Abstract
Previous studies have used analysis of Ca2+ sparks extensively to investigate both normal and pathological Ca2+ regulation in cardiac myocytes. The great majority of these studies used line-scan confocal imaging. In part, this is because the development of open-source software for automatic detection of Ca2+ sparks in line-scan images has greatly simplified data analysis. A disadvantage of line-scan imaging is that data are collected from a single row of pixels, representing only a small fraction of the cell, and in many instances x-y confocal imaging is preferable. However, the limited availability of software for Ca2+ spark analysis in two-dimensional x-y image stacks presents an obstacle to its wider application. This study describes the development and characterization of software to enable automatic detection and analysis of Ca2+ sparks within x-y image stacks, implemented as a plugin within the open-source image analysis platform ImageJ. The program includes methods to enable precise identification of cells within confocal fluorescence images, compensation for changes in background fluorescence, and options that allow exclusion of events based on spatial characteristics.
Introduction
Ca2+ sparks are elementary, localized Ca2+ release events that result from the activation of ryanodine receptor (RyR) clusters within the sarcoplasmic reticulum (SR) of cardiac, skeletal, or smooth muscle (1). In the two decades since their discovery in cardiac myocytes (2), numerous studies have used detailed analysis of sparks to provide fundamental insights into issues such as excitation contraction coupling (3–5), disease mechanisms (6–11), and the actions of therapeutic agents (12).
The great majority of past studies on Ca2+ sparks have involved laser scanning confocal microscopy (LSCM) operating in line-scan mode, where the data are collected from a single line of pixels. In part, this reflects the fact that one can obtain adequate temporal resolution using relatively low-specification confocal systems. In addition, the development of software to automate detection of Ca2+ sparks in line-scan images markedly accelerated and simplified the process of Ca2+ spark detection and analysis (13). Its subsequent implementation as a plugin within the open-source image analysis environment ImageJ was widely adopted (14), providing the additional benefit that data collected by different labs could be compared more readily.
A fundamental limitation of line-scan imaging is that data are collected from only a small fraction of the cell being studied. In circumstances in which the proximity of localized Ca2+ release events is of interest (e.g., in relation to membrane structures, organelles, channels, or transporters), x-y imaging is clearly preferable. However, despite the increasing availability of fast confocal scanning systems, only a few studies have provided limited statistical data on Ca2+ sparks based on analysis of x-y image stacks from either skeletal or cardiac muscle cells (14–17). One reason for this is that descriptions and evaluations of algorithms for automatic detection and analysis of Ca2+ sparks in x-y image stacks are currently limited, and typically such programs operate within specialist software development environments (15–18).
This study describes the development and characterization of software (xySpark) to enable automatic detection and analysis (amplitude, width, frequency, duration, and mass) of Ca2+ sparks in x-y image stacks, implemented as a plugin for ImageJ. xySpark provides an interactive graphical user interface and includes novel (to our knowledge) methods to enable accurate identification of cells within confocal fluorescence images, compensation for slow changes in background fluorescence during data collection, and options that allow exclusion of aberrant events based on spatial characteristics.
Materials and Methods
Myocyte preparation and dye loading
Adult Wistar rats (125–175 g) were sacrificed in accordance with the UK Home Office Guidance on the Operation of Animals (Scientific Procedures) Act of 1986 and the guidelines of the National Institutes of Health for the Care and Use of Laboratory Animals (1996). Ventricular myocytes were isolated by collagenase digestion (Worthington Biochemical, Reading, UK) as described previously (19). Intact myocytes were then perfused with solutions containing (mM) 113 NaCl, 5.4 KCl, 1 MgCl2, 1.0 CaCl2, 0.37 Na2HPO4, 5.5 glucose, 5 HEPES (20–22°C, pH 7.1). Changes in cytosolic [Ca2+] were detected by loading myocytes with fluo-4 AM (5 μM) for 15 min at room temperature (20–22°C). After loading, 1 hr was allowed for dye desertification before the experiments were commenced. Fluo-4 was obtained from Calbiochem (Nottingham, UK) and all other chemicals were obtained from Sigma-Aldrich (Gillingham, UK).
Confocal imaging
The experimental chamber was placed on the stage of a Nikon Diaphot Eclipse TE2000 inverted microscope and cells were viewed using a Nikon 60× water immersion lens (Plan Apo, NA 1.2). Rapid x-y confocal Ca2+ imaging was carried out using an Andor Revolution confocal unit (Belfast, UK) equipped with a Yokogawa CSU X1 spinning disk (10,000 rpm). During imaging, 100% of the light passed via the side port of the camera through a camera mount and lens with a further 2× magnification. Light was detected using an iXon3 897 electron multiplying (EM) CCD camera sensor. The iXon3 897 camera is capable of recording 35 frames per second (fps) at 512 × 512 pixel resolution (i.e., full frame), and this can be increased to ∼170 fps by reading only part of the EMCCD array. Further increases in the fps rate can be achieved by using pixel binning; for example, 2 × 2 binning can allow frame rates between 68 and 313 fps depending on the scan size. In this study, the decrease in resolution associated with 2 × 2 binning was not found to compromise spark detection. Fluo-4 was excited at 488 nm and emitted fluorescence was detected at >500 nm.
Ca2+ spark detection and analysis
Image processing and Ca2+ spark detection were done using the program that forms the basis of this study, xySpark (http://www.fbs.leeds.ac.uk/go/xyspark), which installs as a plugin for ImageJ (http://rsb.info.nih.gov/ij/) or the Fiji distribution of ImageJ (http://fiji.sc/Fiji). ImageJ and xySpark are written in Java, which allows both to run on any platform with a Java Virtual Machine. xySpark requires ImageJ version ≥ 1.47n. All associated files must be located within a folder named xySpark, copied to the plugins folder of ImageJ. The xySpark folder must also contain the ImageAccess class file (http://bigwww.epfl.ch/teaching/iplabsite/tutorial.php). The input data for xySpark is an x-y confocal fluorescence image stack (8, 16, or 32 bit), typically 512 × 512 pixels. In this study we used a PC running 64 bit Windows 7 (ultimate), with an i5 processor and 8GB RAM, with 6GB allocated to ImageJ (see Edit/Options/Memory and Threads). This allowed us to analyze 1000–2000 frame 16 bit image stacks using xySpark. Memory requirements can be reduced by cropping the stacks to exclude a proportion of the pixels outside the cell boundary. However, reducing the bit depth or rotating the image before analysis is not recommended because it may lead to loss of precision or changes in pixel values during interpolation.
The output data from xySpark (frame number, spark coordinates, amplitude, frequency, full width at half-maximum amplitude (FWHM), t1/2 of the descending phase, r2 value for Gaussian fits, and summary statistics) were imported into OriginPro (version 8.4, 64 bit; OriginLab, Silverdale Scientific Ltd., Stoke Mandeville, UK) to allow plotting of histograms and curve fitting. Results are presented as the mean ± SE. Statistical significance was assessed using a t-test. A difference between means was considered significant at p < 0.05.
Results
Our aim in this study was to develop an algorithm to automatically detect and analyze Ca2+ sparks in x-y confocal image stacks obtained from quiescent myocytes, i.e., where the background cell fluorescence is relatively constant and spontaneous or triggered global Ca2+ transients are absent. A detailed summary of the algorithm is provided in Fig. S1 in the Supporting Material and key aspects are described in detail below.
Variations in noise within raw and normalized confocal fluorescence images
We sought to identify Ca2+ sparks by applying a threshold, θ, where θ = the mean background fluorescence within the cell + SD × ε, a user-defined variable (13). Relating θ to the SD of the background noise makes it more likely that a constant value of ε can be applied satisfactorily to data sets with variable noise. However, marked changes in noise were often apparent immediately beyond the cell boundary, making reliable implementation of this approach challenging. Fig. 1 A shows a pixel-averaged (50 frames) confocal image of a fluo-4-loaded cardiac myocyte and a corresponding image (Fig. 1 B), where each pixel is ascribed the value of the SD. In this example, the line profiles show that both the fluorescence and SD are relatively high within the cell and decrease only gradually beyond the cell boundary.
Figure 1.
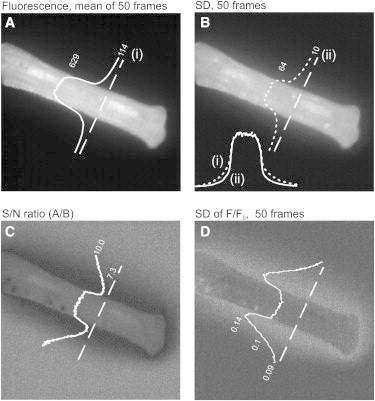
Noise variation in confocal images of myocytes. (A) Image created by pixel averaging of 50 consecutive raw fluorescence images obtained from a fluo-4-loaded ventricular myocyte. (B) Each pixel is the SD of the 50 values at each coordinate position in the raw fluorescence stack. (C) SNR image created by dividing the image of mean pixel values (A) by the image of SD pixel values (B). (D) Each pixel value is the SD of the 50 values at each coordinate position in the F/F0 image stack. In all images, the broken line indicates the orientation of the line profile shown above, superimposed on the cell. In B, the inset shows the line profile from A and B normalized and superimposed.
The gradual decline in fluorescence likely reflects the fact that light from dye trapped within the cell is emitted into the surrounding medium, creating a diffuse halo (see Figure 4 A below). However, the normalized line profiles (Fig. 1 B, inset) show that the mean fluorescence decreases more rapidly beyond the cell boundary than the SD, and as a result, the signal-to-noise ratio (SNR, i.e., image A/B) decreases sharply beyond the cell boundary before increasing to a higher level (Fig. 1 C).
Figure 4.
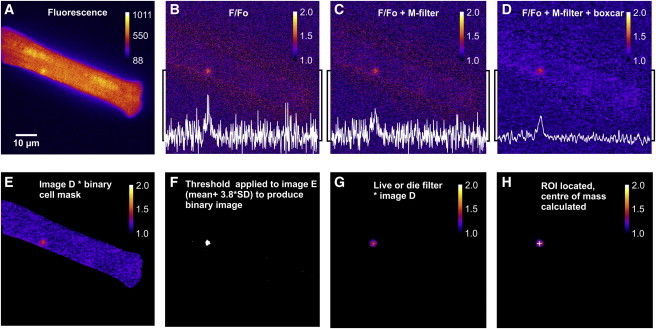
Image processing and spark detection. (A) Original fluorescence image of a fluo-4-loaded myocyte, showing a single spark event close to the lower edge. (B) F/F0 image obtained by dividing image A by the user-selected normalizing image (not shown). Increased noise is apparent immediately outside the cell boundary. The line profile passing through the center of mass of the spark (white trace) shows the amplitude of the spark relative to the background noise. (C) Application of the M filter attenuates noise outliers. (D) The boxcar filter markedly reduces the background noise. (E) Multiplication of the filtered F/F0 image by the binary cell mask (see Fig. 4) sets all pixel values outside the cell boundary to zero, thereby excluding them from subsequent analysis steps. (F) User-defined threshold (θ = mean + ε ×; see Fig. 1) applied to image E to produce a binary image, identifying spark. (G) The live-or-die filter was applied to a speckled binary image to produce a uniform ROI (above threshold = 1; otherwise, 0), and then multiplied by image E to fill the ROI with pixel values. (H) ROI located in the image. Center of spark mass coordinates were calculated and marked with a cross. To see this figure in color, go online.
In practice, this variation in SD has most relevance to the normalized (F/F0) image stack, in which the mean and SD are calculated before detection of Ca2+ sparks. In Fig. 1 D, each pixel value is the SD of the F/F0 values at each coordinate (50 frames), and it is apparent that the SD increases rapidly beyond the cell boundary. This increase in noise immediately beyond the cell boundary in the F/F0 stack has the potential to cause two types of error. First, overestimation of the area occupied by the cell will cause an error in the calculation of the SD and therefore θ, compromising spark detection. Second, noise just beyond the cell boundary might exceed θ, resulting in incorrect identification as a Ca2+ spark. Accurate determination of the cell boundary is therefore essential for reliable spark detection by thresholding.
xySpark input window
The program xySpark is started by selecting it from the Plugins menu within ImageJ (Fig. 2 A). Fig. 2 B shows the Input window, which is presented first upon running xySpark. This allows one to input calibration information about the image stack, including pixel size and the frame interval. It is also necessary to select two normalizing images (lacking sparks), which are used to divide all other frames, and produce the F/F0 image stack. The center of mass tolerance allows for the fact that when a spark appears in successive frames, the calculated center of mass is unlikely to have precisely the same coordinates, and avoids incorrect detection as a new event. The value is set to ±10 pixels by default.
Figure 2.
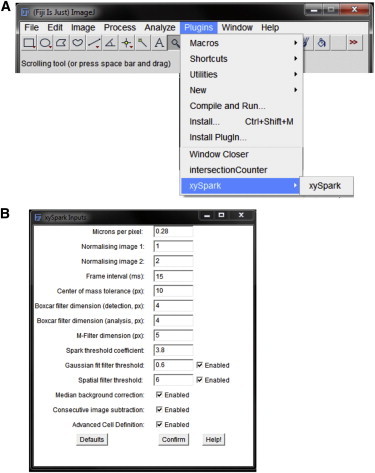
Plugin selection and inputs. (A) xySpark is available via the ImageJ Plugins menu. (B) The Inputs window allows entry of the pixel size, the frame interval, and index numbers of two normalizing frames selected by the user. The user can enter the boxcar filter and M filter dimensions that are applied to the F/F0 image stack before detection, and the dimension of the boxcar filter that is applied before analysis. Events can be excluded based on the r2 value of the Gaussian fit to a line profile passing through the center of mass of each spark at maximum amplitude, or the FWHM (microns) of the spark at maximum amplitude (Spatial filter threshold). It is also possible to select methods for correction of changes in the F/F0 images over time. The Confirm button enters the values into xySpark and begins the next stage. Default values can also be loaded if required. To see this figure in color, go online.
The algorithm is designed such that image processing of the F/F0 stack for optimal detection of Ca2+ sparks is distinct from that used during analysis. Hence, there are dimension settings (in pixels) for the boxcar and M filter used to filter the F/F0 image stack before spark detection, and a separate boxcar filter is used to process regions identified as Ca2+ sparks before analysis.
The Spatial filter allows events greater than a specified FWHM to be excluded. A Gaussian fit filter, based on the r2 value of the curve fitted to a line profile passing through the center of mass of each identified event, is also provided as an option. This was implemented because it was found that regions that were occasionally misidentified as Ca2+ sparks often had highly irregular, non-Gaussian line profiles. The default value of 0.6 (where a perfect fit = 1) only excludes detected events that are very poor fits. The user-defined coefficient ε dictates the threshold for spark detection and in normal use is typically in the range of 3.4–3.8. If necessary, it is also possible to select methods to correct small systematic changes in background fluorescence over time. Finally, the Advanced Cell Definition option implements an interactive algorithm that enables accurate definition of the region of the image occupied by the cell (below). The Confirm button is used to confirm any changes and initiate the next stage. Any changes made by the user are reloaded when xySpark is next run, although default values can also be loaded if required (Defaults button).
Advanced cell definition
A simple approach for defining the region of the image occupied by the cell is to threshold the fluorescence image using the median pixel value, i.e., to set all values greater than the median to 1 (white) and all values below the median to 0 (black) (16). This produces a speckled binary image, with the white pixels being most densely clustered within the cell boundary, where the raw fluorescence is highest. A live-or-die algorithm can then be applied to create a contiguous region of white pixels that defines the cell. However, in this study we found this method to be unreliable for use with cardiac myocytes (see Discussion), and therefore implemented a more accurate two-stage interactive method of defining the cell boundary.
A stack of 20 candidate binary images is first created by applying a threshold = min pixel value + τ (max-min), where τ = 0–1, with increments of 0.05. Values ≥ threshold are set to 1 (white) and those < threshold are set to 0 (black). A live-or-die filter is then applied in 7 × 7 pixel blocks such that if the number of white pixels is ≥12, then all pixels are set to 1 (otherwise, 0). The binary image then has 1 added to each pixel value before multiplication by the original fluorescence image. The effect of this final step is to produce an image in which regions above the threshold are 2× greater (and visibly brighter), whereas all other regions are unaffected (Fig. 3 A).
Figure 3.
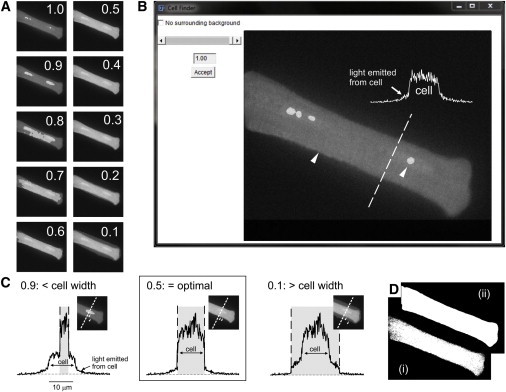
Advanced cell-definition algorithm. (A) Twenty threshold (τ) values are applied to the first image in the raw fluorescence stack (τ = min pixel value + τ (max-min), where τ = 1–0.05, increments of 0.05), followed by a live-or-die filter to produce a binary image where regions above threshold = 1.1 are then added to each pixel before multiplying by the raw fluorescence image to produce 20 candidate images, with the region above threshold highlighted (10 shown). (B) The interactive Cell Finder window allows the user to scroll through the 20 candidate images and select the value of τ that best defines the region occupied by the cell. The arrow on the left indicates the cell boundary, and the arrow on the right indicates the bright region indicating the pixels above threshold at τ =1.0. The line profile (above) from pixels positioned along the broken line shows that beyond the edge of the cell, there is a slow decline in fluorescence due to emitted light. (C) Images at τ = 0.9, 0.5, and 0.1 (insets) and line profiles (below) taken from the regions indicated. The line profiles show that as τ is decreased from 0.9 to 0.5, the region above threshold increases until it aligns with the edge of the cell. At τ = 0.1, the region above threshold extends beyond the cell boundary. (D) With τ = 0.5 having been selected as the optimal value, the image is rethresholded, producing a speckled binary representation of the cell (i). This is followed by application of a live-or-die filter with a strict exclusion criterion (≥30 pixels of 7 × 7 = 1; otherwise, 0), producing a uniform binary cell mask that accurately defines the region occupied by the cell (ii). Because it is sometimes desirable to obtain confocal images from ROIs entirely within a cell, there is also a check box to indicate no surrounding background, in which case the cell-definition algorithm is not applied. To see this figure in color, go online.
This stack of modified fluorescence images is presented in the Cell Finder window, which allows the user to move sequentially through the image stack and visualize expansion of the thresholded region across the cell as τ decreases from 1 to 0.05 (Fig. 3 B). Selected line profiles with τ = 0.9, 0.5, and 0.1 show the elevated (i.e., brighter) regions of the original image that are above the threshold, and how this relates to the cell boundary and the halo of diffuse fluorescence surrounding the cell (Fig. 3 C). The user selects (Accept button) the frame with the highlighted region that corresponds most accurately to the cell. In the second stage, this modified image is rethresholded using min+0.5(max-min) and the resulting speckled binary image (D(i)), filtered using the same live-or-die algorithm, but with the pixel threshold increased from 12 to 30 per 7 × 7 block, which effectively excludes surrounding noise. This two-stage process produces a binary cell mask (D(ii)) that tightly defines the region of the image occupied by the cell.
Image processing and spark detection
The main steps during image processing and detection of Ca2+ sparks are shown in Fig. 4. Each raw fluorescence frame (Fig. 4 A) is divided by a normalizing image lacking Ca2+ sparks (not shown) to produce an F/F0 image (Fig. 4 B). Division by the normalizing image removes large variations in background fluorescence and sets the mean pixel value to 1, both inside and outside the cell. In Fig. 4 B, the increased SD immediately beyond the cell boundary (see Fig. 1 D) appears as a fringe of increased noise.
In this example, a single spark is apparent close to the bottom edge of the cell. The line profile (white trace), which passes horizontally through the spark center of mass, shows that the amplitude is initially small relative to the surrounding noise (Fig. 4 B). However, the image is then filtered, first with a modified 5 × 5 median filter (M filter; Fig. 4 C) and then with a 4 × 4 boxcar filter (Fig. 4 D). The M filter sets the center pixel to the median value if it is 1.5× the interquartile range above or below the median. This removes outliers, but otherwise has no effect on the image.
The combined effect of both filters is to markedly improve the ability to resolve the Ca2+ spark against the background noise (Fig. 4 D). One risk of applying the boxcar filter is that the noise just beyond the edge of the cell is amalgamated into larger contiguous regions, which might then be wrongly identified as a spark. However, this issue can be minimized by multiplying the filtered image by the binary cell mask (see Fig. 3 D), which zeroes all values outside the cell boundary (Fig. 4 E). The user-defined threshold is used to create a binary image, which in this case shows the single Ca2+ spark, correctly identified (Fig. 4 F). A live-or-die filter is then applied (to remove any remaining pixel noise above threshold and set all values within the spark boundary to 1) before multiplying by the F/F0 image (Fig. 4 G). Multiplication by the F/F0 image has the effect of filling regions identified in the binary image as a spark with the corresponding pixel values. An algorithm is then used to find the regions identified as Ca2+ sparks in each image and to calculate the coordinates of the center of mass, indicated with a cross (Fig. 4 H).
The coordinates of the center of mass are used to position a horizontal line profile, which is fitted with a Gaussian function, allowing calculation of the amplitude (ΔF/F0) and FWHM for each event. The data points comprising each line profile, with the corresponding Gaussian curve superimposed, are written to an image stack. This stack initially includes all instances of each spark, i.e., when an event is detected rising and then falling in successive frames. However, for most of the final measurements, the values of interest are those taken from the frame containing the spark at its maximum amplitude. Therefore, the center of mass is used to identify and then exclude submaximal instances of the same event in other frames. At this point, any other user-selected exclusion filters (e.g., the spatial filter) are also applied.
Compensation for changes in background cell fluorescence during data collection
During data collection, small time-dependent decreases (e.g., due to bleaching) or increases (e.g., due to changes in bound dye properties (20)) in cell fluorescence can occur. When present, these changes in raw fluorescence have corresponding effects on the F/F0 stack, which ultimately compromise detection of sparks by thresholding. In this study, we implemented two methods to compensate for changes in background fluorescence. In Fig. 5 A, synthesized data were used to model a rise in F/F0 during a single Ca2+ spark (upper). For illustrative purposes, a very large and rapid increase in the mean pixel value from 1 to 1.1 is shown. The first method involves subtraction of each preceding frame in the F/F0 stack (middle), which corrects effectively the upward drift in F/F0. It also shifts the mean background pixel value to zero, which is reversed by adding 1 to all pixels (not shown). Unfortunately, consecutive image subtraction (CIS) has a significant limitation because the amplitude and other spark parameters actually represent the difference between the amplitudes of the spark detected in successive images. Hence, measurements on images subjected to CIS will underestimate spark amplitude in a manner that depends upon the frame rate. The second method (lower) involves subtraction of the difference between the median pixel value within the cell in the first image and the median pixel value in the nth image (Δ median subtraction: ΔMS). The median is used because it is less affected than the mean by changes in spark frequency.
Figure 5.
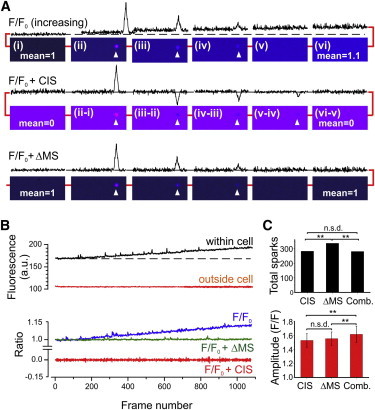
Compensation for changes in background fluorescence. (A) Synthesized data showing sequential (left to right) F/F0 frames containing a single spark event (arrowheads), where the background florescence increases progressively from a mean of 1 to 1.1 (upper). For each frame, a line profile passing through the peak of the spark is shown above. The effect of CIS (middle) or ΔMS (lower) is also shown. (B) Original data obtained by taking the mean of ROIs (20 × 20 pixels) within 1000 sequential confocal images. The background fluorescence increased progressively inside, but not outside, the cell. The progressive increase in fluorescence results in a corresponding increase in F/F0, which can be corrected by using ΔMS or CIS. (C) Mean data from myocytes exhibiting a marked rise in baseline fluorescence, comparing the total number of detected sparks (upper) and the mean speak amplitude (lower) after implementation of CIS, ΔMS, or a combined method (Comb.) in which CIS is used during spark detection and ΔMS is used during analysis (n = 4, ∗∗p < 0.05). To see this figure in color, go online.
Fig. 5 B shows a pronounced time-dependent change in mean fluorescence obtained from a region of interest (ROI) within a myocyte, and the absence of any change distant from the cell. Also shown is the effect of this increase in raw fluorescence on F/F0, and correction of the increase by (i) ΔMS or (ii) CIS. Although this confirms, along with original fluorescence data, that both methods can compensate effectively for a rise in F/F0, their performance during spark detection differed. Fig. 5 C shows the number of detected sparks (upper) and the mean spark amplitude (lower) obtained from four myocytes selected because they exhibited a marked change in background fluorescence. The ΔMS method resulted in detection of more (∼17%) events compared with CIS. Inspection of the raw data revealed that ΔMS was associated with detection of slightly more false-positive events, late in the image sequence (not shown). This is probably because with CIS, subtraction of the preceding image compensates effectively for changes in the pattern of cell background fluorescence over time compared with ΔMS, which simply involves the subtraction of a single value. The detection of small-amplitude false positives may also explain why the mean amplitude of events detected using CIS was not significantly smaller than that detected with ΔMS, which would be expected given that after CIS, the amplitude is the difference between the spark amplitude detected in successive frames.
Based on these findings, xySpark includes an option that enables the combined use of ΔMS and CIS when analyzing data with a significant change in background fluorescence. Specifically, in combined mode, CIS is used only during spark detection and ΔMS is then used to correct background pixel values during analysis, i.e., in a copy of the F/F0 image stack that has not been subjected to CIS. This provides optimal detection of Ca2+ sparks using CIS, and determination of spark amplitude and other parameters using ΔMS correction. As expected, the spark amplitude obtained using the combined method was significantly greater than that obtained with CIS or ΔMS alone (Fig. 5 C).
Analysis and outputs
The main interactive output window shows the original unfiltered fluorescence image stack (Fig. 6 A). The user can advance through the stack image by image. In each image, regions identified as spark peaks are highlighted with a square frame (scaled depending on pixel size) and the corresponding region is shown expanded in the upper right of the window. Where multiple spark peaks occur within a single frame, the box can be advanced sequentially to each event. The amplitude, FWHM, and coordinates of each event are provided at the bottom of the window. As the user moves from one spark to the next, the amplitude, FWHM, and coordinates update automatically.
Figure 6.
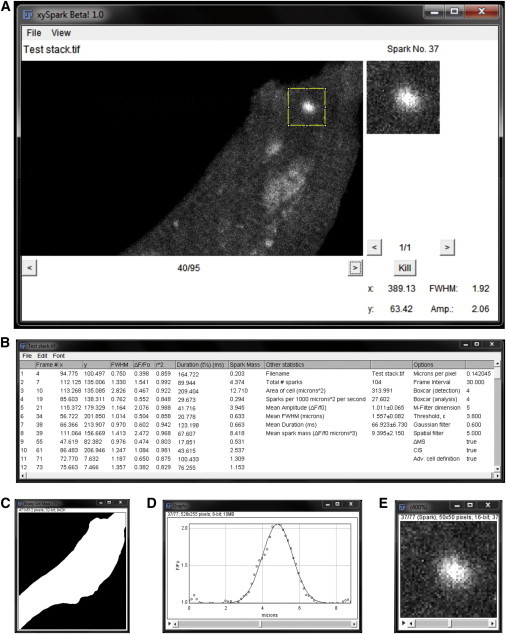
Analysis and outputs. (A) The main output window shows the raw fluorescence image (left) and allows the user to advance through the stack or to jump to a specified frame (option under the File menu). Detected sparks are highlighted with a bounding box in the main image and are also shown on an expanded scale (top right). The coordinates, amplitude, width, and duration for the selected spark are given at the bottom. The Kill button allows individual events to be excluded from the results. (B) The output table (selected via View). (C) The binary cell mask produced via the advanced cell-definition algorithm. (D) Gaussian curves fitted to a line profile passing through the center of mass of each spark (at its maximum amplitude) are available as a stack. (E) A stack containing all of the detected sparks at maximum amplitude is also available (selected via the View menu; see A). To see this figure in color, go online.
The outputs are available in tabular form (Fig. 6 B) and include the coordinates of each spark at its maximum value, the frame number, maximum amplitude (ΔF/F0), FWHM, duration of the descending phase (t1/2), r2 value of each Gaussian fit and summary statistics (cell area, mean ± SE of FWHM, amplitude, width, t1/2 of the descending phase, and event frequency, expressed as sparks/1000 μm2/s). The table can be saved in Excel (.xls) format or copied directly to a spreadsheet for further analysis. Other outputs available via the View menu include the binary cell mask (Fig. 6 C), the Gaussian curve fitted to a line profile passing thorough the center of mass of each spark (Fig. 6 D), and an image stack containing all of the excised sparks, aligned to the center of mass (Fig. 6 E). If an obvious error, e.g., due to a wave, is identified by the user, the Kill button can be used to remove the event from the outputs and the table will then update automatically. The File menu includes options to jump to a specific frame and the ability to save all output files to a user-specified directory, each labeled automatically with the original file name.
Correction of spark amplitudes for noncell background fluorescence
The pixel values outside the cell are typically >0 (e.g., see Fig. 1). Failure to take this into consideration leads to a slight underestimation of spark amplitude, and xySpark includes an option to apply a correction. This involves multiplying the amplitudes (ΔF/F0) determined after running xySpark on the raw images by a correction factor = cell fluorescence/(cell fluorescence-background fluorescence). The background fluorescence (measured using ImageJ before running xySpark) is entered by the user after the initial analysis (via File/Background correction), and xySpark then uses the mean cell fluorescence to calculate the correction factor. The amplitude values in the output table are automatically updated.
Performance of xySpark assessed using synthesized data
To allow comparison with commonly used software for detection and analysis of sparks in line-scan images (21), was evaluated the performance of xySpark using synthesized images containing a cell (mean cell background = 38) and sparks of varying amplitudes, embedded in Gaussian noise (σ = 19, 12.6, or 9.5), as a function of SNR and threshold (Fig. S2). The advantage of using synthesized data is that it is possible to determine with certainty whether an event has been detected correctly at each SNR and amplitude. These data suggest that at SNR = 4, which is typical of that obtained in our confocal images of myocytes, setting ε = 3.4–4.0 provides optimal sensitivity (Fig. S2, upper) and precision (Fig. S2, lower).
Performance of xySpark assessed using confocal image stacks
We analyzed confocal image stacks obtained from fluo-4-loaded rat ventricular myocytes using xySpark. Mean data showing the amplitude, width, and duration of the descending phase (t1/2) at ε = 3, 3.4, 3.8, or 4.2 are shown in Fig. 7 A.
Figure 7.
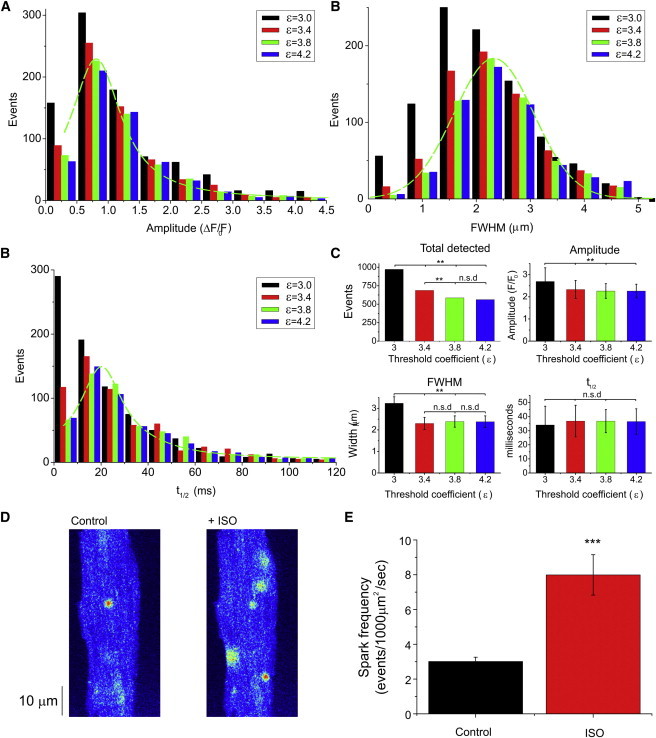
Ca2+ spark data obtained using xySpark. Accumulated data were obtained using xySpark to analyze confocal image stacks obtained from rat ventricular myocytes loaded with fluo-4. (A) Mean data showing the total number of detected events, amplitude, width, and duration (t1/2) at ε = 3, 3.4, 3.8, or 4.2. Also given are histograms of spark amplitude (B), FWHM (C), and the half time (t1/2) of the descending phase (D) at ε = 3, 3.4, 3.8, or 4.2. For clarity, curves (B and D: Poisson; C: Gaussian) are fitted only to data obtained at ε = 3.8 (dashed line). ∗∗p < 0.05, n = 11, n.s.d. indicates no significant difference. Also shown are original (E) and mean (F) data from a second group of cells, showing the effect of 200 nM ISO on spark frequency. ∗∗∗p < 0.01, n = 10. To see this figure in color, go online.
As expected, the total number of detected events increased as ε decreased from 4.2. to 3.0 (Fig. 7 A, upper left). The mean amplitude also decreased significantly between ε = 4.2 and 3.0, and the width decreased significantly as ε was reduced from 3.8 to 3.0 (Fig. 7 A, upper right and lower left). The variation in the t1/2 of the descending phase was larger than the other parameters and the mean values at ε = 4.2–3.0 were not significantly different. However, histograms of the data provide further information about the effect of changing ε. Histograms of the amplitude (Fig. 7 B) and duration (Fig. 7 D) data were fitted with Poisson curves skewed to the left, and FWHM was approximated to a Gaussian distribution (Fig. 7 C). At ε = 3 and to a lesser extent 3.4, the width data diverge markedly from the Gaussian distribution due to an increase in the number of detected events with very small widths. At ε = 3, similar divergence from a Gaussian distribution is apparent in the duration data and to some extent the amplitude data. This is consistent with an increase in the number of small-amplitude events as ε approaches 3, which would be expected to include an increased number of false positives (Fig. S2). Visual examination of individual detected events, the corresponding line profiles, and Gaussian fits (using the Output window) confirmed that ε = 3.4–3.8 produces a good balance of sensitivity and accuracy under the conditions of this study.
Fig. 7, E and F, show original and accumulated data that demonstrate changes in spark frequency detected using xySpark. In this example, spark frequency was increased on average ∼2.7-fold after the introduction of 200 nM isoproterenol (ISO). In the output table, spark frequency (expressed as sparks/1000 μm2/s) is calculated from the total number of detected sparks, the area of the cell derived from the binary cell mask (Fig. 1), and the frame rate.
Discussion
This study describes the development of software to automate detection and analysis of Ca2+ sparks within x-y confocal image stacks. Our assessment of the performance of xySpark using synthetic images (Fig. S2) suggests that its performance is comparable to that of the most commonly used algorithm for spark detection in line-scan images (21). Furthermore, analysis of Ca2+ sparks in confocal image stacks obtained from fluo-4-loaded myocytes produced histogram distributions and mean values for amplitude, FWHM, and t1/2 (Fig. 7) that are broadly consistent with previous studies that used line-scan imaging. Unlike previously described algorithms for spark detection in x-y image stacks, xySpark is implemented in the ImageJ open-source environment and includes a configurable interactive user interface.
Challenges associated with the use of thresholding to detect sparks in x-y image stacks
In common with most methods that employ automatic spark detection in line-scan images, xySpark identifies Ca2+ sparks by applying a threshold to the region of the image occupied by the cell. However, in this study we found that the application of thresholding to identify Ca2+ sparks in x-y image stacks of cardiac cells required new approaches to overcome limitations inherent in previously described methods.
In previous work on skeletal muscle, the cell was identified by thresholding the normalizing image using the median pixel value (16). In this study, identification of myocytes often failed completely using this method because the median was dominated by pixel values outside the cell (not shown). This reflects the fact that cardiac myocytes are markedly smaller than skeletal muscle cells and therefore the surrounding pixels occupy a larger and variable proportion of the image. However, even when the cell was identified correctly, the boundary was not tightly defined, leading to inclusion of surrounding pixel noise (Fig. 4). Overestimation of the cell boundary undermines accurate spark detection due to incorrect calculation of the SD of the background noise within the cell, with consequent effects on θ, and due to false identification of noise outside the cell boundary as Ca2+ sparks. We addressed this by implementing a two-stage interactive algorithm that produces a binary cell mask that accurately defines the cell boundary and excludes surrounding noise (Fig. 3).
Another difficulty with the use of thresholding to detect Ca2+ sparks is that small changes in the background fluorescence within the cell can occur during data collection (20). This compromises reliable spark detection because a constant threshold is being applied to a changing mean background. In the Sparkmaster ImageJ plugin for analysis of line-scan images, it is possible to divide the line-scan image into a number of sections, thereby limiting the influence of a progressive change in baseline (21). However, adaptation of this method for use with xySpark would require the user to identify many normalizing images (lacking sparks) at intervals throughout the stack. Instead, the implemented solution takes advantage of the fact that xySpark treats detection and analysis of sparks as two separate processes. CIS provides an effective method to correct for changes in the mean background fluorescence and any changes in the pattern of background fluorescence, allowing spark peaks to be detected reliably, even late in the image sequence. However, after CIS, the amplitude of each event is actually the difference between the true amplitudes as detected in successive images. Therefore, during analysis, ΔMS is applied to the original F/F0 stack to correct any changes in background fluorescence. This combined approach provides an acceptable balance between accurate detection and subsequent analysis of individual spark properties against a changing background.
Relation to previous studies
Previous studies employed a version of the conventional thresholding algorithm for spark detection in x-y images of skeletal muscle cells (15) and, more recently, cardiac myocytes (14). Although the performance of this algorithm on synthetic sparks at variable SNRs does not appear to have been published, we would expect the algorithm to perform similarly to xySpark in terms of sensitivity and accuracy of detection. However, the ability to separate and fine-tune the filtering characteristics for detection and analysis within xySpark may provide performance benefits. In addition, the algorithm described by Brum and colleagues (15) does not appear to include measurement of spark duration, spatial filters, or automatic correction of changes in background fluorescence, and the program operates only within IDL.
Another study on skeletal muscle also described a modified thresholding algorithm for spark detection in x-y images, implemented with IDL (16). This included the use of thresholding to identify the cell and CIS to compensate for changes in background fluorescence. However, the analysis of spark properties was carried out on images subjected to CIS, which can provide useful data regarding spark frequency and location, but not amplitude or duration. Although xySpark includes an option to use CIS, this is only employed during the spark detection phase, and amplitude and width are measured from the unsubtracted images. In addition, xySpark provides an improved interactive method to define the cell within the image, as well as spatial filtering methods, which help to exclude false-positive events.
Alternative approaches to detect Ca2+ sparks have also been described; for example, methods incorporating the use of wavelet transforms (time-frequency signal decomposition methods) have been described for line-scan imaging (22,23). In addition, a recent study evaluated the performance of a matched-filter detection algorithm, which involves defining a stereotypical Ca2+ spark before searching for instances of the object within the data (18). Both of these approaches may offer improved performance when the SNR is particularly low. However, when the SNR is >2 (which is typical for our x-y confocal data), the improvement appears marginal. The method described by Bankhead and colleagues (23) can, however, be used in situations where the background fluorescence is changing.
Appropriate use of the algorithm and known limitations
Although the xySpark algorithm includes options to allow spark detection despite changes in background fluorescence, this may not always be appropriate, such as when the changes are large, of unknown origin, or due to changes in cytosolic [Ca2+]. Optimal dye loading and decreased laser illumination can reduce changes in background fluorescence (20), which is preferable to image correction. In addition, given the large file sizes inherent in x-y imaging, it may be more practical to acquire a number of shorter image sequences during an experimental protocol, and the reduced laser exposure will also limit any changes in background fluorescence.
The xySpark algorithm has a number of limitations. For example, accurate spark detection is reliant upon the binary cell mask, which is determined by analyzing the first normalizing image in the sequence (Fig. 3). If the cell moves during data collection, false-positive events may be detected outside cell boundary if the mask is unaligned with the cell. Waves must be absent from the image sequence, or at least removed before analysis. Use of the Spatial filter to exclude events with FWHM ≤ 6 μm (Fig. 2) is not recommended because it will artificially attenuate the width distribution (Fig. 7). Similarly, use of the Kill button (Fig. 6) to exclude events could introduce user bias. However, it is justifiable to exclude events that do not comply with predefined criteria, e.g., if examination of detected events in the output window reveals errors caused by propagation of a Ca2+ spark to form a local wave. Occasionally, the performance of the advanced cell-definition algorithm was impaired by the presence of small, very bright regions (e.g., membrane vesicles containing dye) outside the cell boundary, which influenced thresholding. However, given the interactive nature of the cell-definition window (Fig. 3), this is immediately apparent and can be rectified by excluding the object from the original data using built-in ImageJ functions (i.e., encircle the object using the freehand tool and then use Subtract to zero the pixel values).
Another potential limitation is the maximum frame rate attainable during x-y confocal imaging. Measurements obtained by running xySpark on stacks containing synthetic sparks were used to quantify the effects of reduced frame interval (Fig. S3). As expected, there is a progressive underestimation of spark amplitude with increasing frame interval ranging from 2.54 ± 2.7% (n = 50) at a frame interval of 4 ms to 22.1 ± 2.9% (n = 50) at a frame interval of 32 ms. There was no significant effect on mean width, whereas duration increased slightly (by 6.4 ± 2.9%, n = 50) at a frame interval of 32 ms. Typically, line-scan imaging is carried out with a temporal resolution of 1–2 ms. However, some x-y imaging systems are already capable of scanning with frame intervals of <5 ms, where errors relating to spark amplitude are minimal (14). In addition, one of the strengths of x-y imaging is that data can be collected from the entire cell and local signals related to structural components (e.g., organelles, membrane structures, channels, and transporters). If the location and frequency of events are the primary consideration, then a modest frame interval of 30 ms should allow all in-focus sparks to be detected.
Conclusions
We have demonstrated that xySpark can reliably detect and analyze Ca2+ sparks in x-y image stacks. Implementation as a plugin for ImageJ will allow users to apply xySpark within a powerful cross-platform, open-source image analysis environment, complemented by numerous existing analysis functions and plugins. One likely benefit of this approach is that accurate determination of Ca2+ spark coordinates and properties can readily be combined with two-dimensional structural information to advance our understanding of Ca2+ signaling microenvironments. Although the performance of xySpark has been tested in cardiac myocytes, the program should also be of value to researchers studying localized Ca2+ release events in other tissues.
Acknowledgments
The authors thank Dr. Zhaokang Yang and Dr. Hannah Kirton for providing isolated myocytes and for advice during development of xySpark.
This work was supported by the British Heart Foundation and the Wellcome Trust. E.S. began working on xySpark as a work experience student from Harrogate Grammar School, North Yorkshire.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting Material
References
- 1.Cheng H., Lederer W.J. Calcium sparks. Physiol. Rev. 2008;88:1491–1545. doi: 10.1152/physrev.00030.2007. [DOI] [PubMed] [Google Scholar]
- 2.Cheng H., Lederer W.J., Cannell M.B. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 1993;262:740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 3.Kockskämper J., Sheehan K.A., Blatter L.A. Activation and propagation of Ca(2+) release during excitation-contraction coupling in atrial myocytes. Biophys. J. 2001;81:2590–2605. doi: 10.1016/S0006-3495(01)75903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shacklock P.S., Wier W.G., Balke C.W. Local Ca2+ transients (Ca2+ sparks) originate at transverse tubules in rat heart cells. J. Physiol. 1995;487:601–608. doi: 10.1113/jphysiol.1995.sp020903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng H., Cannell M.B., Lederer W.J. Partial inhibition of Ca2+ current by methoxyverapamil (D600) reveals spatial nonuniformities in [Ca2+]i during excitation-contraction coupling in cardiac myocytes. Circ. Res. 1995;76:236–241. doi: 10.1161/01.res.76.2.236. [DOI] [PubMed] [Google Scholar]
- 6.Balke C.W., Shorofsky S.R. Alterations in calcium handling in cardiac hypertrophy and heart failure. Cardiovasc. Res. 1998;37:290–299. doi: 10.1016/s0008-6363(97)00272-1. [DOI] [PubMed] [Google Scholar]
- 7.Lindner M., Brandt M.C., Beuckelmann D.J. Calcium sparks in human ventricular cardiomyocytes from patients with terminal heart failure. Cell Calcium. 2002;31:175–182. doi: 10.1054/ceca.2002.0272. [DOI] [PubMed] [Google Scholar]
- 8.Ward C.W., Reiken S., Lacampagne A. Defects in ryanodine receptor calcium release in skeletal muscle from post-myocardial infarct rats. FASEB J. 2003;17:1517–1519. doi: 10.1096/fj.02-1083fje. [DOI] [PubMed] [Google Scholar]
- 9.Kubalova Z., Terentyev D., Györke S. Abnormal intrastore calcium signaling in chronic heart failure. Proc. Natl. Acad. Sci. USA. 2005;102:14104–14109. doi: 10.1073/pnas.0504298102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voigt N., Li N., Dobrev D. Enhanced sarcoplasmic reticulum Ca2+ leak and increased Na+-Ca2+ exchanger function underlie delayed afterdepolarizations in patients with chronic atrial fibrillation. Circulation. 2012;125:2059–2070. doi: 10.1161/CIRCULATIONAHA.111.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Weisleder N., Ma J. Uncontrolled calcium sparks act as a dystrophic signal for mammalian skeletal muscle. Nat. Cell Biol. 2005;7:525–530. doi: 10.1038/ncb1254. [DOI] [PubMed] [Google Scholar]
- 12.Hilliard F.A., Steele D.S., Knollmann B.C. Flecainide inhibits arrhythmogenic Ca2+ waves by open state block of ryanodine receptor Ca2+ release channels and reduction of Ca2+ spark mass. J. Mol. Cell. Cardiol. 2010;48:293–301. doi: 10.1016/j.yjmcc.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H., Song L.S., Stern M.D. Amplitude distribution of calcium sparks in confocal images: theory and studies with an automatic detection method. Biophys. J. 1999;76:606–617. doi: 10.1016/S0006-3495(99)77229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shkryl V.M., Blatter L.A., Ríos E. Properties of Ca2+ sparks revealed by four-dimensional confocal imaging of cardiac muscle. J. Gen. Physiol. 2012;139:189–207. doi: 10.1085/jgp.201110709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brum G., González A., Ríos E. Fast imaging in two dimensions resolves extensive sources of Ca2+ sparks in frog skeletal muscle. J. Physiol. 2000;528:419–433. doi: 10.1111/j.1469-7793.2000.00419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bányász T., Chen-Izu Y., Izu L.T. A new approach to the detection and statistical classification of Ca2+ sparks. Biophys. J. 2007;92:4458–4465. doi: 10.1529/biophysj.106.103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bray M.A., Geisse N.A., Parker K.K. Multidimensional detection and analysis of Ca2+ sparks in cardiac myocytes. Biophys. J. 2007;92:4433–4443. doi: 10.1529/biophysj.106.089359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong C.H., Soeller C., Cannell M.B. Increasing sensitivity of Ca2+ spark detection in noisy images by application of a matched-filter object detection algorithm. Biophys. J. 2008;95:6016–6024. doi: 10.1529/biophysj.108.135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan D.J., Yang Z., Harrison S.M. TNF-α and IL-1β increase Ca2+ leak from the sarcoplasmic reticulum and susceptibility to arrhythmia in rat ventricular myocytes. Cell Calcium. 2010;47:378–386. doi: 10.1016/j.ceca.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koester H.J., Baur D., Hell S.W. Ca2+ fluorescence imaging with pico- and femtosecond two-photon excitation: signal and photodamage. Biophys. J. 1999;77:2226–2236. doi: 10.1016/S0006-3495(99)77063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picht E., Zima A.V., Bers D.M. SparkMaster: automated calcium spark analysis with ImageJ. Am. J. Physiol. Cell Physiol. 2007;293:C1073–C1081. doi: 10.1152/ajpcell.00586.2006. [DOI] [PubMed] [Google Scholar]
- 22.Szabó L.Z., Vincze J., Szentesi P. Improved spark and ember detection using stationary wavelet transforms. J. Theor. Biol. 2010;264:1279–1292. doi: 10.1016/j.jtbi.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Bankhead P., Scholfield C.N., McGeown J.G. Detecting Ca2+ sparks on stationary and varying baselines. Am. J. Physiol. Cell Physiol. 2011;301:C717–C728. doi: 10.1152/ajpcell.00032.2011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


