Abstract
In order to deduce the molecular mechanisms of biological function, it is necessary to monitor changes in the sub-cellular location, activation and interaction of proteins within living cells in real time. Förster resonance energy transfer (FRET)-based biosensors that incorporate genetically encoded, fluorescent proteins permit high spatial resolution imaging of protein–protein interactions or protein conformational dynamics. However, non-specific fluorescence background often obscures small FRET signal changes, and intensity-based biosensor measurements require careful interpretation and several control experiments. These problems can be overcome by using lanthanide (Tb(III) or Eu(III)) complexes as donors and green fluorescent protein (GFP) or other conventional fluorophores as acceptors. Essential features of this approach are the long-lifetime (~ms) luminescence of Tb(III) complexes and time-gated luminescence microscopy. This allows pulsed excitation followed by a brief delay that eliminates nonspecific fluorescence before detection of Tb(III)-to-GFP emission. The challenges of intracellular delivery, selective protein labeling, and time-gated imaging of lanthanide luminescence are presented, and recent efforts to investigate the cellular uptake of lanthanide probes are reviewed. Data is presented showing that conjugation to arginine-rich, cell penetrating peptides (CPPs) can be used as a general strategy for cellular delivery of membrane impermeable lanthanide complexes. A heterodimer of a luminescent Tb(III) complex, Lumi4, linked to trimethoprim (TMP) and conjugated to nonaarginine via a reducible disulfide linker rapidly (~10 min) translocates into the cytoplasm of Maden Darby canine kidney cells from culture medium. With this reagent, the intracellular interaction between GFP fused to FK506 binding protein 12 (GFP-FKBP12) and the rapamycin binding domain of mTOR fused to Escherichia coli dihydrofolate reductase (FRB-eDHFR) was imaged at high signal-to-noise ratio with fast (1–3 s) image acquisition using a time-gated luminescence microscope. The data reviewed and presented here show that lanthanide biosensors enable fast, sensitive and technically simple imaging of protein-protein interactions in live cells.
INTRODUCTION
Ligand-sensitized complexes of lanthanide cations, especially Tb(III) and Eu(III), have unique photophysical properties that make them particularly advantageous for luminescence-based biological analyses.1–2 Following near-UV (320–400 nm) ligand absorption, Tb(III) and Eu(III) complexes emit at multiple, discrete wavelengths with narrow bandwidths (<10 nm at half-maximum) that span the visible and near-infrared spectral region (Fig. 1a). Most notably, emission lifetimes are long (0.1 – 2 ms), and this allows for time-gated detection (TGD) strategies, where pulsed light is used to excite the sample, and lanthanide emission is detected after a brief delay (~μs) that effectively eliminates scattering and short-lifetime (~ns) fluorescence background signals. The ability to temporally and spectrally isolate lanthanide emission signals makes it possible to detect analytes at small concentrations (pM – nM) in complex matrices, and lanthanide-based assays are routinely used for diagnostics and high throughput screening using commercial plate reader instrumentation.3–4 In recent years, there has been considerable interest in leveraging the inherent sensitivity of TGD with Tb(III) and Eu(III) complexes for applications in live-cell microscopic imaging.1–2 In this context, the chemistry and photophysics of lanthanide complexes must be considered in relation to the workings and limitations of microscopes, the interaction and compatibility of complexes with cells, and the nature of the biological questions that are typically addressed by live-cell imaging experiments.
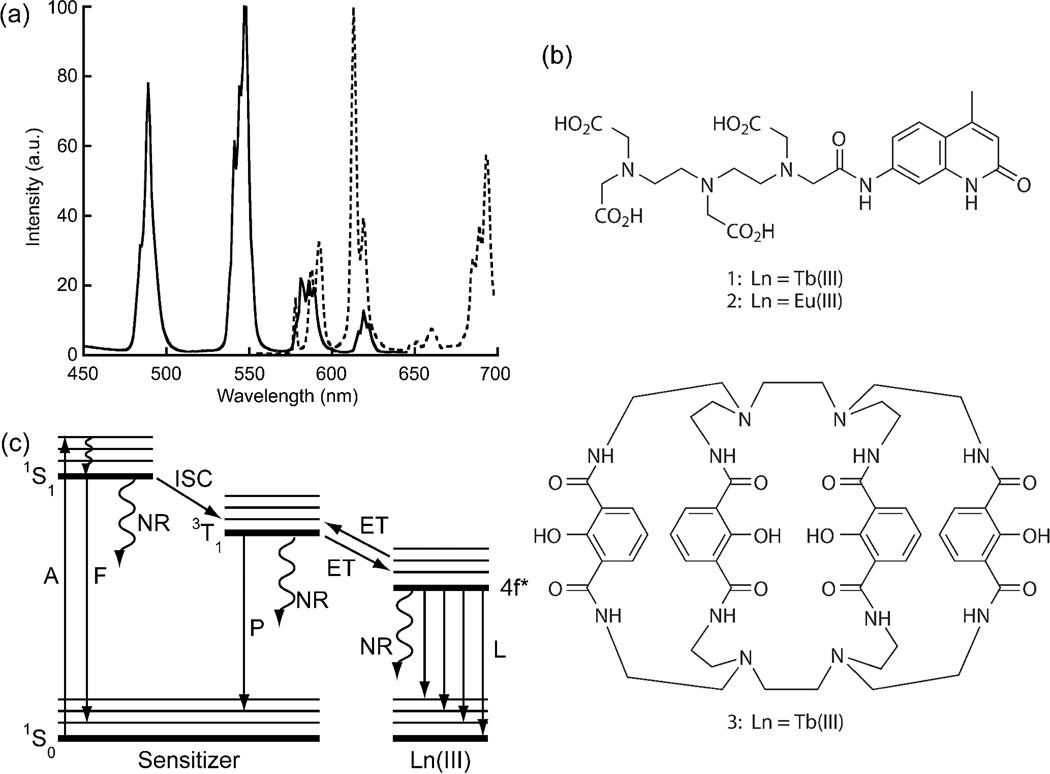
Figure 1. Structure and photophysics of sensitized organic lanthanide complexes.
(a) Typical emission spectra of a luminescent Tb(III) complex (solid) and Eu(III) complex (dotted). (b) Exemplary structures of sensitized lanthanide complexes. With DTPA-cs124, the chelator and sensitizer are separate entities.8 The 7-amino-4-methyl-2(1H)-quinolinone (cs124) moiety effectively sensitizes both Tb(III) (1) and Eu(III) (2) luminescence. With the Lumi4-Tb(III) complex, 3, the 4 hydroxyisophthalamide units serve as both chelators and sensitizers.9 (c) Schematic representation of major energy transitions in a lanthanide complex. S = singlet state, T = triplet state, A = absorption, F = fluorescence, P = phosphorescence, NR = nonradiative, ISC = intersystem crossing, ET = energy transfer, L = metal luminescence.
The photophysics and critical design features of luminescent lanthanide complexes are well established and have been amply described in many reviews.5–7 Because lanthanide f-f transitions are parity forbidden, direct excitation is inefficient, and emissive lanthanide complexes incorporate the metal ion into an organic chelating ligand that contains a sensitizing chromophore with a small singlet-triplet energy gap and a triplet energy at least 1500cm−1 above the receiving Ln(III) level. (Fig. 1b).7–9 Following light absorption by the chromophore, energy is transferred to the lanthanide excited state which then emits. Most commonly, energy transfer proceeds through the ligand triplet state, although transfer through singlet and charge transfer states has been observed (Fig. 1c). For practical use in bioassays, lanthanide complexes require several features: (i) kinetic inertness with respect to metal binding; (ii) a high extinction coefficient (>10,000 M−1 cm−1 ) and quantum yield (>0.1) of emission (i.e., good brightness); (iii) a long-wavelength (>350 nm) ligand absorption maximum; (iv) resistance to photobleaching; and (v) one or more functional groups that can be used for conjugation to biomolecules or targeting moieties.10 While hundreds of luminescent Tb(III) and Eu(III) complexes have been reported, relatively few meet all of the above-mentioned requirements.
For use in live-cell imaging, lanthanide complexes with long-wavelength absorption and good brightness are especially critical. Fluorescence microscopes incorporate high-numerical aperture (NA), aberration-corrected objectives to both deliver excitation light and collect emission signals. Very few of these objectives transmit light at wavelengths below 350 nm. Moreover, resolution and signal-to-noise ratio (SNR) in optical imaging fundamentally depend on the number of photons collected. With ms-scale lifetimes, lanthanides have low photon emission rates (<1000 photons/s). Thus, the brightest possible lanthanide complexes must be used to allow for adequate photon collection within limited exposure times, as we discuss in more detail below.
Fluorescence microscopy is used most effectively to visualize dynamic changes in the concentration, location, interactions or activity of biomolecules directly within the natural environment of the living cell. Central to this technique are luminescent markers; either sensing probes that report, say, localized changes in analyte concentration (e.g., H2O2, Ca2+, Zn2+)11–13 or tags that bind selectively to specific proteins or other macromolecules.14–16 The existing toolbox of organic fluorophores and genetically encoded fluorescent proteins currently enables multi-color fluorescence imaging in live cells with, variously, millisecond time resolution, nanometer spatial precision, single-molecule sensitivity, and absolute biochemical specificity.17–18 Given this context, it is worthwhile to consider the particular biological problems that can be addressed using lanthanide complexes and time-gated imaging that cannot be easily solved using existing technologies. In this Forum Article, we describe the use of lanthanide complexes as donors for time-gated, Förster Resonance Energy Transfer (FRET) microscopy with a particular focus on the imaging of protein-protein interactions in living mammalian cells. Time-gated microscope technology is briefly reviewed and its technical aspects are presented in terms of quantitative measures of image quality including SNR, photon collection efficiency and image acquisition time. Further, we attempt to define benchmark values of these parameters as they relate to obtaining high quality images. We also emphasize, with examples from our group and others, experimental techniques to achieve controlled delivery of lanthanide labels and specific, intracellular labeling of proteins. While the focus is on FRET imaging of protein-protein interactions, the concepts and examples given here are also relevant for lanthanide-based FRET imaging of protein conformational dynamics as well as imaging of lanthanide complex-based sensors, another area of considerable research effort.1–2, 6
FRET IMAGING
In order to deduce the molecular mechanisms by which proteins regulate cellular processes such as signal transduction, it is necessary to monitor changes in both sub-cellular location and protein activity in living cells in real time.19 Often, changes in protein activity are correlated with changes in conformation or propensity to interact with other proteins. FRET microscopy has emerged as a powerful tool for dynamically imaging changes in the level of interaction between two proteins or in the conformation of a single protein.20–22 FRET is the radiationless transfer of energy from an excited fluorescent donor to a nearby (<10 nm) acceptor whose absorption spectrum overlaps with the donor emission spectrum. FRET can be detected as a decrease in the fluorescence intensity, lifetime, or anisotropy of the donor or as an increase in the fluorescence intensity of the acceptor upon donor excitation (if the acceptor is fluorescent).23 As energy transfer efficiency inversely varies with the sixth power of the separation between donor and acceptor, FRET signal changes are highly sensitive to distance changes over the length scale of proteins. FRET-based biosensors for live-cell imaging often incorporate two differently colored fluorescent proteins, usually cyan (CFP) and yellow (YFP) as donors and acceptors to read out changes in protein conformation or interaction (Fig. 2a).22, 24
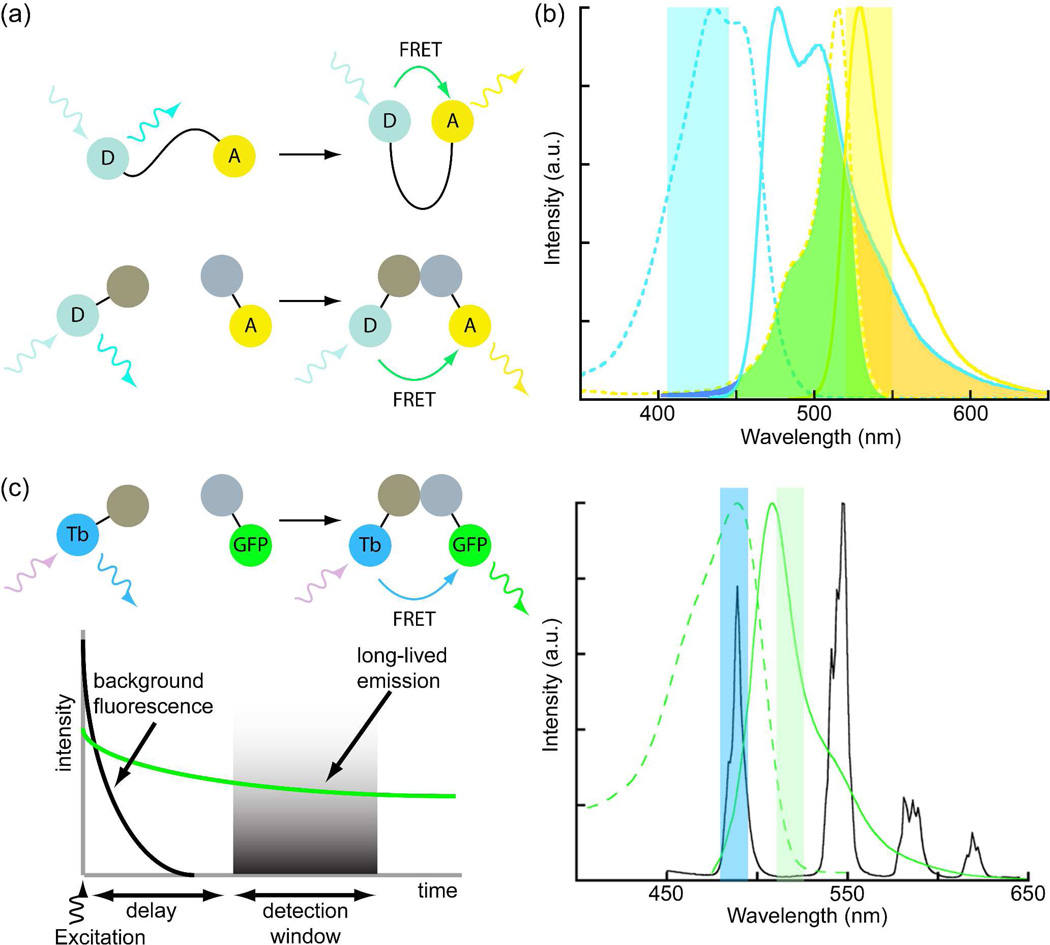
Figure 2. Förster resonance energy transfer (FRET)-based biosensors and effects of donor and acceptor photophysics on detection.
(a) Conformational changes of a single-chain biosensor (top) or interaction of dual chain biosensor (bottom) components bring donor (D) and acceptor (A) fluorophores within FRET distance (<10 nm). (b) Excitation (dotted) and emission spectra (solid) of cyan fluorescent protein (CFP, cyan) and yellow fluorescent protein (YFP, yellow), a common donor-acceptor pair for live cell FRET imaging. Overlap of CFP emission and YFP excitation spectra (green) allows sensitized YFP emission (yellow band) to be detected upon excitation of CFP (cyan band). Crosstalk, or direct excitation of YFP in the CFP band (blue) and bleedthrough of CFP emission into the YFP band (orange) obscure true FRET signals, necessitating multiple measurements with different filter sets. (c) With a long-lifetime Tb(III) donor and short-lifetime, green fluorescent protein (GFP) acceptor, Tb(III) and Tb(III)-sensitized, GFP emission can be separated using narrow-pass emission filters, eliminating bleedthrough. Crosstalk is eliminated by time-gated detection of long-lifetime, Tb(III)-to-GFP FRET.
FRET between two fluorescent proteins is usually imaged in one of three ways: i) detection of donor-sensitized, acceptor emission at steady-state (so-called, “Filter FRET”); ii) detection of donor dequenching upon acceptor photobleaching; and iii) detection of changes in donor emission lifetime.23–24 The practical challenge with any of these techniques is to differentiate biologically relevant FRET signal changes from other sources of fluorescence present in a live cell, and each method has various advantages and disadvantages. At this point in time, Filter FRET is the most commonly used method, can be easily implemented on wide-field, epifluorescence microscopes, and in certain circumstances can be used to measure energy transfer efficiency on a pixel-by-pixel basis in a whole-cell image.24 The acceptor photobleaching method can be used to directly measure FRET efficiency (the fraction of excited donors that transfer energy), but it is destructive of the sample and precludes time-lapse studies.23–24 Fluorescence lifetime imaging microscopy (FLIM) can measure FRET independently of changes in fluorophore concentration or emission intensity across the sample. However, FRET-FLIM measurements in the time domain require expensive instrumentation, long (~min) image acquisition times, and complex data fitting algorithms to resolve nanosecond-scale lifetimes.25 Frequency domain FRET-FLIM instruments that incorporate inexpensive light emitting diode (LED) light sources can achieve relatively fast (~10 s) frame rates.26–27 Recently, so-called phasor or polar plot methods of image analysis have been shown to accurately measure relative FRET changes without having to explicitly determine lifetimes, thereby allowing for faster image acquisition.28–30
When used to image intermolecular interactions between two labeled proteins, i.e., when imaging a dual-chain biosensor, the Filter FRET method is limited by i) crosstalk (direct acceptor excitation by light used to excite the donor); ii) bleedthrough (partial overlap of donor and acceptor emission wavelengths); and iii) non-unitary ratios of donor- and acceptor-labeled proteins (Fig. 2b).23–24, 31 Because of these limitations, at least 3 images must be collected with different filter sets: donor excitation/donor emission; donor excitation/acceptor emission (FRET); and acceptor excitation/acceptor emission. The images are then processed post-acquisition to separate biochemically relevant FRET signals from non-FRET fluorescence background and to normalize the FRET signal to the amounts of donor and/or acceptor.31 Even with these corrections, sensitized emission FRET suffers from low SNR that often lead to false negative results (i.e., interaction occurs but is not seen) and that can prevent accurate quantitative measurements.24 When used to image conformational changes of a single-chain biosensor labeled with both donor and acceptor fluorophores, the image acquisition and processing requirements are less stringent, and two-color, ratiometric imaging (donor excitation/donor emission and donor excitation/acceptor emission) is often sufficient to quantify FRET signal changes.31 The problems of crosstalk and bleedthrough are inherent to the broad excitation and emission spectra of fluorescent proteins, and they present an even greater challenge when one wishes to perform multiplexed imaging of two or more FRET biosensors in a single cell.19, 32
The use of lanthanides, especially Tb(III), as FRET donors along with TGD (often referred to as lanthanide-based FRET, or LRET) offers distinct advantages over conventional FRET with fluorescent proteins for imaging molecular interactions in live cells.5 Firstly, bleedthrough is minimized because the narrow emission bands of Tb(III) can be spectrally isolated from sensitized acceptor emission signals (Fig. 2c). Secondly, TGD eliminates directly excited acceptor fluorescence (crosstalk) and autofluorescence background signals. Thus, Tb(III)-sensitized emission of, for example, green fluorescent protein (GFP), can be detected in a single microscope image by exciting Tb(III) with near-UV light and acquiring GFP emission through a narrow-pass filter centered at 520 nm following a brief delay (~μs) after excitation (Fig. 2c). A third benefit of Tb(III) based FRET imaging is that it affords the possibility of multiplexed FRET imaging, where Tb(III) can sensitize emission of two or more differently colored acceptors.33 Finally, it should be noted that time-gated imaging can be implemented on a conventional wide-field fluorescence microscope in a straightforward manner using commercially available components.34 To fully implement time-gated FRET microscopy of protein-protein interactions, suitably bright and stable lanthanide complexes that can be selectively targeted to proteins in live cells, methods to deliver these complexes into specific cellular compartments without unduly perturbing cellular physiology,2 and a time-gated luminescence microscope are required.
TIME-GATED LUMINESCENCE MICROSCOPY
The potential benefits of TGD for sensitive microscopic imaging of long-lived luminescence were first recognized in the early 1990’s when epifluorescence microscopes equipped with a pulsed xenon flashlamp as excitation source and chopper-gated CCD detectors were used to image inorganic phosphors and europium chelates in immunohistochemical specimens.35–36 Time-gated imaging of Tb(III) or Eu(III) luminescence requires pulsed UV excitation and the ability to turn on or unshutter the detector after the end of the excitation pulse. Various microscope configurations have been described that utilize flashlamps, pulsed lasers or LEDs for excitation and mechanically or electronically gated CCD detectors.37 Most of these systems employ through-objective sample illumination and widefield, array detection, although a time-gated, two-photon scanning microscope was reported.38 Multiphoton excitation of lanthanide complexes in the near-infrared (NIR) could also be used to avoid potentially phototoxic UV illumination when imaging live cells, and some examples of two-photon imaging of lanthanide luminescence in cells have been reported.39–41 However, the inherently low photon emission rates of lanthanide complexes require long pixel dwell times and therefore long image acquisition times.
In our lab, we adapted a conventional, epi-fluorescence microscope for time-gated imaging by incorporating a collimated, UV LED (365 nm) excitation source, an intensified CCD (ICCD) camera, and a pulse generator for synchronized triggering of source and detector.34 With this system, the intensifier component serves both as a shutter and as an emission signal amplifier, and the output of multiple excitation/emission cycles can be integrated on the CCD sensor during a single camera frame. The instrument is capable of rapidly (1–3 s) acquiring images of dim lanthanide specimens including 40 nm nano-spheres (containing ~400 Eu(III) complex molecules), Tb(III) complexes in the cytoplasm of living mammalian cells (cellular concentration, 1–10 μM), and long-lifetime (20–200 μs), non-metal luminescent probes.42 The ability to quickly acquire high-contrast images from a minimal number of emitting molecules is critical for live cell imaging, as overly long exposure times increase phototoxicity and limit temporal resolution. Under such imaging conditions, only a few tens of photons per pixel may be acquired from lanthanide emitters in a single camera frame. Therefore, the gain function of the intensifier is critical for producing images with adequate SNR.
PROTEIN-TARGETED LANTHANIDE COMPLEXES
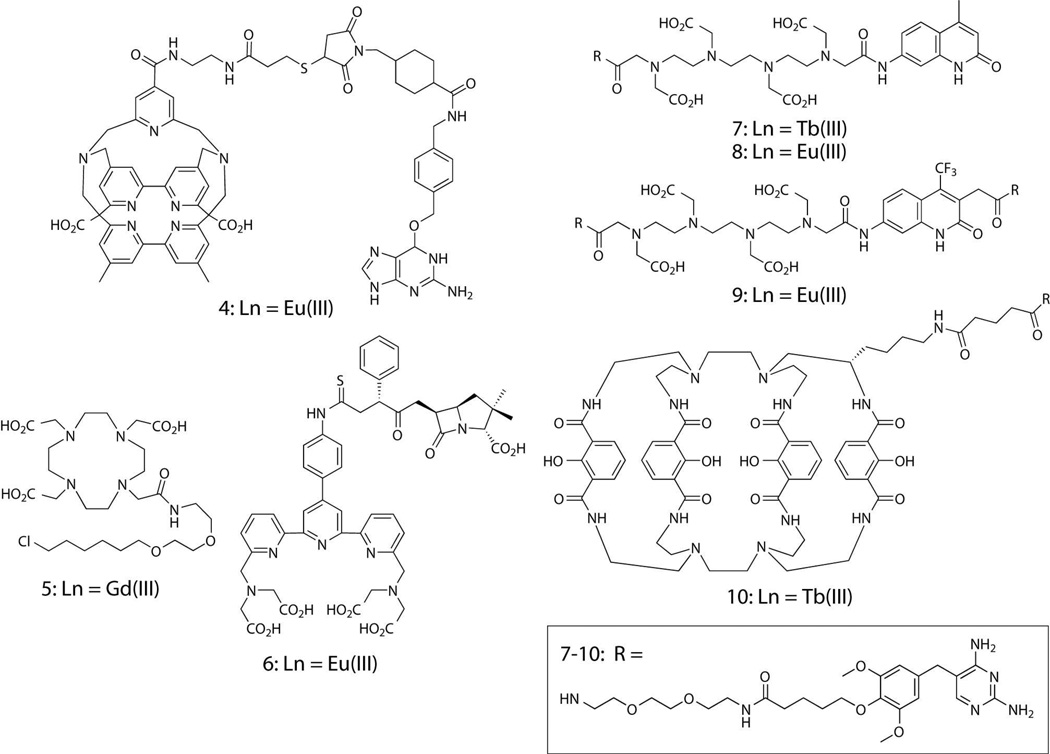
Time-gated imaging studies require a reliable means to selectively and stably label a protein with a lanthanide complex. A variety of hybrid, chemical-genetic labeling methods have been developed that can be used to attach fluorophores or other functional small molecules to proteins in live cells.14–16 These methods generally employ a strategy whereby target proteins are genetically encoded as fusions to a receptor protein, protein domain or peptide sequence. The small molecule probe consists of a receptor-binding ligand coupled to a fluorophore or other functional entity. Note that here, the term ligand refers to the receptor-targeting moiety and not to a metal complexing agent. Ideally, the ligand binds non-covalently or covalently to the recombinantly expressed, receptor protein fusion without appreciable non-specific binding to other cellular targets. Ligand-receptor methods have been used in a few cases to label proteins with luminescent lanthanide species (Fig. 3). Pin and co-workers covalently labeled g-protein coupled receptors (GPCRs) with Eu(III) and Tb(III) complexes for time-gated FRET spectroscopic studies of GPCR oligomerization. Heterodimers of the macrocyclic Tb(III) complex, Lumi4-Tb™, linked to O6-benzylguanine and O2-benzylcytosine as well as an O6-benzylguanine-Eu(III) cryptate analog (4) were used to tag GPCR subunits fused to mutant forms of human alkylguanine alkyltransferase (SNAP-Tag™ and CLIP-Tag™) on the surface of mammalian cells.43–44 The Meade lab reported increased relaxivity of chloroalkyl-derivatized, DOTA-Gd(III) MR contrast agents such as 5 upon binding to an engineered dehalogenase protein (HaloTag™) in vitro.45 Mizukami, et al. reported binding and time-gated luminescence imaging of 6, an ampicillin-terpyridinetetracetate-Eu(III) conjugate to cell-surface TEM-1 beta-lactamase.46
Figure 3. Ligand-lanthanide complex heterodimers that bind selectively to recombinant receptor fusion proteins in vitro or in live cells.
In our lab, we have leveraged the non-covalent interaction between the common antibiotic, trimethoprim (TMP) and the Escherichia coli form of dihydrofolate reductase (eDHFR) to develop protein-targeted lanthanide labels. TMP binds tightly (KD = ~1 nM) to eDHFR but weakly (KD > 1 μM) to mammalian forms of the enzyme, thereby allowing selective labeling of eDHFR fusion proteins in cultured mammalian cells or cell lysates.47–48 Because eDHFR is small (~18 kDa), soluble and monomeric, it can be fused to the N-or C-terminus of target proteins with minimal perturbation of native function. Conjugates of TMP linked to polyaminocarboxylate Tb(III) and Eu(III) complexes (7–9) and to the macrocyclic complex, Lumi4-Tb (10) retain characteristic Tb(III) luminescence and nanomolar affinity for eDHFR fusion proteins in purified preparations and in bacterial lysates.49–50 Moreover, TMP-linked Tb(III) complexes are excellent FRET donors to GFP. In an in vitro assay performed in 96-well plates, we titrated a fusion of GFP to FK506 binding protein 12 (GFP-FKBP12) against a fixed concentration of eDHFR fused to the rapamycin binding domain of mTOR (FRB-eDHFR). Following addition of rapamycin (to induce protein interaction) and TMP-Tb(III) complex 7, we observed a ~3500% increase in long-lifetime, Tb-to-GFP sensitized emission.51 The high dynamic range of Tb(III)-based FRET (>30-fold) greatly exceeds the best dynamic range for non-dimerizing fluorescent proteins (<5-fold) observed in vitro and hints at the potential sensitivity of lanthanide-based FRET microscopy.24, 52
IMAGING PROTEIN-PROTEIN INTERACTIONS: PROOF-OF-PRINCIPLE
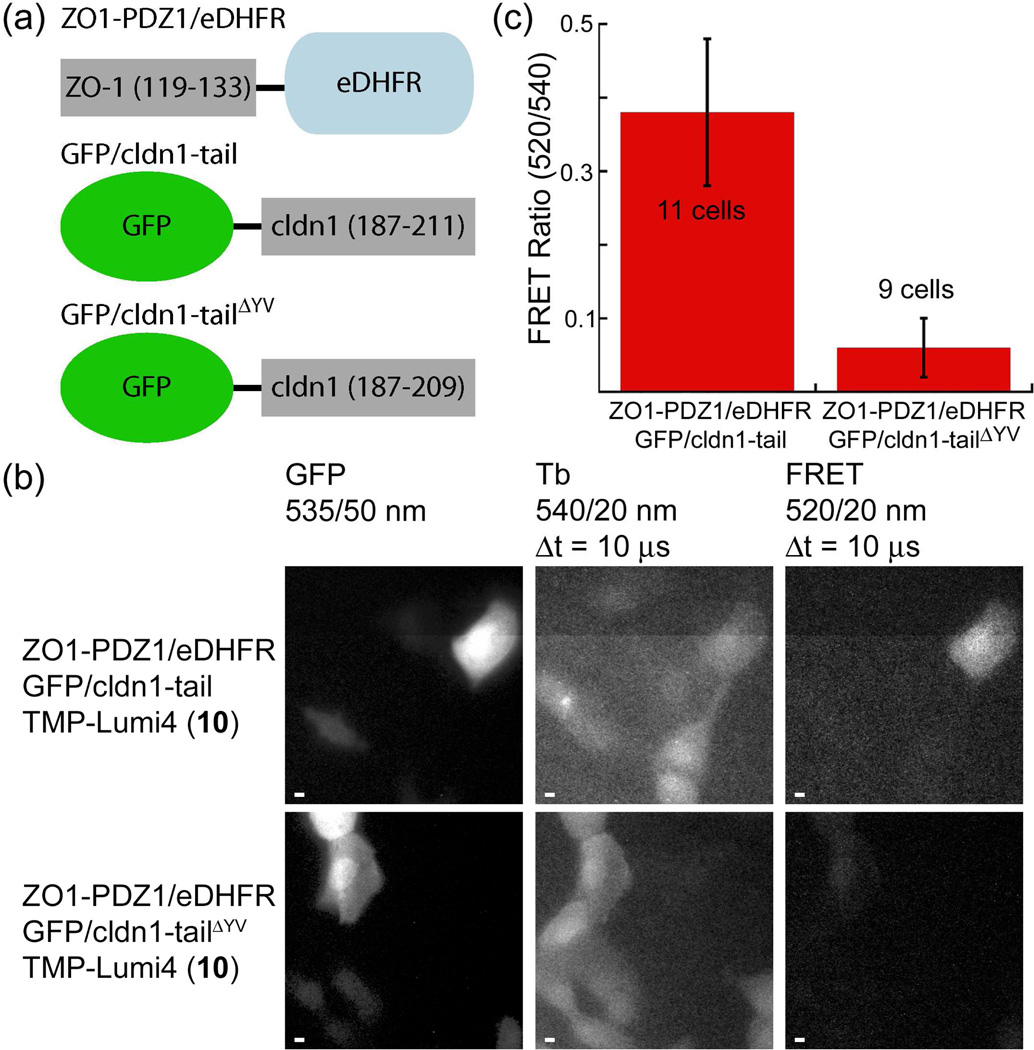
We first demonstrated successful time-gated imaging of protein-protein interactions in living cells using a TMP-Lumi4 heterodimer (10).53 As a model, we imaged the association of the most N-terminal PDZ domain of ZO-1 with the C-terminal binding motif of the transmembrane protein, claudin-1. In epithelia, ZO-1 directs polymerization of claudins which is necessary to form tight junctions,54 and a direct interaction between ZO-1/PDZ-1 and the C-terminal cytoplasmic tail domain of claudin-1 has been demonstrated using recombinant proteins.55 Maden Darby canine kidney (MDCKII) epithelial cells were transiently co-transfected with plasmid DNA encoding a C-terminal fusion of eDHFR to the PDZ1 domain (residues 19–113) of ZO-1 (ZO1-PDZ1/eDHFR) and an N-terminal fusion of EGFP to the C-terminal cytoplasmic domain (residues 187–211) of claudin-1 (GFP/cldn1-tail, see Fig. 4a). We used osmotic lysis of pinosomes to load the otherwise membrane-impermeable 10 into the cytoplasm of transfected cells. Following intracellular delivery, time-gated FRET imaging revealed Tb-sensitized GFP emission only in transfected cells loaded with 10, providing an unambiguous image of the protein-protein interaction (Fig. 4b, top). By contrast, only faint FRET signals were seen in negative control cells that expressed a GFP/cldn1-tail construct lacking the C-terminal YV motif (GFP/cldn1-tailΔYV), which is unable to bind to ZO-1, along with ZO-1/PDZ1-eDHFR (Fig. 4b, bottom). Furthermore, there was a highly significant (P < 10−6), >500% difference between the mean, donor-normalized FRET signal (520 nm/540 nm emission ratio) from cells expressing ZO1-PDZ1/eDHFR and GFP/cldn1-tail and that seen from cells expressing non-interacting, ZO1-PDZ1/eDHFR and GFP/cldn1-tailΔYV (Fig. 4c).
Figure 4. Time-gated FRET microscopy detects protein-protein interactions in live cells with high confidence in single-frame images.
(a) Schematic of fusion proteins. (b) MDCKII cells were loaded with a TMP-Lumi4(TbIII) heterodimer (10) by osmotic lysis of pinosomes. Cells co-expressing indicated fusion proteins exhibited steady-state, GFP fluorescence (left). Time-gated detection of Tb(III) luminescence (middle) revealed cells loaded with 10. Tb(III)-to-GFP FRET (right) is seen in cells co-expressing ZO1-PDZ1/eDHFR and GFP/cldn1-tail and loaded with 10. No FRET signal is visible in cells co-expressing non-interacting ZO1-PDZl/eDHFR and GFP/cldn1-tailΔYV and loaded with 10. Micrographs: GFP fluorescence, λex = 480/40 nm; time-gated luminescence, λex = 365 nm, Δt = delay between excitation and detection. Emission at indicated wavelengths. Scale bars, 5 μm. (c) A significant (P < 10−6), >500% difference in the mean, donor-normalized FRET emission ratio (520/540 nm) was observed between cells expressing interacting and non-interacting fusion proteins. Mean ratios calculated from indicated sample size. Error bars, s.d. Data adapted from Rajapakse, et al.53
The initial, proof-of-principle study yielded three key results that support and guide our ongoing efforts to develop time-gated microscopy with lanthanides as a viable and, in some measures, improved alternative for live-cell FRET imaging. First, compound 10 diffuses freely throughout the cytoplasm and nucleus without detectable nonspecific binding and stably labels eDHFR. To our knowledge, this was the first reported example of selective, intracellular protein labeling with a lanthanide complex, and it suggests that other ligand/receptor tagging methods and improved forms of cytoplasmic probe delivery should prove equally useful. Second, 10 is sufficiently bright to enable single-channel, TGD of intermolecular, Tb(III)-to-GFP sensitized emission with image acquisition times of 1–4 s. These acquisition times are on the order of those seen with muti-channel, fluorescent protein-based, Filter-FRET measurements made on optimized microscopes;24 further improvements may increase the time resolution of live-cell FRET imaging. Moreover, we observed that 10 remains stably luminescent in cells for hours and photobleaches with a lifetime of ~2 min when excited with 365 nm light at the irradiance levels used on our system (~0.5 mW/cm2). As typical exposure times are 1–4 s with the LED pulsing at 50% duty cycle, ~30–100 images may be acquired before significant (>10%) photobleaching occurs. Third, elimination of background fluorescence allows FRET signals from positive (with interacting proteins) and negative (with non-interacting proteins) control cell samples to be distinguished with a high degree of confidence. Thus, lanthanide-based FRET may produce fewer false negative results in comparison to conventional FRET imaging techniques.
CELLULAR INTERNALIZATION AND LOCALIZATION OF LANTHANIDE COMPLEXES
Any exogenous, luminescent probe or label intended for use in live-cell imaging experiments should ideally have the following properties: 1) the molecule should passively diffuse through lipid bilayers, thereby allowing delivery into the cell interior by simple addition to culture medium; 2) the molecule should diffuse freely throughout all intracellular organelles and compartments, making it possible to study the widest range of biological phenomena; 3) the molecule should not bind extensively or tightly to endogenous molecules, therefore allowing it to sense or interact with the target molecule of interest.16, 56 Delivery by passive diffusion makes it possible to control, to a certain extent, the overall amount of fluorophore that accumulates within the cells by varying the extracellular probe concentration and incubation time. If the probe does not bind appreciably to endogenous molecules, the cytoplasmic concentration will reach a steady-state level that never exceeds the extracellular concentration. Minimal levels of non-specific binding are desirable because affinity for large macromolecules can sequester probes away from, say, target proteins or prevent diffusion into certain sub-cellular compartments.56 For example, the nuclear pore complex allows passage of macromolecules <40 kDa; therefore, binding of probes to large, cytoplasmic proteins would prevent entry into the nucleus.57 In practice, very few small molecule fluorophores and, to our knowledge, no lanthanide complexes meet all of the aforementioned criteria.
Membrane permeability, intracellular distribution, and non-specific affinity for endogenous biomolecules are dependent on physico-chemical properties including molecular size, charge, and lipophilicity. As with drug molecules, smaller, uncharged and more lipophilic probes are more likely to be membrane permeable.58 Compounds with ionizable groups (e.g., carboxyl or sulfoxyl groups) are usually impermeant to cell membranes and only enter cells via facilitated diffusion through membrane channels or via endocytosis. Acylation of ionizable oxygens and protection of carboxyl groups with esterase-labile acetoxymethyl or other esters is a common strategy to make probes charge-neutral and thus enhance uptake kinetics.48, 59–60 Charge may also affect intracellular distribution. Certain cationic dyes such as rhodamines partition to respiring mitochondria which exhibit a negative potential across their inner membranes.61 Anionic dyes such as fluorescein generally seem to exhibit less non-specific binding and more even intracellular distribution. The apparent lack of intracellular, non-specific binding seen with 10 may be due to its anionic character (net charge = −1), as most cytoplasmic proteins and DNA are anionic at physiological pH.62
As lanthanide complexes are hydrophilic and often charged, they are unlikely to diffuse passively through lipid membranes. Therefore, effective intracellular delivery requires structural modifications that favor alternative uptake pathways (e.g., endocytosis) or experimental loading techniques that breach the plasma membrane. Direct microinjection of complexes into cells is a straightforward way to achieve cytoplasmic delivery,63 but this method requires expensive apparatus and considerable technical expertise. We have successfully adopted established biochemical methods of cytoplasmic delivery to enable intracellular protein labeling with the TMP-Lumi4 heterodimer, 10, including osmotic lysis of pinocytic vesicles64 and reversible membrane permeabilization with the bacterial toxin, streptolysin O (SLO).65 While we showed that both loading techniques can be applied without compromising cell viability,53 their use requires careful optimization to achieve consistently uniform and high loading levels. Conjugation of lanthanide complexes to cell-surface receptor-targeting ligands or hydrophobic molecules has been shown to enhance cellular uptake. Bornhop and coworkers linked a cyclen-based chelator to PK11195, an isoquinoline carboxamide that binds selectively to the peripheral benzodiazepine receptor (PBR).66 Cellular uptake and multimodal (magnetic resonance and luminescence) imaging of C6 glioblastoma cells was achieved following administration of a cocktail of PBR-targeted Eu(III) and Gd(III) complexes. The Meade group reported intracellular accumulation and progesterone-mediated transcription of a reporter gene following exposure of T47D breast cancer epithelial cells to progesterone-DOTA-Gd(III) conjugates.67 More recently, Nagano reported enhanced cellular uptake of DOTA-Gd(III) conjugated to hydrophobic dyes including Cy7 and boron dipyrromethene.68
Parker and co-workers have done extensive work to explore the effects of probe structure on the cellular uptake and compartmentalization behavior of more than 60 cyclen-based Tb(III) and Eu(III) complexes.69 The majority (~80%) of compounds studied were microscopically observed to reside in endosomes or lysosomes following uptake into cells.70 Similar cellular distributions were observed for helical, dinuclear Eu(III) complexes reported by Bünzli.71–73 Smaller subsets of Parker’s compounds distributed in mitochondria,74 or ribosomes and nucleoli.75–77 From this series of work, it was found that the nature of the heteroaromatic sensitizing chromophore and its mode of attachment to the cyclen chelator, and not complex charge or lipophilicity, was the primary determinant of uptake kinetics and intracellular trafficking. Additional studies by Parker showed substantial variation in uptake kinetics, sub-cellular distribution and cytotoxicity of 11 Tb(III) complexes variable only at a single position on the sensitizing chromophore.39, 78 These observations are consistent with follow-up studies that implicated macropinocytosis as the cell uptake mechanism for cyclen-based complexes.79–80 Macropinocytosis involves non-specific (i.e., receptor-independent) invagination and trapping of extracellular fluid and solutes within large (0.5–5 μm) cytoplasmic vesicles that subsequently fuse with endosomes or lysosomes.81 Once taken into cells, probes may escape from inherently leaky macropinocytotic vesicles into the cytoplasm. There, they may be expected to traffic to cellular substructures in a manner dependent on their propensity to bind to endogenous membranes or biomolecules.
CELL PENETRATING PEPTIDES FOR LANTHANIDE COMPLEX DELIVERY
Besides direct structural modification or conjugation to receptor-targeting ligands, covalent coupling to cell penetrating peptides (CPPs) offers another means to effect enhanced cellular uptake of lanthanide complexes. CPPs are a class of short (10–30 amino acids), cationic peptides that have been shown in numerous studies to traverse biomembranes.82–87 Considerable efforts have been made to characterize and optimize CPPs as molecular transporters that facilitate the passage of nucleic acids, proteins, therapeutics and imaging agents into cells.86–87 CPPs and their attached cargo can enter cells by two distinct routes: endocytosis and direct translocation through the lipid bilayer.82–85 The extent to which either mechanism is operative depends on numerous factors including the CPP sequence, the size and type of cargo, cell type, and the composition of the culture medium. Arginine-rich peptides such as oligoarginines or a fragment of the HIV-1 trans-activating transcriptional activator (Tat, residues 49–57) are a class of CPPs that exhibit high transduction efficiency.85 Studies using fluorophore-labeled oligoarginine and Tat reveal that these peptides utilize both endocytic and non-endocytic pathways. At lower micromolar concentrations, uptake has been observed to occur via macropinocytosis, clathrin-mediated endocytosis and caveolae/lipid-raft-mediated endocytosis. However, above a threshold concentration (5–10 μM), uptake occurs via an energy-independent, non-endocytic pathway that leads to diffuse cytosolic and nuclear distribution of the peptides.82–84
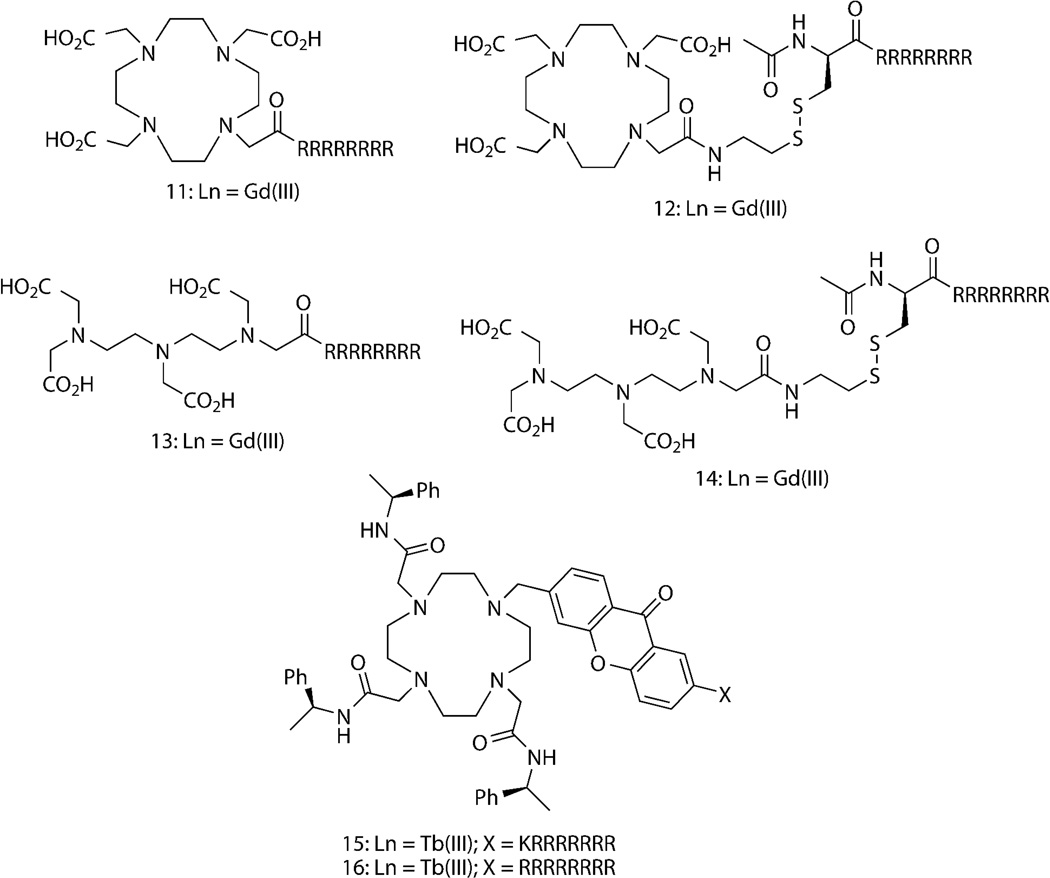
CPPs have been covalently attached to lanthanide complexes to enhance cell uptake and retention (Fig. 5). Both DOTA and DTPA Gd(III) chelates have been covalently linked to Tat and oligoarginine peptides to aid cell uptake.88–89 Meade and coworkers have extensively explored oligoarginine as a delivery vehicle for magnetic resonance contrast agents.90–94 Conjugation to a peptide containing at least eight arginines, as shown for DOTA and DTPA analogues, 11 and 13 was found to be necessary for sufficient cell uptake.91 Compounds 12 and 14, which incoroporate a disulfide bond between DOTA-Gd(III) and DTPA-Gd(III) and octaarginine exhibited enhanced retention in NIH/3T3 fibroblasts because the disulfide was cleaved in the reducing environment of the cell, presumably preventing peptide-mediated export.93 Kielar, et al. observed rapid (<5 min) internalization and endosomal localization of cyclen-based Tb(III) chelates that contained an N-coordinated, azaxanthone sensitizer conjugated to oligoarginines (compounds, 15 and 16 ).39, 78 A recent study by Thielemann, et al. reported cellular uptake of pentadecanuclear Tb(III) and Eu(III) clusters linked to multiple, cell penetrating peptoid monomers. Confocal microscopy revealed distribution of the luminescent clusters within endosomes, cytoplasm and nucleus of HeLa cells, suggesting endocytosis and subsequent escape from endosomes/lysosomes as the mechansism of cell entry.95
Figure 5. Reported lanthanide complexes conjugated to oligoarginine, cell penetrating peptides (CPPs) that exhibit enhanced uptake into mammalian cells.
Abbreviations: X = peptide sequence; capital letters, L-amino acids.
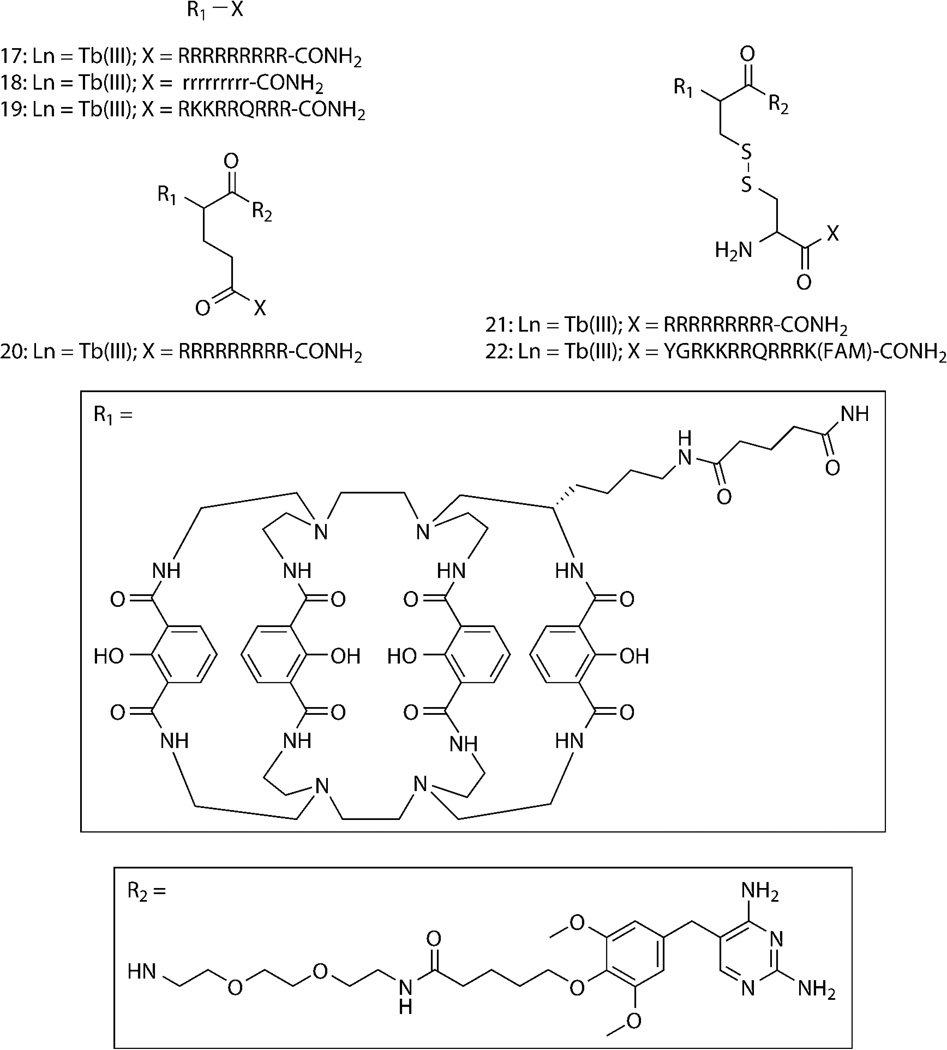
We recently reported that covalent coupling to CPPs mediates cellular delivery of Lumi4-Tb and TMP–Lumi4 heterodimers and specific labeling of nucleus-localized eDHFR fusion proteins.96 Several conjugates were prepared as N-terminal fusions of either Lumi4 or a heterodimer of Lumi4 and TMP to variations of nonaarginine (R9) or Tat (Fig. 6). We used TGD of Tb(III) luminescence or Tb(III)-mediated FRET signals to assess cell uptake, sub-cellular peptide distribution and specific intracellular protein labeling. For both Tat and R9 conjugates, the mechanism of cell uptake and the resultant cellular distribution depended on the effective extracellular peptide concentration (Fig. 7a). At low concentrations, Tb(III) conjugates enter cells via an endocytic pathway, resulting in a punctate cellular distribution. Above a threshold concentration, uptake occurs via direct translocation from culture medium to cytoplasm, as evidenced by a diffuse distribution of Tb(III) luminescence throughout the cell. Incubation in medium lacking fetal bovine serum lowered the threshold concentration for observing diffuse staining (e.g., from 20 μM to 5μM for 17, Fig. 7a), presumably because R9 binds to serum proteins, thus lowering its effective concentration in complete medium.84 Observation of diffuse luminescence in MDCKII cells incubated at 4°C in serum-free medium containing 17 (10 μM) provided further evidence that the uptake at higher effective concentrations occurs through an energy-independent, non-endocytic translocation mechanism (Fig. 7a). We were able to identify experimental conditions that enabled cytoplasmic delivery of all the prepared conjugates into a variety of cell types including NIH/3T3, HeLa, and HEK293 cells.
Figure 6. Structures of Lumi4-Tb(III) and TMP-Lumi4 heterodimers linked to nonaarginine and Tat-derived CPPs.
Abbreviations: R1, linker-functionalized derivative of Lumi4; R2, triethyleneglycolamino derivative of TMP; capital letters, L-amino acids; small letters, D-amino acids; FAM, 5,6-carboxyfluorescein.
Figure 7. Arginine-rich, cell penetrating peptides (CPPs) conjugated to Lumi4-Tb(III) analogs directly translocate from culture medium to cytoplasm and TMP-Lumi4-Tb(III) peptide conjugates bind to eDHFR following cytoplasmic delivery.
(a) Micrographs of time-gated luminescence (delay = Δt = 10 μs, λex = 365 nm, λem = 540/20 nm); scale bars, 5 μm. MDCKII cells were incubated for 30 min at indicated temperatures in Dulbecco’s modified Eagle’s medium (DMEM) with (+) or without (−) fetal bovine serum (FBS, 10% v/v) that contained indicated concentrations of 17, a conjugate of Lumi4-Tb(III) to (L)-R9. Incubation in DMEM with FBS containing low (5 μM) concentration of 17 results in punctate Tb(III) luminescence (top, left) whereas incubation in DMEM with FBS above a threshold concentration (20 μM) results in diffuse distribution of Tb(III) luminescence throughout the cytoplasm and nucleus (top, right). Incubation in DMEM without FBS lowers the threshold concentration (5 μM) for cytoplasmic delivery (bottom, left). Incubation at 4 °C results in a diffuse staining pattern (bottom, right), suggesting an energy-independent, direct translocation uptake mechanism. (b) MDCKII cells transiently expressing H2B–TagRFPT–eDHFR were incubated for 30 min at 37 °C in DMEM without FBS containing TMP-Lumi4-Tb(III) linked to the N-terminus of (L)-R9 (20, 10 μM). A steady-state fluorescence (λex = 545/30 nm, λem = 610/70 nm) image reveals nucleus-localized TagRFP-T fluorescence in expressing cells (top left). Time-gated images (delay = Δt = 10 μs, λex = 365 nm, λem as indicated) of Tb(III) luminescence (top, right) and Tb(III)-sensitized, TagRFP-T emission (bottom, left) show that nucleus-localized FRET signals occur in expressing cells loaded with 20. The sensitized emission signal disappears when TMP (final concentration = 100 μM) was added to medium (bottom, right); scale bars, 5 μm. Data adapted from Mohandessi, et al.96
Time-gated FRET imaging was used to directly visualize binding of CPP-conjugated, Lumi4-TMP heterodimers to an eDHFR fusion protein in live cells. We observed specific labeling of a three-component protein chimera consisting of histone 2B linked to the red fluorescent protein TagRFP-T97 and eDHFR (H2B–TagRFPT–eDHFR). Following incubation of MDCKII cells in medium containing 20, time-gated imaging of Tb(III)-to-TagRFP-T sensitized emission revealed nuclear luminescence that coincided with the steady-state fluorescence of TagRFP-T in expressing cells (Fig. 7b). When unconjugated TMP was added to the imaging medium, it diffused into cells, competed with 20 for eDHFR binding, and eliminated the FRET signal (Fig. 7b), providing further evidence of a direct interaction between 20 and H2B-TagRFPT-eDHFR. Similar results were seen with 21, a cysteine amide-linked heterodimer of Lumi4 and TMP conjugated to CR9 via a reducible disulfide bond. Intracellular reduction and cleavage of disulfide bond was experimentally confirmed using compound 22, a cysteine amide-linked heterodimer of Lumi4 and TMP via a disulfide bond to the Tat-derived peptide CYGRKKRRQRRRK(FAM). The intramolecular FRET signal between Tb(III) and a fluorescein moiety linked to the lysine side chain was observed to diminish within 30 min to 2 h following cell loading.
The ability to directly deliver protein-targeted, Tb(III) complexes into the cytoplasm of live cells with technically simple, mix-wash-image experimental protocols will enable time-gated FRET imaging of a wide variety of target molecules. Furthermore, our results suggest that conjugation to the N-terminus of R9 or Tat peptides via amide or disulfide linkages may offer a general strategy for the controlled cellular delivery of a wide variety of membrane-impermeable probes and protein labels. We are currently assessing the cell uptake performance of other R9-linked probes including benzyl guanineand benzyl cytosine-Lumi4-Tb(III) heterodimers that bind to SNAP and CLIP fusion proteins,98 polyaminocarboxylate Eu(III) complexes, and cyanine dyes. Additionally, we are working to quantify the extent to which the intracellular concentration of CPP conjugates can be controlled by varying experimental labeling conditions.
QUANTITATIVE TIME-GATED MICROSCOPY WITH LANTHANIDES
The quality of fluorescence microscope images, as indicated by perceived contrast, resolution and SNR, is a function of the number of photons acquired by the imaging system.99 Photon acquisition, in turn, depends on the microscope optics (magnification, numerical aperture, transmittance), detector quantum efficiency, specimen properties (thickness, transparency), luminescent probe characteristics (brightness, lifetime, concentration), and image acquisition parameters (excitation light intensity, exposure time). In practice, live cell imaging requires adequate photon acquisition from a limited number of fluorophores. For example, over-expressed fluorescent proteins and exogenous fluorescent sensors are typically imaged in mammalian cells at concentrations of 1–10 μM (106–107 copies per cell).100–101 These cellular concentration levels reflect a need to balance detectability with minimal perturbation of physiological function. Assuming a diffuse cytoplasmic distribution, a diffraction limited volume element (~1 fL) may contain only ~100 fluorescent molecules. Furthermore, exposure times are limited by the need to resolve dynamic biological processes and to limit photobleaching of probes and photodamage to cells. Therefore, there is always a trade-off between image quality and temporal resolution.
When imaging biosensors in living cells, the goal is often to measure, as precisely as possible, dynamic changes in emission intensities over a range that reflects biochemically relevant changes in biosensor activity (e.g., changes in level of interaction between two proteins).19 The precision of intensity measurements is characterized by SNR which, for a Poissonian process, cannot exceed the square root of the number of photons/pixel in the image.99 In practice, it is desirable to utilize the full dynamic range of a biosensor by imaging under conditions where the maximum emission intensity approaches detector saturation. The presence of background light from, for example, cellular autofluorescence, increases photon noise and reduces the effective capacity of the detector, thus reducing both SNR and dynamic range. Maximal SNR is particularly important for Filter-FRET or ratiometric imaging where two or more raw images are processed post-acquisition to quantify signal changes. When mathematical operations are performed on raw images, the stochastic noise adds to the total noise levels in the resulting ratio image.31
The ability to eliminate cellular autofluorescence and, for FRET imaging, directly excited acceptor fluorescence would seem to favor time-gated microscopy with lanthanide probes over steady-state fluorescence imaging with conventional organic fluorophores or fluorescent proteins. However, with long emission lifetimes and commensurately low photon emission rates, it is not clear whether the SNR achievable with lanthanides is high enough to quantitatively image biosensor activity under biologically relevant conditions. The Hahn lab is at the forefront of biosensor design and application, and their genetically encoded, FRET-based sensors of Rho-family GTPase activity represent a performance benchmark against which other sensor designs may be compared.102–103 Hahn has shown that single-chain and dual-chain biosensor activity can be quantitatively imaged in the cytoplasm of live cells at effective cellular concentrations of a few μM with exposure times of 0.3—0.9 s per image.103 They used a Filter FRET-based approach, collecting two (for single-chain sensor) or three (for dual-chain sensor) images at successive time-points, yielding total image acquisition times up to several seconds on a microscope with motorized filter changers. Each image typically has SNR of at least 3 in the darkest regions. With these performance characteristics as a point of comparison, we have initiated studies to quantitatively assess lanthanide-based FRET imaging.
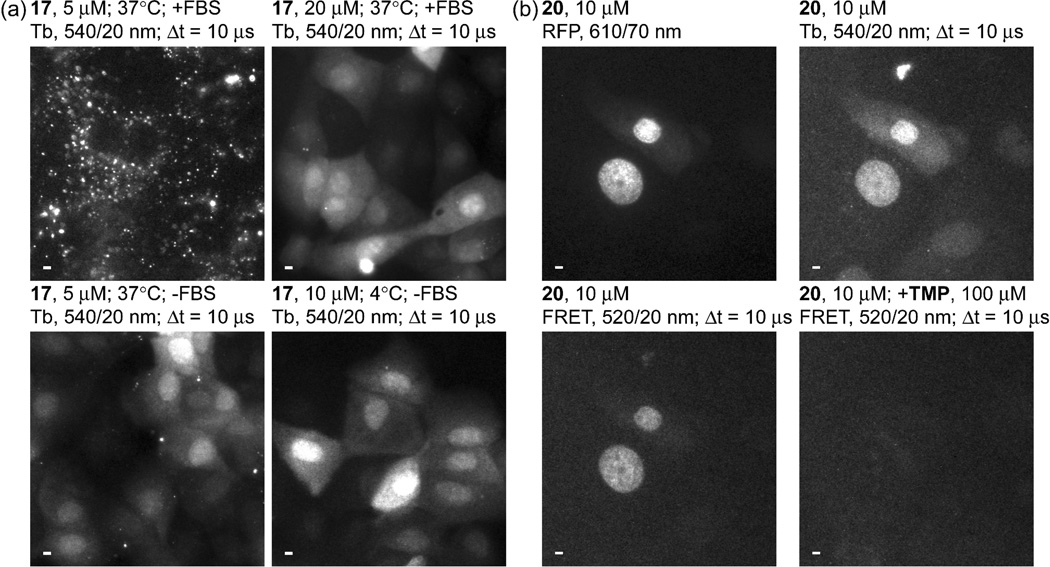
The rapamycin-induced interaction between FKBP12 and FRB51, 104 presents an ideal model system for quantifying the detection limit, SNR, dynamic range and other measures of image quality that may be observed with time-gated FRET imaging of protein-protein interactions. Because both proteins diffuse throughout the cytoplasm and nucleus when expressed in mammalian cells,105 cytoplasmic signal levels are proportional to total cellular protein concentration. Moreover, the degree of interaction can be controlled by titrating rapamycin levels, allowing for direct measurement of FRET dynamic range. In a preliminary experiment, we transiently co-transfected MDCKII cells with DNA encoding GFP-FKBP12 and FRB-eDHFR. Following induction with rapamycin (200 nM, 1 h), cells were loaded with compound 21, a cell-permeable conjugate of Lumi4-TMP linked to CR9. Time-gated imaging revealed Tb-to-GFP, FRET-sensitized emission only in expressing cells that were loaded with probe (Fig. 8a).
Figure 8. Interaction between GFP-FKBP12 and FRB-eDHFR imaged at high signal-to-noise ratio (SNR).
(a) MDCKII cells transiently co-expressing GFP-FKBP12 and FRB-eDHFR were exposed to rapamycin (200 nM, 1 h) and subsequently incubated for 10 min at 37 °C in DMEM without serum containing 21 (7.5 μM), a Lumi4-CysTMP heterodimer linked via a disulfide bond to CR9. The five images shown represent a single field of view acquired under different conditions: BF, bright field; GFP, steady-state fluorescence reveals expressing cells; Tb, time-gated emission (494 nm) shows which cells were loaded with 21; FRET, rightmost 2 images show Tb-to-GFP FRET signal observed in cells that both express fusion proteins and contain 21. Time-gated Tb luminescence and FRET micrographs (delay = Δt = 10 μs, λex = 365 nm, λem as indicated) were generated by summing 4 frames of indicated length to yield single, 12-bit images. SNR and photons/pixel were calculated for a region of interest in the cytoplasm (e.g., square in right-most image). SNR was calculated as the mean, background-subtracted pixel gray value in the ROI divided by its standard deviation. Values given are range for a 10-cell sample. FRET images are represented at identical contrast levels. Scale bars, 5 μm. (b) Bar chart represents the mean, donor-normalized FRET emission ratio (520/540 nm) observed in FRET positive (+rapamycin) and FRET negative (-rapamycin) cells imaged at 0.667 s frame lengths. Error bars, s.d. (c) Relative MDCKII cell metabolic activity assessed with MTT assay. Bar chart shows mean absorbance at 550 nm as a percentage of positive control (DMEM only) following incubation with 21 (30 min) at indicated concentrations (3 replicates for each condition). Error bars, s.d.
Time-gated FRET images were acquired at frame lengths of 0.333 s and 0.667 s, and 4 frames were summed to increase image SNR, yielding total acquisition times of 1.33 s and 2.67 s, respectively. We measured the mean, background-subtracted pixel gray values for ROIs in the cytoplasm and calculated the GFP/Tb emission ratio, number of photons/pixel and SNR (defined as the mean gray value divided by the standard deviation within the ROI) for a 9-cell sample. There was a highly significant (P = 0.0005), ~10-fold difference between the mean, cytoplasmic GFP/Tb emission ratio from FRET-positive cells imaged at 0.667 s frame lengths (+rapamycin, 1.4 ± 0.5) and FRET-negative cells (−rapamycin, 0.14 ± 0.10; mean ± s.d., Fig. 8b). The mean number of photons/pixel observed in the cytoplasm ROIs ranged from ~50–180 for Tb(III) luminescence and Tb-to-GFP FRET under the indicated imaging conditions, corresponding to SNRs of ~6–11. An MTT assay was used to assess MDCKII cell redox activity following exposure to 21. No difference in MTT reduction was observed in cells exposed to 21 (5 uM, 20 uM) for 30 min relative to that observed in positive control cells (Fig. 8c), suggesting that CPP-linked Lumi4 analogs are non-toxic under typical exposure conditions used for these probes.
These results suggest that Tb(III)-mediated FRET microscopy can be used to quantitatively image protein-protein interactions in live cells. For accurate quantification of time-gated FRET signal changes, it should be sufficient to take a ratio of the FRET emission over the donor emission, necessitating only two images at each time point. Importantly, the SNR observed here exceeded the minimum value (5) considered acceptable for pixel-wise, two-color ratiometric FRET imaging,106 even at acquisition times of only ~1.3 s. Two cameras can be mounted via a beamsplitter to simultaneously acquire FRET and donor images, thereby enabling single-frame imaging. Such an approach would improve temporal resolution in comparison to conventional Filter FRET measurements that require 3 images to accurately quantify bimolecular interactions. Further studies are underway in our lab using FKBP12/FRB and other model systems to quantify dynamic range and detection limits for time-gated FRET imaging. As transient transfection yields widely varying expression levels of each fusion protein in different cells, we are preparing stable cell lines where GFP-FKBP12 is constitutively expressed at modest levels and FRB-eDHFR expression is under control of an inducible promoter. Using this construct, we will be able to obtain statistically robust measurements of image quality and relate these measurements to the total amount of protein expressed. We are also developing image analysis protocols that will allow for Tb(III)-mediated FRET quantification on a pixel-wise basis, which is the standard for live cell biosensor imaging.103
PHOTOXICITY OF UV EXCITATION LIGHT USED IN LANTHANIDE IMAGING
All Tb(III) complexes and most Eu(III) complexes with sufficient brightness and photostability for cellular imaging require near-UV (<380 nm) light for single-photon excitation. Because UV light is considerably more cytotoxic than visible or NIR radiation, lanthanide complexes have more limited applicability in live cells than conventional fluorophores. For chemists working in the field of lanthanide microscopy, it is helpful to understand the mechanisms of UV-mediated photoxicity and the extent to which it poses a problem for live-cell studies. Such understanding can guide the design of novel probes or experimental approaches that overcome photoxicity limitations and expand the usefulness of lanthanide-based imaging methods.
Phototoxicity results from the production of reactive oxygen species (ROS) including superoxide (•O2−), hydroxyl radical (HO•), various peroxides and, especially, singlet oxygen (1O2). ROS react with a variety of easily oxidizable cellular components, such as proteins, nucleic acids, and membrane lipids.107 Native fluorescent species or exogenous fluorescent labels act as photosensitizers, reacting in their triplet excited state with dioxygen (3O2) to produce singlet oxygen (1O2). The magnitude of observed phototoxic effects, then, depends largely on the level of singlet oxygen production which, in turn, depends on several variables including the concentration, sub-cellular location and photochemical nature of sensitizing fluorophores, and the intensity and wavelength of the excitation light. For a given excitation intensity, UV light is more toxic than visible or near-infrared (NIR) light because there are a variety of endogenous fluorescent species present in cells that can absorb UV light and sensitize ROS production, including flavins, NAD(P)H and porphyrins.108 For example, Gorgidze et al. observed wavelength-dependent, mitotic inhibition in pig kidney embryo cells with near-UV (360 nm) light being 5 times more inhibitory than blue light (423–488 nm) and >50 times more inhibitory than green light (>500 nm).109
When the mechanism of UV-induced photodamage is orthogonal to the biological process being investigated, or when UV exposure is limited, reliable experimental results can be obtained. For example, UV microscopy has been successfully used to image a number of cellular processes, including calcium signaling110–111 and cholesterol distribution.112 UV illumination is also commonly used for the photolytic activation of caged compounds.113–114 Because our lanthanide imaging methodology requires only low exposures (~500 mW/cm2) of short duration (1–3 s), it should be possible to obtain reliable results for many important cellular processes before damage accumulates, including cytoskeletal dynamics, golgi dynamics, or signaling molecule interactions that occur on time scales of seconds to a few minutes. As with any biological study, sufficient controls will be needed to ensure that experimental conditions do not affect results. Moreover, addition of oxygen scavengers (e.g., OxyFuor) or antioxidants such as ascorbic acid can be added to imaging media to minimize photodamage and prolong imaging experiments.115 Therefore, lanthanide imaging with existing probe and microscope technologies should be immediately useful for a variety of biological applications.
Lanthanide probes that can be excited by visible light or by two-photon absorption in the NIR offer another means to overcome the limitations imposed by UV, single-photon excitation. A number of Eu(III) complexes with appreciable absorptivity at ~400 nm have been developed using acridone, coumarin or azathiaxanthone as sensitizers.76, 116–118 The previously mentioned pentadecanuclear Eu(III) clusters reported by Thielemann, et al. also absorb in this wavelength range.95 Recent work by Maury and co-workers shows that push-pull, aryl-alkynyl chromophores can sensitize Eu(III) emission from a charge transfer excited state, extending absorbance maxima well into the visible spectrum.119 Such chromophores were recently used to develop very bright (εϕem > 30,000 M−1cm−1), stable, triazacyclononane Eu(III) complexes,120–122 albeit with shorter wavelength absorption (λmax = ~330 nm). Moreover, the charge transfer character of the ligands can be tuned by varying donor substituents, allowing high two-photon excitation cross-sections (>700 GM at 740 nm) and effective sensitization of Eu(III) luminescence.123–124 At the present time, time-gated, multiphoton excitation with standard beam-scanning microscopeis likely to be impractical for live-cell imaging because the long emission times of lanthanide complexes would require prohibitively long exposure times.38 However, the ongoing development of fast-acquisition, two-photon microscopes,125–127 coupled with further development of multiphoton-excited lanthanide bioprobes opens up a new frontier for lanthanide-based cell imaging.
CONCLUSIONS AND FUTURE PERSPECTIVES
While TGD with ms-lifetime Tb(III) and Eu(III) lumiphores is firmly established for high-throughput screening and in vitro assays, the development of time-gated microscopy and lanthanide probe technologies for live-cell imaging has begun to accelerate. Increased understanding about the mechansisms by which lanthanide complexes interact with and enter cells will guide future development of lanthanide sensors and protein labels. The ability to directly load luminescent Tb(III) complexes into the cytoplasm of live cells by conjugating them to CPPs and to selectively label intracellular fusion proteins (Fig. 7) will enhance the development of lanthanide-based biosensors and time-gated FRET imaging of protein interactions and activity. The sensitivity of time-gated microscopes based on UV LED light sources and gated, ICCD cameras makes it possible to image lanthanide luminescence and lanthanide-mediated FRET signals of protein-protein interactions at levels of protein expression and temporal resolution that are comparable to conventional fluorescence imaging (Fig. 8). Further improvements in probe design, CPP-mediated delivery and quantitative time-gated microscopy will serve to make lanthanide-based imaging a routine tool for biological investigations.
EXPERIMENTAL SECTION
Materials and Reagents
Materials listed here and protocols given below pertain to data presented in Figure 8. Experimental methods for previously reported data (Figs. 4, 7) may be found in the cited references. Enzymes and cloning reagents were obtained from New England Biolabs, Inc. Cell culture reagents were purchased from Life Technologies, Inc. Plasmid vectors pRSETB-EGFP-FKBP12 and pRSETB-FRB-eDHFR were described previously.51 MDCKII cells were obtained from the American Type Culture Collection (cat. no. CRL-2936). Synthesis and characterization of compound 21 was described in Mohandessi, et al.96
Plasmids
A 318 bp fragment encoding FRB was amplified by PCR from pRSETB-FRB-eDHFR using the primers 5'-C GAC GCT CGA GAT AAG ATG CCA ATC CTC TGG C-3' (XhoI, coding strand) and 5'-CC AGA TCC CGA ATT CAC CTT TGA GAT TCG TCG G-3' (EcoRI, non-coding strand). This fragment was inserted between the XhoI and EcoRI sites in pLL-1 (Active Motif, Inc.) to generate pLL-1-FRB-eDHFR, encoding FRB-eDHFR. A 360 bp fragment encoding FKBP12 was amplified by PCR from pRSETB-EGFP-FKBP12 using the primers 5'-GGA AGT GCT CGA GGT GGA GTG CAG GTG G -3' (XhoI, coding strand) and 5'-GCA GCC GGA TCC AGC TTC AGC TTA TTC CAG TTT TAG AAG C-3' (BamHI, non-coding strand). This fragment was inserted between the XhoI and BamHI sites in pEGFP-C1 (Clontech, Inc.) to generate pEGFP-FKBP12, encoding GFP-FKBP12. Plasmid integrity was confirmed by direct sequencing.
Cell culture, transfection and labeling
MDCKII cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 unit/mL penicillin and 100 μg/mL of streptomycin (DMEM+) at 37 °C and 5% CO2. Cells were seeded at 105 cells per well into a 6-well plate. After ~18 h incubation at 37 °C and 5% CO2, adherent cells (~80% confluent) were co-transfected with 1 μg each of pLL1-FRB-eDHFR and pEGFP-FKBP12 using Lipofectamine2000™ (Invitrogen, Inc.) according to manufacturer’s instructions. Approximately 6 h after transfection, cells were trypsinized and reseeded at 15,000 cells/well into 8-well chambered coverglasses (Nunc™, 12-565-470) and incubated at 37 °C and 5% CO2 overnight.
Transfected cells growing in DMEM in 8-well chambered coverglasses were washed 1X with phosphate buffered saline (PBS), re-immersed in DMEM without FBS at 37 °C containing 21 (10 μM) and incubated for 10 min. Cells were washed 2X in PBS and re-immersed in DMEM supplemented with 1 mM Patent Blue™ dye to quench extracellular luminescence of 21 bound non-specifically to cells or coverslips. Following incubation and washing, cells were maintained (<1 h) at 37 °C and 5% CO2 until imaging. The GFP-FKBP12/FRB-eDHFR interaction was induced by incubating cells in DMEM containing rapamycin (200 nM) at 37 °C for 1 h. Rapamycin induction was performed prior to loading cells with 21.
MTT assay
Cellular metabolic activity following exposure to 21 was assessed using MTT (Life Technologies, cat. no. M6494) according to manufacturer’s instructions. MDCKII cells were seeded into clear-bottom, 96-well, tissue culture-treated plates (30,000 cells/well) and incubated in DMEM(+) at 37 °C, 5% CO2 overnight. Cells were washed with 100 μL PBS and re-immersed in culture medium containing the following components (3 wells for each condition): positive control, DMEM(+) without phenol red (50 μL); negative control, EtOH (50 μL); DMEM(+) without phenol red containing 5 μM 21 (50 μL); DMEM(+) without phenol red containing 20 μM 21 (50 μL). Following 30 min incubation at 37 °C, 5% CO2, the cells were washed 1X with 100 μL PBS, and 100 μL DMEM(+) without phenol red was added. MTT (1μL of 12 mM stock solution in PBS) was added to each well and mixed by pipetting. The cells were then incubated for 4 h at 37 °C, 5% CO2. Then, 85 μL of medium was removed from each well and replaced with 50 μL DMSO to solubilize MTT reduction product, the cells were incubated for 10 min, and absorbance at 550 nm was measured on a plate reader (Perkin Elmer, Victor 3V). Absorbance of a second negative control consisting of DMEM(+) without phenol red (100 μL), MTT (10 μL) and DMSO (50 μL) was also measured. The mean absorbance (~0.05 for negative controls, ~0.5 for positive control) for all conditions (three replicates) was plotted as a percentage of absorbance measured for the positive control. Error bars represent s.d.
Microscopy, image processing and analysis
Imaging of adherent live cells was performed using a previously described, modified epifluorescence microscope (Axiovert 200, Carl Zeiss, Inc.).34 All images were acquired using a EC Plan Neofluar, 63X, 1.25 N.A. objective (Carl Zeiss, Inc.) Filter cubes containing the appropriate excitation and emission filters and dichroics allowed for wavelength selection. Continuous wave fluorescence images were acquired using an Axiocam MRM CCD camera (Carl Zeiss, Inc.). For time-gated luminescence, pulsed excitation light from a UV LED (λem = 365 nm, illumination intensity = ~0.5 W/cm2 at the sample plane) was synchronized with the intensifier component of an intensified CCD camera (ICCD, Mega-10EX, Stanford Photonics, Inc.) such that a 10 μs delay was inserted between the end of the LED pulse and the intensifier start-time. For each acquisition, the signal from multiple excitation/emission events was accumulated on the ICCD sensor and read out at the end of the camera frame. The source/camera timing parameters and intensifier gain voltage settings were the same for all of the time-gated images and data presented: excitation pulse width = 1500 μs, pulse period = 3000 μs, delay time = 10 μs, intensifier on-time = 1390 μs, gain = 778 V. The sensitivity of time-gated imaging is dependent on the number of excitation/detection events integrated on the CCD during a single camera frame and on the intensifier gain voltage. For frame lengths of 0.067 s, 0.333 s and 0.667 s, the number of excitation/detection events equaled 22, 110 and 220, respectively. Frame summing was used to increase SNR and to remove ion-feedback noise from the intensifier. Each frame summed effectively increases the bit depth of the resulting image in increments of 1024 (i.e., 1 frame yields bit depth equal to 1024, 2 frames, 2048, etc.). Images presented in Figure 8 are composites of 4 frames with resulting bit-depths of 4096.
ICCD images (tagged image file format, .TIFF) were captured with Piper control software (v2.4.05, Stanford Photonics, Inc.) and Axiocam images (.ZVI) were captured with Zeiss AxioVision software (v4.6). All images were cropped, adjusted for contrast and analyzed using NIH ImageJ (v1.42q). For quantitative analysis of time-gated microscope images, the emission signal intensity was calculated according to the equation: S = μsignal − μbckg, where, μsignal is equal to the mean pixel gray value in a region of interest (ROI) drawn within the cytoplasm, and μbckg is equal to the mean pixel gray value in a similarly sized ROI in a nearby part of the image with no cells. SNR was defined as S/σS, where . The mean number of photons/pixel was calculated from a proportionality factor that relates pixel gray value to the apparent number of incident photons. The proportionality factor was measured from a graph of variance versus mean intensity for a set of images, as described in Gahlaut and Miller.34 The donor normalized FRET signal was defined as the ratio (S520/S540) of mean gray values from corresponding ROIs in each image pair. Cells were selected for analysis that exhibited both GFP expression and loading of 21 as determined by examining corresponding continuous wave fluorescence images (λex = 480/40 nm, λem = 535/50 nm) and time-resolved images of terbium emission (λex = 365 nm, λem = 494/20 nm). The mean and standard deviation of FRET ratios was determined for a 10-cell sample each of cells expressing interacting (+rapamycin) and non-interacting (-rapamycin) fusion proteins. P-value was determined from a two-tailed, two-sample, unequal variance t-test of each sample.
ACKNOWLEDGMENTS
We thank Dr. Darren Magda at Lumiphore, Inc. for ongoing material and intellectual support. This study was supported by the National Institutes of Health (National Institute of General Medical Sciences Grant R01GM081030-01A1) and by the National Science Foundation (1013776). Lumi4 is a trademark of Lumiphore, Inc.
REFERENCES
- 1.Bunzli JC. Chem. Rev. 2010;110:2729–2755. doi: 10.1021/cr900362e. [DOI] [PubMed] [Google Scholar]
- 2.New EJ, Parker D, Smith DG, Walton JW. Curr. Opin. Chem. Biol. 2010;14:238–246. doi: 10.1016/j.cbpa.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Ghose S, Trinquet E, Laget M, Bazin H, Mathis G. J. Alloys Compd. 2008;451:35–37. [Google Scholar]
- 4.Degorce F, Card A, Soh S, Trinquet E, Knapik GP, Xie B. Curr. Chem. Genomics. 2009;3:22–32. doi: 10.2174/1875397300903010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selvin PR. Annu. Rev. Biophys. Biomol. Struct. 2002;31:275–302. doi: 10.1146/annurev.biophys.31.101101.140927. [DOI] [PubMed] [Google Scholar]
- 6.Hemmila I, Laitala V. J. Fluoresc. 2005;15:529–542. doi: 10.1007/s10895-005-2826-6. [DOI] [PubMed] [Google Scholar]
- 7.Bunzli JC, Eliseeva SV. Basics of lanthanide photophysics. In: Hanninen P, Harma H, editors. Lanthanide luminescence: Photophysical, analytical and biological aspects. Vol. 7. Berlin Heidelberg: Springer-Verlag; 2011. pp. 1–47. [Google Scholar]
- 8.Li M, Selvin PR. J. Am. Chem. Soc. 1995;117:8132–8138. [Google Scholar]
- 9.Xu J, Corneillie TM, Moore EG, Law GL, Butlin NG, Raymond KN. J. Am. Chem. Soc. 2011;133:19900–19910. doi: 10.1021/ja2079898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hovinen J, Guy PM. Bioconjugate Chem. 2009;20:404–421. doi: 10.1021/bc800370s. [DOI] [PubMed] [Google Scholar]
- 11.Terai T, Nagano T. Curr. Opin. Chem. Biol. 2008;12:515–521. doi: 10.1016/j.cbpa.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Nolan EM, Lippard SJ. Acc. Chem. Res. 2009;42:193–203. doi: 10.1021/ar8001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J, Dodani SC, Chang CJ. Nat. Chem. 2012;4:973–984. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller LW, Cornish VW. Curr. Opin. Chem. Biol. 2005;9:56–61. doi: 10.1016/j.cbpa.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Marks KM, Nolan GP. Nat. Methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 16.Jing C, Cornish VW. Acc. Chem. Res. 2011;44:784–792. doi: 10.1021/ar200099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 18.Xie XS, Yu J, Yang WY. Science. 2006;312:228–230. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 19.Welch CM, Elliott H, Danuser G, Hahn KM. Nat. Rev. Mol. Cell Biol. 2011;12:749–756. doi: 10.1038/nrm3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Campbell RE, Ting AY, Tsien RY. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 21.Jares-Erijman EA, Jovin TM. Curr. Opin. Chem. Biol. 2006;10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Miyawaki A. Annu. Rev. Biochem. 2011;80:357–373. doi: 10.1146/annurev-biochem-072909-094736. [DOI] [PubMed] [Google Scholar]
- 23.Jares-Erijman EA, Jovin TM. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 24.Piston DW, Kremers GJ. Trends Biochem. Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Day RN, Periasamy A. Nat. Protoc. 2011;6:1324–1340. doi: 10.1038/nprot.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buranachai C, Kamiyama D, Chiba A, Williams BD, Clegg RM. J. Fluoresc. 2008;18:929–942. doi: 10.1007/s10895-008-0332-3. [DOI] [PubMed] [Google Scholar]
- 27.Hum JM, Siegel AP, Pavalko FM, Day RN. Int. J. Mol. Sci. 2012;13:14385–14400. doi: 10.3390/ijms131114385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redford GI, Clegg RM. J. Fluoresc. 2005;15:805–815. doi: 10.1007/s10895-005-2990-8. [DOI] [PubMed] [Google Scholar]
- 29.Colyer RA, Lee C, Gratton E. Microsc. Res. Tech. 2008;71:201–213. doi: 10.1002/jemt.20540. [DOI] [PubMed] [Google Scholar]
- 30.Hinde E, Digman MA, Welch C, Hahn KM, Gratton E. Microsc. Res. Tech. 2012;75:271–281. doi: 10.1002/jemt.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berney C, Danuser G. Biophys. J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galperin E, Verkhusha VV, Sorkin A. Nat. Methods. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 33.Geissler D, Stufler S, Lohmannsroben HG, Hildebrandt N. J. Am. Chem. Soc. 2013;135:1102–1109. doi: 10.1021/ja310317n. [DOI] [PubMed] [Google Scholar]
- 34.Gahlaut N, Miller LW. Cytometry A. 2010;77:1113–1125. doi: 10.1002/cyto.a.20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beverloo HB, van Schadewijk A, van Gelderen-Boele S, Tanke HJ. Cytometry. 1990;11:784–792. doi: 10.1002/cyto.990110704. [DOI] [PubMed] [Google Scholar]
- 36.Seveus L, Vaisala M, Syrjanen S, Sandberg M, Kuusisto A, Harju R, Salo J, Hemmila I, Kojola H, Soini E. Cytometry. 1992;13:329–338. doi: 10.1002/cyto.990130402. [DOI] [PubMed] [Google Scholar]
- 37.Connally RE, Piper JA. Ann. N Y Acad. Sci. 2008;1130:106–116. doi: 10.1196/annals.1430.032. [DOI] [PubMed] [Google Scholar]
- 38.Ramshesh VK, Lemasters JJ. J. Biomed. Opt. 2008;13:064001. doi: 10.1117/1.3027503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kielar F, Congreve A, Law GL, New EJ, Parker D, Wong KL, Castreno P, de Mendoza J. Chem. Commun. (Cambridge, UK) 2008:2435–2437. doi: 10.1039/b803864c. [DOI] [PubMed] [Google Scholar]
- 40.Law GL, Wong KL, Man CW, Wong WT, Tsao SW, Lam MH, Lam PK. J. Am. Chem. Soc. 2008;130:3714–3715. doi: 10.1021/ja710418d. [DOI] [PubMed] [Google Scholar]
- 41.Eliseeva SV, Aubock G, van Mourik F, Cannizzo A, Song B, Deiters E, Chauvin AS, Chergui M, Bunzli JC. J. Phys. Chem. B. 2010;114:2932–2937. doi: 10.1021/jp9090206. [DOI] [PubMed] [Google Scholar]
- 42.Vaasa A, Ligi K, Mohandessi S, Enkvist E, Uri A, Miller LW. Chem. Commun. (Cambridge, UK) 2012;48:8595–8597. doi: 10.1039/c2cc33565d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurel D, Comps-Agrar L, Brock C, Rives ML, Bourrier E, Ayoub MA, Bazin H, Tinel N, Durroux T, Prezeau L, Trinquet E, Pin JP. Nat. Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doumazane E, Scholler P, Zwier JM, Trinquet E, Rondard P, Pin JP. FASEB J. 2011;25:66–77. doi: 10.1096/fj.10-163147. [DOI] [PubMed] [Google Scholar]
- 45.Strauch RC, Mastarone DJ, Sukerkar PA, Song Y, Ipsaro JJ, Meade TJ. J. Am. Chem. Soc. 2011;133:16346–16349. doi: 10.1021/ja206134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizukami S, Yamamoto T, Yoshimura A, Watanabe S, Kikuchi K. Angew. Chem. Int. Ed. Engl. 2011;50:8750–8752. doi: 10.1002/anie.201103775. [DOI] [PubMed] [Google Scholar]
- 47.Miller LW, Cai YF, Sheetz MP, Cornish VW. Nat. Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 48.Calloway NT, Choob M, Sanz A, Sheetz MP, Miller LW, Cornish VW. Chembiochem. 2007;8:767–774. doi: 10.1002/cbic.200600414. [DOI] [PubMed] [Google Scholar]
- 49.Rajapakse HE, Reddy DR, Mohandessi S, Butlin NG, Miller LW. Angew. Chem. Int. Ed. Engl. 2009;48:4990–4992. doi: 10.1002/anie.200900858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy DR, Pedro Rosa LE, Miller LW. Bioconjugate Chem. 2011;22:1402–1409. doi: 10.1021/bc200131k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yapici E, Reddy DR, Miller LW. Chembiochem. 2012;13:553–558. doi: 10.1002/cbic.201100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen AW, Daugherty PS. Nat. Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 53.Rajapakse HE, Gahlaut N, Mohandessi S, Yu D, Turner JR, Miller LW. Proc. Natl. Acad. Sci. U S A. 2010;107:13582–13587. doi: 10.1073/pnas.1002025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 55.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. J. Cell. Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunningham CW, Mukhopadhyay A, Lushington GH, Blagg BS, Prisinzano TE, Krise JP. Mol. Pharm. 2010;7:1301–1310. doi: 10.1021/mp100089k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nigg EA. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 58.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug. Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 59.Tsien RY. Nature. 1981;290:527–528. doi: 10.1038/290527a0. [DOI] [PubMed] [Google Scholar]
- 60.Minta A, Kao JP, Tsien RY. J. Biol. Chem. 1989;264:8171–8178. [PubMed] [Google Scholar]
- 61.Baracca A, Sgarbi G, Solaini G, Lenaz G. Biochim. Biophys. Acta. 2003;1606:137–146. doi: 10.1016/s0005-2728(03)00110-5. [DOI] [PubMed] [Google Scholar]
- 62.Schwartz R, Ting CS, King J. Genome Res. 2001;11:703–709. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- 63.Hanaoka K, Kikuchi K, Kobayashi S, Nagano T. J. Am. Chem. Soc. 2007;129:13502–13509. doi: 10.1021/ja073392j. [DOI] [PubMed] [Google Scholar]
- 64.Okada CY, Rechsteiner M. Cell. 1982;29:33–41. doi: 10.1016/0092-8674(82)90087-3. [DOI] [PubMed] [Google Scholar]
- 65.Walev I, Bhakdi SC, Hofmann F, Djonder N, Valeva A, Aktories K, Bhakdi S. Proc. Natl. Acad. Sci. U S A. 2001;98:3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manning HC, Goebel T, Thompson RC, Price RR, Lee H, Bornhop DJ. Bioconjugate Chem. 2004;15:1488–1495. doi: 10.1021/bc049904q. [DOI] [PubMed] [Google Scholar]
- 67.Lee J, Burdette JE, MacRenaris KW, Mustafi D, Woodruff TK, Meade TJ. Chem. Biol. 2007;14:824–834. doi: 10.1016/j.chembiol.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 68.Yamane T, Hanaoka K, Muramatsu Y, Tamura K, Adachi Y, Miyashita Y, Hirata Y, Nagano T. Bioconjugate Chem. 2011;22:2227–2236. doi: 10.1021/bc200127t. [DOI] [PubMed] [Google Scholar]
- 69.Montgomery CP, Murray BS, New EJ, Pal R, Parker D. Acc. Chem. Res. 2009;42:925–937. doi: 10.1021/ar800174z. [DOI] [PubMed] [Google Scholar]
- 70.Frias JC, Bobba G, Cann MJ, Hutchison CJ, Parker D. Org. Biomol. Chem. 2003;1:905–917. doi: 10.1039/b301385p. [DOI] [PubMed] [Google Scholar]
- 71.Chauvin AS, Comby S, Song B, Vandevyver CD, Thomas F, Bunzli JC. Chem. -- Eur. J. 2007;13:9515–9526. doi: 10.1002/chem.200700883. [DOI] [PubMed] [Google Scholar]
- 72.Chauvin AS, Comby S, Song B, Vandevyver CD, Bunzli JC. Chem. -- Eur. J. 2008;14:1726–1739. doi: 10.1002/chem.200701357. [DOI] [PubMed] [Google Scholar]
- 73.Chauvin AS, Thomas F, Song B, Vandevyver CDB, Bunzli JCG. Philos. Trans. R. Soc. A. 2013;371:20120295. doi: 10.1098/rsta.2012.0295. [DOI] [PubMed] [Google Scholar]
- 74.Murray BS, New EJ, Pal R, Parker D. Org. Biomol. Chem. 2008;6:2085–2094. doi: 10.1039/b803895c. [DOI] [PubMed] [Google Scholar]
- 75.Yu J, Parker D, Pal R, Poole RA, Cann MJ. J. Am. Chem. Soc. 2006;128:2294–2299. doi: 10.1021/ja056303g. [DOI] [PubMed] [Google Scholar]
- 76.Pal R, Parker D. Chem. Commun. (Cambridge, UK) 2007:474–476. doi: 10.1039/b616665b. [DOI] [PubMed] [Google Scholar]
- 77.Pal R, Parker D. Org. Biomol. Chem. 2008;6:1020–1033. doi: 10.1039/b718993a. [DOI] [PubMed] [Google Scholar]
- 78.Kielar F, Law GL, New EJ, Parker D. Org. Biomol. Chem. 2008;6:2256–2268. doi: 10.1039/b804369h. [DOI] [PubMed] [Google Scholar]
- 79.New EJ, Parker D. Org. Biomol. Chem. 2009;7:851–855. doi: 10.1039/b822145f. [DOI] [PubMed] [Google Scholar]
- 80.New EJ, Congreve A, Parker D. Chem. Sci. 2010;1:111–118. [Google Scholar]
- 81.Falcone S, Cocucci E, Podini P, Kirchhausen T, Clementi E, Meldolesi J. J. Cell Sci. 2006;119:4758–4769. doi: 10.1242/jcs.03238. [DOI] [PubMed] [Google Scholar]
- 82.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 83.Fretz MM, Penning NA, Al-Taei S, Futaki S, Takeuchi T, Nakase I, Storm G, Jones AT. Biochem. J. 2007;403:335–342. doi: 10.1042/BJ20061808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kosuge M, Takeuchi T, Nakase I, Jones AT, Futaki S. Bioconjugate Chem. 2008;19:656–664. doi: 10.1021/bc700289w. [DOI] [PubMed] [Google Scholar]
- 85.Nakase I, Takeuchi T, Tanaka G, Futaki S. Adv. Drug. Deliv. Rev. 2008;60:598–607. doi: 10.1016/j.addr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 86.Stewart KM, Horton KL, Kelley SO. Org. Biomol. Chem. 2008;6:2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
- 87.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. Adv. Drug Deliv. Rev. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhorade R, Weissleder R, Nakakoshi T, Moore A, Tung CH. Bioconjugate Chem. 2000;11:301–305. doi: 10.1021/bc990168d. [DOI] [PubMed] [Google Scholar]
- 89.Liu M, Guo YM, Wu QF, Yang JL, Wang P, Wang SC, Guo XJ, Qiang YQ, Duan XY. Biochem. Biophys. Res. Commun. 2006;347:133–140. doi: 10.1016/j.bbrc.2006.06.081. [DOI] [PubMed] [Google Scholar]
- 90.Allen MJ, Meade TJ. J. Biol. Inorg. Chem. 2003;8:746–750. doi: 10.1007/s00775-003-0475-2. [DOI] [PubMed] [Google Scholar]
- 91.Allen MJ, MacRenaris KW, Venkatasubramanian PN, Meade TJ. Chem. Biol. 2004;11:301–307. doi: 10.1016/j.chembiol.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 92.Endres PJ, Macrenaris KW, Vogt S, Allen MJ, Meade TJ. Mol. Imaging. 2006;5:485–497. [PubMed] [Google Scholar]
- 93.Endres PJ, MacRenaris KW, Vogt S, Meade TJ. Bioconjugate Chem. 2008;19:2049–2059. doi: 10.1021/bc8002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Major JL, Meade TJ. Acc. Chem. Res. 2009;42:893–903. doi: 10.1021/ar800245h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thielemann DT, Wagner AT, Rosch E, Kolmel DK, Heck JG, Rudat B, Neumaier M, Feldmann C, Schepers U, Brase S, Roesky PW. J. Am. Chem. Soc. 2013;135:7454–7457. doi: 10.1021/ja403539t. [DOI] [PubMed] [Google Scholar]
- 96.Mohandessi S, Rajendran M, Magda D, Miller LW. Chem. -- Eur. J. 2012;18:10825–10829. doi: 10.1002/chem.201201805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 99.Murray JM, Appleton PL, Swedlow JR, Waters JC. J. Microsc. 2007;228:390–405. doi: 10.1111/j.1365-2818.2007.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sala F, Hernandez-Cruz A. Biophys. J. 1990;57:313–324. doi: 10.1016/S0006-3495(90)82533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tsien RY. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 102.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hodgson L, Shen F, Hahn K. Curr. Protoc. Cell Biol. 2010:14.11.1–14.11.26. doi: 10.1002/0471143030.cb1411s46. Chapter 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 105.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. Nat. Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang YL. J. Microsc. 2007;228:123–131. doi: 10.1111/j.1365-2818.2007.01834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dailey M, Manders E, Soll D, Terasaki M. Confocal Microscopy of Living Cells. In: Pawley JB, editor. Handbook Of Biological Confocal Microscopy. US: Springer; 2006. pp. 381–403. [Google Scholar]
- 108.Konig K, Krasieva T, Bauer E, Fiedler U, Berns MW, Tromberg BJ, Greulich KO. J. Biomed. Opt. 1996;1:217–222. doi: 10.1117/12.233373. [DOI] [PubMed] [Google Scholar]
- 109.Gorgidze LA, Oshemkova SA, Vorobjev IA. Biosci. Rep. 1998;18:215–224. doi: 10.1023/a:1020104914726. [DOI] [PubMed] [Google Scholar]
- 110.Sako Y, Sekihata A, Yanagisawa Y, Yamamoto M, Shimada Y, Ozaki K, Kusumi A. J. Microsc. 1997;185:9–20. doi: 10.1046/j.1365-2818.1997.1480707.x. [DOI] [PubMed] [Google Scholar]
- 111.Stricker SA, Whitaker M. Microsc. Res. Tech. 1999;46:356–369. doi: 10.1002/(SICI)1097-0029(19990915)46:6<356::AID-JEMT4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 112.Mukherjee S, Zha X, Tabas I, Maxfield FR. Biophys. J. 1998;75:1915–1925. doi: 10.1016/S0006-3495(98)77632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mitchison TJ, Sawin KE, Theriot JA, Gee K, Mallavarapu A. Methods Enzymol. 1998;291:63–78. doi: 10.1016/s0076-6879(98)91007-2. [DOI] [PubMed] [Google Scholar]
- 114.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 115.Knight MM, Roberts SR, Lee DA, Bader DL. Am. J. Physiol. Cell Physiol. 2003;284:C1083–C1089. doi: 10.1152/ajpcell.00276.2002. [DOI] [PubMed] [Google Scholar]
- 116.Dadabhoy A, Faulkner S, Sammes PG. J. Chem. Soc. Perkin Trans. 2002;2:348–357. [Google Scholar]
- 117.Bretonniere Y, Cann MJ, Parker D, Slater R. Org. Biomol. Chem. 2004;2:1624–1632. doi: 10.1039/b400734b. [DOI] [PubMed] [Google Scholar]
- 118.Szijjarto C, Pershagen E, Ilchenko NO, Borbas KE. Chem. -- Eur. J. 2013;19:3099–3109. doi: 10.1002/chem.201203957. [DOI] [PubMed] [Google Scholar]
- 119.D'Aleo A, Picot A, Beeby A, Gareth Williams JA, Le Guennic B, Andraud C, Maury O. Inorg. Chem. 2008;47:10258–10268. doi: 10.1021/ic8012969. [DOI] [PubMed] [Google Scholar]
- 120.Butler SJ, McMahon BK, Pal R, Parker D, Walton JW. Chem. -- Eur. J. 2013;19:9511–9517. doi: 10.1002/chem.201301273. [DOI] [PubMed] [Google Scholar]
- 121.McMahon BK, Pal R, Parker D. Chem. Commun. (Cambridge, UK) 2013;49:5363–5365. doi: 10.1039/c3cc42308e. [DOI] [PubMed] [Google Scholar]
- 122.Walton JW, Bourdolle A, Butler SJ, Soulie M, Delbianco M, McMahon BK, Pal R, Puschmann H, Zwier JM, Lamarque L, Maury O, Andraud C, Parker D. Chem. Commun. (Cambridge, UK) 2013;49:1600–1602. doi: 10.1039/c2cc35247h. [DOI] [PubMed] [Google Scholar]
- 123.D'Aleo A, Picot A, Baldeck PL, Andraud C, Maury O. Inorg. Chem. 2008;47:10269–10279. doi: 10.1021/ic8012975. [DOI] [PubMed] [Google Scholar]
- 124.Picot A, D'Aleo A, Baldeck PL, Grichine A, Duperray A, Andraud C, Maury O. J. Am. Chem. Soc. 2008;130:1532–1533. doi: 10.1021/ja076837c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Nat. Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Nat. Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 127.Choi H, Tzeranis DS, Cha JW, Clemenceau P, de Jong SJG, van Geest LK, Moon JH, Yannas IV, So PTC. Opt. Express. 2012;20:26219–26235. doi: 10.1364/OE.20.026219. [DOI] [PMC free article] [PubMed] [Google Scholar]