Abstract
NOTCH1 mutations have been reported to occur in 10 to 15% of head and neck squamous cell carcinomas (HNSCC). To determine the significance of these mutations, we embarked upon a comprehensive study of NOTCH signaling in a cohort of 44 HNSCC tumors and 25 normal mucosal samples through a set of expression, copy number, methylation and mutation analyses. Copy number increases were identified in NOTCH pathway genes including the NOTCH ligand JAG1. Gene set analysis defined a differential expression of the NOTCH signaling pathway in HNSCC relative to normal tissues. Analysis of individual pathway-related genes revealed overexpression of ligands JAG1 and JAG2 and receptor NOTCH3. In 32% of the HNSCC examined, activation of the downstream NOTCH effectors HES1/HEY1 was documented. Notably, exomic sequencing identified 5 novel inactivating NOTCH1 mutations in 4/37 of the tumors analyzed, with none of these tumors exhibiting HES1/HEY1 overexpression. Our results revealed a bimodal pattern of NOTCH pathway alterations in HNSCC, with a smaller subset exhibiting inactivating NOTCH1 receptors mutations but a larger subset exhibiting other NOTCH1 pathway alterations, including increases in expression or gene copy number of the receptor or ligands as well as downstream pathway activation. Our results imply that therapies that target the NOTCH pathway may be more widely suitable for HNSCC treatment than appreciated currently.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a disease with significant morbidity and mortality. More than 50,000 new cases of HNSCC are diagnosed in the United States yearly, with a mortality rate of 12,000 annually. As with lung cancer, this malignancy is also predominantly related to smoking with alcohol as a co-carcinogen, although infection with the human papillomavirus has also been associated with the majority of oropharynx cancers (1). Despite significant progress in therapeutic interventions, including surgery, radiotherapy, and chemotherapy, there have been only modest improvements in survival of patients with HNSCC in the past 30 years.
HNSCC, like other solid tumors, develops through a prolonged multistage process involving the accumulation of genetic and epigenetic alterations. Investigators have uncovered several critical genes and pathways important in the tumorigenesis of HNSCC. These include TP53 (2), CDKN2A(3), Cyclin D1 (4), PIK3CA (5), HRAS, and EGFR (6). However, these molecular alterations do not fully recapitulate the pathogenesis of HNSCC. To gain a comprehensive view of the genetic alteration in HNSCC, Agrawal et al. (7) and Stransky et al. (8) used a high-throughput next-generation sequencing technique to analyze the HNSCC genome. Both groups sequenced the exons of all known human genes in tumor DNA and compared the sequence to that of the corresponding normal DNA from the identical patient. In total, the genomic landscapes of 32 and 74 tumors were examined. Mutations were confirmed in genes that had been previously known to play a role in HNSCC, such as TP53, CDKN2A, PIK3CA, PTEN, and HRAS. Both research groups reported novel mutations in NOTCH1. In both studies, inactivating mutations of NOTCH1 were found in 10 to 15% of the HNSCC tumors, making NOTCH1 the second most frequently mutated gene after TP53. In several tumors, both alleles harbored mutations in NOTCH1. More recently, Pickering et al. have discovered changes in gene copy number and expression for some other NOTCH pathway genes including increased copy number changes in NOTCH receptor ligands JAG1 and JAG2 and in NUMB that were associated with elevated mRNA (9).
NOTCH signaling pathway has been linked to multiple biological functions, including regulation of self-renewal capacity, cell cycle exit, and survival. The pathway is initiated when one cell expressing the appropriate ligand (Jagged or Delta) interacts with another cell expressing a NOTCH receptor (NOTCH1-4). Upon ligand binding, the transmembrane NOTCH receptor is subsequently cleaved by ADAM metalloprotease and γ-secretase complex. The cleaved product, intracellular fragment of NOTCH (NICD), translocates into the nucleus where it interacts with the nuclear DNA-binding factors, CSL/CBF1/RBPjk), and recruits co-activators (MAML proteins) to turn on transcription factors of target genes. The most prominent targets of the NOTCH pathway include a set of basic-helix-loop factors of the Hes and Hey families (10, 11). Several studies suggest that NOTCH mutation can have either an oncogenic or a tumor-suppressive effect. In T cell acute lymphoblastic leukemia/lymphoma, NOTCH signaling had previously implicated as pro-tumorigenic by activating mutations and translocations observed in the genes for NOTCH receptors or their regulators (12, 13); whereas in chronic myelomonocytic leukemia, cutaneous, lung and HNSCC tumors, several of the NOTCH family mutations in HNSCC encode inactivating mutations, suggesting a tumor suppressor function (14–16).
Due to the discovery of NOTCH mutations in HNSCC, a high priority is placed on a more comprehensive understanding of the complex molecular alterations of NOTCH signaling pathway in HNSCC. In this study we examined the comprehensive genetic, epigenetic and transcriptional alterations of the NOTCH signaling pathway in a cohort of primary HNSCC, and report a systematic dysregulation of the NOTCH signaling pathway in HNSCC. Further analysis revealed that the NOTCH signaling pathway was activated in a subset of HNSCC tumors, and this pathway activation was independent of the NOTCH1 mutation status. These findings provide important new insights of NOTCH signaling pathway into the pathogenesis of HNSCC and highlight the NOTCH pathway as a target for therapeutic development.
Methods
A more detailed description of our methods and statistical procedures is available in Supplemental Methods.
Human tissue samples
The tumor tissue samples from patients with HNSCC and normal samples were obtained from patients surgically treated in the Department of Otolaryngology-Head and Neck Surgery at Johns Hopkins Medical Institutions, Baltimore, using appropriate informed consent obtained after institutional review board approval. Microdissection of frozen tumor tissue was done to assure that >80% of tissue contained HNSCC. The normal tissues consisted of tissues obtained from non-cancer affected control patients that underwent uvulopalatopharyngoplasty (UPPP). After review by a pathologist, a section of dissected mucosal layer from discarded UPPP specimens was immediately frozen in liquid nitrogen. All specimens were stored at −80 °C until processing. For microarray analysis, a cohort including 44 HNSCC tumors and 25 normal mucosa tissues was used (Supplementary Table I). For quantitative real-time PCR analysis, in addition to the above cohort used for microarray analysis, two separate cohorts with one comprising of 31 HNSCC tumor tissues and 17 normal mucosa tissues and the other 63 HNSCC tumor tissues and 30 normal mucosa were also used. The application of different cohorts was based on the sample cohort availability.
DNA extraction and RNA isolation
DNA was extracted from all samples by digestion with 50 μg/mL proteinase K (Boehringer, Mannheim, Germany) in the presence of 1% SDS at 48 °C overnight, followed by phenol/chloroform extraction and ethanol precipitation (17). Total RNA was isolated with trizol reagent according to the manufacturer’s instructions, and then purified with an RNeasy Kit (Qiagen, Germantown, MD).
HPV analysis
The HPV status was determined as described previously (18). In brief, specific primers and probes have been designed to amplify the E6, E7 regions of HPV16. Their sequences are available in a previous publication. All the samples were run by quantitative Taqman PCR in duplicate. Primers and probes to a housekeeping gene (β-actin) were run in duplicate and parallel to normalize input DNA. HPV copy number more than 1 copy per cell for tumor samples were regarded as positive.
Copy Number Analysis
The cohort of 44 HNSCC tumor tissues and 25 normal mucosa samples were run on Affymetrix SNP6.0 arrays for copy number analysis. All arrays were run according to the manufacturers’ instructions. DNA processing, preparation, hybridization and chip scanning were performed by the Johns Hopkins Microarray Core Facility. The data was normalized using the crlmm package and genome build HG18 annotations (19). The 25 normal mucosa samples and 44 HNSCC samples were then analyzed using the Tibshirani and Hastie outlier statistical approach (20) and compared to 1.74 million values generated by permutation of sample labels to generate an empirical p-value using the approximation of Smyth and Phipson (21). Outlier samples were defined in the standard way.
Methylation Array Analysis
Bisulfite conversion of genomic DNA from the above cohort was done with the EZ DNA methylation Kit (Zymo Research, D5002) by following manufacturer’s protocol with modifications for Illumina Infinium Methylation Assay in the Johns Hopkins microarray core. We have applied the same statistical analysis used for copy number looking for hypomethylation outliers in NOTCH pathway genes defined in the Kyoto Encyclopedia of Genes and Genomes (22). NOTCH genes with a P value <0.05 were considered significant.
Gene expression profiling
RNA isolated from the cohort of 44 HNSCC tumor tissues and 25 normal mucosa samples were run on Affymetrix HuEx 1.0 GeneChips for expression analysis. All arrays were run according to the manufacturers’ instructions (23–25). To look at the expression of NOTCH pathway genes on a tumor sample basis, we compared the expression level of each gene in each individual tumor to the distribution of expression levels in normal mucosa samples assuming a normal distribution. We declared a gene as significantly differentially expressed if the tumor expression level was 2 σs away from the mean normal expression level, effectively setting a threshold of α = 0.05.
Heat map
Heat maps of differential gene expression were created using the R-package gplots. The visualized data comprised gene by sample expression levels, and expression was normalized to Z-score on a gene-by-gene basis to convert all genes to the same scale.
Gene set analysis
Gene Set Analysis was performed using a mean rank gene set enrichment test (26), as provided in the limma R package (27). Initial gene ranks were generated using an Empirical Bayes statistic. Significance of the NOTCH signaling pathway gene set (KEGG database) or Nguyen_Target_Set (Molecular Signature Database from the Broad Institute) (28) were measured. Recently, Nguyen identified 85 differentially expressed genes as a gene set (Named as Nguyen_NOTCH1_Targets in Molecular Signatures Database of the Broad Institute) of NOTCH downstream targets that are concomitantly modulated by activated NOTCH1 in mouse and human primary keratinocytes (28). With this gene set, we also assessed the expression status of NOTCH signaling pathway in the above cohort of HNSCC tumors.
Exome sequencing of NOTCH1
The exome sequencing of NOTCH1 was performed by Asuragen (Austin, Texas) (29–31). We retained only the high coverage regions for analysis, high coverage being defined by having greater than 10% of the sample median coverage and above 100 reads. We also flagged the loci that are known to have frequent false positive calls in our library. For each set of matched samples, we filtered out variants that were present in the lymphocytes as putative germline variants, or as sample specific systematic error. In our analysis, if the matched normal loci is found to have greater than 1% variant reads, or the variant score difference is within the 99.5% percentile of all pairwise differences for non-annotated loci across the matched pair, the background is considered high and the variant is removed from the list. Variants were annotated using gene structure from the NCBI RefSeq transcript set. Coding base substitutions were classified as missense, nonsense, splice site, or silent. In addition to NOTCH1, we also performed targeted sequencing on 51 genes selected from the COSMIC mutation database; those data will be reported separately.
Quantitative real-time RT-PCR
The same RNA samples used for microarray analysis, the RNA samples from an independent HNSCC cohort as described above, as well as RNA extracted from cell lines were assessed for NOTCH1, HES1 and HEY1 expression levels using quantitative real-time RT-PCR (Taqman). Reverse transcription was performed with random hexamer primers and Superscript II reverse transcriptase (Invitrogen Corp.) as described previously (17). Quantitative RT-PCR was then carried out on the Applied Biosystems 7900 Sequence Detection Instrument (Applied Biosystems, Carlsbad, CA). The primers and probes (Integrated DNA technologies, San Diego, CA, USA) used in the analysis are available upon request. In our analysis, serial dilutions of cDNA product for each gene of interest were used for constructing the calibration curves on each plate.
Immunohistochemistry
56 HNSCC and 11 non-cancer formalin-fixed and paraffin-embedded samples were obtained from the Head and Neck Tissue Bank at Johns Hopkins and were used to construct a tissue microarray under JHU-IRB approved protocols. The protocol incorporated heat-induced antigen retrieval with citrate buffer (pH 6.0) followed by peroxide-blocking step and primary antibody incubation for 15 minutes with rabbit polyclonal antibody against HES1 (Millipore, AB5702, dilution 1: 200), for 30 minutes with rabbit polyclonal antibody against HEY1(Abcam, ab22614, dilution 1: 200) or 30 minutes with goat polyclonal antibody against cleaved/activated NOTCH1 (Santa Cruz Biotechnology, sc-6014, dilution1: 400). The tissues were analyzed using Aperio software. The staining was categorizes as strong, moderate or none/weak for each individual tissue and averaged for tissue quadruplicates.
Cell Culture
Human HNSCC cell lines UPCI-SCC090 (090) and SCC61 were used in experiments. 090 was received from Dr. Susanne Gollin, University of Pittsburgh. SCC61 was received from Dr. Ralph Weichselbaum (University of Chicago). In our study, each cell line was authenticated using a short tandem repeat analysis kit, identifier (Applied Biosystems, Foster City, CA), as directed at the Johns Hopkins Genetic Resources Core Facility. The results were shown in Supplementary Table II. Cell growth conditions were maintained at 37°C in an atmosphere of 5% CO2.
Transient Transfection, NOTCH inhibition and Cell Proliferation Assay
ON-TARGETplus Pool of siRNAs against NOTCH1, HEY1 non-targeting Pool of siRNA (Thermo Scientific) was used to downregulate the expression of NOTCH1, HEY1 or used as control. Cells were seeded in 96-well plates and allowed to grow until the cells were approximately 70% confluent. Cells were transfected with siRNA using Lipofectamine RNAiMAX Reagent (Invitrogen). NOTCH1 was inhibited by Gamma Secretase Inhibitor XXI, Compound E (GSI-XXI, EMD Millipore). Cell metabolic activity was determined every 24 hours using the CCK-8 colorimetric assay (Dojindo). Values are mean ± SEM for pentaplicates of cultured cells. The transfection efficiency was confirmed by qRT-PCR as described above for primary tissues with primers and probes for NOTCH1, or HEY1 and normalized to GAPDH at 48 hours time point.
Analysis of the TCGA HNSCC Data
The TCGA datasets, including DNA copy number datasets from 288 HNSCC tumor tissues, RNA-seq expression datasets from 279 HNSCC tumor tissues and 37 adjacent normal tissues, and NOTCH1 mutation data sets from 281 HNSCC tumor tissues, were obtained from the public access data portal (32). The DNA copy number, RNA expression, and NOTCH1 mutation in HNSCCs were assayed using Affymetrix 6.0 SNP arrays, RNAseq, and Whole genome sequencing respectively. Among these data sets, 279 HNSCC tumor tissues had full DNA copy number, RNA-seq expression, and NOTCH1 mutation data available, and hereby were used in our study.
Statistical Analysis
All RT-PCR analyses utilized an unpaired Student’s t test to determine statistical significance between experimental variables. All statistical tests were two sided without multiple testing correction. A p value threshold of α = 0.05 was used to indicate statistical significance. All computations were done in R 2.15.2.
Results
DNA Copy Number and promoter methylation analysis of NOTCH signaling pathway genes in HNSCC vs normal mucosa
Recently, our laboratory and others have reported that NOTCH1 is a putative tumor suppressor gene mutated and inactivated at a significant frequency (~15%) in HNSCC (7). NOTCH1 encodes a member of NOTCH signaling pathway, a pathway comprised of 47 genes according to the KEGG database annotation (Supplementary Table III). The goal of this study was to explore the comprehensive alterations of NOTCH signaling pathway in HNSCC and their potential involvement in HNSCC development. To achieve this goal, we analyzed the DNA copy number, methylation and expression array data generated from 44 HNSCC tumor tissues and 25 normal mucosa tissues in our laboratory. The microarray data was uploaded to GEO with an accession number “33205”. The demographic and clinical characteristics of this cohort are presented in Supplementary Table I.
We firstly performed DNA copy number array data analysis on 38 of the 47 NOTCH signaling pathway genes from the above cohort, as these genes had data available on Affymetrix SNP 6.0 arrays. Outlier sums were used to identify those NOTCH signaling pathway genes with significant copy number gains, highlighting 8 NOTCH signaling pathway genes, including NOTCH ligand JAG1 (P=0.040), MAML2 (P=0.026), MAML3 (P=0.006), NCOR2 (P=0.003), NUMB (P=0.039), NUMBL (P=0.020), PSEN1 (P=0.010), and NSCTN (P=0.049) (Figure 1A). Our analysis identified a number of samples exhibiting copy number gains in one or more of these 8 genes. These comprised 5, 5, 5, 9, 6, 4, 7, and 6 out of the 44 HNSCC tumors for copy number gains of JAG1, MAML2, MAML3, NCOR2, NUMB, NUMBL, PSEN1, and NSCTC, respectively (Figure 1A and 1B).
Figure 1. Copy number analysis of NOTCH signaling pathway genes in the cohort of 44 HNSCC tumors and 25 normal mucosa.
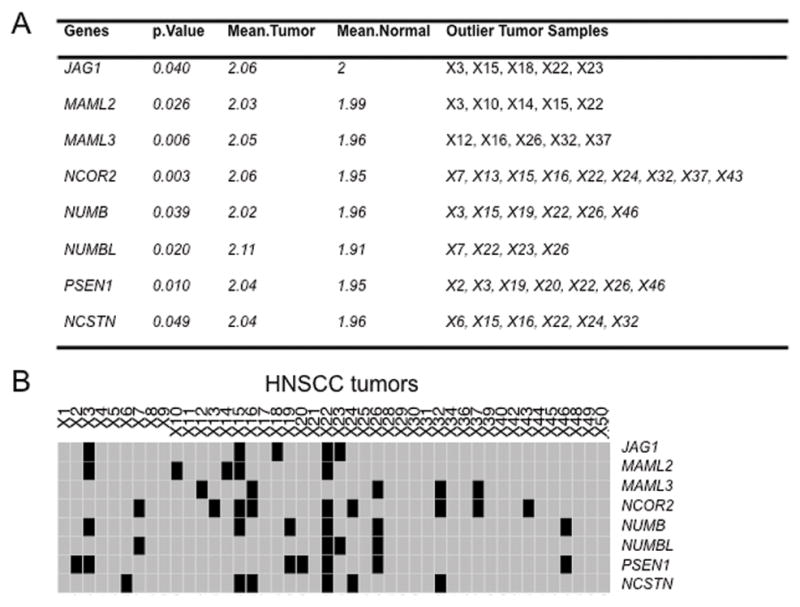
A, NOTCH signaling pathway genes showing significant copy number gains in HNSCC tumor tissues vs normal mucosa. The copy number levels in 44 HNSCC tumors and 25 normal mucosa were analyzed with the Tibshiani and Hastie statistic, and the P-values were generated by permutation of sample labels to generate an empirical P-value using the approximation of Smyth and Phipson as described in methods. P<0.05 was considered significant. B, Copy number status of the 8 NOTCH pathway genes in A) in the 44 HNSCC tumors. Gray denotes no copy-number gain, Black, copy number gain and also the outlier HNSCC tumor for given gene.
We then analyzed the promoter methylation status of NOTCH signaling pathway genes using the DNA methylation array data from the cohort of 44 HNSCC tumor tissues and 25 normal mucosal tissues. Preliminary analysis was conducted using the 12, 023 probes on the arrays that included ≥3 CpG islands probe using empirical Bayes comparisons for all tissue types. Given that we were interested in NOTCH pathway genes that are promoter hypomethylated and overexpressed in HNSCC tumors, the outlier sums were applied in the analysis to identify those NOTCH pathways exhibiting left-tail outliers, indicating promoter hypomethylation in HNSCC tumors in comparison to that in normal mucosa. With this analysis, we found none of the NOTCH pathway genes is significantly promoter hypomethylated in HNSCC tumors versus normal mucosa. However, PSEN2 and PSENEN trended toward promoter hypomethylation with p-values of 0.052 and 0.072 respectively (data not shown).
Gene Expression Analysis of NOTCH signaling pathway genes in HNSCC tumors vs normal mucosa
We then extended our analysis on the transcriptional levels of the NOTCH pathway genes using the expression array data from the same cohort described above. Forty-three of the 47 NOTCH pathway genes illustrated by KEGG database are available on the Affymetrix HuEx 1.0 GeneChip (Supplementary Table III). When the differential expression of these individual genes was analyzed, we found 15 genes showing a significant differential expression (P<0.05). Among the 15 genes, the mRNA levels of JAG1, JAG2, NOTCH3, NCSTN, ADAM17, DTX3L, DVL3, HES1, HDAC2, NCOR2, and NUMBL were significantly higher in primary HNSCC tumors than in normal mucosa whereas the mRNA levels of KAT2B, MAML3, DTX1 and MFNG significantly lower in HNSCC tumors than in normal mucosa (Supplementary Table IV). Of note, JAG1 and JAG2 ligands and NOTCH3 receptor were among the NOTCH pathway genes significantly overexpressed in tumors (JAG1, log2 mean, 9.4; JAG2, log2 mean, 7.9; NOTCH3, log2 mean, 8.3) compared to those in normal mucosa (JAG1, log2 mean, 8.5, P<0.001; JAG2, log2 mean, 7.4, P<0.001; NOTCH3, log2 mean, 8.0, P=0.007) (Figure 2A).
Figure 2. Expression array analysis of NOTCH signaling pathway genes in the cohort of 44 HNSCC tumors and 25 normal mucosa.

A, JAG1, JAG2 and NOTCH3 are overexpressed in HNSCC tumors vs. normal mucosa. Expression of JAG1, JAG2, and NOTCH3 were assessed by Affymetrix HuEx 1.0 GeneChip platform in a cohort of 44 HNSCC tumors and 25 normal tissues. Dots, relative expression levels in different tissue samples. Boxes represent the interquartile range (25th–75th percentile) and horizontal lines inside the boxes indicate median. Whiskers indicate the minimum and maximum values. P value was calculated by using t test. N, normal mucosa; T, tumors. B, Heat map of gene expression of NOTCH signaling pathway and its downstream targets in the cohort of 44 HNSCC tumor tissues and 25 normal tissues. All HNSCC tumor samples are labeled black and normal tissues gray. Genes in heat map are shown in rows; each individual sample is shown in one column. For each gene, the scale bar shows color-coded differential expression from the mean gene expression level of normal tissues in standard deviation units. Red and green indicate over- or under-expression, respectively.
We constructed a heat map depicting the differential expression of the NOTCH Signaling pathway genes in the cohort of 44 HNSCC tumors and 25 normal mucosa (Figure 2B). Importantly, we found 29.5%(13/44), 34.1%(15/44), and 18.2% (8/44) of HNSCC tumors showed overexpression of JAG1, JAG2, and NOTCH3 when compared with the normal mucosa. Of note, in our gene-set analysis, we observed a marginal statistical trend (P=0.067) that NOTCH signaling pathway genes tended to be differentially expressed. We also found that there was significant differential expression of the gene set of Nguyen_NOTCH1_Targets (P<0.001) in the HNSCC tumor cohort, suggesting that NOTCH signaling pathway is dysregulated on a pathway basis in HNSCC. Of note, the value of the Nguyen_NOTCH1_Targets set in evaluation of the NOTCH signaling pathway status in HNSCC tumors may be limited, given that the gene set was identified in primary keratinocytes and that it may differ from the NOTCH downstream targets in HNSCCs.
In addition, we compared the gene expression levels of the eight NOTCH pathway genes (exhibiting copy number gains; Figure 1) among the tumors with increased copy number gains, the tumors without increased copy number gains, and normal tissues of our study cohort. Among these genes, our results showed the significant overexpression of JAG1 (P=0.014) and PSEN1 (P=0.015) in the tumors with increased copy number gains in comparison to that in the normal tissues (Supplementary Figure 1).
NOTCH signaling pathway activation in HNSCC tumors
Given the extensive NOTCH signaling pathway alterations identified in HNSCC tumors and in order to determine the functional status of NOTCH signaling, we next determined whether the NOTCH signaling pathway is activated in HNSCC. It is known that the NOTCH pathway signals through transcriptional activation of targets genes, HES1 and HEY1. We therefore analyzed the mRNA expression of HES1 and HEY1 using expression array data from the above cohort. We found that the mRNA expression of HES1 and HEY1 was significantly higher in HNSCC tumors than (HES1, log2 mean, 8.1; HEY1, log2 mean, 6.8) in normal mucosa (HES1, log2 mean, 7.8, P=0.017; HEY1, log2 mean, 6.7, P<0.001) (Figure 3A). Using the same threshold as defined above to declare a gene was overexpressed in an individual sample, we found that 14% (6/44) and 25% (11/44) of HNSCC tumors showed overexpression of HES1 and HEY1 when compared with the normal mucosa (Figure 3B). In total, we found 31.8%(14/44) of HNSCC tumors showed overexpression of HES1 and/or HEY1. Given the likelihood of low expression status of HES1 and HEY1 as transcription factors in certain tumor samples, we also analyzed their downstream target genes as additional indicators of HES1 or HEY1 overexpression. Based on literature report, genes downregulated by HES1 included NEUROG3 (33), GAA (34), CDKN1B (35), RCOR1 and CFD; genes downregulated by HEY1, GATA4 and GATA6 (36). Our results revealed that all those HNSCC tumors showing downregulation of one or more HES1/HEY1 target genes demonstrate overexpression of HES1 or HEY1. Of note, the mRNA expression of GATA4 and NEUROG3 were found significantly lower in HNSCC tumors with HES1/HEY1 overexpression compared with those HNSCC tumors without HES1/HEY1 overexpression (P=0.008) and compared with normal mucosa (P=0.02), respectively; The mRNA expression of NEUROG3 showed significant downregulation in HNSCC tumors with HES1/HEY1 overexpression compared to those HNSCC tumors without HES1/HEY1 overexpression and showed a trend toward downregulation (P=0.03) when compared with the normal mucosa (P=0.067) (Figure 3C). Taken together, these results suggested that NOTCH signaling pathway is activated in 31.8% of HNSCC tumors.
Figure 3. Microarray Expression analysis of downstream activation of NOTCH signaling pathway in the cohort of 44 HNSCC tumors and 25 normal mucosa.

A, NOTCH downstream targets HES1 and HEY1 are overexpressed in HNSCC tumors. Expression of HES1 and HEY1 were assessed by Affymetrix HuEx 1.0 GeneChip platform in a cohort of 44 HNSCC tumors and 25 normal tissues. N, normal mucosa; T, tumors. Boxes represent the interquartile range (25th–75th percentile) and horizontal lines inside the boxes indicate median. Whiskers indicate the minimum and maximum values. P value was calculated by using t test. B, Heat map expression of downstream target genes (HES1 and HEY1) for NOTCH signaling pathway in the cohort of 44 HNSCC tumor tissues and 25 normal tissues. As proposed by literature, expression of NEUROG3, GAA, CDKN18, RCOR1, and CFD are regulated by HES1; expression of GATA4 and GATA6 are regulated by HEY1. Thus these genes were used to assist the estimation of the HES1/HEY1 expression. All HNSCC tumor samples are labeled black and normal tissues gray. Genes in heat map are shown in rows; each individual sample is shown in one column. For each gene, the scale bar shows color-coded differential expression from the mean gene expression level of normal tissues in standard deviation units. Red and green indicate over- or under-expression, respectively. C, GATA4 and NEUROG3 had decreased expression in HNSCC tumors with HES1/HEY1 overexpression in comparison to those without HES1/HEY1 overexpression. N, normal mucosa; T-HES1/HEY1, HNSCC tumors without HES1/HEY1 overexpression; T+HES1/HEY1, HNSCC tumors with HES1/HEY1 overexpression. Boxes represent the interquartile range (25th–75th percentile) and horizontal lines inside the boxes indicate median. Whiskers indicate the minimum and maximum values. P value was calculated by using t test.
In addition, to compare the pattern and intensity of HES1/HEY1 in tumor versus normal epithelium and correlate these with NOTCH1 at the protein levels, we constructed a tissue microarray comprising 56 primary HNSCC and 11 normal control mucosa, and performed immunohistochemistry analysis using commercially available antibody for HES1, HEY1, as well as activated NOTCH1. Due the availability of the samples, we were able to include 5 HNSCC tumors on the tissue microarrays that originated from the same HNSCC cohort used for our microarray genomic analysis. The patient demographic and clinical variables were similar to the original 44 tumors and 25 normal tissues. Our results showed that that 1) NOTCH1 protein is significantly overexpressed in tumor samples, as compared to normal tissues (P=0.02); 2) HES1 and HEY1 were overexpressed in 17.9% (10/56) and 14.3% (8/56) tumor samples; either HES1 or HEY1 were overexpressed in 26.8% (15/56) of HNSCC samples which is comparable to that in our expression array data (31.8%); 3) 10 HNSCC tumors with elevated levels of NOTCH1 have overexpressed HES1 or HEY1; 4) among the five tumor samples originating from the same patient samples used for our microarray analysis, two (X7 and X11) demonstrated strong correlation of RNA and protein for HES1, HEY1 and NOTCH1 expression (Supplementary Figure 2).
Quantitative RTPCR Validation of NOTCH ligand, receptor, and target overexpression HES1, HEY1 in primary HNSCC
To validate the reliability of HES1 and HEY1 overexpression as assessed by expression microarrays in the cohort of 44 HNSCC tumors and 25 normal mucosa, we performed quantitative RT-PCR analysis on the same cohort. Our quantitative RT-PCR results confirmed the significant overexpression of HES1 and HEY1 in HNSCC tumors versus normal mucosa. The mean log10 mRNA levels for HES1 and HEY1 in primary HNSCCs were 1.1 (log10 range, 0.4 to 2.0) and 1.0 (log10 range, 0.1 to 1.8); in normal mucosa, the mean log10 mRNA levels for HES1 and HEY1 were 0.8 (log10 range, 0.1 to 1.4, P=0.013) and 0.8 (log10 range, 0.4 to 1.1, P=0.043), showing that Hes1 and Hey1 was significantly overexpressed (Figure 4A).
Figure 4. Quantitative RT-PCR analysis validation of HES1, HEY1, JAG1 and JAG2 expression in different cohorts of HNSCC tumors and normal mucosa.

A, Quantitative RT-PCR analysis of HES1 and HEY1 expression in the original study cohort of 44 HNSCC tumors and 25 normal mucosa. Expression of HES1 and HEY1 were significantly higher in HNSCC tumor tissues in comparison to normal tissues. Experiments were performed by Taqman realtime RT-PCR in triplicate and the data were normalized to ACTIN per sample. B, C and D, Quantitative RT-PCR analysis of HES1 and HEY1 (B), and JAG1 (C) expression in a separate cohort of 31 HNSCC tumors and 17 normal mucosa, and JAG2 expression in a additional cohort of 63 HNSCC tumors and 30 normal mucosa (D). Expression of HES1, HEY1, JAG1, and JAG2 were significantly higher in HNSCC tumor tissues in comparison to normal tissues. Experiments were performed by Taqman realtime RT-PCR in triplicate and the data were normalized to ACTIN or GAPDH per sample.
We further validated the overexpression of HES1 and HEY1 in an independent HNSCC cohort with 31 HNSCC tumors and 17 normal mucosa in non-cancer controls by quantitative RT-PCR. As expected, significantly increased expression levels of Hes1 and Hey1 were found in HNSCC (HES1, log10 mean, 1.0; HEY1, log10 mean, 0.9) versus normal mucosal tissues (HES1, log 10 mean, 0.7, P<0.01; HEY1, log10 mean, 0.5, P<0.01) (Figure 4B). We also validated the overexpression of JAG1 in this independent cohort. Our quantitative RT-PCR assay showed significantly increased levels of JAG1 expression in HNSCC tumor tissues (log10 mean, 1.1) in comparison with normal mucosa (log10 mean, 0.6, P<0.001) (Figure 4C). Further, we validated the overexpression JAG2 in a cohort of 63 HNSCC tumors vs 30 normal mucosa. The mRNA levels of JAG2 were significantly increased in HNSCC tumors (log10 mean, 1.1) in comparison with that in normal mucosa (log10 mean, 0.9, P<0.001) (Figure 4D). The application of different cohorts for quantitative RTPCR validation showing similar alterations validates the initial findings in our discovery cohort.
A subset of wildtype NOTCH1 HNSCC tumors has increased HES1/HEY1 expression
To determine the potential relevance of NOTCH1 genetic mutations to the transcriptional alterations in the NOTCH signaling pathway, we performed selected exome sequencing on 37 HNSCC tumors from the above cohort (6 samples including X14, X15, X20, X26, X30 and X37 were eliminated for sequencing due to inadequate DNA; an additional sample X3 was eliminated for not passing quality control criteria) and their paired normal lymphocytes. A total of 5 NOTCH1 mutations were identified in 4 of 37 (10.8%) HNSCC tumors. Consistent with our previous study, next to TP53, NOTCH1 was found the most frequently mutated gene found in the 37 HNSCC tumors (Figure 5A). Also the NOTCH1 mutation rate in this study was found similar to that in our former reported cohort. Four of the NOTCH1 mutations were predicted to truncate the protein product, whereas 1 was missense. Among the four nonsense mutations (R56*, Q154*, E1294*, W1843*), one mutation of W1843* was clustered in the RBP-Jk-associated module (RAM) domain, whereas the other 3 mutations next to or in the N-terminal epidermal growth factor (EGF)-like ligand binding domain. Similarly, the 1 missense mutation (T1138I) was also clustered in the N-terminal EGF-like ligand binding domain (Figure 5B). Whereas NOTCH1 contains activating mutations in T cell acute lymphoblastic leukemia and chronic lymphocytic leukemia (mainly lying at the heterodimerizaion (HD) domain and C-terminal polypeptide-enriched proline, glutamate, serine, and threonine (PEST) domain of NOTCH1) (13), the mutations we found in the study cohort appeared to be loss-of-function mutations, consistent with those recently described in myeloid leukemia and HNSCC tumors (7, 8, 15).
Figure 5. NOTCH1 mutations identified in the 37 HNSCC tumor tissues from the initial cohort of 44 HNSCC tumors and 25 normal mucosa.
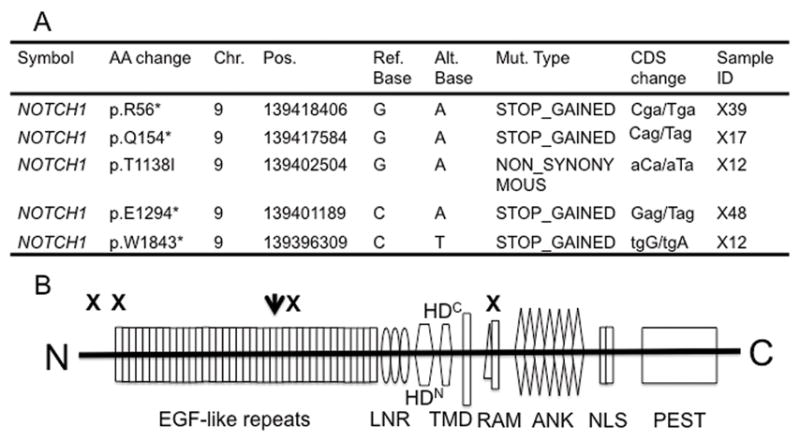
A. NOTCH1 mutations in the 37 HNSCC tumor tissues identified by whole exome sequencing. B. Schematic depiction of mutations in NOTCH1. EGF, epidermal growth factor; LNR, Lin12-NOTCH repeats; HDN, N-terminal heterodimerization domain; HDC, C-terminal heterodimerization domain; TMD, transmembrane domain; RAM, recombination signal-binding protein 1 for J-κ (RBPjκ) association module; NLS, nuclear localization signal; PEST, proline, glutamic acid, serine/threonine-rich motifs. Black arrow (missense mutation) and “X” (truncating mutation) depict mutations observed in 4 out of the 37 HNSCC tumors observed in this study by targeted exome sequencing.
We then compared downstream activation of NOTCH signaling pathway driven by transcriptional changes between the HNSCC tumors with and without NOTCH1 mutations. We examined the association of NOTCH1 mutations and HES1/HEY1 expression status. We found the significant lower expression of HES1 (P=0.013) and/or HEY1 (P=0.003) in HNSCC tumors with mutant type NOTCH1 than those with wild type NOTCH1 (Figure 6A). Of note, the mRNA levels of HES1 (P=0.104) or HEY1 (P=0.970) in HNSCC tumors with NOTCH1 mutant were similar to those in the normal mucosa, and none of these 4 HNSCC tumors with NOTCH1 mutant exhibited HES1/HEY1 overexpression, consistent with the loss-of-function of NOTCH1 mutations described above. Intriguingly, we found that among the 33 HNSCC tumors with wild type status, 10 (30.3%, 10/33) exhibited HES1/HEY1 overexpression, implicating NOTCH pathway activation in these HNSCC tumors. Thus, by comparison of the pattern of copy number alteration, transcriptional alteration and downstream activation of NOTCH pathway activation in wildtype NOTCH1 tumors, we found in the wildtype NOTCH1 HNSCC tumors there exist a group of NOTCH1 pathway activated HNSCC tumors (Figure 6B).
Figure 6. A subset of wildtype NOTCH1 HNSCC tumors has increased HES1/HEY1 expression.
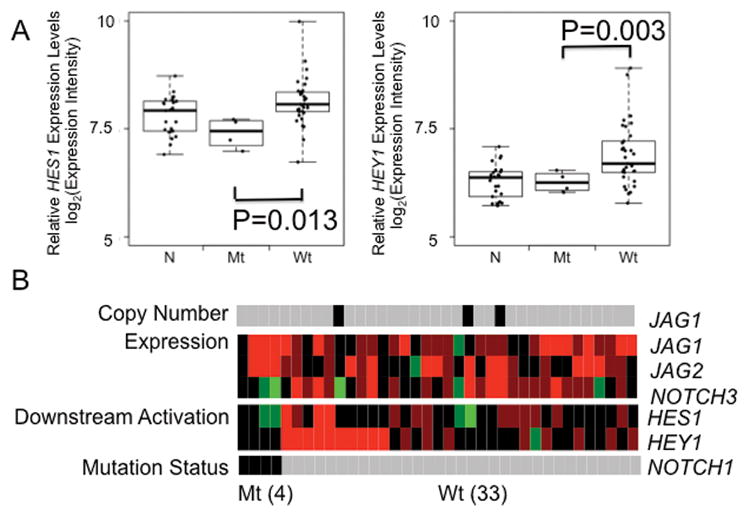
A. Comparison of HES1/HEY1 expression among normal mucosa, NOTCH1 mutant and wild type HNSCC tumors. NOTCH1 mutant HNSCC tumors had decreased HES1/HEY1 expression in comparison to those in NOTCH1 wildtype HNSCC tumors, and were similar to those in normal mucosa. N, normal mucosa, Mt, NOTCH1 mutant HNSCC tumors, Wt, NOTCH1 wild type HNSCC tumors. Boxes represent the interquartile range (25th–75th percentile) and horizontal lines inside the boxes indicate median. Whiskers indicate the minimum and maximum values. P value was calculated by using t test. B. Heat-map display of copy number alteration (JAG1), transcriptional alteration (JAG1, JAG2 and NOTCH3) and downstream activations (HES1 and HEY1) of NOTCH pathway in 4 NOTCH1mutant and 33 NOTCH1 wild type HNSCC tumors as defined in Figure 5. Gray denotes HNSCC tumor with wild type NOTCH1, black, HNSCC tumor with mutant type NOTCH1. Red and green indicate over- or under-expression, respectively.
In the 44 HNSCC from our initial study cohort, 13 were HPV-positive. We hereby determined the association between HPV status and NOTCH1 mutation status, as well as the mRNA levels of HES1 and HEY1. We found that the association between HPV status and NOTCH1 status was not statistically significant (P=0.55, Fisher’s exact test); No statistically significant differences in mRNA levels of HES1 (P=0.37, student’s t test) and HEY1 (P=0.96, student’s t test) between HPV-positive and HPV-negative HNSCC tumors were observed (data not shown). Therefore, we did not perform the NOTCH signaling pathway analyses in HPV-positive or HPV-negative tumors separately in this study.
The subset of wild-type NOTCH1 HNSCC tumors with increased HES1/HEY1 expression is validated in TGCA HNSCC cohort
To validate the pattern of NOTCH1 pathway alterations revealed in our HNSCC cohort, we further analyzed the available NOTCH1 mutation, copy number, and expression data of NOTCH pathway in the TCGA HNSCC cohort (including NOTCH1 mutation, copy number and expression data sets from 279 HNSCC tumors and expression data set from 37 adjacent normal tissues). Similar to the alterations revealed in our initial HNSCC cohorts, increased expression of JAG1 (P<0.001), JAG2 (P<0.001) and NOTCH3 (P=0.004) were found in HNSCC tumors vs adjacent normal tissues in the TCGA HNSCC cohort (Supplementary Figure 3A). The significant overexpression of JAG1 (P<0.001) and PSEN1 (P<0.001) in HNSCC tumors with increased copy number gains compared with that in normal tissues were also validated in the TCGA HNSCC cohort. Moreover, in the TCGA HNSCC cohort, we found that JAG1 and PSEN1 were overexpressed in the tumors with increased copy number gains in comparison to those tumors without increased copy number gains. (Supplementary Figure 3B).
In the TCGA cohort, a total of 67 mutations in NOTCH1, comprised of 14 frame shift dels, 1 in frame del, 51 nonsense mutations, and 1 splice site-associated mutation, were found in 52 HNSCC tumors. We observed that decreased expression of HES1 had a borderline significance (P=0.064) in NOTCH1 mutant type vs wild type HNSCC tumors whereas increased expression of HEY1 has a statistically significant difference (P=0.012) (Figure 7A). It should be of note that in the TGCA HNSCC cohort, we were not able to validate the overexpression of HES1 in HNSCC tumors vs adjacent normal tissues although a borderline significance trend was shown; this may be due to the fact that the adjacent normal tissues used in the TCGA cohort were collected from patients with HNSCC and include non mucosal tissues, whereas the normal mucosal tissues in our initial HNSCC cohort include only mucosal tissue from non-cancer patients. Accordingly, overexpression of HEY1 instead of HES1 was used as an indication of the NOTCH pathway activation in this TCGA cohort. Of interest, we found that among the 227 NOTCH1 wild-type HNSCC tumors, 50 (22.0%) exhibited overexpression of HEY1, confirming a subset of NOTCH pathway activation, wild-type NOTCH1 HNSCC primary tumors (Figure 7B).
Figure 7. Validation of the subset of wildtype NOTCH1 HNSCC tumors with increased HES1/HEY1 expression in TCGA HNSCC cohort of 279 tumor tissues and 37 adjacent normal tissues.
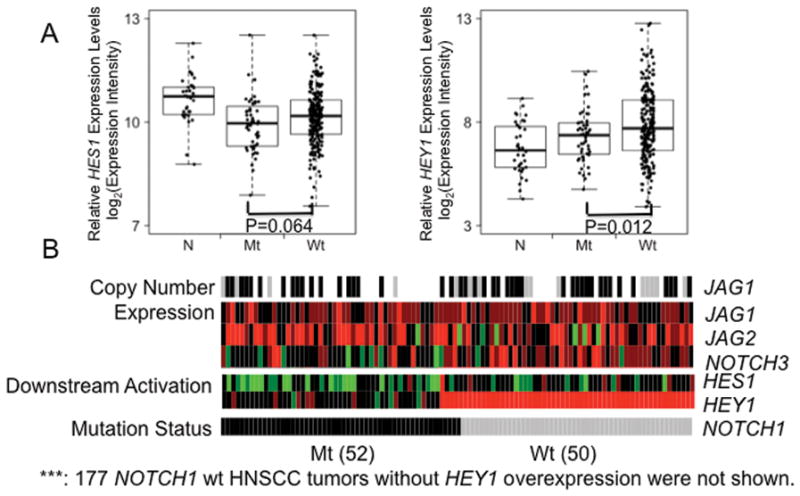
A. Comparison of HES1/HEY1 expression among adjacent normal mucosa, NOTCH1 mutant and wild type HNSCC tumors. NOTCH1 mutant HNSCC tumors have decreased HES1/HEY1 expression in comparison to those in NOTCH1 wildtype HNSCC tumors. N, adjacent normal mucosa, Mt, NOTCH1 mutant HNSCC tumors, Wt, NOTCH1 wild type HNSCC tumors. Boxes represent the interquartile range (25th–75th percentile) and horizontal lines inside the boxes indicate median. Whiskers indicate the minimum and maximum values. P value was calculated by using t test. B. Heat-map display of copy number alteration (JAG1), transcriptional alteration (JAG1, JAG2 and NOTCH3) and downstream activations (HES1 and HEY1) of NOTCH pathway in 52 NOTCH1 mutant and 50 NOTCH1 wild type HNSCC tumors (with HEY1 overexpression) from the TCGA HNSCC cohort. Gray denotes HNSCC tumor with wild type NOTCH1, black, HNSCC tumor with mutant type NOTCH1. Red and green indicate over- or under-expression, respectively.
To test the feasibility of the NOTCH pathway as target for future directed therapy, we explored the functional consequences when NOTCH1 is inhibited by siRNA in NOTCH1 wildtype cells (090 and SCC61). As shown in Supplementary Figure 4, following transfection with NOTCH1 siRNAs, NOTCH1 wildtype 090 and SCC61 cells showed dramatic and modest decrease in cell proliferation respectively. To validate the different functional consequences mediated by inhibition with NOTCH1 siRNAs in NOTCH1 wildtype cells, we then examined the growth effects upon siRNA inhibition of HEY1. Consistent with the results showing by siRNA inhibition of NOTCH1, we noted a significant decrease of cell growth in NOTCH1 wildtype 090 cells inhibited with HEY1 siRNAs (Supplementary Figure 5). Similarly, when NOTCH pathway inhibitor GSI-XXI was applied, the cell growth in NOTCH1 wildtype 090 cells was modestly inhibited (Supplementary Figure 6).
Discussion
The NOTCH signaling pathway regulates many facets of cancer biology, including stem cell renewal, proliferation, tumor angiogenesis, and metastasis (37). The NOTCH signaling pathway has been implicated in the tumorigenesis of several solid tumor malignancies including, but not limited to, non-small cell lung adenocarcinoma (38), melanoma, and ovarian carcinoma (39). However, the molecular alterations of the NOTCH signaling pathway in HNSCC are less well defined. We have previously reported that NOTCH1 mutations occur in ~15% of HNSCC patients, implicating a critical role of NOTCH signaling pathways in HNSCC tumors (7). Here we extend our study to analyze the comprehensive molecular alterations of NOTCH signaling pathway genes and the NOTCH signaling pathway downstream activation status in HNSCC tumors. To better define the alterations of NOTCH signaling pathway, we have conducted DNA copy number, methylation, expression and mutation analysis from a cohort of 44 HNSCC tumors and 25 normal mucosa.
Our data demonstrate the frequent occurrence of aberrant DNA copy number gains of NOTCH signaling pathway genes in HNSCC tumors compared to that in normal mucosa. We reported that 8 NOTCH pathways genes exhibited increased DNA copy numbers in HNSCC tumors versus normal mucosa based on outlier analysis. We observed that there are outlier HNSCC tumors with copy number gains of these NOTCH pathway genes. The broad copy number alterations identified in this study imply that in addition to NOTCH1 mutations, copy number gains of NOTCH signaling pathway genes may contribute to HNSCC tumor development. Among these genes featuring copy number gains in HNSCC tumors, the identification of NOTCH ligand JAG1 is intriguing. In support of the findings of the copy number increase of JAG1 in this study, previously studies from our laboratory and others using SNP6.0 microarrays or high-resolution microarray comparative genomic hybridization showed that chromosome 20p, a region containing JAG1, is amplified at high frequency in HNSCC tumors (40). Moreover, our expression array analysis demonstrated that JAG1 was significantly overexpressed (30%) in HNSCC tumors compared to that in normal mucosa. Our study suggests that genomic amplification in part contributes to the overexpression of JAG1 in HNSCC (Pearson correlation coefficient between copy number levels and mRNA expression: 0.6, data not shown). Recently, studies on the function significance of JAG1 in cancer development have been published. For example, the upregulation of JAG1 in breast cancer has been implicated metastatic disease and correlated with poor prognosis (41) and JAG1 plays a critical role in the promotion of bone metastatic outgrowth of breast cancer (11). In addition, we note that 4 out of 5 patients with JAG1 gains also have a gain in either NUMB or NUMBL, which are know inhibitors of the NOTCH pathway. In this respect, it remains unclear whether expressing the NOTCH ligand JAG1 and NOTCH regulating genes such as NUMB and NUMBL within the same tumors would actually contribute to NOTCH signaling or instead result in cis-inhibition of the pathway.
To identify the potential epigenetic alterations of NOTCH signaling pathway genes, in particular promoter methylation in HNSCC tumors, we performed methylation analysis using Illumina’s Infinium methylation data generated from the above cohort. We report that none of the NOTCH pathway genes present in on the methylation array exhibit promoter hypomethylation in HNSCC tumors compared with that in normal mucosa although two genes (PSEN2 and PSENSEN) with borderline significance were noted, implicating that methylation alterations may play a less common role in regulating Notch pathway genes during HNSCC tumor development.
We next examined the transcriptional alterations of NOTCH signaling pathways genes in HNSCC tumors using the expression array data from the cohort of 44 HNSCC tumors and 25 normal mucosa. These include genes involved in the molecular and biochemical events occur during of ligand-receptor recognition, receptor activation, intramembrane proteolysis, and target gene selection (42). Our result revealed that 11 genes, including JAG1, JAG2, NOTCH3, NCSTN, DTX3L, ADAM17, DVL3, HES1, HDAC2, NCOR2 and NUMBL, were significantly upregulated, and 4, including KAT2B, MAML3, DTX1 and MFNG, downregulated. Of the overexpressed genes, JAG2 was found to promote lung adenocarcinoma metastatis through a miR-200-dependent pathway in mice (43). In nasopharyngeal carcinoma, Man CH et al’s studies indicat that activation of NOTCH3 pathway is a critical oncogenic event in nasopharygeal carcinoma development (44). NCSTN, encoding a γ-secretase component, was recently shown to be a potential driver genes in human liver carcinoma by genome wide screening (45). DTX3L, a member of the deltex family of proteins that function as E3 ligases and modify NOTCH signaling, has been shown amplification and overexpression in cervical cancer (46); and consisted with our result, ADAM17 (ADAM metallopeptidase domain 17) was reported an elevated expression in malignant cells in HNSCC (47). In addition, we observed a marginal statistical trend that NOTCH pathway genes tend to be differentially expressed. Taken together, these results suggest that NOTCH signaling pathway genes are altered at the transcriptional levels in HNSCC tumors.
Given the prevalence of the transcriptional alterations of NOTCH signaling pathway we observed in HNSCCs, we further investigated the activation of the NOTCH signaling pathway in HNSCC. To date, the best characterized gene targets indicating NOTCH signaling pathway activation are members of the hairy and enhancer of split (HES) and the Hes related-repressor protein (HERP) families of bHLH transcriptional repressors such as HES1 and HEY1 (42, 48). In our study, we found that HES1 and HEY1 were significantly overexpressed in HNSCCs in comparison to that in normal mucosa. Our results revealed ~31.8% (14/44) of HNSCC tumors exhibited overexpression of HES1 and/or HEY1. Notably, the overexpression of HES1 and/or HEY1 was supported by the downregulation of their downstream responsive genes. The findings that HES1 and HEY1 were overexpressed in our HNSCC cohort shown in expression microarray were also validated by quantitative real-time RTPCR analysis in the same HNSCC cohort as well as an additional validation cohort. In addition, we found a significant differential expression of Nguyen_NOTCH1_Target gene set (differentially expressed genes concomitantly modulated by activated NOTCH1 in mouse and human primary keratinocytes) in 44 HNSCC tumor tissues (28). Although the estimating power of Nguyen gene set in evaluated the NOTCH signaling pathway status maybe moderate given the evidence that this gene set originated from primary keratinocyte rather than head and neck epithelial cells, this result support the notion that NOTCH signaling pathway is dysregulated in HNSCCs.
To gain a more complete understanding of the NOTCH1 mutations relevant to the transcriptional alterations of NOTCH signaling pathway and to the NOTCH signaling pathway activation, we performed selected exome sequencing using the massive parallel next generation sequencing techniques on 37 HNSCC tumors used for the above microarray expression analysis. We observed 5 NOTCH1 mutations occurring in ~10.8% (4/37) HNSCC tumors. Of these NOTCH1 mutations, 4 predicted to result in loss of the majority of amino acids from the translated protein; the remaining 1 missense mutation located in N-terminal EGF-like ligand-binding domain. A dual biological of NOTCH1 as either cancer promoting or tumor suppressing has been highlighted. It is reported that gain-of-function NOTCH1 mutations found in leukemia’s cluster in the negative regulatory region and the C-terminal PEST domain (13), whereas the loss-of-function NOTCH mutations in cutaneous and lung squamous cell carcinoma span ectodomain and the N-terminal portion of the intracellular domains that include nonsense mutations leading to receptor truncations (14). Thus it is likely that gain- and loss-of-function mutations occur at different regions of the NOTCH1 gene. Our and others’ study of sequence analysis and in HNSCC tumors demonstrated that that the majority of NOTCH1 mutations in HNSCC affect either the EGF-like ligand-binding domain or the NICD domain, suggesting loss of function (7, 8). Our data support the notion that the NOTCH1 mutations found in this study are loss-of-function mutations and play a tumor-suppressive role during HNSCC development.
Moreover, our result showed that the HNSCC tumors with NOTCH1 mutation exhibited decreased HES1/HEY1 expression in comparison to that in HNSCC tumors with NOTCH1 wild type. However, we reported that the mRNA levels of HES1/HEY1 in HNSCC tumors with NOTCH1 mutant is similar to that in the normal mucosa and overexpression of HES1/HEY1 was found in none of the HNSCC tumors with NOTCH1 mutant. This data is in agreement with a lack of NOTCH activation in NOTCH1 mutant HNSCC tumors. It is notable that, although the absence of elevated HES1/HEY1 correlated with NOTCH1 mutations, these 4 patient samples also appear to have lower NOTCH3 compared to many of the samples.
Interestingly, our data also suggest that a large subset of HNSCC tumors with NOTCH1 wild type sequence exhibit NOTCH pathway copy number increase, increased expression and downstream activation (Figure 6 and Figure 7). In our HNSCC initial cohort, a subset of NOTCH1 wild type (~30.3%) HNSCC tumors had HES1/HEY1 overexpression (downstream activation). This subset of HNSCC tumors featuring wild type NOTCH1 status and HES1/HEY1 overexpression was also validated in TCGA HNSCC cohort. In support of this notion, our initial functional results showed that in the NOTCH1 wildtype 090 HNSCC cells, siRNA inhibition of NOTCH1 or HEY1 significantly decreased cell growth; and the application of GSI-XXI, a NOTCH pathway inhibitor to 090 HNSCC cells also inhibited cell growth. It is possible that one explanation for the the differential response of two wildtype cell lines (090 and SCC61) to NOTCH pathway inhibition may be the presence of HPV (090, HPV positive; SCC61, HPV negative), however, other concurrent pathway alterations may also explain this response.
In summary, the present study demonstrated systematic transcriptional alterations of NOTCH signaling pathway in HNSCC tumors by using a large cohort comprised of HNSCC tumors and normal mucosas. The study provides strong evidence supporting the prevalent activation of NOTCH signaling pathway in HNSCC tumors and an overall bimodal pattern in HNSCC. In HNSCC tumors with NOTCH1mutant, there is a lack of NOTCH pathway activation due to the loss-of-function mutations; whereas in HNSCC tumors with NOTCH1 wild type, there is a larger subset of HNSCC tumors exhibit ligand receptor copy number increase, increased expression and downstream pathway activation. Our studies may suggest that in a subset of HNSCC tumors, the NOTCH pathway may be driving the growth is a potential therapeutic target; yet it is unclear if in other HNSCC tumors, the signaling may actually impair the growth of wildtype tumors to some degree. The findings reported in the present study may have important implications for development of NOTCH pathway targeting therapies, in particular for those NOTCH wildtype, NOTCH pathway activated HNSCC tumors that may be targeted by NOTCH inhibiting therapies.
Supplementary Material
Acknowledgments
Grant Support: The work was supported by National Institute of Dental and Craniofacial Research (NIDCR) and NIH Challenge Grant RC1DE020324 and RC2DE020789, and NCI P50 DE 019032 Head and Neck Cancer SPORE.
The manuscript/analysis of this article is based on a web database application provided by Research Information Technology Systems (RITS)–https://www.rits.onc.jhmi.edu/.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nature reviews Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Poeta ML, Manola J, Goldenberg D, Forastiere A, Califano JA, Ridge JA, et al. The Ligamp TP53 Assay for Detection of Minimal Residual Disease in Head and Neck Squamous Cell Carcinoma Surgical Margins. Clin Cancer Res. 2009;15:7658–65. doi: 10.1158/1078-0432.CCR-09-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demokan S, Chuang A, Suoglu Y, Ulusan M, Yalniz Z, Califano JA, et al. Promoter methylation and loss of p16(INK4a) gene expression in head and neck cancer. Head Neck. 34:1470–5. doi: 10.1002/hed.21949. [DOI] [PubMed] [Google Scholar]
- 4.Papadimitrakopoulou VA, Izzo J, Mao L, Keck J, Hamilton D, Shin DM, et al. Cyclin D1 and p16 alterations in advanced premalignant lesions of the upper aerodigestive tract: role in response to chemoprevention and cancer development. Clin Cancer Res. 2001;7:3127–34. [PubMed] [Google Scholar]
- 5.Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. International journal of oncology. 2008;32:101–11. [PubMed] [Google Scholar]
- 6.Bonner JA, Harari PM, Giralt J, Cohen RB, Jones CU, Sur RK, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 11:21–8. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer discovery. 2013 doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalaitzidis D, Armstrong SA. Cancer: The flipside of Notch. Nature. 473:159–60. doi: 10.1038/473159a. [DOI] [PubMed] [Google Scholar]
- 11.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 14.Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17761–6. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–3. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun W, Zaboli D, Wang H, Liu Y, Arnaoutakis D, Khan T, et al. Detection of TIMP3 promoter hypermethylation in salivary rinse as an independent predictor of local recurrence-free survival in head and neck cancer. Clin Cancer Res. 18:1082–91. doi: 10.1158/1078-0432.CCR-11-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho AL, Henrique R, Jeronimo C, Nayak CS, Reddy AN, Hoque MO, et al. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:4782–9. doi: 10.1158/1078-0432.CCR-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scharpf RB, Irizarry RA, Ritchie ME, Carvalho B, Ruczinski I. Using the R Package crlmm for Genotyping and Copy Number Estimation. J Stat Softw. 2011;40:1–32. [PMC free article] [PubMed] [Google Scholar]
- 20.Tibshirani R, Hastie T. Outlier sums for differential gene expression analysis. Biostatistics (Oxford, England) 2007;8:2–8. doi: 10.1093/biostatistics/kxl005. [DOI] [PubMed] [Google Scholar]
- 21.Phipson B, Smyth GK. Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol. 2010;9:Article39. doi: 10.2202/1544-6115.1585. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic acids research. 2004;32:D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 26:2363–7. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber W, Gentleman R. matchprobes: a Bioconductor package for the sequence-matching of microarray probe elements. Bioinformatics. 2004;20:1651–2. doi: 10.1093/bioinformatics/bth133. [DOI] [PubMed] [Google Scholar]
- 25.Scharpf RB, Irizarry RA, Ritchie ME, Carvalho B, Ruczinski I. Using the R Package crlmm for Genotyping and Copy Number Estimation. J Stat Softw. 40:1–32. [PMC free article] [PubMed] [Google Scholar]
- 26.Michaud J, Simpson KM, Escher R, Buchet-Poyau K, Beissbarth T, Carmichael C, et al. Integrative analysis of RUNX1 downstream pathways and target genes. BMC genomics. 2008;9:363. doi: 10.1186/1471-2164-9-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Dudoit SVC, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 28.Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–42. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27:1025–31. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.TCGA Sees Heterogeneity in Head and Neck Cancers. Cancer Discov. 2013;3:475–6. [Google Scholar]
- 33.Lee JC, Smith SB, Watada H, Lin J, Scheel D, Wang J, et al. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–36. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- 34.Yan B, Raben N, Plotz PH. Hes-1, a known transcriptional repressor, acts as a transcriptional activator for the human acid alpha-glucosidase gene in human fibroblast cells. Biochemical and biophysical research communications. 2002;291:582–7. doi: 10.1006/bbrc.2002.6483. [DOI] [PubMed] [Google Scholar]
- 35.Murata K, Hattori M, Hirai N, Shinozuka Y, Hirata H, Kageyama R, et al. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Molecular and cellular biology. 2005;25:4262–71. doi: 10.1128/MCB.25.10.4262-4271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer A, Klattig J, Kneitz B, Diez H, Maier M, Holtmann B, et al. Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Molecular and cellular biology. 2005;25:8960–70. doi: 10.1128/MCB.25.20.8960-8970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiuza UM, Arias AM. Cell and molecular biology of Notch. The Journal of endocrinology. 2007;194:459–74. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 38.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. Journal of the National Cancer Institute. 2000;92:1355–7. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 39.Bedogni B, Warneke JA, Nickoloff BJ, Giaccia AJ, Powell MB. Notch1 is an effector of Akt and hypoxia in melanoma development. J Clin Invest. 2008;118:3660–70. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smeets SJ, Braakhuis BJ, Abbas S, Snijders PJ, Ylstra B, van de Wiel MA, et al. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25:2558–64. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 41.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 42.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121:1373–85. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Man CH, Wei-Man Lun S, Wai-Ying Hui J, To KF, Choy KW, Wing-Hung Chan A, et al. Inhibition of NOTCH3 signalling significantly enhances sensitivity to cisplatin in EBV-associated nasopharyngeal carcinoma. The Journal of pathology. 2012;226:471–81. doi: 10.1002/path.2997. [DOI] [PubMed] [Google Scholar]
- 45.Woo HG, Park ES, Lee JS, Lee YH, Ishikawa T, Kim YJ, et al. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–66. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilting SM, de Wilde J, Meijer CJ, Berkhof J, Yi Y, van Wieringen WN, et al. Integrated genomic and transcriptional profiling identifies chromosomal loci with altered gene expression in cervical cancer. Genes, chromosomes & cancer. 2008;47:890–905. doi: 10.1002/gcc.20590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stokes A, Joutsa J, Ala-Aho R, Pitchers M, Pennington CJ, Martin C, et al. Expression profiles and clinical correlations of degradome components in the tumor microenvironment of head and neck squamous cell carcinoma. Clin Cancer Res. 2010;16:2022–35. doi: 10.1158/1078-0432.CCR-09-2525. [DOI] [PubMed] [Google Scholar]
- 48.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. Journal of cellular physiology. 2003;194:237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


