Abstract
Two novel trialkyl-substituted aromatic acids, solwaric acids A and B, were isolated from a marine Solwaraspora sp. cultivated from the ascidian Trididemnum orbiculatum. Solwaric acids A and B were isotopically labeled with U-13C glucose, and analysis of a 13C–13C COSY allowed for unambiguous determination of the location of the phenyl methyl group. The two novel compounds demonstrated antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA).
Keywords: Trididemnum orbiculatum, Solwaraspora, methicillin-resistant Staphylococcus aureus
1. Introduction
Infectious disease continues to be a major issue, as it is the second leading cause of death worldwide [1]. Complicating the treatment of infectious disease is antibiotic resistance, which results in about $20 billion in health service costs and at least 23,000 deaths in the United States per year [2,3]. Among antibiotic resistant bacteria, methicillin-resistant Staphylococcus aureus (MRSA) has become a major concern for human health [4,5,6,7,8]. While the mortality percentages in methicillin-susceptible Staphylococcus aureus (MSSA) patients range from 5% to 28%, the mortality percentages for MRSA patients tend to be even higher, ranging from 10% to 64% [5,6,7,8]. Although ranging between studies, one study encompassing data from different European countries found an additional 80% excess mortality rate at day 30 after infection was contributed by methicillin resistance in S. aureus [5,6,8]. Therefore, antibiotic resistance escalates the importance for discovering new and effective antibiotics in an efficient manner.
In our pursuit of novel antibacterial compounds, we isolated two novel aromatic acids that were named solwaric acids A (1) and B (2) and the known 2,4,6-triphenyl-1-hexene (3) [9,10,11], from a marine Solwaraspora sp. (Strain WMMB329) cultivated from the ascidian Trididemnum orbiculatum (Van Name, 1902) [12]. To our knowledge, these are the first novel compounds reported from a Solwaraspora sp., although strain WMMB329 also showed 99% 16S rDNA sequence similarity to some Micromonospora spp. Solwaric acids A (1) and B (2) demonstrated antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA). Aromatic acids are somewhat rare in natural products, with most being produced by actinomycetes [13,14,15,16].
Although the solwaric acids did not present elucidation challenges typical of some natural products, such as few protons, the aromatic ring was substituted only with carbon-based substituents. As a result, the chemical shifts of the protons presented challenges with respect to unambiguously determining the location of the methyl group. HMBC correlations were not conclusive and 13C calculations using density functional theory were questionable due to the small differences between two potential structures. Therefore, solwaric acids A (1) and B (2) were isotopically enriched with U-13C glucose, which allowed for acquisition of a 13C–13C COSY and unambiguous assignment of the aromatic methyl group location. While the gCOSY requires isotopic enrichment, the method is still economically feasible, and can be made even more so by the use of 13C-optimized cryoprobes, which drastically reduce the amount of compound needed and therefore the amount of U-13C glucose required. We have more broadly explored the 13C gCOSY as a rapid route for establishing carbon connectivity, and it appears broadly useful for a broad range of bacterially produced natural products. Overall, we have found this method to be highly promising.
2. Results and Discussion
2.1. Bacterial Strain Selection and Structure Elucidation
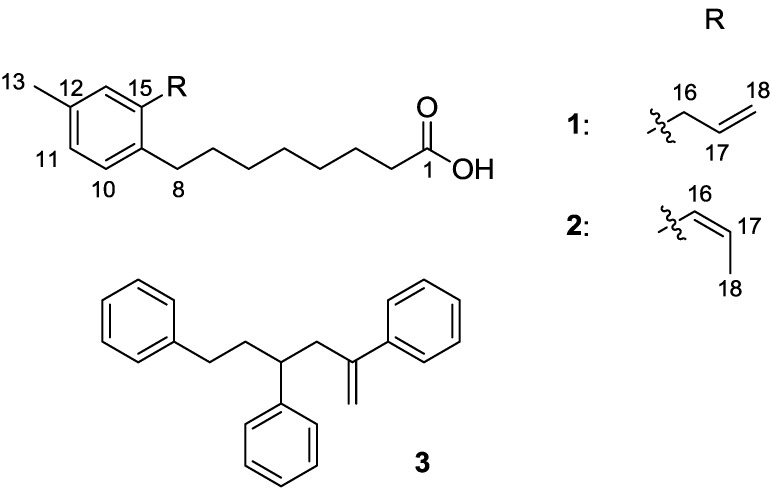
Strain WMMB329 was investigated due to the lack of chemistry described in the literature from Solwaraspora spp. as well as antibacterial activity from initial screening of a crude extract. After fermentation and isolation of the bioactive compounds, WMMB329 was found to produce solwaric acids A (1) and B (2), as well as the known 2,4,6-triphenyl-1-hexene (3) (Figure 1).
Figure 1.
Structures of 1–3.
HRMS supported the molecular formula of C18H26O2 for both solwaric acid A (1) and B (2). The structures of solwaric acids A (1) and B (2) were determined by analysis of 1H and 13C NMR data (Table 1, Supplementary Figures S1–S16). Since the assignment of solwaric acid A (1) and B (2) was confirmed by direct 13C–13C correlation experiments, the discussion of elucidation has been eliminated with the exception of the olefin at C-16 and C-17 in solwaric acid B (2), which was determined to be Z based on the vicinal coupling constant (3JH 11.4) [17]. The presence of a methyl group (C-13) attached to the phenyl ring was indicated by HMBC correlations to three aromatic carbon atoms, but it could not be determined unambiguously relative to the other aromatic carbons. The major problem was that two protons in the aromatic ring (H-11 and H-14) were at nearly identical chemical shifts. Additionally, C-9 and C-15 were at nearly identical chemical shifts. Therefore, the HMBC correlations supported two possible arrangements for C-10 through C-14 relative to positions 9 and 15. To further complicate matters, the close chemical shifts of H-11 and H-14 made it difficult to justify assignment based on NOE. Finally, DFT calculations showed differences in the calculated 13C shifts that were within what we have observed as typical experimental error (i.e., not overly conclusive). Direct 13C–13C correlations, however, would solve the problem. Consequently, we evaluated the use of 13C-labeling of the two compounds using U-13C glucose and subsequent acquisition of a 13C–13C gCOSY to address this issue.
Table 1.
1H and 13C NMR data for 1 and 2 (600 MHz for 1H, 150 MHz for 13C, CDCl3).
| Position | 1 | 2 | COSY | HMBC | |||
|---|---|---|---|---|---|---|---|
| δC, mult. | δH (J in Hz) | δC, mult. | δH (J in Hz) | ||||
| 1 | 180.1, C | 180.5, C | |||||
| 1-OH | 9.34, s | ||||||
| 2 | 34.4, CH2 | 2.33, t (6.6) | 34.8, CH2 | 2.29, t (6.6) | 3 | 3, 4 | |
| 3 | 24.9, CH2 | 1.60, m | 25.0, CH2 | 1.59, m | 2, 4 | 2, 4 | |
| 4 | 29.2, CH2 | 1.30, m | 29.3, CH2 | 1.30, m | 3 | ||
| 5 | 29.4, CH2 | 1.30, m | 29.4, CH2 | 1.30, m | |||
| 6 | 29.9, CH2 | 1.30, m | 29.6, CH2 | 1.30, m | 7 | ||
| 7 | 31.3, CH2 | 1.51, m | 30.9, CH2 | 1.48, m | 8 | 9 | |
| 8 | 32.5, CH2 | 2.52, t (7.6) | 33.2, CH2 | 2.50, t (7.6) | 7 | 6, 9, 10 | |
| 9 | 137.9, C | 138.1, C | |||||
| 10 | 129.4, CH | 7.02, d (7.2) | 129.0, CH | 7.04, d (7.2) | 11 | 8, 12 | |
| 11 | 127.2, CH | 6.95, d (7.2) | 127.6, CH | 6.96, d (7.2) | 10 | 14 | |
| 12 | 135.5, C | 134.7, C | |||||
| 13 | 21.2, CH3 | 2.27, s | 21.2, CH3 | 2.30, s | 11, 12, 14 | ||
| 14 | 130.4, CH | 6.94, s | 130.3, CH | 6.97, s | 9, 13 | ||
| 15 | 137.5, C | 136.1, C | |||||
| 16 | 37.2, CH2 | 3.33, br d (6.4) | 129.1, CH | 6.47, br d (11.4) | 17 | 12, 14, 18 | |
| 17 | 137.7, CH | 5.93, tdd (6.4, 10.1, 16.9) |
126.9, CH | 5.77, dq (11.4, 7.0) |
16, 18 | 18 | |
| 18 | 115.7, CH2 | 5.04, tdd (1.7, 1.7, 10.1) |
14.6, CH3 | 1.71, dd (1.8, 7.0) | 17 | 16, 17 | |
| 5.00, tdd (1.7, 1.7, 16.9) |
|||||||
2.2. 13C–13C gCOSY
NMR experiments, such as the 2D INADEQUATE, have been used for direct determination of carbon-carbon connectivity within natural products [18,19]. Even with cryoprobes, the INADEQUATE can present difficulties due to sample solubility since it requires a fairly concentrated sample. Therefore, we decided to evaluate a 13C–13C gCOSY, which would require a shorter minimum phase cycle than the INADEQUATE, could be easily implemented, and could be easily interpreted. In particular, calculations suggested that the standard two-pulse gCOSY implemented on NMR spectrometers for 1H experiments could be used for 13C–13C with no modifications to the pulse program. In fact, after adjusting for 13C and spectral widths, a theoretical sampling of coupling constants in the range 30 to 60 Hz would produce maximum polarization transfer at 33 to 17 ms, respectively. Additionally, S/N for JCC = 60 Hz would be maximized with 450 increments—though smaller values (e.g., 256) could be used without much loss in sensitivity—resulting in the 2D resolution of 0.55 ppm (Supplementary Figure S17). The combination of easily optimizing for a range of 13C–13C coupling constants combined with the fact that the gCOSY does not require 13C inversion pulses and that the experiment could be immediately implemented by others with no requirement for pulse programming made the simple gCOSY an encouraging prospect (Supplementary Figures S18 and S19). The only drawback was that the 13C–13C COSY experiment requires isotopic enrichment. To address this problem, we tested the feasibility of 13C incorporation of microbial-derived natural products in conjunction with evaluating the feasibility of the 13C–13C gCOSY.
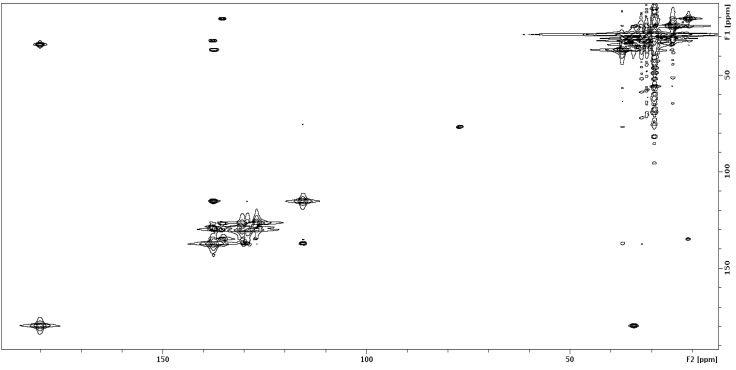
To increase the 13C abundance, fermentation of WMMB329 in 1 L ASW-A using uniformly 13C-labeled glucose (10 g/L ASW-A) and subsequent purification yielded solwaric acids A (1) and B (2) with about 35% 13C incorporation (as evidenced by MS isotopic distribution). The 13C–13C gCOSY (Figure 2) was acquired in two hours on 18 µmoles (105 µM) solwaric acid A (1) and allowed for the methyl group (C-13) to be unambiguously assigned as attached to C-12; likewise, 13C–13C COSY correlations were seen between C-12 and C-14, as well as C-14 and C-15. Analysis of the 13C–13C COSY also confirmed the carbon connectivity throughout the rest of the structure (Supplementary Figure S20). While the 13C–13C gCOSY was acquired on 5 mg of solwaric acid A (1), the experiment can be effectively acquired at considerably lower concentrations. The exact concentration is dependent on the level of 13C incorporation and the spectrometer used, but our initial studies suggest that a 13C–13C gCOSY can be effectively acquired on low µM concentrations in less than 24 h.
Figure 2.
13C–13C COSY of 13C-labeled 1.
We have also tested to see if this approach would be more broadly applicable to other types of natural products. For example, we have produced two novel 13C-labeled peptides from marine-derived bacteria, the structures of which will be reported in a subsequent publication. For complicated structures, this method will drastically reduce elucidation time, which is currently a bottleneck in natural products discovery. Also, recent work by Williamson and Martin has shown that 13C–13C coupling constants can aid in establishing configuration [20]. Even modest levels of 13C incorporation would be helpful to apply 13C–13C coupling constants for help in structure elucidation.
Additionally, 13C-labeling and acquisition of a 13C–13C COSY can be useful for analyzing crude extracts or mixtures of compounds. We have demonstrated this approach by acquiring a 13C–13C COSY on the extract from the CHCl3 partition of WMMB329 (Supplementary Figure S21). The CHCl3 partition was the first step of purification of the crude extract, and therefore, contained a mixture of compounds. Acquisition of the 13C–13C COSY on 13C-labeled extract allows for separation of the chemistry on a large sweep width (~200 ppm) and elimination of most media components—which are unlabeled—from the spectrum. Although there may still be a mixture of several labeled compounds, this method could rapidly provide carbon-carbon connectivity for portions of the structures, which could be helpful for dereplication. Therefore, 13C-labeling of natural products can be a valuable tool for structure elucidation, as well as other areas of natural product research.
2.3. Biological Activity
Solwaric acids A (1) and B (2) showed antibacterial activity and were more potent against gram positive bacteria (Table 2). Vancomycin and gentamicin were used as a positive control in assays with gram positive and gram negative bacteria, respectively. The known 2,4,6-triphenyl-1-hexene (3) (Supplementary Figures S22–S24) demonstrated no antibacterial activity.
Table 2.
Minimum Inhibitory Concentration (µg/mL) of 1–3.
| MRSA | MSSA | E. coli | P. aeruginosa | |
|---|---|---|---|---|
| 1 | 32 | 64 | 128 | 128 |
| 2 | 32 | 64 | 128 | 128 |
| 3 | >128 | >128 | >128 | >128 |
3. Experimental Section
3.1. General Experimental Procedures
Optical rotations were measured on a Perkin–Elmer 241 Polarimeter. UV spectra were recorded on an Aminco/OLIS UV-Vis spectrophotometer. IR spectra were measured with a Bruker Equinox 55/S FT–IR spectrophotometer. NMR spectra were obtained in CDCl3 with a Bruker Avance 600 MHz spectrometer equipped with a 1.7 mm 1H{13C/15N} cryoprobe, a Bruker Avance 500 MHz spectrometer equipped with a 13C/15N{1H} cryoprobe, and a Varian Unity-Inova 500 MHz spectrometer. HRMS data were acquired with a Bruker MaXis™ 4G QTOF mass spectrometer. RP HPLC was performed using a Shimadzu Prominence HPLC system and a Phenomenex Luna C18 column (250 × 10 mm, 5 µm), as well as a Gilson Preparative HPLC and Phenomenex Gemini C18 column (250 × 30 mm, 5 µm).
3.2. Biological Material
Ascidian specimens were collected on 11 October 2010, in the Florida Keys (24°37.4873′, 81°27.443′). Identification was confirmed by Shirley Parker-Nance. A voucher specimen (FLK10-5-1) for Trididemnum orbiculatum (Van Name, 1902) [12] is housed at the University of Wisconsin-Madison. For cultivation, a sample of ascidian (1 cm3) was rinsed with sterile seawater, macerated using a sterile pestle in a micro-centrifuge tube, and dilutions were made in sterile seawater, with vortexing between steps to separate bacteria from heavier tissues. Dilutions were separately plated on three media: ISP2 supplemented with artificial seawater [21], R2A [22], and M4 [23]. Each medium was supplemented with 50 µg/mL cycloheximide and 25 µg/mL nalidixic acid. Plates were incubated at 28 °C for at least 28 days, and strain WMMB329 was purified from an M4 isolation plate.
3.3. Sequencing
16S rDNA sequencing was conducted as previously described [24]. WMMB329 was identified as a Solwaraspora sp. and demonstrated 99% sequence similarity to Solwaraspora sp. UMM486 (accession number AY552774) and 99% sequence similarity to Micromonospora sp. S3-1 (accession number AB645957). The 16S sequence for WMMB329 was deposited in GenBank (accession number KC856821).
3.4. Fermentation, Extraction, and Isolation
One 10 mL seed culture (25 × 150 mm tube) in medium modified ASW-A (5 g soluble starch, 10 g glucose, 5 g peptone, 5 g yeast extract per liter of 50%/50% artificial seawater/diH2O) were inoculated with strain WMMB329 and shaken (200 RPM, 28 °C) for fifteen days. For fermentation, 500 mL baffled flasks (2 × 100 mL) containing ASW-A (20 g soluble starch, 10 g glucose, 5 g peptone, 5 g yeast extract, 5 g CaCO3 per liter of artificial seawater) were inoculated with 4 mL seed culture and were incubated (200 RPM, 28 °C) for fourteen days. Two-liter flasks (15 × 500 mL) containing medium ASW-A with Diaion HP20 (7% by weight) were inoculated with 5 mL from the 100 mL culture and shaken (200 RPM, 28 °C) for ten days. Filtered HP20 and cells were washed with H2O and extracted with acetone. The acetone extract (7.1 g) was subjected to liquid-liquid partitioning using 90%/10% MeOH/H2O and hexanes (1:1) followed by liquid-liquid partitioning of the previous aqueous layer using 70%/30% MeOH/H2O and CHCl3 (1:1). The hexanes partition (287 mg) was fractionated by Sephadex LH20 column chromatography (2.5 × 40 cm, CHCl3:MeOH, 1:1). Fractions containing 1–3 were subjected to RP HPLC (70/30% to 90/10% ACN/H2O with H2O containing 0.1% acetic acid, 25 min, 25 mL/min followed by 90/10% to 100/0% of same solvents, 3 min, 25 mL/min, and a hold at 100/0% of same solvents) using a Phenomenex Gemini C18 column (250 × 30 mm, 5 µm), yielding 1 (15.4 mg, tR 18.0 min), 2 (13.8 mg, tR 19.5 min), and 3 (4.9 mg, tR 31.5 min). For 13C incorporation, the same procedure was used with two-liter flasks (2 × 500 mL) containing medium ASW-A (U-13C-glucose substituted for unlabeled glucose). Yields for solwaric acids A (1) and (2) were approximately the same during fermentation with U-13C glucose as with unlabeled glucose.
Solwaric acid A (1). colorless solid; UV (MeOH) λmax (log ε) 204 (2.77), 218 (2.49), 271 (1.27) nm; IR (ATR) νmax 2928, 2856, 1707, 1412, 1217, 994, 911, 819, 755 cm−1; 1H and 13C NMR (Table 1); HRMS [M + Na]+m/z 297.1836 (calcd. for C18H26O2Na+, 297.1825).
Solwaric acid B (2). colorless solid; UV (MeOH) λmax (log ε) 208 (2.79), 238 (2.48), 281 (1.29) nm; IR (ATR) νmax 2928, 2856, 1708, 1410, 1217, 820, 755, 707, 667 cm−1; 1H and 13C NMR (Table 1); HRMS [M + Na]+m/z 297.1838 (calcd. for C18H26O2Na+, 297.1825).
3.5. Antibacterial Assay
Solwaric acid A (1) and B (2) and 2,4,6-triphenyl-1-hexene (3) were tested for antibacterial activity against MSSA (ATCC #29213), MRSA (ATCC #33591), E. coli (ATCC #25922), and P. aeruginosa (ATCC #27853), and MICs were determined using a dilution antimicrobial susceptibility test for aerobic bacteria [25]. Solwaric acids A (1) and B (2) and 2,4,6-triphenyl-1-hexene (3) were dissolved in DMSO, serially diluted to 10 concentrations (0.25–128 µg/mL), and tested in a 96-well plate. Vancomycin was used as a control and exhibited an MIC of 1 µg/mL against MSSA and 1 µg/mL against MRSA. Gentamicin was used as a control and exhibited an MIC of 4 µg/mL against E. coli and 4 µg/mL against P. aeruginosa. Solwaric acids A (1) and B (2) and 2,4,6-triphenyl-1-hexene (3) were tested in triplicate, and vancomycin and gentamicin was tested in triplicate. Six untreated media controls were included on each plate. The plates were incubated at 33 °C for 18 h. The MIC was determined as the lowest concentration that inhibited visible growth of bacteria.
4. Conclusions
We reported the isolation and structure elucidation of two novel trialkyl-substitued aromatic acids, solwaric acids A (1) and B (2), and the known 2,4,6-triphenyl-1-hexene (3). The novel compounds demonstrated antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA). Solwaric acid A (1) and B (2) were enriched with 13C-labeled glucose that allowed for the acquisition of a 13C–13C COSY and unambiguous assignment of the methyl group location on the phenyl ring. While this example utilized 13C-labeling to determine carbon connectivity for one challenging portion of the structure, this method could be valuable for molecules with multiple tetrasubstituted centers, which make structure elucidation by standard NMR experiments more challenging. Hence, 13C incorporation and subsequent acquisition of a 13C–13C COSY—aided by the increasing sensitivity of NMR spectrometers—could drastically reduce the time for structure determination of microbial-derived natural products, including peptides and terpenes. Compared to the cost and time involved with other methods such as computer assisted structure determination, labeling microbial natural products offers a cost effective solution while providing high confidence in the proposed structure.
Acknowledgments
This work was supported by funding from the University of Wisconsin-Madison School of Pharmacy, the Graduate School at the University of Wisconsin, and the UW College of Agriculture and Life Sciences. This work was also funded by the NIH, NIGMS Grant R01 GM092009 and by R01 GM104192. We would like to thank the Analytical Instrumentation Center at the University of Wisconsin-Madison for the facilities to acquire spectroscopic data, as well as the University of Wisconsin-Madison Chemistry Instrument Center (The purchase of the Waters (Micromass) Autospec® in 1994 was partially funded by NSF Award #CHE-9304546 to the Department of Chemistry). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grants P41RR02301 (BRTP/NCRR) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. We would like to thank Don Demaria for assistance with collection and Shirley Parker-Nance for taxonomy.
Supplementary Files
Supplementary Information (PDF, 668 KB)
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.World Health Organization . Deaths by Cause, Sex, and Mortality Stratum in WHO Regions, Estimates for 2002: World Health Report—2004. World Health Organization; Geneva, Switzerland: 2004. p. 1. [Google Scholar]
- 2.Smith R., Coast J. The true cost of antimicrobial resistance. Br. Med. J. 2013;346 doi: 10.1136/bmj.f1493. [DOI] [PubMed] [Google Scholar]
- 3.Center for Disease Control and Prevention. [(accessed on 13 December 2013)]. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/
- 4.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Jr., Gilbert D., Rice L.B., Scheld M., Spellberg B., Bartlett J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 5.Stefani S., Chung D.R., Lindsay J.A., Friedrich A.W., Kearns A.M., Westh H., MacKenzie F.M. Meticillin-Resistant Staphylococcus aureus (MRSA): Global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents. 2012;39:273–282. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Köck R., Becker K., Cookson B., van Gemert-Pijnen J.E., Harbarth S., Kluytmans J., Mielke M., Peters G., Skov R.L., Struelens M.J., et al. Methicillin-Resistant Staphylococcus aureus (MRSA): Burden of disease and control challenges in Europe. Euro Surveill. 2010;15:19688–19696. doi: 10.2807/ese.15.41.19688-en. [DOI] [PubMed] [Google Scholar]
- 7.Klevens R.M., Morrison M.A., Nadle J., Petit S., Gershman K., Ray S., Harrison L.H., Lynfield R., Dumyati G., Townes J.M., et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 8.de Kraker M.E.A., Wolkewitz M., Davey P.G., Koller W., Berger J., Nagler J., Icket C., Kalenic S., Horvatic J., Seifert H., et al. Clinical impact of antimicrobial resistance in European hospitals: Excess mortality and length of hospital stay related to methicillin-resistant Staphylococcus aureus bloodstream infections. Antimicrob. Agents Chemother. 2011;55:1598–1605. doi: 10.1128/AAC.01157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staudinger H., Steinhofer A. Über hochpolymere verbindungen. 107. Mitteilung. Beiträge zur kenntnis der polystyrole. Justus Liebigs Ann. Chem. 1935;517:35–53. doi: 10.1002/jlac.19355170104. [DOI] [Google Scholar]
- 10.Mayo F.R. The dimerization of styrene. J. Am. Chem. Soc. 1968;90:1289–1295. doi: 10.1021/ja01007a032. [DOI] [Google Scholar]
- 11.Ayer W.A., Muir D.J., Chakravarty P. Phenolic and other metabolites of Phellinus pini, a fungus pathogenic to pine. Phytochemistry. 1996;42:1321–1324. doi: 10.1016/0031-9422(96)00125-2. [DOI] [Google Scholar]
- 12.Van Name W.G. The ascidians of the Bermuda Islands. Trans. Connecticut Acad. Sci. 1902;11:325–412. [Google Scholar]
- 13.Iwata F., Sato S., Mukai T., Yamada S., Takeo J., Abe A., Okita T., Kawahara H. Lorneic acids, trialkyl-substituted aromatic acids from a marine-derived actinomycete. J. Nat. Prod. 2009;72:2046–2048. doi: 10.1021/np900353y. [DOI] [PubMed] [Google Scholar]
- 14.Wenzel S.C., Bode H.B. Novel polyene carboxylic acids from Streptomyces. J. Nat. Prod. 2004;67:1631–1633. doi: 10.1021/np049852t. [DOI] [PubMed] [Google Scholar]
- 15.Mehnaz S., Saleem R.S., Yameen B., Pianet I., Schnakenbur G., Pietraszkiewicz H., Valeriote F., Josten M., Sahl H.G., Franzblau S.G., et al. Lahorenoic acids A–C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013;76:135–141. doi: 10.1021/np3005166. [DOI] [PubMed] [Google Scholar]
- 16.Raju R., Gromyko O., Fedorenko V., Luzhetskyy A., Müller R. Lorneic acids C and D, new trialkyl-substituted aromatic acids isolated from a terrestrial Streptomyces sp. J. Antibiot. 2013;66:347–349. doi: 10.1038/ja.2013.18. [DOI] [PubMed] [Google Scholar]
- 17.Williams D.H., Fleming I. Spectroscopic Methods in Organic Chemistry. 4th ed. McGraw-Hill Book Company Limited; London, UK: 1987. pp. 143–146. [Google Scholar]
- 18.Bugni T.S., Bernan V.S., Greenstein M., Janso J.E., Maiese W.M., Mayne C.L., Ireland C.M. Brocaenols A–C: Novel polyketides from a marine-derived Penicillium brocae. J. Org. Chem. 2003;68:2014–2017. doi: 10.1021/jo020597w. [DOI] [PubMed] [Google Scholar]
- 19.Meyer S.W., Köck M. NMR studies of phakellins and isophakellins. J. Nat. Prod. 2008;71:1524–1529. doi: 10.1021/np8000706. [DOI] [PubMed] [Google Scholar]
- 20.Bifulco G., Riccio R., Martin G.E., Buevich A.V., Williamson R.T. Quantum chemical calculations of 1JCC coupling constants for the stereochemical determination of organic compounds. Org. Lett. 2013;15:654–657. doi: 10.1021/ol3034803. [DOI] [PubMed] [Google Scholar]
- 21.Harrison P.J., Waters R.E., Taylor F.J.R. A broad spectrum artificial seawater medium for coastal and open ocean phytoplankton. J. Phycol. 1980;16:28–35. doi: 10.1111/j.1529-8817.1980.tb00724.x. [DOI] [Google Scholar]
- 22.Reasoner D.J., Geldreich E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldonado L.A., Fragoso-Yáñez D., Pérez-García A., Rosellón-Druker J., Quintana E.T. Antinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek. 2009;95:111–120. doi: 10.1007/s10482-008-9294-3. [DOI] [PubMed] [Google Scholar]
- 24.Wyche T.P., Hou Y., Braun D., Cohen H.C., Xiong M.P., Bugni T.S. First natural analogs of the cytotoxic thiodepsipeptide thiocoraline A from a marine Verrucosispora sp. J. Org. Chem. 2011;76:6542–6547. doi: 10.1021/jo200661n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. 7th ed. NCCLS; Villanova, PA, USA: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (PDF, 668 KB)