3.
Amindination(%) of lysine residues of the BMV CP.
| Residues | Location | B1B | B1W | B2.3/4B | B2.3/4W |
|---|---|---|---|---|---|
| K8 | Internal | 100.0 | 100.0 | 34.3 | 0.0 |
| K41 | Internal | 51.5 | 99.8 | 81.5 | 96.1 |
| K44 | Internal | 99.3 | 99.9 | 97.3 | 96.1 |
| K53 | Inter-subunit | 100.0 | 78.1 | 90.1 | 100.0 |
| K64 | External | 38.1 | 100.0 | 63.1 | 66.6 |
| K81 | Inter-subunit | 79.3 | 100.0 | 100.0 | 85.4 |
| K83 | External | 79.3 | 100.0 | 100.0 | 79.8 |
| K86 | Internal | 3.6 | 99.9 | N/D | 40.7 |
| K105 | External | 97.8 | 98.2 | 59.7 | 100.0 |
| K111 | External | 33.6 | 30.9 | 53.4 | 44.7 |
| K130 | Inter-subunit | 58.3 | 71.2 | 72.3 | 14.6 |
| K165 | External | N/D | N/D | N/D | N/D |
RNA sequences that contact the capsid do not differ as a function of the host
To determine whether the host can influence the conformations of the encapsidated RNAs, we used a modified version of the UV crosslinking immunoprecipitation and high throughput DNA sequencing (CLIP-seq) assay.22 CLIP-seq combines UV crosslinking of the capsid and RNA, fragmentation of the RNA, immunoprecipitation of the CP and CP-RNA complexes, cDNA synthesis and next-generation DNA sequencing to identify the RNA fragments that contact CP. Assays performed with and without immunoprecipitation of the BMV CP demonstrate that the majority of the RNA identified were immunoprecipitated with the CP (sFIg. 1). The average lengths of the RNAs that contacted the capsids of BMVB, BMVW and BMVN, was ca. 50-nt and all of the libraries yielded over one million reads each that mapped to the BMV genome (Fig. 5B). The RNA fragments from all three virion preparations had virtually identical sequences that co-immunoprecipitated with the capsid (Fig. 5B). In the assignment of the reads, we cannot distinguish those that are from RNA4 from those of the second half of RNA3. However, this does not affect the interpretation of the results. These results show that the host did not have detectable effects on RNA structure within the virions. The effects of the host are primarily on the capsid.
BMV CP has host-specific covalent modifications
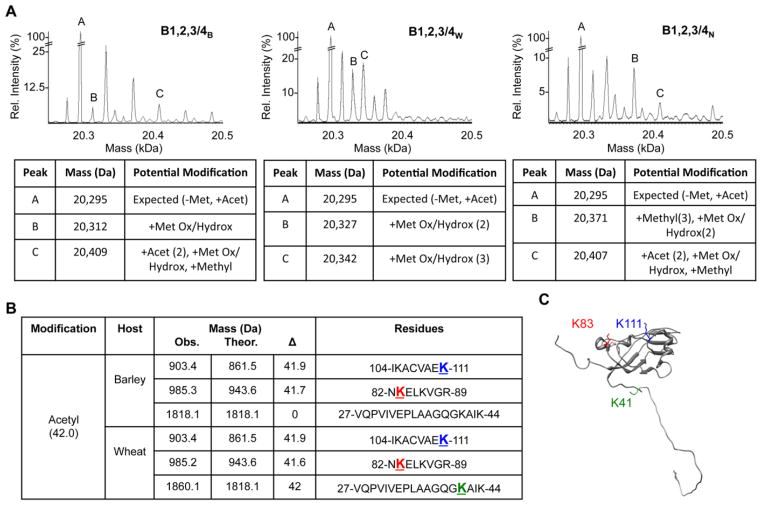
To explain the heterogeneity of BMV virions as a function of host, mass spectrometry was used to examine whether the capsid of BMVB, BMVW, and BMVN possess post-translational modifications. The mass spectra of the CPs from all three hosts revealed several discrete forms. The most abundant peak has a mass of 20,295 Da, which corresponds to the mass of the BMV CP that has the N-terminal methionine removed and acetylation of the serine residue (Fig. 6A). Protein N-terminal methionine excision is an essential activity for all kingdoms of life and N-terminal acetylations is known for the vast majority of eukaryotic proteins and has been reported for the BMV CP.23–25 Therefore, the BMV CP of 20,295 Da should be considered the BMV CP that lacks internal modifications. Importantly, several discrete peaks that correspond to masses larger than 20,295 Da were observed in BMV virions from all three hosts (Fig. 6A). Furthermore, some peaks have masses that are unique for each host. While the identities of the specific modifications need to be validated experimentally, the masses of several of the CPs are consistent with a combinations of oxidation, acetylations, hydroxylation, phosphorylations and methylation in each of the three BMV preparations (Fig. 6A).
Fig. 6. The BMV capsid contains post-translational modifications.
A) Mass spectrometry spectra showing modified forms of the BMV CP from virions purified from barley, wheat and N. benthamiana. Below each spectrum are summaries of the potential modifications predicted to exist on the three CPs labeled within each spectrum. Abbreviations: Met: methionine, Acet: acetylation, Met Ox: methionine oxidation, Hydrox: hydroxylation, ΔMethyl: removal of the N-terminal methionine. B) Comparison of lysine acetylation in tryptic fragments of the CPs from BMVB and BMVW. The mass of the observed peptides, the theoretical mass, and the mass differences are shown. The residue most likely to be modified due to predictions are shown in color and underlined. C) The location of the acetylated lysine residues in a CP subunit. The colors of labeled residues correspond to those in panel B.
To better resolve the locations of the CP modifications, virions were digested by trypsin and the fragmented peptides were analyzed. A large number of CP peptides with masses consistent with modified residues were observed. An acetylated K111 was found in BMVW, BMVB, and BMVN, and an acetylated K83 was present in BMVW and BMVB, but not in BMVN. Interestingly, BMVW had an acetylated K41 that is located in the N-terminal tail, but BMVB did not (Fig. 6B, 6C). It is also worth noting that K83 is adjacent to E84, which coordinates a Mg2+ ion within the CP trimer to stabilize the BMV capsid (Fig. 6C).12 The post-translational modifications of the BMV CP as a function of the plant host should account for at least some of the observed differences in the physiochemical properties and capsid-RNA contacts of the BMV virions (Fig. 3 to 5).
BMV is a population of highly heterogeneous particles
Thus far we demonstrated that BMV produced from barley and wheat have distinct physiochemical properties that could be attributed to the differences in capsid-RNA interactions. However, the fact that the ratio of RNA3 and RNA4 was not equal suggests that a greater degree of heterogeneity in the virions exist (Fig. 1A). Our ability to separate the RNA1 virions from the other two virions by manipulating the CsCl gradients suggests that the density gradient could bias the virions we analyzed.12 This notion prompted us to compare the virions before and after the CsCl purification.
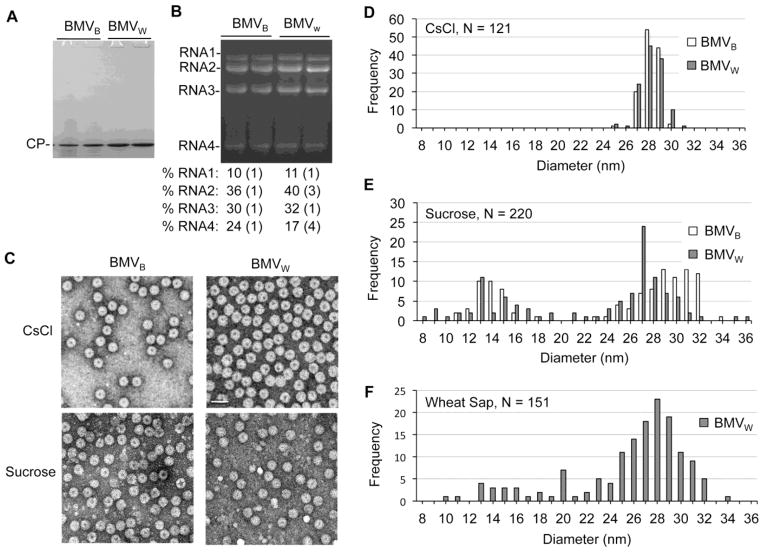
BMV virions are extracted from leaf homogenates by phase-separating the lysate in 10% chloroform to remove chlorophyll, pelleting the virions through a 10% sucrose cushion prior to CsCl density gradients. We examined the BMV enriched with the sucrose cushion and found that BMV CP was the predominant protein present in the sucrose cushion-pelleted material (Fig. 7A). Unlike the CsCl-purified virions, the relative abundances of the four viral RNAs extracted from BMVB and BMVW purified by sucrose cushions were highly similar (Fig. 7B, Table 1). In both BMVB and BMVW, RNA2 was the most abundant and RNA3 was present in higher amounts than RNA4 (Fig. 7B). These RNAs were encapsidated since free BMV RNA did not pellet through the sucrose cushion (data not shown).
Fig. 7. Variation in the diameters of the BMV particles.
A) The CP is the most abundant protein from BMVW and BMVB purified by sucrose density cushions. 2 μg of total protein determined by optical density was loaded per lane in the SDS-PAGE. The gel was stained with Coomassie blue. B) Relative abundances of the virion RNAs from BMVW and BMVB enriched with sucrose cushions. RNAs was visualized by staining with ethidium bromide. The quantification of the relative amount of the RNAs were from three independent experiments. C) Negative-stained electron micrograph of BMVW and BMVB purified from CsCl gradients or the sucrose cushion. D) The size distribution of CsCl purified BMVW and BMVB. The outer diameters of the viral particles were measured from multiple micrographs to ensure random sampling. E) The size distribution of the BMVW and BMVB particles enriched through the sucrose cushion. G) The size distribution of BMVW for clarified wheat sap.
Electron microscopy was used to examine virions purified with CsCl density gradients and with the sucrose cushions. The CsCl-purified BMVW and BMVB were homogeneous with mean diameters of ca. 28 nm, which is widely reported for BMV (Fig. 7C, 7D).12,26 Virions pelleted by the sucrose cushion, however, exist in two broad populations, one of 8 to 19 nm in diameter and a second of 21 to 34 nm (Fig. 7C, 7E).
To ensure that sucrose cushion did not eliminate any subset of the BMV virions, BMV in leaf lysates clarified only with a chloroform extraction and a 5000 g centrifugation to remove insoluble materials was examined. BMVW prepared this way exhibited a bimodal distribution similar to that of virions purified through sucrose cushions (Fig. 7F). BMVB also exhibited a wide range of diameters, but the number of virions recovered was lower (data not shown). These results support the notion that BMV forms more diverse virions than was previously assumed.7,12
Finally, to demonstrate that the CsCl gradient selected for BMV virions of ca. 28 nm, the protein and RNA content of the entire gradient was examined by Western blots or Northern blot. The BMV CP was present in the majority of the ten fractions for both BMVB and BMVW (Fig. 8, top images). Virion RNAs extracted from the fractions also revealed different relative abundances of the four BMV RNAs, including unequal amounts of RNA3 and RNA4 (Fig. 8, bottom images). These results confirmed that BMV virions have a greater heterogeneity during infection than previously believed and that virions can encapsidate RNA3 and RNA4 separately.
Fig. 8. BMV virions exhibit a range of densities in CsCl density gradients.
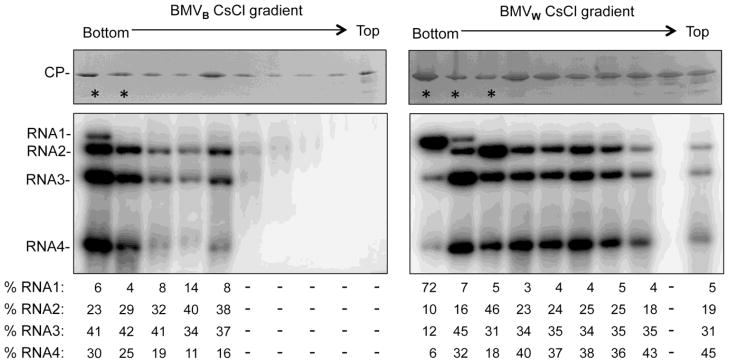
BMVW or BMVB enriched by sedimentation through a sucrose cushion were further centrifuged in a 45% cesium chloride gradient for 20 h at 50,000 g. Fractions were then collected following puncturing the bottom of the tube. The stars denote the fractions that appear in the opalescent white band. The upper images show the presence of the CP in each fraction. The SDS-PAGE gel was stained with Commassie blue. The lower images show the detection of viral RNAs by Northern blot. The quantification show the relative amount of the four viral RNAs in each fraction. Approximately equal amount of virions or viral RNAs from each fraction were used for the visualization, except for the fractions that contained significantly less viral RNAs.
DISCUSSION
In nature, BMV has been found in monocot plants.4 In the laboratory, a variety of hosts are used to study its infection process, including S. cerevisiae, where BMV can replicate and transcribe its RNAs.5,27–29 These systems have provided valuable insight into the roles of host factors in viral RNAs replication for this well-studied model positive-strand RNA virus.30 However, relatively little is known about how the host influences virion production. In this study, we found that the relative abundances of the four encapsidated RNAs varied for BMV produced in different hosts. While the host species is associated with different levels of BMV RNA accumulation and may also promote RNA recombination, the difference in RNA accumulation cannot account for the distinct relative abundances of the encapsidated RNAs. Instead, two additional factors also impact the progeny production: 1) altered interaction between the BMV capsid and the RNA that is a function of the host from which BMV was purified, and 2) a biased recovery of virions due to the purification protocol that is standard in the field for nonenveloped icosahedral viruses.
BMV virions from wheat and barley exhibit differences in the regions of the CP that contact the RNA. These changes have been mapped using RCAP and amidination interference assays with compatible results (Table 2 and 3, Fig. 5A). Distinct capsid-RNA contacts and local conformation of the capsid are also associated with differences in protease sensitivity, isoelectric points and the thermodenaturation of virions purified from different hosts (Fig. 3B, 3C, and 4). In Vaughan et al. we documented that the replication of RNA1 is related to its specific interaction with the capsid when the infection was launched from virion.17 Given that capsid-RNA interactions can change as a function of the host and that viral infection process is in a carefully timed race with host innate immune responses,31,32 virions from host species should impact the outcome of subsequent infections.
Post-translational modifications of the BMV capsid likely contribute to the distinct capsid-RNA contacts and the physiochemical properties of the BMV virions from different hosts. Post-translational modification can induce changes to protein structure.33,34 These structural changes may impact the interaction between capsid subunits and between the capsid subunits and viral RNA. In a set of viruses, phosphorylation of capsid protein has been reported to affect CP-RNA interaction and have consequences in the uncoating, replication, encapsidation and trafficking of the viral RNAs.35–37 While our mass spectrometric analyses indicate that virions from barley, wheat and N. benthamiana all contain modified forms of the CP (Fig. 6A), it needs to be followed by future study to establish the specific roles of these host-dependent modifications.
One portion of the CP that exhibits significant host-specific difference in function is the N-terminal tail of the CP (Table 2).12,38–41 The N-terminal tail from the CP of Cowpea chlorotic mottle virus (closely related to BMV) and Cucumber necrotic virus (CNV, a tombusvirus) can transit from the inside to the outside of the virion.42,43 With CNV, the structurally flexible region of the CP is phosphorylated to affect trafficking of the CNV to plastids in the cell.44 The N-terminal tail of the BMV CP appears to share both the structural flexibility, as shown by the sensitivity to protease digestion (Fig. 4) and the CP is post-translationally modified (Fig. 6B). It is likely that these properties will affect the infection process. The BMV CP is known to affect translation and can bind to the core promoter for minus-strand RNA synthesis.45,46 Additional analysis of the effects of post-translational modifications on the infection process will be a different study.
Curiously, results from the CLIP-seq assay demonstrated that the capsid-contacting regions in the encapsidated RNA were indistinguishable for BMV produced from different hosts (Fig. 5B and Table 4). This indicates that the conformation of the encapsidated RNA is more conserved when compared to that of the capsid (Table 3). Furthermore, the major factor in host-dependent differences in the BMV virions are likely due to modifications of the capsid (Table 2 and Fig. 5A). This observation agrees with the hypothesis that in virion assembly the BMV RNA folds into a core structure that is defined by secondary and tertiary nucleic acid interaction and then the capsid forms around it.12,47 Perhaps the extensive secondary and tertiary folding of the BMV RNAs renders the virion RNA to be more resistant to effects of different hosts.
Our understanding of viral RNA encapsidation is significantly impacted by the virion purification protocol used. The homogeneous virions prepared by collecting a band from a density gradient allows for biochemical and structural analyses. However, virus prepared in this manner affected interpretations for virion assembly such as the sequential co-encapsidation of RNA3 and RNA4.48 While these rules will apply to a subset or even a majority of the viral particles, it is important to consider that BMV RNA encapsidation can be more of a stochastic process than previously conceived. The diameter of the BMV virion is influenced by the size of the encapsidated RNA or other artificial cargo.26,49 Instead of packaging viral RNA(s) with a length close to 3 kb, the small virions in the sucrose cushion pelleted BMV may encapsidate individual RNA3 or RNA4, or the novel subgenomic RNA3a derived from the 5′ half of RNA3.50 The possibility of this scenario is supported by the unbalanced amount of the RNA3 and RNA4 in the sucrose cushion pelleted virus and in the fractions of the CsCl gradient density (Fig. 7B, 8). The high degree of heterogeneity in the virions could influence the timing and/or the outcome of the infection process. We believe that it is reasonable to expect a higher degree of heterogeneity in other viral species.
It is widely accepted that the genomes of RNA viruses have a high degree of heterogeneity that could impact fitness.51 We observed that after the transfer of BMVN to two monocot hosts, RNA1 recombined with RNA2 or RNA3. This event is absent in N. benthamiana, suggesting that viral RNA recombination is impacted by host factors. This observation is consistent with the notion that both host and viral factors are involved in viral RNA replication.52 Similar host-dependent effect on viral RNA recombination has been shown in other (+)-stand RNA plant viruses, including tomato bushy stunt virus (TBSV) and cumcumber mosaic virus (CMV).53,54 In-depth characterization of the recombination of TBSV in the model host yeast further demonstrated that the viral RNA recombination is regulated by a complex network of host factors.55 Since recombination of viral RNA serves as a mechanism to guard viral genome integrity and to increase the genome variability, the higher recombination frequency in specific hosts may enhance the fitness of the virus and facilitate the evasion of host defense responses.56 Interestingly, the recombination of the tRNA-like region introduced nucleotide changes to motifs required for viral replicase assembly and initiation of minus-strand RNA synthesis in RNA1.14,42 These changes could account for the higher levels of BMV RNA accumulation in the monocot plants. Furthermore, the 1a protein encoded by RNA1 is needed in higher amounts than the polymerase to form and translocate the replication factory.58,59 The accumulation of the recombination events likely reflects the specialized role for RNA1 in BMV RNA replication.
MATERIALS AND METHODS
Virion preparation
The barley and wheat plants used for infection were 7 or 8 days old and N. benthamiana plants were 4 weeks old. The inoculum BMVN was produced by in N. benthamiana 7 days after the agroinfiltration, as described by Gopinath et al. (2007).28 Subsequent passage in barley, wheat or N. benthamiana were conducted by gently rubbing leaves with carborundum dusted BMV (0.05 mg/ml).
Infected leaves were typically harvested 7 days post infection. The leaves were homogenized in virus buffer (250 mM NaOAc, 10 mM MgCl2, pH 5,2) followed by the removal of insoluble debris by a 5000 × g centrifugation. The lysate containing the virions was clarified by phase separation with 10% vol/vol of chloroform. The virions in the supernatant was layered on a 10% sucrose cushion and centrifuged for 3 h at 28,000 rpm using a Beckman SW32 rotor to pellet the virus. CsCl gradients used the pellet from the sucrose cushion dissolved overnight in VB and centrifuged using a Beckman TLA110 rotor. The opalescent white band of virus was collected and dialyzed with three changes of VB. Virions containing RNA1 and those containing either RNA2 and RNA3/4 were dissolved in VB with a 45% (w/v) CsCl and banded by centrifugation for 24 h at 50,000 rpm using a Beckman TLA110 rotor. The upper and lower bands were collected and reconstituted into a 45% (w/v) CsCl for a second round of centrifugation for 20 to 24 h at 50,000 rpm. The separated two bands were collected and exchanged into VB for analysis.
Transmission electron microscopy
Virus samples were applied to glow-discharged carbon coated copper grids and stained with 1% uranyl acetate for electron microscopy imaging. The virions were visualized by a magnification of 60,000 on a JEOL JEM 1010 electron microscope operating at a voltage of 80 kV. Images were collected on a 4K × 4K CCD Gatan Camera. Particle measurements were taken from the images using a measurement tool in the Gatan software.
RNA analysis
Total plant RNA was isolated from infected plant leaves at the indicated time using Trizol reagent (Life Science Technologies). The RNA was treated with glyoxal, electrophoresed on 1.2% agarose gel and transformed to nylon membrane for Northern blot. An in vitro-transcribed riboprobe was used to detect the tRNA-like region common to the positive-strand BMV RNAs.60 Northern blots were quantified using ImageQuant software. Virion RNAs were isolated from purified virus using Trizol and analyzed by Northern blot as described above.
DNA Sequencing
RNAs isolated from purified virion were used to template the first-strand cDNA synthesis by M-MLV reverse transcriptase (New England Biolab) with a primer complementary to the common 3′ terminal sequence. The first cDNA strand was amplified by PCR using pairs of primers that generate a series of ~1 kb overlapping DNA fragments that covered the entire BMV genome. The cDNA population from PCR was gel purified and sequenced using specific primers. The cDNA may also be cloned into pGEM-T Easy Vector (Promega) for sequencing of individual copy of cDNA.
Isoelectric focusing
Capillary isoelectric focusing (cIEF) used a Convergent Bioscience iCE280 Analyzer with a Prince autosampler (Protein Simple). The samples were prepared in a 200 μl reaction containing 0.35% methylcellulose, 8 μl pharmalytes of pH 5–8 (Sigma), 1 μl of each pI marker (pI 5.12 and 7.90, Protein Simple), and 40 μg of the BMV virion. Absorbance of the virions was monitored at 280 nm with a sample injection time of 210s and a injection pressure of 1000 mbar. All samples were pre-focused for 1 min using a voltage of 1500 V followed by 8 min at 3000 V. After each run, the capillary was rinsed with 0.5% MC for 180 s. Each reaction was performed in triplicate to ensure cIEF reproducibility.
Differential scanning fluorimetry
DSF was performed as described in Ni et al.38 The temperature with the fastest change in fluorescence intensity was extrapolated to be the apparent melting temperature of the capsid. The difference in the apparent melting temperature between samples were taken as ΔTmApp.
Partial proteolysis assay
Partial proteolysis of the purified virions was performed in ammonium bicarbonate solution (100 mM, pH 7.0). Trypsin was added to solution to 2% weight ratio of the CP at 37°C. Aliquots removed at each indicated time point was added to the Laemmli sample buffer to terminate the reaction and the samples stored on ice until SDS-PAGE. The SDS-PAGE gel was stained with silver.
Partial proteolysis monitored by mass spectrometry
MALDI-ToF analysis of peptides derived from BMV CP used a 0.5% weight ratio of trypsin to the CP in the digestion. The reaction was also amended with 4 μM trypsin-resistant Bradykinin fragment (Sigma) that will serve as an ionization control. Aliquots were stored in 0.1% trifluoroacetic acid until all samples were collected. The peptides were purified and concentrated for analyzing by MALDI-TOF mass spectrometer in reflection mode. Peptide quantification used a summation of the fragmented CP peptide ion intensities after normalization to the Bradykinin fragment ionization intensity.
Database search was carried out using Mascot (Matrix Science, Boston, MA) against the BMV protein sequence database from NCBI. Label-free peptide quantification was performed using the ProteinQuant Suite,61 and molecular modeling used the crystal structure of BMV virion (Protein Data Bank code 1JS9).
LCT analysis and modification prediction
Nominal mass measurements of BMV CP was performed using a Waters/Micromass LCT classic system equipped with a Waters CapLC autosampler as its inlet. Multiply charged spectra were then analyzed using MaxEnt, a software package designed to deconvolve mass spectra using a maximum-entropy approach.62 Modifications in the BMV CP were predicted using FindMod ( http://web.expasy.org). Methylation and acetylations in the CP were predicted with the PSKAcePred with a threshold of 0.5 and Phosphorylation was predicted using NetPhosK 1.0 with a threshold of 0.5.63,64
Reversible crosslinking–peptide fingerprinting (RCAP) assay
BMV particles were treated with formaldehyde and analyzed using the RCAP assay as previously described.18,19 Samples processed in parallel without formaldehyde treatment served as a negative control. After digestion and reversal of crosslinking, the samples were passed through a Ziptip column (Millipore) and analyzed by MALDI-TOF (Bruker Autoflex III, Agilent Technologies) in positive ion mode. Assigned peaks all corresponded to within 0.5 Da of the theoretical masses of peptides from BMV coat protein.
Amidination protection assay
Differential amindination of lysines was analyzed as described previously.21 Briefly, 160 pmol BMV virions were diluted into a 50 μl reaction containing 500 mM HEPES (pH 7.0), 20 mM MgCl2, and pre-incubated for 5 min. An equal volume of 200 mM SMTA dissolved in the same buffer was then added and the mixture incubated for 1 h. After exchanging the buffer with 100 mM NH4HCO3, trypsin was added at a ratio of 1:20 (w/w) and incubated overnight at 37°C. Digested samples were then treated with S-methyl-thiopropionate to normalize peptide ionization efficiency.19 The samples were analyzed using a LTQ linear ion trap mass spectrometer (Thermo Scientific). The tryptic digests were injected onto the C18 column (Waters NonaAcquity UPLC(r) BEH130 C18 column of 100 μm × 100 mm, 1.7 μm) and eluted with a linear gradient of 1 to 45% acetonitrile (in water with 0.1% FA). The column was developed with a flow rate was 500 nl/min, and the effluent was electro-sprayed into the LTQ mass spectrometer. MS/MS spectra were obtained by collision-induced dissociation. Database search was carried out using Mascot program, as described above.
CLIP-seq
Virion in VB was irradiated at 0.4 mJ/cm2 by 254 nm UV-light for three sessions. The virion was then pelleted by ultracentrifuge and incubated in the RNA fragmentation buffer (100 mM Tris-HCl, pH 7.5 and 10 mM ZnCl2) at 70°C for 15 minutes. The reaction was quenched by 20 mM EDTA. Immunoprecipitation used anti-CP serum and protein A/G magnetic Beads (Pierce) according to the manufacturer’s protocol. Materials on the beads were digested with 2 μg/ml proteinase K at 37°C for 2 h to release the crosslinked RNA fragments. The eluted RNA was subsequently purified by phenol-chloroform extraction and ethanol precipitation. To construct the library for Illumina MiSeq sequencing, the RNA was treated by Protein kinase R (New England Biolab) to remove a 3′ phosphate and add a 5′ phosphate. The pre-treated RNA was then prepared using Illumina Truseq small RNA preparation kit. After amplification of the cDNA, the PCR product was purified by elution from 1.8% agarose TAE gel. The Libraries from different virus samples with distinct barcodes were pooled at equimolar ratios for sequencing using the Miseq personal sequencer.
The Miseq reads for each library was cleaned by trimming the 3′ adaptor sequences and by removing any base with a quality score lower than 20 using FASTX-toolkits (http://hannonlab.cshl.edu/fastx_toolkits/). Alignment of the cleaned reads to reference genome was performed by Bowtie2 under the default local alignment settings.65 The alignment results of each library were analyzed by the SAMtools to obtain the actual coverage of each position in the reference RNA.66 To adjust for the difference in the depth of sequencing, the actual coverage was divided by the computed average coverage of the corresponding RNA to obtain the normalized coverage.
Highlights.
How the host species affect RNA virion structure is not understood
The host was found to affect the ratios of RNAs encapsidated by Brome mosaic virus
Virions from different hosts had distinct physiochemical properties
Virions from different hosts also differed in modifications of the capsid
A wide range of viral particles could impact the infection process
Acknowledgments
We thank the Indiana University Cereal Killers for their helpful discussions and support during this work, especially J. Ford, R. Qi and B. Fan for help with the CLIP-seq experiment, and the IU Mass spectrometry facility for the use of spectrometers. This work was supported by the NIH grant 1R01AI090280 to C.K.
Abbreviations used
- BMV
Brome mosaic virus
- CP
coat protein
- dpi
days post-inoculation
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE CITED
- 1.Levine B, Hardwick JM, Griffin DE. Persistence of alphaviruses in vertebrate hosts. Trends Microbiol. 1994;2:25–28. doi: 10.1016/0966-842x(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 2.Hayes EB, Komar N, Nasci RS, Montgomery SP, O’Leary DR, Campbell GL. Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao AL. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 4.Lane LC. The bromoviruses. Adv Virus Res. 1974;19:151–220. doi: 10.1016/s0065-3527(08)60660-0. [DOI] [PubMed] [Google Scholar]
- 5.Rao A, Grantham G. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology. 1995;211:42–52. doi: 10.1006/viro.1995.1377. [DOI] [PubMed] [Google Scholar]
- 6.Knepper C, Day B. From perception to activation: the molecular-genetic and biochemical landscape of disease resistance signaling in plants. The Arabidopsis book / American Society of Plant Biologists. 2010;8:e012. doi: 10.1199/tab.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kao CC, Sivakumaran K. Brome mosaic virus, good for an RNA virologist’s basic needs. Mol Plant Pathol. 2000;1:91–97. doi: 10.1046/j.1364-3703.2000.00017.x. [DOI] [PubMed] [Google Scholar]
- 8.Subba-Reddy CV, Tragesser B, Xu Z, Stein B, Ranjith-Kumar CT, Kao CC. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase in human cells reveals requirements for de novo initiation and protein-protein interaction. J Virol. 2012;86:4317–27. doi: 10.1128/JVI.00069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller WA, Dreher TW, Hall TC. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 10.Siegel RW, Adkins S, Kao CC. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–43. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane LC, Kaesberg P. Multiple genetic components in bromegrass mosaic virus. Nat New Biol. 1971;232:40–43. doi: 10.1038/newbio232040a0. [DOI] [PubMed] [Google Scholar]
- 12.Lucas RW, Larson SB, McPherson A. The crystallographic structure of brome mosaic virus. J Mol Biol. 2002;317:95–108. doi: 10.1006/jmbi.2001.5389. [DOI] [PubMed] [Google Scholar]
- 13.Ahlquist P, Dasgupta R, Kaesberg P. Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J Mol Biol. 1984;172:369–83. doi: 10.1016/s0022-2836(84)80012-1. [DOI] [PubMed] [Google Scholar]
- 14.Chapman MR, Kao CC. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J Mol Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 15.Kim C, Kao CC, IT RNA motifs that determine specificity between a viral replicase and its promoter. Nat Struct Mol Biol. 2000;7:521. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 16.Figlerowicz M, Bujarski JJ. RNA recombination in brome mosaic virus, a model plus strand RNA virus. Acta Biochim Pol. 1998;45:847–868. [PubMed] [Google Scholar]
- 17.Vaughan R, Tragesser B, Ni P, Ma X, Dragnea B, Kao CC. The tripartite virions of the brome mosaic virus have distinct physical properties that affect the timing of the infection process. PLoS Pathog. 2013 doi: 10.1128/JVI.00377-14. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hema M, Murali A, Ni P, Vaughan RC, Fujisaki K, Tsvetkova I, Dragnea B, Kao CC. Effects of amino-acid substitutions in the Brome mosaic virus capsid protein on RNA encapsidation. Mol Plant-Microbe Interact : MPMI. 2010;23:1433–47. doi: 10.1094/MPMI-05-10-0118. [DOI] [PubMed] [Google Scholar]
- 19.Vaughan R, Running WE, Qi R, Kao C. Mapping protein-RNA interactions. Virus Adaptation and Treatment. 2012;4:29–41. [Google Scholar]
- 20.Yi G, Vaughan RC, Yarbrough I, Dharmaiah S, Kao CC. RNA binding by the brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J Mol Biol. 2009;391:314–26. doi: 10.1016/j.jmb.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Running WE, Ni P, Kao CC, Reilly JP. Chemical reactivity of brome mosaic virus capsid protein. J Mol Biol. 2012;423:79–95. doi: 10.1016/j.jmb.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 23.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell Mol Life Sci. 2004;61:1455–1474. doi: 10.1007/s00018-004-3466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polevoda B, Sherman F. Nalpha -terminal acetylation of eukaryotic proteins. J Biol Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 26.Sun J, DuFort C, Daniel MC, Murali A, Chen C, Gopinath K, Stein B, De M, Rotello VM, Holzenburg A, Kao CC, Dragnea B. Core-controlled polymorphism in virus-like particles. Proc Natl Acad Sci USA. 2007;104:1354–9. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–70. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 28.Gopinath K, Dragnea B, Kao C. Interaction between Brome Mosaic Virus Proteins and RNAs : Effects on RNA Replication, Protein Expr ession, and RNA Stability. J Virol. 2005;79:14222–14234. doi: 10.1128/JVI.79.22.14222-14234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Jong W, Ahlquist P. Host-specific alterations in viral RNA accumulation and infection spread in a brome mosaic virus isolate with an expanded host range. J Virol. 1995;69:1485–1492. doi: 10.1128/jvi.69.3.1485-1492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- 31.Morel JB, Dangl JL. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997;4:671–683. doi: 10.1038/sj.cdd.4400309. [DOI] [PubMed] [Google Scholar]
- 32.Bendahmane M, Szecsi J, Chen I, Berg RH, Beachy RN. Characterization of mutant tobacco mosaic virus coat protein that interferes with virus cell-to-cell movement. Proc Natl Acad Sci U S A. 2002;99:3645–3650. doi: 10.1073/pnas.062041499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groban ES, Narayanan A, Jacobson MP. Conformational changes in protein loops and helices induced by post-translational phosphorylation. PLoS Comput Biol. 2006;2:e32. doi: 10.1371/journal.pcbi.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xin F, Radivojac P. Post-translational modifications induce significant yet not extreme changes to protein structure. Bioinformatics. 2012;28:2905–2913. doi: 10.1093/bioinformatics/bts541. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov KI, Puustinen P, Gabrenaite R, Vihinen H, Ronnstrand L, Valmu L, Kalkkinen N, Makinen K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell. 2003;15:2124–2139. doi: 10.1105/tpc.012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law L, Everitt J, Beatch M. Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. J Virol. 2003;77:1764–1771. doi: 10.1128/JVI.77.3.1764-1771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Wang JC, Zlotnick A. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog. 2011;7:e1002388. doi: 10.1371/journal.ppat.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni P, Wang Z, Ma X, Das NC, Sokol P, Chiu W, Dragnea B, Hagan M, Kao CC. An examination of the electrostatic interactions between the N-terminal tail of the Brome Mosaic Virus coat protein and encapsidated RNAs. J Mol Biol. 2012;419:284–300. doi: 10.1016/j.jmb.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calhoun SL, Speir Ja, Rao aLN. In vivo particle polymorphism results from deletion of a N-terminal peptide molecular switch in brome mosaic virus capsid protein. Virology. 2007;364:407–21. doi: 10.1016/j.virol.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi YG, Rao aL. Molecular studies on bromovirus capsid protein. VII. Selective packaging on BMV RNA4 by specific N-terminal arginine residuals. Virology. 2000;275:207–17. doi: 10.1006/viro.2000.0513. [DOI] [PubMed] [Google Scholar]
- 41.Rao ALN, Grantham GL. Molecular studies on bromovirus capsid protein II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology. 1996;305:294–305. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- 42.Speir JA, Bothner B, Qu C, Willits DA, Young MJ, Johnson JE. Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J Virol. 2006;80:3582–91. doi: 10.1128/JVI.80.7.3582-3591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakani K, Reade R, Rochon D. Evidence that vector transmission of a plant virus requires conformational change in virus particles. J Mol Biol. 2004;338:507–17. doi: 10.1016/j.jmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Xiang Y, Kakani K, Reade R, Hui E, Rochon D. A 38-amino-acid sequence encompassing the arm domain of the cucumber necrosis virus coat protein functions as a chloroplast transit Peptide in infected plants. J Virol. 2006;80:7952–7964. doi: 10.1128/JVI.00153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi G, Letteney E, Kim CH, Kao CC. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA. 2009;15:615–26. doi: 10.1261/rna.1375509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Gopinath K, Murali A, Yi G, Hayward SD, Zhu H, Kao C. RNA-binding proteins that inhibit RNA virus infection. Proc Natl Acad Sci USA. 2007;104:3129–34. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuznetsov YG, Daijogo S, Zhou J, Semler BL, McPherson A. Atomic force microscopy analysis of icosahedral virus RNA. J Mol Biol. 2005;347:41–52. doi: 10.1016/j.jmb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Choi YG, Rao ALN. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J Virol. 2003;77:9750–9757. doi: 10.1128/JVI.77.18.9750-9757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krol Ma, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc Natl Acad Sci USA. 1999;96:13650–5. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wierzchoslawski R, Urbanowicz A, Dzianott A, Figlerowicz M, Bujarski JJ. Characterization of a novel 5′ subgenomic RNA3a derived from RNA3 of Brome mosaic bromovirus. J Virol. 2006;80:12357–66. doi: 10.1128/JVI.01207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahlquist P, Noueiry AO, Lee W, David B, Dye BT, Kushner DB. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desvoyes B, Scholthof HB. Host-dependent recombination of a Tomato bushy stunt virus coat protein mutant yields truncated capsid subunits that form virus-like complexes which benefit systemic spread. Virology. 2002;304:434–442. doi: 10.1006/viro.2002.1714. [DOI] [PubMed] [Google Scholar]
- 54.Chen YK, Goldbach R, Prins M. Inter- and intramolecular recombinations in the cucumber mosaic virus genome related to adaptation to alstroemeria. J Virol. 2002;76:4119–4124. doi: 10.1128/JVI.76.8.4119-4124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A. 2005;102:10545–10550. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagy PD. The roles of host factors in tombusvirus RNA recombination. Adv Virus Res. 2011;81:63–84. doi: 10.1016/B978-0-12-385885-6.00008-0. [DOI] [PubMed] [Google Scholar]
- 57.Choi S, Hema M, Gopinath K, Santos J, Kao C. Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells. J Virol. 2004;78:13420–13429. doi: 10.1128/JVI.78.24.13420-13429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz M, Chen J, Janda M, Sullivan M, Den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Molecular cell. 2002;9:505–14. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 59.Chen J, Noueiry A, Ahlquist P. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J Virol. 2001;75:3207–3219. doi: 10.1128/JVI.75.7.3207-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hema M, Kao CC. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J Virol. 2004;78:1169–1180. doi: 10.1128/JVI.78.3.1169-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mann B, Madera M, Sheng Q, Tang H, Mechref Y, Novotny MV. ProteinQuant Suite: a bundle of automated software tools for label-free quantitative proteomics. Rapid Commun Mass Spectrom. 2008;22:3823–3834. doi: 10.1002/rcm.3781. [DOI] [PubMed] [Google Scholar]
- 62.Phillips S, Anderson R, Schapire R. Maximum entropy modeling of species geographic distributions. Ecological modelling. 2006;190:231–259. [Google Scholar]
- 63.Suo SB, Qiu JD, Shi SP, Sun XY, Huang SY, Chen X, Liang RP. Position-specific analysis and prediction for protein lysine acetylation based on multiple features. PLoS One. 2012;7:e49108. doi: 10.1371/journal.pone.0049108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Felden B, Florentz C, Giege R, Westhof E. Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J Mol Biol. 1994;235:508–531. doi: 10.1006/jmbi.1994.1010. [DOI] [PubMed] [Google Scholar]