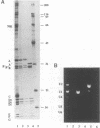
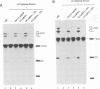
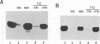Abstract
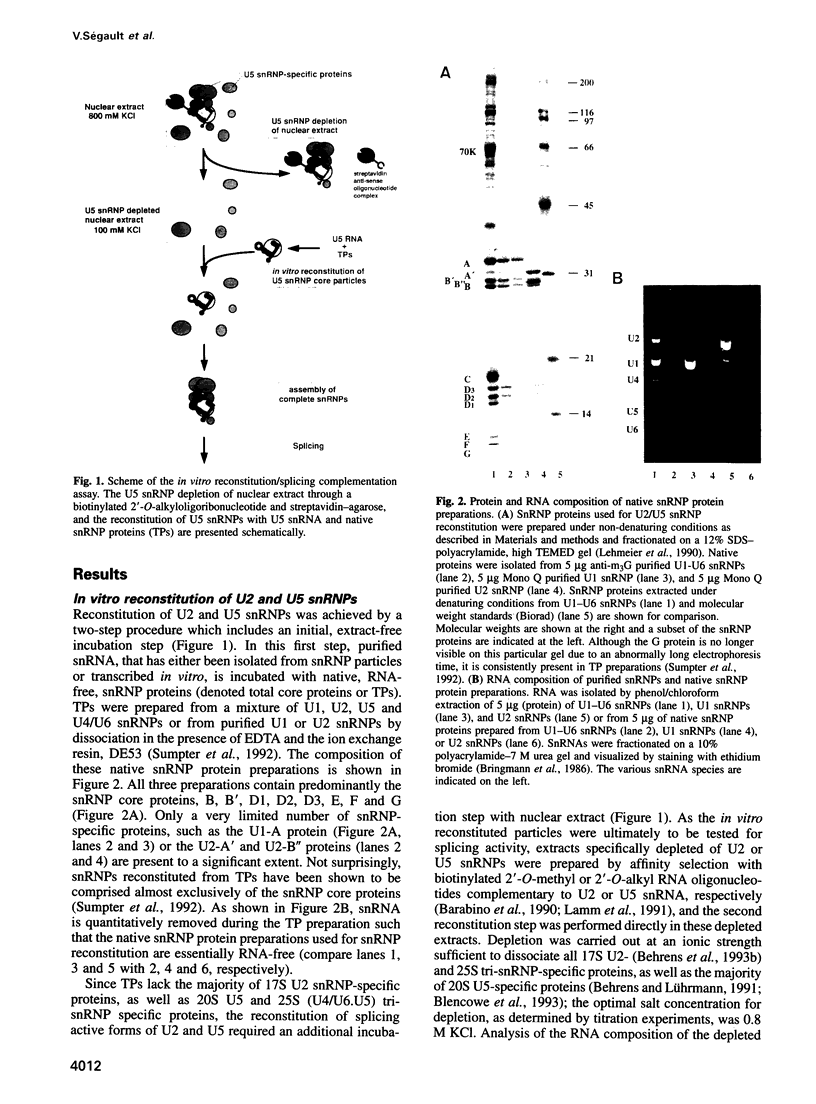
An in vitro reconstitution/splicing complementation system has been developed which has allowed the investigation of the role of mammalian U2 and U5 snRNP components in splicing. U2 or U5 snRNP cores are first reconstituted from purified native snRNP core proteins and snRNA in the absence of cellular extract and are subsequently added to splicing extracts depleted of either U2 or U5 snRNP. When snRNPs reconstituted with HeLa U2 or U5 snRNA were added to U2- or U5-depleted nuclear extract, splicing was complemented. Addition of naked snRNA, on the other hand, did not restore splicing, demonstrating that the core proteins are essential for both U2 and U5 snRNP functions in splicing. Hybrid U2 or U5 snRNPs, reconstituted with core proteins isolated from U1 or U2 snRNPs, were equally active in splicing complementation, indicating that the snRNP core proteins are functionally interchangeable. U5 snRNPs reconstituted from in vitro transcribed U5 snRNA restored splicing to a level identical to that observed with particles reconstituted from authentic HeLa U5 snRNA. In contrast, splicing could not be restored to U2-depleted extract by the addition of snRNPs reconstituted from synthetic U2 snRNA, suggesting that U2 snRNA base modifications are essential for U2 snRNP function.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach M., Bringmann P., Lührmann R. Purification of small nuclear ribonucleoprotein particles with antibodies against modified nucleosides of small nuclear RNAs. Methods Enzymol. 1990;181:232–257. doi: 10.1016/0076-6879(90)81125-e. [DOI] [PubMed] [Google Scholar]
- Bach M., Winkelmann G., Lührmann R. 20S small nuclear ribonucleoprotein U5 shows a surprisingly complex protein composition. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6038–6042. doi: 10.1073/pnas.86.16.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Beggs J. D. Yeast protein splicing factors involved in nuclear pre-mRNA splicing. Mol Biol Rep. 1993 Aug;18(2):99–103. doi: 10.1007/BF00986763. [DOI] [PubMed] [Google Scholar]
- Behrens S. E., Galisson F., Legrain P., Lührmann R. Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8229–8233. doi: 10.1073/pnas.90.17.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S. E., Lührmann R. Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes Dev. 1991 Aug;5(8):1439–1452. doi: 10.1101/gad.5.8.1439. [DOI] [PubMed] [Google Scholar]
- Behrens S. E., Tyc K., Kastner B., Reichelt J., Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993 Jan;13(1):307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J., Carmo-Fonseca M., Behrens S. E., Lührmann R., Lamond A. I. Interaction of the human autoantigen p150 with splicing snRNPs. J Cell Sci. 1993 Jul;105(Pt 3):685–697. doi: 10.1242/jcs.105.3.685. [DOI] [PubMed] [Google Scholar]
- Bochnig P., Reuter R., Bringmann P., Lührmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem. 1987 Oct 15;168(2):461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Ebel J. P., Lazar E., Haendler B., Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1(10):1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann P., Lührmann R. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J. 1986 Dec 20;5(13):3509–3516. doi: 10.1002/j.1460-2075.1986.tb04676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R., Gröning K., Behrens S. E., Lührmann R., Krämer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993 Oct 1;262(5130):102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Datta B., Weiner A. M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991 Aug 29;352(6338):821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., McPheeters D. S., Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989 Dec;3(12B):2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- Fischer U., Darzynkiewicz E., Tahara S. M., Dathan N. A., Lührmann R., Mattaj I. W. Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J Cell Biol. 1991 May;113(4):705–714. doi: 10.1083/jcb.113.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990 Aug 17;249(4970):786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- Fischer U., Sumpter V., Sekine M., Satoh T., Lührmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993 Feb;12(2):573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Patton J. G., Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994 Jul 15;13(14):3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Hackl W., Fischer U., Lührmann R. A 69-kD protein that associates reversibly with the Sm core domain of several spliceosomal snRNP species. J Cell Biol. 1994 Feb;124(3):261–272. doi: 10.1083/jcb.124.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Darzynkiewicz E., Tahara S. M., Mattaj I. W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990 Aug 10;62(3):569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Mattaj I. W. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989 Oct 6;59(1):159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Scherly D., Mattaj I. W. Multiple domains of U1 snRNA, including U1 specific protein binding sites, are required for splicing. EMBO J. 1990 Apr;9(4):1237–1244. doi: 10.1002/j.1460-2075.1990.tb08231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann H., Fabrizio P., Raker V. A., Foulaki K., Hornig H., Brahms H., Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995 May 1;14(9):2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinterberger M., Pettersson I., Steitz J. A. Isolation of small nuclear ribonucleoproteins containing U1, U2, U4, U5, and U6 RNAs. J Biol Chem. 1983 Feb 25;258(4):2604–2613. [PubMed] [Google Scholar]
- Jacobson M. R., Rhoadhouse M., Pederson T. U2 small nuclear RNA 3' end formation is directed by a critical internal structure distinct from the processing site. Mol Cell Biol. 1993 Feb;13(2):1119–1129. doi: 10.1128/mcb.13.2.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Mattaj I. W. The determinants for Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO J. 1993 Jan;12(1):223–232. doi: 10.1002/j.1460-2075.1993.tb05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B., Bach M., Lührmann R. Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1710–1714. doi: 10.1073/pnas.87.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm G. M., Blencowe B. J., Sproat B. S., Iribarren A. M., Ryder U., Lamond A. I. Antisense probes containing 2-aminoadenosine allow efficient depletion of U5 snRNP from HeLa splicing extracts. Nucleic Acids Res. 1991 Jun 25;19(12):3193–3198. doi: 10.1093/nar/19.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmeier T., Foulaki K., Lührmann R. Evidence for three distinct D proteins, which react differentially with anti-Sm autoantibodies, in the cores of the major snRNPs U1, U2, U4/U6 and U5. Nucleic Acids Res. 1990 Nov 25;18(22):6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührmann R., Kastner B., Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990 Nov 30;1087(3):265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992 Nov 27;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986 Sep 12;46(6):905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., De Robertis E. M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985 Jan;40(1):111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- McPheeters D. S., Fabrizio P., Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 1989 Dec;3(12B):2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- Nelissen R. L., Will C. L., van Venrooij W. J., Lührmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994 Sep 1;13(17):4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Pan Z. Q., Prives C. Assembly of functional U1 and U2 human-amphibian hybrid snRNPs in Xenopus laevis oocytes. Science. 1988 Sep 9;241(4871):1328–1331. doi: 10.1126/science.2970672. [DOI] [PubMed] [Google Scholar]
- Pan Z. Q., Prives C. U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev. 1989 Dec;3(12A):1887–1898. doi: 10.1101/gad.3.12a.1887. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Patton J. R. Formation of pseudouridine in U5 small nuclear RNA. Biochemistry. 1994 Aug 30;33(34):10423–10427. doi: 10.1021/bi00200a025. [DOI] [PubMed] [Google Scholar]
- Patton J. R., Jacobson M. R., Pederson T. Pseudouridine formation in U2 small nuclear RNA. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3324–3328. doi: 10.1073/pnas.91.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J. R. Pseudouridine modification of U5 RNA in ribonucleoprotein particles assembled in vitro. Mol Cell Biol. 1991 Dec;11(12):5998–6006. doi: 10.1128/mcb.11.12.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C. W., Bindereif A., Green M. R. In vitro reconstitution of snRNPs: a reconstituted U4/U6 snRNP participates in splicing complex formation. Genes Dev. 1989 Apr;3(4):479–487. doi: 10.1101/gad.3.4.479. [DOI] [PubMed] [Google Scholar]
- Plessel G., Fischer U., Lührmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol. 1994 Jun;14(6):4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond B. C., Rokeach L. A., Hoch S. O. Human snRNP polypeptide D1 promotes pre-mRNA splicing in yeast and defines nonessential yeast Smd1p sequences. Nucleic Acids Res. 1993 Jul 25;21(15):3501–3505. doi: 10.1093/nar/21.15.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkukalek A., Myslinski E., Mougin A., Luhrmann R., Branlant C. Phylogenetic conservation of modified nucleotides in the terminal loop 1 of the spliceosomal U5 snRNA. Biochimie. 1995;77(1-2):16–21. doi: 10.1016/0300-9084(96)88099-0. [DOI] [PubMed] [Google Scholar]
- Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995 May 1;14(9):2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Wells S. E., Ares M., Jr Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Sep;14(9):6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersig C., Bindereif A. Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res. 1990 Nov 11;18(21):6223–6229. doi: 10.1093/nar/18.21.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersig C., Bindereif A. Reconstitution of functional mammalian U4 small nuclear ribonucleoprotein: Sm protein binding is not essential for splicing in vitro. Mol Cell Biol. 1992 Apr;12(4):1460–1468. doi: 10.1128/mcb.12.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C. L., Behrens S. E., Lührmann R. Protein composition of mammalian spliceosomal snRNPs. Mol Biol Rep. 1993 Aug;18(2):121–126. doi: 10.1007/BF00986766. [DOI] [PubMed] [Google Scholar]
- Wolff T., Bindereif A. Conformational changes of U6 RNA during the spliceosome cycle: an intramolecular helix is essential both for initiating the U4-U6 interaction and for the first step of slicing. Genes Dev. 1993 Jul;7(7B):1377–1389. doi: 10.1101/gad.7.7b.1377. [DOI] [PubMed] [Google Scholar]
- Wolff T., Bindereif A. Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J. 1992 Jan;11(1):345–359. doi: 10.1002/j.1460-2075.1992.tb05057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. A., Manley J. L. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991 Aug 29;352(6338):818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zavanelli M. I., Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991 Dec;5(12B):2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- Zavanelli M. I., Britton J. S., Igel A. H., Ares M., Jr Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol Cell Biol. 1994 Mar;14(3):1689–1697. doi: 10.1128/mcb.14.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Zapp M. L., Berget S. M. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol. 1988 Feb;8(2):814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]