Abstract
The exopeptidases of family S46 are exceptional, as the closest homologs of these enzymes are the endopeptidases of clan PA. The three-dimensional structure of S46 enzymes is unknown and only one of the catalytic residues, the serine, has been identified. The catalytic histidine and aspartate residues are not experimentally identified. Here we present phylogenetic and experimental data that identify all residues of the catalytic triad of S46 peptidase, dipeptidyl aminopeptidase BII (DAP BII) from Pseudoxanthomonas mexicana WO24. Phylogenetic comparison with the protein and S46 peptidases, revealed His-86, Ser-657, and five aspartate residues as possible catalytic residues. Mutation studies identified the catalytic triad of DAP BII as His-86, Asp-224, and Ser-657, while secondary structure analysis predicted an extended alpha-helical domain in between Asp-224 and Ser-657. This domain is unique for family S46 exopeptidases and its absence from the endopeptidases of clan PA might be key to their different hydrolysis activities.
Peptidases are a type of hydrolases, which hydrolyze primarily peptide bonds in proteins and peptides to release amino acids or shorter peptides. Some enzymes are also able to hydrolyze imido bonds. Peptidases are classified into peptidase families on the basis of sequence homology. These families are further subdivided into peptidase clans on the basis of similarities in three-dimensional structures, their amino acid sequence around the catalytic amino acids, or the order of catalytic residues in the amino acid sequence. Clan PA contains both serine and cysteine peptidases whose peptidase unit exhibits a double beta barrel fold that is constituted from closed and crossed antiparallel beta barrel domains and contains catalytic residues serine (or cysteine), histidine and aspartate. Clan PA contains over seven hundred identified peptidases, mainly endopeptidases such as trypsin and chymotrypsin, the best known of all peptidases, and forms the largest member of all peptidase clans.
Pseudoxanthomonas is a bacterial genus, first classified in the year 2000 based on the characteristic of related group bacteria to reduce nitrite, but not nitrate, with the production of nitrous oxide (N2O)1,2. Pseudoxanthomonas species have been isolated from soil, compost, midgut, plant tissue, coral reef and sludge of paper and cotton mills and a dairy factory1,2,3,4,5,6,7. Species related to Pseudoxanthomonas are plant pathogens, like Xanthomonas axonopodis, Xanthomonas campestris and Xylella fastidiosa8,9,10, while the multidrug-resistant opportunistic pathogen Stenotrophomonas maltophilia, can form a risk for immunocompromised patients11. Pseudomonas sp. strain WO24, in this study reclassified as Pseudoxanthomonas mexicana WO24, was originally isolated from wastewater of a bean curd (tofu) factory when screening for hydrolytic activities that would release the N-terminal dipeptide Gly-Phe from Gly-Phe-p-nitroanilide (Gly-Phe-pNA)12.
We have reported several dipeptidyl peptidases (DPP), such as dipeptidyl aminopeptidase BI12 (DAP BI), DAP BII13, DAP BIII13 and DAP IV14, from P. mexicana WO24. DPP (EC 3. 4. 14), which we call DAP in this study, is an exopeptidase that releases dipeptides from the free amino-terminus. DAPs are present in all organisms, from bacteria to mammals, and the enzyme contributes to several important physiological functions such as the absorption of nutrients, immunity and the ability of pathogens to invade host cells15,16,17. DAP BI18 and DAP IV19 show amino acid sequence homology and similar substrate specificity to enzymes that belong to the peptidase family S920 in clan SC, such as oligopeptidase B and DPP IV. In contrast, no homologs have been identified for DAP BII and DAP BIII. DAP BI and DAP BIII possess both endo- and exopeptidase activity, while DAP BII has only an exo activity. It specifically acts on dipeptide p-nitroanilide substrates with aliphatic and aromatic residues and can remove many residues of bioactive oligopeptides such as angiotensin I and neuromedin N. Apart from peptide bonds, DAP BII is able to hydrolyze imido bonds.
DAP BII is a putative serine protease that is sensitive to diisopropyl fluorophosphate, a major serine peptidase inhibitor. Substrate specificity similar to that of DAP BII is only found for dipeptidyl peptidase 7 (PgDPP7), a serine peptidase exhibiting broad substrate specificity from the periodontal pathogen Porphyromonas gingivalis. PgDPP7 has been assigned to the peptidase family S46 in clan PA21,22,23,24. Family S46 of exopeptidases is unique for clan PA, as all other members of this clan are endopeptidases. Preliminary examination of purified DAP BII using peptide sequencing and a homology search by blastp suggested that DAP BII could be assigned to family S46. Other enzymes that belong to this family are, apart from PgDPP7, the dipeptidyl peptidases 11 from P. gingivalis and Shewanella putrefaciens (PgDPP11 and SpDPP11, respectively)22,23. DPP11 was first discovered in a pathogen of human periapical periodontitis, Porphyromonas endodontalis (PeDPP11)22. Much attention has recently been paid to P. gingivalis because of its close relationship with periodontal and ischemic diseases, including cardiovascular disorders, decreased kidney function and rheumatoid arthritis25,26. P. gingivalis is known to utilize dipeptides, instead of free amino acids, as the preferential source of energy and cellular material. DAPs providing dipeptides are crucial for the metabolism of this bacterium; and hence DPP7 and DPP11 are studied extensively with regards to their mechanism of substrate recognition. Investigation of substrate recognition by members of family S46 showed that Gly-666 of PgDPP7, Arg-673 of PgDPP11 and Ser-684 of SpDPP11 discriminate the P1 residue of the substrate23 and Phe-664 of PgDPP7 and Phe-671 of PgDPP11 recognize the residue at the P2 position of the substrate24. Based on sequence comparison among the conserved region containing the serine of the catalytic His, Asp, Ser triad, the S46 family peptidases have been classified into a phylogenetic tree of gram-negative bacteria that branches into five clusters of Bacteroides and three clusters of Proteobacteria23. For each cluster, peptidases were differentiated into three classes according to their type of specificity for the P1 position in the substrate: a) DPP7-type broad substrate specificity, b) PgDPP11-type acidic residue specificity, or c) SpDPP11-type specificity for an acidic residue or proline. Although a three-dimensional structure has not yet been determined for a peptidase of this family, the family S46 is assigned to clan PA since a contiguous region surrounding the catalytic serine of DPP7 and DPP11 is homologous to the equivalent region around the catalytic Ser-237 of glutamyl peptidase I (V8 protease) from Staphylococcus aureus, which belongs to clan PA27,28,29. For PgDPP11, Ser-655, corresponding to Ser-648 in PgDPP7, has been experimentally identified as the catalytic serine22. The other two residues of the catalytic triad, histidine and aspartate, have, however, not yet been experimentally identified for family S46 peptidases. Bioinformatic data analyses show that family S46 peptidases contain a number of highly conserved Asp-residues which could be the aspartate of the catalytic triad. In this study, we provide bioinformatic and experimental evidence for the identification of the catalytic His and Asp in DAP BII, a peptidase belonging to the family S46. The predicted secondary structure of family S46 peptidases is marked by a unique, extended helical region, which suggests that the tertiary structure of this family of enzymes will be different from that of other clan PA peptidases. Elucidation of the structure-function relationship in DAP BII may be key to understanding the difference between the endo- and exo activities of peptidases in clan PA.
Results
Reclassification of Pseudomonas sp. strain WO24 as Pseudoxanthomonas mexicana
In this study we aimed to determine the catalytic residues in the dipeptidyl aminopeptidase DAP BII, which we purified from a strain that was isolated from the effluent of a tofu factory and had been tentatively identified as a Pseudomonas species12,13. By obtaining the complete sequence for its 16S rRNA gene (DDBJ Acc. no AB375392), we were able to assess the origins of this bacterium more precisely. Phylogenetic tree analysis with 16S rRNA gene sequences retrieved from the GenBank database (NCBI) (Supplemental Fig. S1) showed very high levels of similarity to 16S rRNA genes from Pseudoxanthomonas japonensis and Pseudoxanthomonas mexicana, both of which were for 99.7% identical by pairwise alignment to that of Pseudomonas sp. strain WO24. The genomic DNA relatedness of these three species was confirmed by analyzing the extent in which biotin-labeled DNA hybridized to genomic DNAs that had been immobilized on filters (Supplemental Table S1). The results indicated that P. japonensis appears less closely related to Pseudomonas sp. strain WO24 than P. mexicana. We therefore propose to reclassify Pseudomonas sp. strain WO24 as a strain of P. mexicana. A morphological and biochemical analysis of Pseudoxanthomonas mexicana strain WO24 is presented in Supplemental Table S2.
Cloning and expression of DAP BII gene
To obtain sequence information that would facilitate the cloning of the dapb2 gene that codes for DAP BII, we purified the native protein from P. mexicana WO24 and determined the sequence of an N-terminal peptide (GEGMWVPQQLPEIAGPLKKA) and of three internal amino acid sequences (ETGQDPFDSPK, YAATMAGWNNTSK, LPADQRVAAVDAWLGGNDAAAVK). The underlined residues were used to design degenerate primers with which a probe could be synthesized that enabled the isolation of dapb2 from a genomic library. The cloned dapb2 contained a 2166 bp open reading frame that coded for a polypeptide of 722 amino acids (aa) with a calculated molecular mass of 78.7 kDa which included the sequenced peptides (DDBJ acc. no. AB889525). The N-terminal signal peptide, with the deduced sequence M1RPNLLAAAIAVPLSLLAAQIAQA24, conformed to the definition of a type I signal peptide of gram-negative bacteria30,31. The SignalP program32 predicted that the gene product would localize to the periplasm. The putative mature protein would be composed of 698 aa with a calculated molecular mass of about 76 kDa which is very close to the molecular mass of 73 kDa for native DAP BII as determined by SDS-PAGE13.
The deduced amino acid sequence of DAP BII exhibited a very high degree of homology with family S46 peptidases, as shown by a BLASTP search of the NCBI database. Pairwise-alignment between DAP BII and known family S46 peptidases PgDPP7, PgDPP11 or SpDPP11 showed that these proteins were identical for, respectively, 31.4%, 33.0% or 43.7% and similar for 48.2%, 47.3% or 60.2% (Fig. 1). For the characterization of DAP BII and the identification of its catalytic residues we tried to heterologously express dapb2 in E. coli hosts such as DH5α or JM109, but had no success, even when expression was controlled from a lac or tac promoter (data not shown). In the sequence coding for the signal peptide (ATG CGT CCG AAC CTG CTC GCC GCC GCC ATC GCG GTC CCG TTG TCC CTG CTC GCC GCC CAG ATC GCC CAG GCG), multiple occurrences of the same codon for Ala (GCC) might, however, cause a problem with translation. As DAP BI, DAP BIII (unpublished result) and DAP IV19 from P. mexicana WO24 could be expressed in E. coli we constructed a chimeric gene that coded for the fusion of the mature DAP BII polypeptide with the signal peptide of DAP BIII. We expressed this fusion gene by means of the lac promoter in JM109 cells and determined the substrate specificity of the enzyme that was purified from cell-free extracts by ammonium sulfate precipitation. The specific activities of DAP BII for Gly-Phe-pNA and Ala-Ala-pNA were 3.4 ± 0.1 U/mg (1 U is defined as the conversion of 1 μmol substrate per min) and 10.0 ± 0.2 U/mg, which were similar to those of the native enzyme, 3.3 ± 0.1 U/mg and 9.5 ± 0.2 U/mg, respectively13. Therefore, the E. coli expressed DAP BII and native DAP BII have the same substrate specificity.
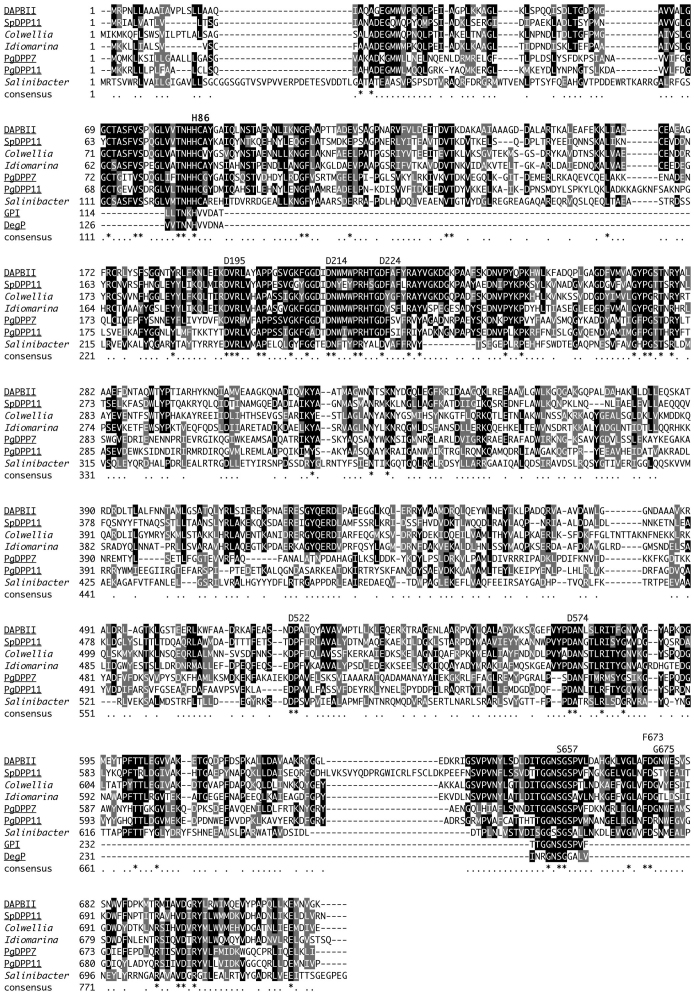
Figure 1. Alignment of DAP BII amino acid sequence with family S46 peptidases and clan PA endopeptidases.
Known peptidases are underlined: DAP BII amino acid sequence (AB889525) was obtained in this study; PgDPP7 (MEROPS ID: MER014366); PgDPP11 (MER034628); SpDPP11 (MER096825); GPI, Glutamyl peptidase I from Staphylococcus aureus (MER00264); DegP from E. coli (MER00266). Other sequences are DAP BII homologs: Colwellia, YP_267442 from Colwellia psychrerythraea 34H; Idiomarina, YP_156891 from Idiomarina loihiensis L2TR; Salinibacter, YP_446097 from Salinibacter ruber DSM 13855. Putative catalytic triad (His-86, Asp-195, Asp-214, Asp-224, Asp-522, Asp-574 and Ser-657) and residues recognizing the P1 position (Gly-675) and the P2 position (Phe-673) in the substrate are shown.
The phylogenetic tree of family S46 exopeptidases
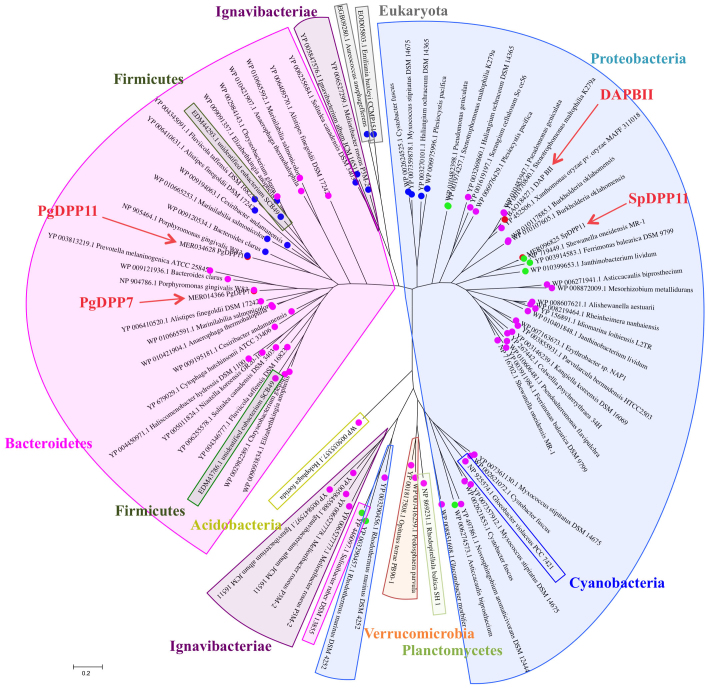
The catalytic residues in serine peptidases are highly conserved. In order to map the conserved residues in DAP BII, we performed an exhaustive homology search using Position-Specific Iterated BLAST (PSI-BLAST) which identified 661 amino acid sequences from 345 strains. These homologs of family S46 peptidases were found in 306 species of bacteria, the large majority being Bacteroidetes and Proteobacteria as reported before23, but also in Acidobacteria, Cyanobacteria, Firmicutes, Ignavibacteriae33, Planctomycetes and Verrucomicrobia. A phylogenetic tree of peptidase family S46 was constructed based on a set of 81 peptidase sequences from 48 independent phylogenetic families that were arbitrarily selected from all family S46 peptidases (Fig. 2). The enzymes from Bacteroidetes and Proteobacteria clustered into large groups bisecting the phylogenetic tree, with a few exceptions, such as the protein YP_446097.1 from Salinibacter ruber DSM 13855. The peptidases from Firmicutes and Cyanobacteria were grouped among the Proteobacteria. Ignavibacteriae33, Acidobacteria, Verrucomicrobia, Planctomycetes and Eukaryota were clustered as groups independent of Bacteroidetes and Proteobacteria. Our phylogenetic analysis indicates that family S46 peptidases are ubiquitous in bacteria.
Figure 2. Phylogenetic tree of family S46 peptidases.
The phylogenetic tree with DAP BII, PgDPP7, PgDPP11, and SpDPP11 was derived on the basis of a sequence set containing 81 peptidase sequences from 48 independent phylogenetic families by MUSCLE multiple sequence alignment and maximum likelihood analysis. The figure was generated using the MEGA program; enclosed clusters are phyla of source organisms and colored circles indicate known family S46 peptidases that differ with respect to the residue that is predicted to recognize the P1 residue of the substrate: glycine (magenta), arginine (blue), or serine (green). In DAP BII, the putative P1-interactive residue is glycine (red).
The predicted catalytic unit of DAP BII suggests a novel classification of clan PA peptidases
No three-dimensional structure has been made available for any member of family S46 yet and the only experimental proof for a catalytic residue has been obtained, by site-specific mutagenesis of PgDPP1122, for the serine but not the histidine or the aspartate residues that together form the catalytic triad of serine peptidases. On account of contiguous homology between regions surrounding the catalytic serines of PgDPP7 and PgDPP11 with their counterparts from enzymes belonging to clan PA, such as glutamyl peptidase I from Staphylococcus aureus, the family S46 peptidases have been assigned to the same clan28. However, no significant sequence conservation was found that could have revealed the catalytic histidine and aspartate residues of family S46 peptidases.
We aimed to identify all residues of the catalytic triad in DAP BII and, therefore, first examined the regions that showed the highest degree of conservation in the multiple sequence alignments of family S46 homologs. The alignment included PgDPP7, PgDPP11 and known clan PA peptidases such as glutamyl peptidase I from Staphylococcus aureus and DegP from E. coli, which belongs to clan PA family S134 (Fig. 1). Conservation between DAP BII regions and those of DegP containing the known catalytic residues His-131 and Ser-236 showed that residues His-86 and Ser-657 of DAP BII were part of its peptidase unit. On the other hand, five aspartate residues (Asp-195, Asp-214, Asp-224, Asp-522 and Asp-574 in DAP BII) turned out to be highly conserved among family S46 peptidases, which precluded unambiguous assignment of the catalytic aspartate by comparison with other clan PA enzymes.
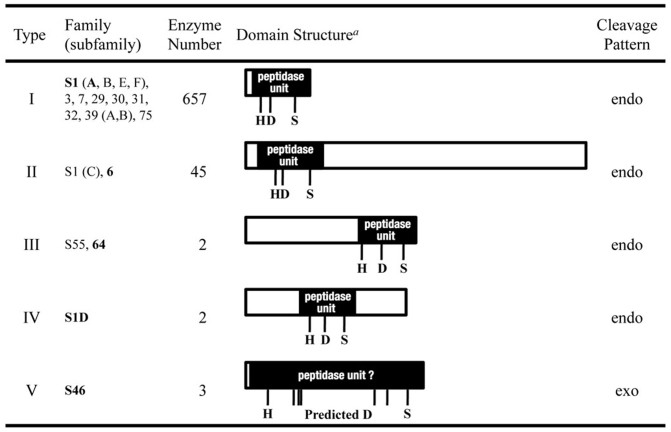
In order to obtain clues on which aspartate residue would be part of the catalytic triad, we classified clan PA peptidases by the position of the peptidase unit containing the catalytic residues within the enzyme35 (Table 1, Fig. 3). By this approach we could distinguish five types of clan PA peptidases. The peptidase unit of type I peptidases is compact and takes up almost the complete protein and are often derived from a virus. Glutamyl peptidase I28 and DegP34, belonging to subfamilies S1B and S1C, respectively, are classified as type I peptidases. The type II, III and IV peptidases have a peptidase unit that is located in either the carboxy-terminal, amino-terminal or central portion of their amino acid sequence, respectively. Like the type I peptidases, their peptidase units are comprised of about 150 to 300 aa. In contrast, the peptidase unit in type V enzymes, which all belong to clan PA family S46, appears dispersed over the enzymes, with the histidine and serine residues of the catalytic triad separated by about 700 aa. Thus, the enzymes of family S46 seem to contain a very large peptidase unit, which makes them, apart from being exopeptidases, again a unique group in clan PA.
Table 1. Clan PA Serine peptidases.
| Familya | seq.b | Ident.c | Strc.d | MEROPS ID | Typical peptidase | Source organisms | Total sizee | peptidase unit sizef | endo/exo | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Stert | End | size | |||||||||
| S1A | 13448 | 613 | 86 | S01.001 | Chymotrypsin A | cattle | 263 | 34 | 263 | 230 | endo |
| S1B | 1038 | 19 | 7 | S01.269 | Glutamyl peptidase I | Staphylococcus aureus | 336 | 69 | 336 | 268 | endo |
| S1C | 3698 | 34 | 5 | S01.273 | Peptidase Do (DegP peptidase) | Escherichia coli | 474 | 114 | 269 | 156 | endo |
| S1D | 98 | 2 | 1 | S01.280 | Lysyl peptidase (bacteria) | Achromobacter lyticus | 653 | 226 | 447 | 222 | endo |
| S1E | 336 | 12 | 7 | S01.261 | Streptogrisin A | Streptomyces griseus | 297 | 116 | 297 | 182 | endo |
| S1F | 66 | 1 | 1 | S01.109 | Astrovirus serine peptidase | human astrovirus | 927 | 447 | 578 | 132 | endo |
| S3 | 81 | 1 | 2 | S03.001 | Togavirin | Sindbis virus | 1245 | 1 | 264 | 264 | endo |
| S6 | 293 | 11 | 1 | S06.003 | Tsh peptidase | Escherichia coli | 1377 | 53 | 308 | 256 | endo |
| S7 | 162 | 1 | 1 | S07.001 | Flavivirin (yellow fever virus) | West Nile virus | 3411 | 1485 | 1666 | 182 | endo |
| S29 | 344 | 2 | 1 | S29.001 | Hepacivirin (NS3) | hepatitis C virus | 3011 | 152 | endo | ||
| S30 | 91 | 2 | 0 | S30.001 | Potyvirus P1 peptidase | plum pox virus | 3141 | 1 | 301 | 301 | endo |
| S31 | 29 | 1 | 0 | S31.001 | Pestivirus NS3 polyprotein peptidase | bovine viral diarrhea virus | 3898 | 1590 | 1800 | 211 | endo |
| S32 | 33 | 2 | 2 | S32.001 | Equine arteritis virus serine peptidase | equine arteritis virus | 3175 | 1065 | 1261 | 197 | endo |
| S39A | 23 | 1 | 1 | S39.001 | Sobemovirus peptidase | cocksfoot mottle virus | 568 | 131 | 319 | 189 | endo |
| S39B | 42 | 1 | 0 | S39.002 | Luteovirus peptidase | potato leaf roll luteovirus | 639 | 204 | 399 | 196 | endo |
| S46 | 506 | 2 | 0 | Dipeptidyl aminopeptidase BII | P. mexicana | 722 | 25 | 722 | 698 | exo | |
| S55 | 347 | 1 | 0 | S55.001 | SpoIVB peptidase | Bacillus subtilis | 426 | 188 | 426 | 239 | endo |
| S64 | 37 | 1 | 0 | S64.001 | Ssy5 peptidase | Saccharomyces cerevisiae | 687 | 459 | 687 | 229 | endo |
| S65 | 3 | 1 | 0 | S65.001 | Picornain-like cysteine peptidase | Breda-1 torovirus | 4445 | 3131 | 3395 | 265 | endo |
| S75 | 1 | 1 | 0 | S75.001 | White bream virus serine peptidase | White bream virus | 6872 | 3449 | 3709 | 261 | endo |
All data were abstracted from MEROPS;
aPeptidase family or subfamily;
bNumber of registered peptidases;
cNumber of identified peptidases;
dNumber of disclosed PDB file;
eFull-length size (in aa) of the typical peptidase;
fPosition and size of the peptidase unit in the typical peptidase.
Figure 3. Typing of peptidase families from clan PA by peptidase unit position.
(a) Type I, Family S1A Chymotrypsin A from Bos taurus (Length: 263 aa, Catalytic triad: H57-D102-S195). Type II, Family S6 Tsh peptidase from E. coli (1377 aa, H125-D153-S259). Type III, Family S64 Ssy5 peptidase from Saccharomyces cerevisiae (687 aa, H465-D545-S640). Type IV, Family S1D Lysyl peptidase from Achromobacter lyticus (653 aa, H262-D318-S399). Type V, DAP BII from P. mexicana (722 aa, H86-D195, D214, D224, D522 or D574-S657).
Identification of the catalytic triad
We decided to mutate the conserved five aspartate residues in order to establish which one is essential for catalytic activity and successfully produced 12 DAP BII-mutants with alanine substitutions in His-86, Asp-195, Asp-214, Asp-224, Asp-522, Asp-574 and Ser-657 or with an asparagine substituting one of the five aspartates, a sterically neutral mutation. The variants were expressed from a T7 promoter after induction with isopropyl-β-D-thiogalactopyranoside (IPTG) and purified. In the case of the D195N mutant, however, only trace amounts could be isolated, although some enzyme activity could be detected (Data not shown). To ascertain that the mutant proteins were not misfolded, circular dichroism (CD) spectra were collected for the wild-type protein and the variants (Supplemental Fig. S2A). The spectra of all variants were almost the same as the spectrum obtained for wild-type DAP BII, indicating that the secondary structure of the mutant proteins was not affected by the amino-acid change.
We noticed that the spectrum of wild-type DAP BII exhibited a positive band in the 190–195 nm region and two negative bands around 208 and 222 nm (Supplemental Fig. S2B), which is characteristic for an alpha-helical conformation. By analyzing the spectrum at 222 nm using an equation described by Chen et al.36, a high alpha-helical content of about 47% was predicted for DAP BII. The DAP BII spectrum was strikingly different from that for alpha-chymotrypsin (214 residues), which is for 9% alpha-helical and for 34% beta-sheet37. This protein is characteristic for the all-beta proteins of clan PA in endopeptidase family S1. The CD spectrum for alpha-chymotrypsin had a strong negative band near 200 nm plus a weak band around 230 nm (Supplemental Fig. S2B). Despite its resemblance to CD spectra of unfolded proteins, such a spectrum has been found for some all-beta globular proteins38. The CD spectra informed us that the two- and three-dimensional structure of DAP BII will be completely different from that of clan PA endopeptidases, especially with respect to the number of alpha-helices.
The specific activity and kinetic parameters of purified mutant enzymes were determined with Gly-Phe-pNA as the substrate (Table 2). Compared to the wild-type protein, the single substitution mutants H86A and S657A caused an 107-fold decrease in relative enzyme activity, indicating that His-86 and Ser-657 are essential residues for the catalytic activity of DAP BII, especially as the remaining activity was too low to allow the determination of kinetic parameters. Of the aspartate substitutions, the change of Asp-224 to either alanine or asparagine caused the largest decrease of relative enzyme activity (to 0.026% and 0.15%, respectively) in comparison to the substitutions of Asp-195 (to 23%), Asp-214 (to 1.5–3%), Asp-522 (to 16–32%) and Asp-574 (to 21–83%). The remaining activity (and catalytic efficiency) of mutants with substitutions in Asp-195, Asp-522 and Asp-574 indicated that these residues are not directly involved in catalysis. Substitution of Asp-214 to asparagine reduced the relative activity but the value for specificity constant kcat/Km remained at 5% of the value determined for wild-type DAP BII, indicating that this residue is not a catalytic residue. Mutation of Asp-224 lead to a dramatic, 103 to 104-fold decrease in specific activity. Asp-224 is, like the catalytic aspartate of clan PA endopeptidases, located outwith the surrounding beta-sheets, whereas Asp-195 is embedded within a beta-sheet (Fig. 4). The experimental results and the predicted difference in structural context for these residues argue that Asp-224 is the catalytic aspartate. We conclude that the catalytic triad of DAP BII consists of His-86, Asp-224, and Ser-657.
Table 2. Relative activities and kinetic parameters of DAP BII variants.
| Relative activity (%) | kcat (/sec) | Km (mM) | kcat/Km (/sec/mM) | |
|---|---|---|---|---|
| WT | 100 | 40.0 ± 1.8 | 1.400 ± 0.063 | 29.0 ± 1.3 |
| H86A | < 10−5 | -b | -b | -b |
| S657A | < 10−5 | -b | -b | -b |
| D195A | 23.0 ± 0.3 | -c | -c | -c |
| D195N | -a | -a | -a | -a |
| D214A | 1.5 ± 0.2 | -c | -c | -c |
| D214N | 3.0 ± 0.1 | 13.00 ± 0.05 | 10.00 ± 0.03 | 1.300 ± 0.005 |
| D224A | 0.026 ± 0.001 | -b | -b | -b |
| D224N | 0.150 ± 0.001 | -b | -b | -b |
| D522A | 32.0 ± 0.4 | 10.0 ± 0.1 | 0.150 ± 0.001 | 67.0 ± 0.5 |
| D522N | 16.0 ± 0.3 | 2.80 ± 0.01 | 0.053 ± 0.003 | 53.0 ± 0.3 |
| D574A | 83.0 ± 5.6 | 17.00 ± 0.04 | 0.53 ± 0.01 | 32.00 ± 0.07 |
| D574N | 21.0 ± 0.7 | 11.0 ± 0.1 | 1.10 ± 0.01 | 10.00 ± 0.09 |
Enzyme activities were assayed by hydrolysis of Gly-Phe-pNA; WT, wild type enzyme; catalytic residues are underlined;
aOnly trace amounts of the mutant were expressed;
bKinetic parameters could not be identified due to low activity of the mutants;
cKinetic parameters of the mutant were not determined.
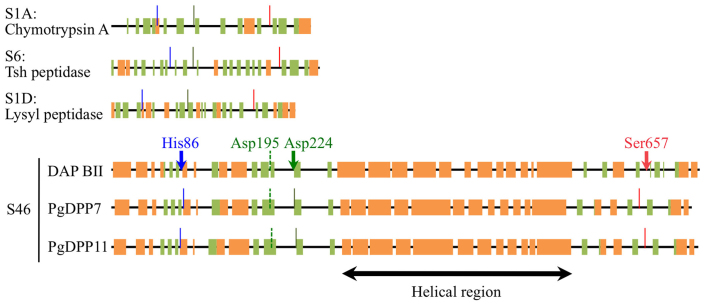
Figure 4. Secondary structure prediction of family S46 peptidases.
Secondary structures of family S46 peptidases as predicted by Jpred compared to those derived for chymotrypsin A from Bos taurus, Tsh peptidase from E. coli, and lysyl peptidase from Achromobacter lyticus. Regions that form a beta-sheet (green) or an alpha-helix (orange) are indicated by boxes, while solid lines and arrows show the His (blue), Asp (green) and Ser (red) residues of the catalytic triads. Arrows indicate the active residues identified in this study, while the broken green lines mark Asp residues that are part of the catalytic triad according to Ohara-Nemoto, Y. et al.22.
Secondary structure of family S46 peptidases
As a test for our assignment of the catalytic residues of DAP BII, we determined the putative secondary structure of family S46 peptidases and compared these to the folding of known clan PA peptidases (Fig. 4). We noticed that, in general, the catalytic residues reside in stretches rich in beta-sheets, while, in particular, the reactive aspartates and histidines are in regions with identical secondary structure in case of the family S46 enzymes, which supported our assignment of these residues as being involved in catalysis. Interestingly, a large alpha-helical domain was predicted to separate the catalytic aspartate and serine residues (Asp-224 and Ser-657 in DAP BII) in family S46 peptidases. This domain is unique for this family of clan PA enzymes and might be key for their mode of action as exopeptidases.
Discussion
In this study, we identified the catalytic residues of DAP BII by means of phylogenetic analysis and experimental examination of the effect of their substitution on relative enzyme activity. The catalytic mutants of DAP BII had lost their peptidase activity to a comparable extent. A trace of enzyme activity could be detected for DAP BII in which His-86 or Ser-657 were mutated, while the activities in the case of the Asp-224 mutants were reduced 103 to 104-fold. The catalytic mechanism and the contribution of the catalytic triad to peptide hydrolysis have been investigated for numerous clan PA peptidases39. During catalysis, the hydroxyl group of the reactive serine, activated by the catalytic histidine and aspartate residues, attacks the peptide bond in the substrate and a tetrahedral intermediate is formed which turns over into an acyl-enzyme intermediate. Attack by a water molecule, which is assisted by the catalytic histidine and aspartate residues, releases the acyl-group. In trypsin from Fusarium oxysporum, alanyl-substitution of the catalytic His-57 and Ser-195 caused a 105-fold decrease of the kcat/Km, while substitution of the catalytic Asp-102 with asparagine lead to a 104-fold reduction40. Thus, in the catalytic triad of some clan PA peptidases the aspartate contributes less to catalysis than the serine and histidine residues. The remaining activities of the trypsin mutants were due to some catalytic effect of the substituting residue or indirect catalytic mechanisms, such as transition state stabilization and proximity effects, which would also occur in case of the DAP BII substitution-mutants. In previous studies, the catalytic serine of family S46 peptidases was determined by alanine substitution, while the catalytic histidine and aspartate residues were predicted on the basis of bioinformatic analysis22,23,24. In PgDPP7, PgDPP11 and SpDPP11, the aspartate residue (Asp-196, Asp-198 and Asp-186, respectively) that corresponded to Asp-195 of DAP BII was thought to be essential for catalysis. We have shown, however, that mutation of this aspartate to alanine in DAP BII only resulted in a 4-fold reduction of relative enzyme activity. In the predicted secondary structures and unlike the catalytic aspartate of clan PA peptidases, Asp-195 of DAP BII, Asp-196 of PgDPP7 and Asp198 of PgDPP11 are located within beta-sheets, suggesting that these residues are not catalytic. Therefore, we concluded that Asp-224 of DAP BII is part of the catalytic triad, together with His-86 and Ser-657, which would imply that catalysis by PgDPP7, PgDPP11 or SpDPP11 relies on homologous triads (His-89, Asp-215, Ser-648), (His-85, Asp-217, Ser-655) or (His-80, Asp-215, Ser-666), respectively.
In order to examine the structure-activity relationship of family S46 peptidases, we compared their predicted secondary structures which revealed a region that covered about 47% of the protein was predicted to have an alpha-helical secondary structure41 (Fig. 4) which was supported by CD spectra of DAP BII which indicated an alpha-helical content of about 47%36 (Supplemental Fig. S2). The helical region is located between the catalytic aspartate and serine residues of family S46 exopeptidases and does not exist in clan PA endopeptidases such as alpha-chymotrypsin which has an alpha-helical content of 9%37, suggesting that this unique structural feature might be required for the exo hydrolase activity. The catalytic triad of clan PA peptidases is brought together by a double beta barrel fold consisting of a N- and a C-terminal beta barrel. The N-terminal beta barrel is constituted from six beta strands in which the catalytic histidine is located between the third and fourth beta strand while the catalytic aspartate lies between the fifth and sixth beta strand. The catalytic serine is located between the fourth and fifth beta strand of the C-terminal beta barrel (Fig. 4). Comparably, the catalytic residues of family S46 peptidases are predicted to be located in beta strand rich regions (Fig. 4). The catalytic triad of DAP BII, (His-86, Asp-224, Ser-657), is dispersed in between beta-sheets as in clan PA endopeptidases. A three-dimensional structure of the C terminal region of PgDPP7 (Asp-566 to Met-695) and PgDPP11 (Asp-572 to Val-702) was predicted with a homology modeling program based on the structure of glutamyl peptidase I24, which is in agreement with our putative secondary structure (Data not shown).
Studies of substrate recognition by family S46 peptidases showed that residues Gly-666 of PgDPP7, Arg-673 of PgDPP11 and Ser-684 of SpDPP11 recognize the P1 position in the substrate23, and Phe-664 of PgDPP7 and Phe-671 of PgDPP11 the P2 position24. Similar to PgDPP7, a glycine in DAP BII (Gly-675) recognizes the P1 position, leading to a broad substrate specificity (Fig. 1, Table 3). PgDPP11 and SpDPP11 show specificity for acidic amino acid residues (Asp, Glu), while SpDPP11 also recognizes alanine and proline at the P1 position. DAP BII is closely related to SpDPP11 in amino acid sequence homology but has a very different substrate specificity due to the glycine residue, instead of a serine, that recognizes the P1 position of the substrate (Fig. 1).
Table 3. Substrate specificities of family S46 peptidases.
| Enzyme | Hydrolysed substrate | Un- hydrolysed substrate |
|---|---|---|
| DAP BII13 | Asp-Arg -|- Val-Tyr -|- Ile-His -|- Pro-Phe -|- His-Leu | |
| Lys-Ile -|- Pro-Tyr -|- Ile-Leu | ||
| Phe-Leu -|- Pro-Leu -|- Ile-Leu-Gly-Lys-Leu-Val-Lys-Gly-Leu-Leu | ||
| Ala-Ala -|- pNA, Lys-Ala -|- MCA | Gly-Arg-pNA, Arg-Arg-MNA | |
| Gly-Phe -|- pNA | Gly-Pro-MNA | |
| Ser-Tyr -|- βNA | ||
| PgDPP721,23,24 | Trp-Ala -|- Gly-Gly-Asp-Ala-Ser-Gly-Glu | Trp-His-Trp-Leu-Glu-Leu-Lys-Pro-Gly-Glu-Pro-Met-Tyr |
| Ile-Ala -|- Arg-Arg-His-Pro-Tyr-Phe-Leu | Ser-Pro-Tyr-Ser-Ser-Glu-Thr-Thr | |
| Lys-Ile -|- Ala-Gly-Tyr-His-Leu-Glu-Leu | Ala-Pro-Val-Arg-Ser-Leu | |
| Phe-Leu -|- Arg-Glu-Pro-Val-Ile-Phe-Leu | Gln-Lys-Gln-Met-Ser-Asp-Arg-Arg-Glu | |
| Ala-Ala -|- pNA, Lys-Ala -|- MCA | Ala-Gly-pNA, Gly-Gly-pNA, Gly-Gly-MCA | |
| Ala-Phe -|- pNA, Gly-Phe -|- pNA | His-Asp-MCA | |
| Met-Leu -|- MCA | Gly-Glu-pNA | |
| Leu-Asp -|- MCA, Val-Asp -|- MCA | Gly-Arg-pNA | |
| Thr-Asp -|- MCA, Ile-Asp -|- MCA | Gly-Lys-pNA | |
| Leu-Glu -|- MCA | Lys-Lys-MCA | |
| Leu-Arg -|- MCA, Val-Arg -|- MCA | Gly-Pro-MCA | |
| Arg-Arg -|- MCA, Gly-Arg -|- MCA | Ala-Pro-pNA | |
| Leu-Lys -|- MCA | ||
| Leu-Gln -|- MCA, Ala-Asn -|- MCA | ||
| Thr-Ser -|- MCA, Ser-Tyr -|- MCA | ||
| PgDPP1122,23 | Leu-Asp -|- MCA, Ile-Asp -|- MCA | ac-Leu-Asp-MCA |
| Leu-Glu -|- MCA | Lys-Ala-MCA | |
| Gly-Gly-MCA | ||
| Met-Leu-MCA | ||
| Leu-Arg-MCA, Val-Arg-MCA, Gly-Arg-MCA | ||
| Leu-Lys-MCA, Lys-Lys-MCA | ||
| Gly-Pro-MCA | ||
| SpDPP1123 | Lys-Ala -|- MCA | Gly-Gly-MCA |
| Lys-Asp -|- MCA | Met-Leu-MCA | |
| Lys-Glu -|- MCA | Gly-Arg-MCA, Leu-Arg-MCA, Val-Arg-MCA | |
| Gly-Pro -|- MCA | Leu-Lys-MCA, Lys-Lys-MCA |
Hydrolytic cleavage bond, -|-; uncleaved bond, -; MCA, 7-methoxycoumarin; MNA, 4-methoxy-2-naphthylamide; βNA, β-naphthylamide.
DAPs from various kinds of organisms show different substrate specificities, making them important targets for industrial application, such as drug design42. Mammalian DPP IV, also called adenosine deaminase complexing protein 2 or CD26, cleaves X-proline dipeptides from the N-terminus of various polypeptides, such as chemokines, neuropeptides, and peptide hormones. In recent years, DPP IV has attracted interest for the many roles it is implicated in, such as immune regulation, hypertension, or hyperglycemia16,43.
Family S46 peptidases are found in diverse organisms (Fig. 2), such as Prevotella melaninogenica which can be isolated from severe anaerobic infections44. All known family S46 peptidases have been studied, but their physiological roles, including those from the pathogenic bacteria P. gingivalis and related Pseudoxanthomonas species, remain unclear. For P. mexicana WO24, our preliminary examination suggests that this bacterium prefers oligopeptides as a carbon - and nitrogen source, which could indicate that DAP BII plays a physiological role in feeding (unpublished result).
In this study, the catalytic triad of the peptidase DAP BII, belonging to family S46, has been identified as His-86, Asp-224, Ser-657. The catalytic histidine and aspartate residues have been experimentally identified for the first time for a family S46 peptidase and this is also the first report of the complete catalytic triad in an exopeptidase of clan PA. A unique helical region was discovered in the region between the catalytic aspartate and serine residues and this region is found exclusively in family S46 exopeptidases and absent from clan PA endopeptidases. Further analysis of DAP BII may unravel the mechanism that underlies endo/exo cleavage selection.
Methods
16S rRNA gene sequence, phylogenetic analysis and DNA-DNA hybridization
Using primers recognizing universally conserved regions, 16S ribosomal gene fragments were amplified from Pseudomonas sp. strain WO2412 genomic DNA, cloned and sequenced. This provided the information to determine the complete 1523 nt sequence of its 16S rRNA gene. Phylogenetic tree analysis of 16S rRNA sequences was carried out with the MEGA program45. DNA-DNA hybridization was done using the membrane filter method described by Ezaki et al.46 with biotin-labeled chromosomal DNA from Pseudomonas sp. strain WO2412, P. mexicana JCM11524 and P. japonensis JCM11525. Extent of hybridization was determined by ELISA using streptavidin-HRP and ABTS (E2,2′-azinobis[3-ethylbenzothiazolin-6-sulfonic acid]).
Physiology of Pseudomonas sp. strain WO24
Morphological and physiological properties were determined as described by Komagata et al.47.
DAP BII peptide sequence analysis
DAP BII was purified from P. mexicana WO24 as described previously13 and the sequence of peptides derived from the N-terminus and of internal fragments was obtained by peptide sequence analysis on a PPSQ-21 (Shimadzu). Degenerate primers were designed with which an ~800 bp fragment was amplified and used as a probe against dapb2 after labeling with [α-32P]dCTP (GE Healthcare) using a BcaBEST Labeling Kit (Takara Bio).
Cloning of dapb2
For the construction of a genomic library of P. mexicana WO24, chromosomal DNA was prepared by a modified CTAB method14 and partially digested with Sau3AI. Fragments of 9 to 20 kb, obtained after fractionation of the digest by ultracentrifugation through a 10 to 40% sucrose gradient (rotor SW28.1, 22 000 rpm, 20°C, 20 h), were inserted into BamHI site of lambda EMBL3 DNA (Stratagene). The recombinant lambda DNA was packaged into phage coat proteins using Gigapack II Plus Packaging Extract and transfected to E. coli XL1-Blue MRA (P2)(Stratagene).
To obtain a full-length clone of dapb2, the library was screened by plaque hybridization. Southern hybridization of EcoRI digested DNA from a positive lambda clone located dapb2 to a 2.9 kb-fragment which was cloned into pBluescript II KS (+) yielding plasmid pDAP BII.
Heterologous expression of DAP BII
For heterologous expression of P. mexicana DAP BII in E. coli, expression vector pKSb2m was constructed in which, because highly repetitive codons in the signal sequence of dapb2 might inhibit translation in E.coli, a chimeric gene consisting of the signal sequence from dapb3 (DDBJ acc. no. AB889526) and the coding region of dapb2 (DDBJ acc. no. AB889525) was placed under the control of the lac promoter. Regions from both genes were cloned into a vector used for the heterologous expression of DAP IV, pKF18-k-219. First, a 2.1 kb KpnI-BamHI fragment from pDAP BII with a part of the coding region of dapb2 and some upstream sequence, was ligated into pKF18k-2, yielding pK8b2. For subclone pK8b3, an 1.5 kb EcoRI-SalI fragment was cloned into pKF18k-2 containing the signal-sequence region of dapb3 SacI sites, introduced by site-directed mutagenesis in pK8b2 and pK8b3, were digested and blunted, and the linearized DNA digested with EcoRI. The generated pK8b2 vector-fragment was recombined with an 0.7 kb insert from pK8b3 and into the resulting plasmid an 0.85 kb ApaI fragment from pDAP BII was inserted with the remainder of dapb2 and some downstream sequence. The chimeric expression cassette in the resulting pKSb2m was verified by sequencing.
Pre-cultures of JM109 transformed with pKSb2m were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) containing 100 μg/ml ampicillin at 37°C for 12 h and diluted 25-fold into 2 × YT medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl) containing 100 μg/ml ampicillin and further incubated at 37°C for 20 h. Cells were harvested by centrifugation, washed with cold saline and suspended in cold 50 mM Tris–HCl (pH 8.0) containing 1 mM dithiothreitol. The suspended cells were disrupted with a Biospec bead beater after which cell debris was removed by centrifugation at 20,000 × g for 30 min. DAP BII was precipitated from the cell-free extracts with 35–70% ammonium sulfate and redissolved in 50 mM Tris–HCl (pH 9.0).
Purification of DAP BII
For the purification of DAP BII from E. coli, overexpression vector pETb2m was constructed, which is pET22b with the fusion gene from pKSb2m. A NdeI site was introduced into pKSb2m by site-directed mutagenesis using the PrimeSTAR Mutagenesis Basal Kit (TaKaRa Bio) at the starting codon of the chimeric gene which was transferred on a NdeI-SphI (blunted) fragment to pET22b. The resulting pETb2m has the chimeric gene under the control of an IPTG-inducible T7 promoter. Site-directed mutagenesis was carried out with a Mutan-Super Express Km (TaKaRa Bio) or PrimeSTAR Mutagenesis Basal Kit (TaKaRa Bio) using primers listed in Supplemental Table S3.
Rosetta2(DE3)pLacI cells transformed with pETb2m (or derived mutants) were pregrown in LB containing 100 μg/ml ampicillin at 37°C for 12 hr, diluted 100-fold into 2 × YT medium containing 100 μg/ml ampicillin, and incubated at 30°C to an A600 of approximately 0.4. Expression of recombinant DAP BII was induced by the addition of IPTG to a final concentration of 0.4 mM. After an 8 h incubation at 30°C cells were harvested by centrifugation, washed with cold saline, resuspended in BugBuster Protein Extraction Reagent (Novagen) and incubated on ice with shaking for 1 hr. Cell-free extracts were obtained by centrifugation at 20,000 × g for 30 min and diluted with an equal amount of cold 100 mM Tris–HCl (pH 8.0). The 35–70% ammonium sulfate precipitates obtained from the cell-free extracts were dissolved in 50 mM Tris–HCl (pH 9.0) and separated on an HiPrep Butyl FF 16/10 column, connected to an AKTApurifier (GE Healthcare), with a linear 30–0% gradient of ammonium sulfate in 50 mM Tris–HCl (pH 9.0). Fractions with DAP BII activity were pooled and equilibrated to 20 mM Tris–HCl (pH 9.0) by column chromatography using HiPrep 26/10 Desalting columns (GE Healthcare). The desalted samples were further purified by ion exchange chromatography over 6-ml Resource Q (GE Healthcare) columns with a linear gradient of 0–0.3 M NaCl in 20 mM Tris–HCl (pH 9.0). The fractions with DAP BII activity were pooled, concentrated and desalted using Vivaspin 20–10 K (GE Healthcare) centrifugal concentrators.
Purified DAP BII and mutants were analyzed by circular dichroism spectroscopy using a Jasco J-805.
Enzyme assay and kinetics
Enzyme activity was assayed by the hydrolysis of pNA substrates in 1 ml reactions, containing enzyme, 0.3 mM pNA substrate and 50 mM Tris-HCl (pH 8.0). After an incubation at 30°C for 20 min, the reaction was stopped by addition of 50 μl 100% trichloroacetic acid, then centrifuged at 20, 000 × g for 5 min and the initial rates of hydrolysis were measured at A385.
Values for kcat and Km were obtained from Lineweaver-Burk plots. For this, enzyme activities were measured under substrate concentrations varying from 0.05 to 0.3 mM, and expressed as the means of three different experiments.
Bioinformatic analysis
Homologs of DAP BII, PgDPP7, PgDPP11 and SpDPP11, were found by PSI-BLAST analysis in the database of non-redundant protein sequences at the National Center for Biotechnology Information. Subsequent analyses were performed by the MEGA program42. In total, 661 candidates for family S46 peptidases were filtered from a MUSCLE multiple sequence alignment48,49, based on the presence of conserved His and Ser residues that form part of the catalytic triad. A phylogenetic tree was constructed for a selected set of family S46 peptidases by maximum likelihood analysis using a WAG +F +G +I substitution model50 which was selected from fits for 48 different amino acid substitution models.
Secondary structures of family S46 peptidases were predicted using Jpred37 (http://www.compbio.dundee.ac.uk/www-jpred/), while secondary structures for known Clan PA peptidases were deduced from tertiary structures deposited in the Protein Data Bank (PDB; http://www.rcsb.org/pdb/home); i.e. for Chymotrypsin A from Bos taurus (1YPH.pdb) with catalytic triad H-75, D-120, S-213, Tsh peptidase from E. coli, (1WXR.pdb) with triad H-125, D-153, S-259, and lysyl peptidase from Achromobacter lyticus (1ARB.pdb) with catalytic triad H-262, D-318, S-399.
Author Contributions
Y.Su. and W.O. conceived and designed the project and experiments. Y.Su. performed the experiments and carried out data analysis. Y.Sa. performed the CD measurements. Y.Su., Y.Sa., N.T., H.O., Y.M. and W.O. discussed about the obtained results. Y.Su. wrote the paper. W.O. edited the paper.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr. N. Suzuki and Prof. T. Senda for their help with the CD spectrum measurements at the Structural Biology Research Center, High Energy Accelerator Research Organization (KEK).
References
- Finkmann W., Altendorf K., Stackebrandt E. & Lipski A. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 50 Pt 1, 273–282 (2000). [DOI] [PubMed] [Google Scholar]
- Thierry S. et al. Pseudoxanthomonas mexicana sp. nov. and Pseudoxanthomonas japonensis sp. nov., isolated from diverse environments, and emended descriptions of the genus Pseudoxanthomonas Finkmann et al. 2000 and of its type species. Int. J. Syst. Evol. Microbiol. 54, 2245–2255, 10.1099/ijs.0.02810-0 (2004). [DOI] [PubMed] [Google Scholar]
- Chen M. Y. et al. Pseudoxanthomonas taiwanensis sp. nov., a novel thermophilic, N2O-producing species isolated from hot springs. Int. J. Syst. Evol. Microbiol. 52, 2155–2161 (2002). [DOI] [PubMed] [Google Scholar]
- Harada R. M., Campbell S. & Li Q. X. Pseudoxanthomonas kalamensis sp. nov., a novel gammaproteobacterium isolated from Johnston Atoll, North Pacific Ocean. Int. J. Syst. Evol. Microbiol. 56, 1103–1107, 10.1099/ijs.0.63556-0 (2006). [DOI] [PubMed] [Google Scholar]
- Weon H. Y. et al. Pseudoxanthomonas suwonensis sp. nov., isolated from cotton waste composts. Int. J. Syst. Evol. Microbiol. 56, 659–662, 10.1099/ijs.0.63749-0 (2006). [DOI] [PubMed] [Google Scholar]
- Yang D. C., Im W. T., Kim M. K. & Lee S. T. Pseudoxanthomonas koreensis sp. nov. and Pseudoxanthomonas daejeonensis sp. nov. Int. J. Syst. Evol. Microbiol. 55, 787–791, 10.1099/ijs.0.63210-0 (2005). [DOI] [PubMed] [Google Scholar]
- Yoo S. H. et al. Pseudoxanthomonas yeongjuensis sp. nov., isolated from soil cultivated with Korean ginseng. Int. J. Syst. Evol. Microbiol. 57, 646–649, 10.1099/ijs.0.64427-0 (2007). [DOI] [PubMed] [Google Scholar]
- Tim S., Schubert S. A. R., Xiaoan, Sun, Tim Gottwald R., James H. Graham & Wayne, N. Dixon. Meeting the challenge of Eradicating Citrus Canker in Florida—Again. Plant Disease 85, 340–356 (2001). [DOI] [PubMed] [Google Scholar]
- Purcell D. L. H. a. A. H. Xylella fastidiosa: Cause of Pierce's Disease of Grapevine and Other Emergent Diseases. Plant Disease 86, 1056–1066 (2002). [DOI] [PubMed] [Google Scholar]
- Chung-Jan Chang, Ruth Donaldson, Phil Brannen, Gerard Krewer & Boland a. R. Bacterial Leaf Scorch, a New Blueberry Disease Caused by Xylella fastidiosa. HortScience 44, 413–417 (2009). [Google Scholar]
- Brooke J. S. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25, 2–41, 10.1128/cmr.00019-11 10.1128/cmr.00019-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara W. et al. A novel dipeptidyl aminopeptidase from Pseudomonas sp. strain WO24. J. Bacteriol. 178, 1283–1288 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara W., Kobayashi G., Okada H. & Morikawa Y. Two types of novel dipeptidyl aminopeptidases from Pseudomonas sp. strain WO24. J. Bacteriol. 178, 6288–6295 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara W., Ogawa Y., Yano K., Okada H. & Morikawa Y. Dipeptidyl aminopeptidase IV from Pseudomonas sp. WO24. Biosci. Biotechnol. Biochem. 60, 2032–2037 (1996). [DOI] [PubMed] [Google Scholar]
- Mierau I. et al. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J. Bacteriol. 178, 2794–2803 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrell M. D., Gysbers V. & McCaughan G. W. CD26: a multifunctional integral membrane and secreted protein of activated lymphocytes. Scand. J. Immunol. 54, 249–264 (2001). [DOI] [PubMed] [Google Scholar]
- Kumagai Y., Yagishita H., Yajima A., Okamoto T. & Konishi K. Molecular mechanism for connective tissue destruction by dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis. Infect. Immun. 73, 2655–2664, 10.1128/iai.73.5.2655-2664.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara W., Kobayashi G., Ishimaru S., Okada H. & Morikawa Y. The gene encoding dipeptidyl aminopeptidase BI from Pseudomonas sp. WO24: cloning, sequencing and expression in Escherichia coli. Gene 206, 229–236 (1998). [DOI] [PubMed] [Google Scholar]
- Ogasawara W. et al. Isoforms of dipeptidyl aminopeptidase IV from Pseudomonas sp. WO24: role of the signal sequence and overexpression in Escherichia coli. Protein Expr. Purif. 41, 241–251, 10.1016/j.pep.2004.10.027 (2005). [DOI] [PubMed] [Google Scholar]
- Polgar L. The prolyl oligopeptidase family. Cell. Mol. Life. Sci. 59, 349–362 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banbula A. et al. Porphyromonas gingivalis DPP-7 represents a novel type of dipeptidylpeptidase. J. Biol. Chem. 276, 6299–6305, 10.1074/jbc.M008789200 (2001). [DOI] [PubMed] [Google Scholar]
- Ohara-Nemoto Y. et al. Asp- and Glu-specific novel dipeptidyl peptidase 11 of Porphyromonas gingivalis ensures utilization of proteinaceous energy sources. J. Biol. Chem. 286, 38115–38127, 10.1074/jbc.M111.278572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouf S. M. et al. Discrimination based on Gly and Arg/Ser at position 673 between dipeptidyl-peptidase (DPP) 7 and DPP11, widely distributed DPPs in pathogenic and environmental gram-negative bacteria. Biochimie 95, 824–832, 10.1016/j.biochi.2012.11.019 (2013). [DOI] [PubMed] [Google Scholar]
- Rouf S. M. et al. Phenylalanine 664 of dipeptidyl peptidase (DPP) 7 and Phenylalanine 671 of DPP11 mediate preference for P2-position hydrophobic residues of a substrate. FEBS open bio 3, 177–183, 10.1016/j.fob.2013.03.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai T. et al. Oral bacteria in the occluded arteries of patients with Buerger disease. J. Vasc. Surg. 42, 107–115, 10.1016/j.jvs.2005.03.016 (2005). [DOI] [PubMed] [Google Scholar]
- Detert J., Pischon N., Burmester G. R. & Buttgereit F. The association between rheumatoid arthritis and periodontal disease. Arthritis. Res. Ther. 12, 218, 10.1186/ar3106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona C. & Gray G. L. Nucleotide sequence of the serine protease gene of Staphylococcus aureus, strain V8. Nucleic Acids Res. 15, 6757 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad L., Leduc Y., Hayakawa K. & Delbaere L. T. The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus. Acta Crystallogr. D Biol. Crystallogr. 60, 256–259, 10.1107/s090744490302599x (2004). [DOI] [PubMed] [Google Scholar]
- Nemoto T. K. et al. Characterization of the glutamyl endopeptidase from Staphylococcus aureus expressed in Escherichia coli. FEBS J. 275, 573–587, 10.1111/j.1742-4658.2007.06224.x (2008). [DOI] [PubMed] [Google Scholar]
- Paetzel M., Karla A., Strynadka N. C. & Dalbey R. E. Signal peptidases. Chem. Rev. 102, 4549–4580 (2002). [DOI] [PubMed] [Google Scholar]
- Auclair S. M., Bhanu M. K. & Kendall D. A. Signal peptidase I: cleaving the way to mature proteins. Protein Sci. 21, 13–25, 10.1002/pro.757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786, 10.1038/nmeth.1701 (2011). [DOI] [PubMed] [Google Scholar]
- Podosokorskaya O. A. et al. Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ. Microbiol. 15, 1759–1771, 10.1111/1462-2920.12067 (2013). [DOI] [PubMed] [Google Scholar]
- Krojer T., Garrido-Franco M., Huber R., Ehrmann M. & Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416, 455–459, 10.1038/416455a (2002). [DOI] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J. & Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–350, 10.1093/nar/gkr987 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T. & Martinez H. M. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry 11, 4120–4131 (1972). [DOI] [PubMed] [Google Scholar]
- Birktoft J. J. & Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl-chymotrypsin at 2 A resolution. J. Mol. Biol. 68, 187–240 (1972). [DOI] [PubMed] [Google Scholar]
- Wu J., Yang J. T. & Wu C. S. Beta-II conformation of all-beta proteins can be distinguished from unordered form by circular dichroism. Anal. Biochem. 200, 359–64 (1992). [DOI] [PubMed] [Google Scholar]
- Polgar L. The catalytic triad of serine peptidases. Cell. Mol. Life. Sci. 62, 2161–2172, 10.1007/s00018-005-5160-x (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R. & Corey C. S. C. An investigation into the minimum requirements for peptide hydrolysis by mutation of the catalytic triad of trypsin. J. Am. Chem. Soc. 114, 1784–1790, 10.1021/ja00031a037 (1992). [Google Scholar]
- Cole C., Barber J. D. & Barton G. J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–201, 10.1093/nar/gkn238 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysant S. G. & Chrysant G. S. Clinical implications of cardiovascular preventing pleiotropic effects of dipeptidyl peptidase-4 inhibitors. Am. J. Cardiol. 109, 1681–1685, 10.1016/j.amjcard.2012.01.398 (2012). [DOI] [PubMed] [Google Scholar]
- Varga T. et al. Higher serum DPP-4 enzyme activity and decreased lymphocyte CD26 expression in type 1 diabetes. Pathol. Oncol. Res. 17, 925–930, 10.1007/s12253-011-9404-9 (2011). [DOI] [PubMed] [Google Scholar]
- Shah H. N. & Collins D. M. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int. J. Syst. Bacteriol. 40, 205–208 (1990). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739, 10.1093/molbev/msr121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki T., Hashimoto Y. & Yabuuchi E. Fluorometric Deoxyribonucleic Acid-Deoxyribonucleic Acid Hybridization in Microdilution Wells as an Alternative to Membrane Filter Hybridization in which Radioisotopes Are Used To Determine Genetic Relatedness among Bacterial Strains. Int. J. Syst. Evol. Microbiol. 39, 224–229, 10.1099/00207713-39-3-224 (1989). [Google Scholar]
- Komagata K. in Classification and Identification of Microorganisms. Vol. 2 (ed Hasegawa, T.) 99–161 (Tokyo: Gakkai Shuppan, 1985).
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797, 10.1093/nar/gkh340 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113, 10.1186/1471-2105-5-113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S. & Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information