Abstract

d-Cycloserine is a second-line drug approved for use in the treatment of patients infected with Mycobacterium tuberculosis, the etiologic agent of tuberculosis. The unique mechanism of action of d-cycloserine, compared with those of other clinically employed antimycobacterial agents, represents an untapped and exploitable resource for future rational drug design programs. Here, we show that d-cycloserine is a slow-onset inhibitor of MtDdl and that this behavior is specific to the M. tuberculosis enzyme orthologue. Furthermore, evidence is presented that indicates d-cycloserine binds exclusively to the C-terminal d-alanine binding site, even in the absence of bound d-alanine at the N-terminal binding site. Together, these results led us to propose a new model of d-alanine:d-alanine ligase inhibition by d-cycloserine and suggest new opportunities for rational drug design against an essential, clinically validated mycobacterial target.
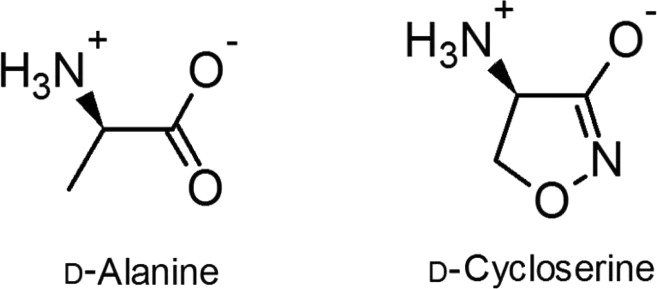
d-Cycloserine [DCS, (R)-4-amino-1,2-oxazolidin-3-one (Scheme 1)] is an antibiotic currently employed as the cornerstone of treatment options for multiresistant and extensively drug resistant tuberculosis.1 DCS is a structural analogue of d-alanine [d-Ala (Scheme 1)] and blocks bacterial growth by inhibiting two enzymes involved in d-Ala metabolism and peptidoglycan biosynthesis: alanine racemase and d-alanine:d-alanine ligase (Ddl).2 The molecular targets and mechanism of action of DCS are unique among all classes of antibiotics that are currently known, and congruently, DCS displays no cross-resistance with any other front- or second-line antitubercular drugs.1 This feature of DCS, along with low reported levels of clinical resistance,1 makes DCS a valuable asset in the antituberculosis compound arsenal. Host toxicity (principally neuronal NMDA receptor partial agonism), however, remains a serious and dose-limiting side effect of DCS treatment.3 Nonetheless, understanding the inhibitory mechanisms and molecular interactions between DCS and its microbial targets could aid future endeavors in the rational design of improved, next-generation antimycobacterial compounds.
Scheme 1. Structures of d-Alanine and d-Cycloserine.
We have previously characterized the steady-state kinetics of DCS inhibition of Mycobacterium tuberculosis Ddl (MtDdl).4 Ddl catalyzes the ATP-dependent formation of the amide bond between two d-alanine molecules, forming the peptidoglycan precursor dipeptide d-alanyl-d-alanine (d-Ala-d-Ala). Ddl catalysis proceeds through two half-reactions, and the two participating d-Ala molecules occupy distinct regions of the active site (N- and C-termini binding sites).5 As such, DCS can potentially inhibit binding of either d-Ala molecule to Ddl, and steady-state kinetic data from us and others support this hypothesis.4,6
During routine spectrophotometric analysis of MtDdl activity, we noticed that individual time courses of DCS-containing reactions exhibited transient biphasic curvature suggestive of slow-onset inhibition.7 Although this phenomenon was disregarded for our initial steady-state kinetic analysis of DCS inhibition, the potential significance of this observation motivated us to investigate it further; our findings are reported herein.
Materials and Methods
Materials
All chemicals were reagent or analytical grade and sourced as previously described.4 Solutions of DCS were prepared in CHES buffer (pH 9.0) and aliquots stored indefinitely at −80 °C. Aliquots were thawed directly before being used and discarded after 24 h.
Protein Purification
Recombinant MtDdl was overexpressed and purified as a hexahistidine tag fusion protein, as previously described.4 The gene encoding Escherichia coli DdlB (EcDdl) was amplified via polymerase chain reaction from E. coli BL21 genomic DNA and cloned into the NdeI and SacI sites of pET28a+. E. coli BL21 was transformed with the resulting vector, and the recombinant enzyme was overexpressed and purified in a manner identical to that of MtDdl. Proteins were stored indefinitely at −20 °C in 20 mM TEA (pH 7.8) containing 50% (v/v) glycerol. Proteins were >95% pure as judged by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (results not shown).
Kinetic Measurements
Steady-state kinetics and equilibrium binding were performed as previously described.4 Time courses for the measurement of slow-onset inhibition were collected on an SX20 stopped-flow spectrometer (Applied Photophysics) at 37 °C, using a 10 mm path-length cell. Data points were recorded every 0.1 s. Catalysis was detected by the same coupled enzyme mechanism as used previously for steady-state kinetic measurements.4 All reaction mixtures contained 50 mM HEPES (pH 7.3), 10 mM MgCl2, 80 mM KCl, 3 mM ATP, 0.25 mM NADH, 1.5 mM PEP, and a 40 μL/mL pyruvate kinase/lactate dehydrogenase enzyme solution (PK-LDH; stock solution of 600–1000 units/mL PK and 900–1400 units/mL LDH). MtDdl was employed at concentrations from 60 to 290 nM and EcDdl at a final concentration of 30 nM. DCS and d-Ala concentrations were varied as required. Reaction components were sorted into two syringes, with rapid mixing of 60 μL from each starting each reaction. For all assays, both syringes contained HEPES, MgCl2, KCl, NADH, PEP, and PK-LDH. MtDdl and d-Ala were always stored in separate syringes. For analysis of slow-onset inhibition at the C-terminal d-Ala binding site (i.e., data for Figure 1), DCS and Mt/EcDdl were stored in separate reaction mixtures prior to the commencement of the reaction. For analysis of slow-onset inhibition at the N-terminal d-Ala site (i.e., data for Figure 2), MtDdl and DCS (with or without ATP) were stored in the same reaction mixture (for at least 4 min) prior to the commencement of the reaction.
Figure 1.
Representative reaction progress curves for (A) MtDdl and (B) EcDdl at multiple concentrations of DCS (4 and 0.75 mM d-Ala, respectively). Gray lines depict data obtained by averaging three injections, and black lines are fittings of individual data sets using eq 1. Insets show replots of kobs vs DCS. Symbols are data, and the solid line is a fit of the data using eq 2.
Figure 2.
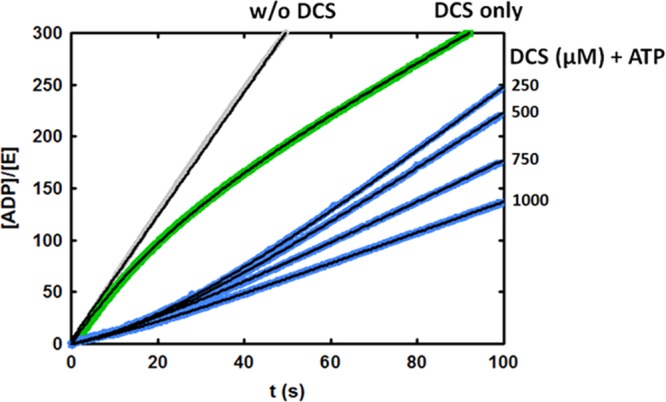
Representative reaction progress curves for MtDdl (final concentrations of 40 mM d-Ala and 3 mM ATP) following preincubation of MtDdl for 5 min with DCS alone (0.5 mM, green curve) or with ATP and multiple concentrations of DCS (blue curves). The progress curve for reaction in the absence of DCS is also displayed (gray). Gray, green, and blue lines depict data, obtained by averaging three injections, and thin black lines are fits of individual data sets using the appropriate equation.
Individual time course data were fit using eq 1
| 1 |
where [P]t is the product formed at time t, [E] is the total MtDdl concentration, vi is the initial rate, vf is the final (steady-state) rate, and kobs is the apparent rate constant for approach to the steady state.7 Time courses were performed in triplicate and kobs values averaged for each DCS and d-Ala concentration tested. Secondary replots of kobs versus DCS concentration were fit using eq 2
| 2 |
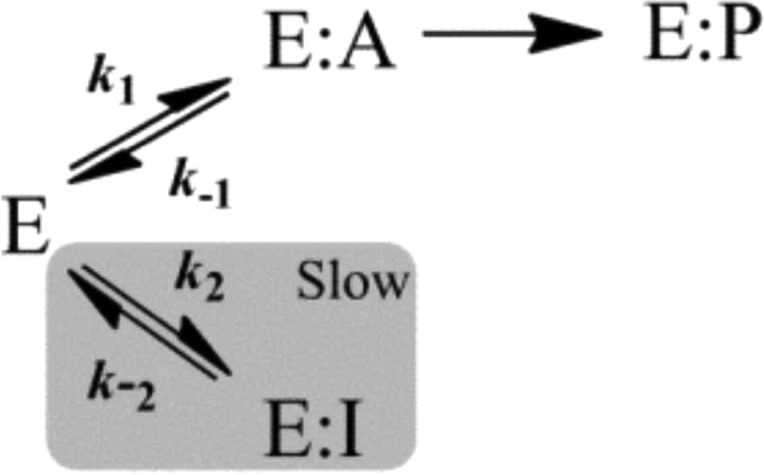
where [I] is the DCS concentration, [S] is the d-Ala concentration, and k2 and k–2 are rate constants as described in Scheme 2. The Km value was that for the C-terminal site (4 mM), as previously reported.4
Scheme 2. Minimal Kinetic Mechanism Describing Slow-Onset Inhibition without Slow Conformational Changes.
pH Dependence
Ki,DCS2 and Kd,DCS1 pH profile data were fit using eq 3, describing the involvement of two acidic (unresolvable pKa values) and two basic (unresolvable pKa values) residues in ligand binding:
| 3 |
where pKx is the negative log10 of the parameter being measured (Ki,DCS2 or Kd,DCS1), pKx0 is the pH-independent value of the parameter being measured, H is the proton concentration, and Ka1 and Ka2 are acid dissociation constants for enzyme or ligand ionizable groups. One-dimensional NMR data indicated that DCS was stable and consisted of a single chemical species across all pHs tested over the time frame of the experiment (data not shown).
Software
All data analysis, curve fitting, and graph design were performed with SigmaPlot 12.0. Reported values and errors were derived from software-assisted regression analyses of averaged replicate (at least triplicate) data sets.
Results and Discussion
Using stopped-flow spectrophotometry to guarantee sufficient sampling and the accuracy of the crucial early time points (first 0–20 s), we generated high-resolution time course data for MtDdl activity at multiple concentrations of DCS and d-Ala (Figure 1A). To simplify the kinetic analysis, all d-Ala concentrations tested were saturating for the N-terminal d-Ala binding site (≫Km,d-Ala1). It was therefore assumed that all DCS binding events involved the MtDdl–ATP–d-Ala1 complex, i.e., competitive with binding of the C-terminal d-Ala.4 Consistent with our previous observations, reaction progress curves displayed biphasic curvature only when DCS was present in the reaction mixture (Figure 1A).
Equation 1 was applied to individual time course data to permit estimation of the rate constants for the approach to steady state, or kobs. The dependence of kobs on inhibitor concentration is diagnostic of the slow-onset inhibitory mechanism being observed.7 Replots of kobs versus DCS concentration (at each d-Ala concentration) correlated linearly (inset Figure 1A), implicating a mechanism as illustrated in Scheme 2. In this case, association and/or dissociation of the enzyme–inhibitor complex occurs slowly (relative to the rate of catalysis), and enzyme–inhibitor complex isomerization steps do not contribute noticeably to the rate of inhibition onset.7 Individual replots of kobs versus DCS concentration, at each d-Ala concentration tested, were then fit to eq 2 to calculate estimates of the individual rate constants for DCS binding and dissociation (k2 and k–2, respectively, in Scheme 2). Our results indicated that the d-Ala concentration employed (from 0.4 to 40 mM) did not affect the magnitudes of the calculated rate constants (results not shown), and therefore, values were averaged across all d-Ala concentrations tested and are listed in Table 1.
Table 1. Kinetic Parameters for Slow-Onset Inhibition of MtDdl and EcDdlB by DCSa.
| preincubationb | parameterc | MtDdl | EcDdld |
|---|---|---|---|
| ATP | k2 (M–1 s–1) | 888 ± 36 | 2970 ± 360 |
| k–2 (s–1) | 0.014 ± 0.003 | 0.13 ± 0.03 | |
| t1/2 (s) | 48 | 5 | |
| ATP with DCS | k2 (M–1 s–1) | 373 ± 11 | |
| k–2 (s–1) | 0.028 ± 0.001 | ||
| t1/2 (s) | 25 |
Values and errors were derived from regression analyses of averaged replicate (at least triplicate) data sets.
Components with which the enzyme was preincubated prior to the commencement of the reaction.
Reaction composition listed in Materials and Methods.
Only 0.75 mM d-Ala (≈Km,d-Ala2) tested for EcDdl
Although our data indicate that isomerization events do not significantly contribute to the rate of inhibition onset, it is possible that rapid enzyme-dependent modifications of DCS could lead to formation of new species with differing affinities for MtDdl. Similarly, other resonant or dimerized forms of DCS may exist under certain conditions and influence inhibition parameters (Scheme S1 of the Supporting Information).8 We therefore checked the stability of DCS by one-dimensional and 13C HSQC NMR in the presence and absence of enzyme (and other buffer components) and found no spectral changes between the two conditions (Figures S1 and S2 of the Supporting Information). Furthermore, liquid chromatography–mass spectrometry analysis of reaction products of MtDdl, d-Ala, DCS, and ATP failed to reveal MtDdl-dependent DCS modifications (data not shown). We therefore conclude that a single, stable DCS species is responsible for the slow-onset behavior observed with MtDdl.
Slow-onset inhibition has not previously been reported for DCS with any Ddl orthologue.2,6,9−12 Therefore, to test if the behavior we observed is unique to MtDdl, or if similar behavior in prior studies had been missed, we overexpressed, purified, and assessed the activity of recombinant E. coli DdlB (EcDdl), an enzyme for which DCS inhibition data are available.12 Steady-state kinetic analysis revealed kcat, Km,d-Ala2, and Ki,DCS2 values consistent with previous reports (Table S1 of the Supporting Information).12 More importantly, progress curves for reaction mixtures containing DCS were nonlinear, although with a substantially smaller curvature (Figure 1B). Accordingly, rate constants calculated from these data (Table 1) were considerably different from those of MtDdl; EcDdl displayed a >3-fold faster rate of association of the enzyme–inhibitor complex (k2) and a 10-fold faster rate of dissociation of the enzyme–inhibitor complex (k–2). These results suggest that although DCS displays slow-onset inhibition against both EcDdl and MtDdl, the response is substantially more prolonged, and therefore more mechanistically significant, for MtDdl.
On the basis of the magnitude of the dissociative half-life alone (48 s), it is difficult to conceive a strong physiological effect for the slow-onset behavior of DCS against MtDdl; however, theoretical calculations provide a DCS–MtDdl complex residence time up to 8 orders of magnitude longer than that for the d-Ala–MtDdl complex (Supporting Information and Figure S3), suggesting a significant and substantial contribution of slow-onset binding to enzyme activity in its physiological setting, and may indeed partially account for the observed narrow-spectrum activity of DCS against M. tuberculosis. With an increasing significance of drug–enzyme residence times in drug discovery ventures,13−15 these results emphasize the importance of DCS as a functional scaffold for future MtDdl-targeted drug design experiments.
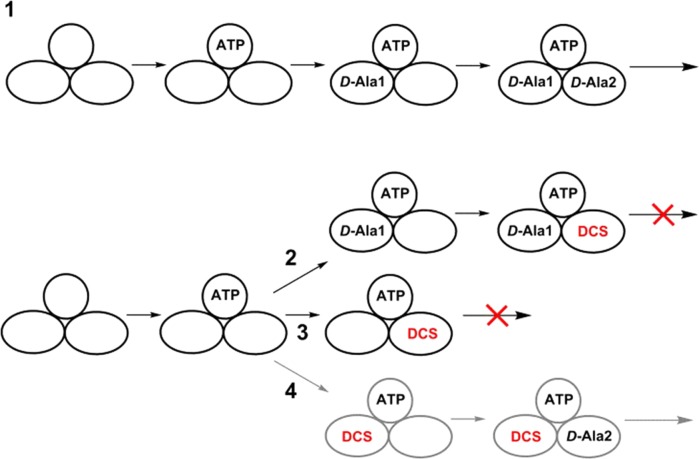
A complementary technique for confirming slow-onset inhibition is to preincubate enzyme with inhibitor prior to the commencement of the reaction. Time-dependent dissociation of the enzyme–inhibitor complex produces a characteristic “lag” phase at the beginning of the time course before equilibration to a steady-state rate.7 This method is impossible to apply to the study of slow-onset inhibition of DCS at the C-terminal d-Ala binding site of MtDdl, as preincubation would require both ATP and d-Ala (the substrate for the N-terminal site), generating a kinetically competent enzyme complex in the process. Instead, by preincubating MtDdl with only ATP and DCS prior to the commencement of the reaction, we were able to investigate whether slow-onset inhibition occurs at the N-terminal d-Ala binding site. Figure 2 shows time courses of MtDdl activity following preincubation with DCS and ATP, or with just DCS. A lag period was evident when both ATP and DCS were included in the preincubation mixture, but not when ATP was absent, reiterating the importance of ATP in DCS binding.4 In conclusion, binding and dissociation of DCS with both the MtDdl–ATP and the MtDdl–ATP–d-Ala complexes can be described by a slow-onset mechanism. Equations 1 and 2 were used on relevant progress curve data to calculate rate constants for DCS binding and dissociation from the MtDdl–ATP complex (Table 1 and Figure S4 of the Supporting Information). We were surprised at the similarity of these values to those obtained for DCS inhibition at the C-terminal d-Ala binding site (2-fold difference between k2 and k–2 values), especially when compared with the differences in active site architecture, d-Ala affinities, and substrate specificities between the two distinct substrate binding pockets.4,16−18 Together with our previous data demonstrating similar (also unexpected) DCS affinity at each binding site (Ki,DCS1 and Ki,DCS2 of 15 and 25 μM, respectively), and in the absence of any published Ddl–DCS cocrystal structure to claim otherwise, we now propose a novel mechanism of DCS inhibition whereby only the C-terminal d-Ala binding site of Ddl is capable of binding DCS but can be occupied either in the presence or in the absence of bound N-terminal d-Ala (Scheme 3). This theory is entirely consistent with previous steady-state kinetic data from our group and other research groups,4,6 as such studies are able to distinguish only between enzyme forms (as per the definition of a competitive inhibitor19); no positional information is derived. It can also explain the inability of two DCS molecules to simultaneously occupy each binding site, as we have previously shown.4 Although structural data are required to definitively assess the true binding mechanism, we were able to lend further support to the proposed mechanism by comparing pH profiles of dissociation constants of the MtDdl–ATP complex and DCS (using intrinsic tryptophan fluorescence quenching) with those of Ki,DCS2 (binding or inhibition of DCS to the MtDdl–ATP–d-Ala complex). As the active site residues that constitute the chemically distinct binding pockets do not share the same pKa values,20 the almost superimposing pH profile curves shown in Figure 3 refute a model involving separate binding sites for the two DCS binding events.
Scheme 3. Representation of the Uninhibited Kinetic Mechanism (1), the DCS-Inhibited Forms Proposed To Exist with MtDdl (2 and 3), and Forms That Are Not Likely To Exist (4).
Figure 3.
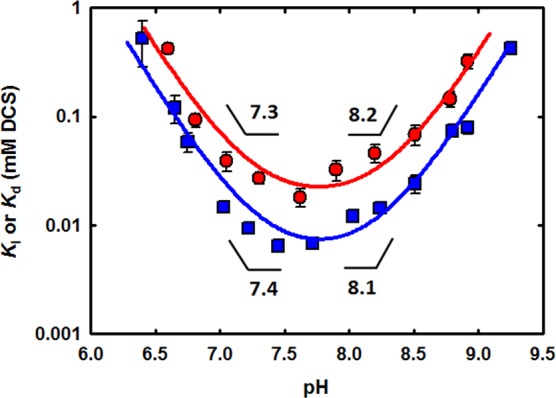
Comparison of the pH dependence on Ki,DCS2 (red circles) and Kd,DCS1 (blue squares) for MtDdl. Ki,DCS2 values were taken from previous work.4Kd,DCS1 values were determined as described in Materials and Methods. Symbols are data, and solid lines are fits of each data set using eq 3. Calculated pKa values are indicated.
In summary, our DCS inhibition results indicate that MtDdl binds DCS slowly, resulting in slow-onset inhibition kinetics, contrary to what has been observed for other orthologues. More importantly, the magnitude of the rate constants obtained in the presence and absence of d-Ala and the similarities observed in pH studies of DCS dissociation and inhibition strongly suggest that DCS inhibition is attained by DCS binding solely at the C-terminal d-Ala site. This information is of paramount importance for the rational development of improved analogues of DCS that can be used to treat human tuberculosis. In particular, enhanced drug target residence time, the specific and high affinity for the C-terminal d-Ala site, and negligible off-target host interactions (by counterscreening against the human NMDA receptor, for example) should all be factored into future MtDdl-targeted drug discovery ventures.
Acknowledgments
We thank the National Institute for Medical Research Large Scale Laboratory for E. coli growth and Dr. Gerald Larrouy-Maumus for initial help with the stopped-flow spectrophotometry. We thank Dr. Geoff Kelly (MRC Biomedical NMR Centre) for conducting NMR experiments.
Glossary
Abbreviations
- DCS
d-cyloserine
- Ddl
d-alanine:d-alanine ligase
- d-Ala-d-Ala
d-alanyl-d-alanine
- Mt
M. tuberculosis
- Ec
E. coli.
Supporting Information Available
Table S1, Scheme S1, and Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was funded by the Medical Research Council (MC_UP_A253_1111).
Supplementary Material
References
- Caminero J. A.; Sotgiu G.; Zumla A.; Migliori G. B. (2010) Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect. Dis. 10, 621–629. [DOI] [PubMed] [Google Scholar]
- Strominger J. L.; Ito E.; Threnn R. H. (1960) Competitive inhibition of enzymatic reactions by oxamycin. J. Am. Chem. Soc. 82, 998–999. [Google Scholar]
- Yew W. W.; Wong C. F.; Wong P. C.; Lee J.; Chau C. H. (1993) Adverse neurological reactions in patients with multidrug-resistant pulmonary tuberculosis after coadministration of cycloserine and ofloxacin. Clin. Infect. Dis. 17, 288–289. [DOI] [PubMed] [Google Scholar]
- Prosser G. A.; de Carvalho L. P. (2013) Kinetic mechanism and inhibition of Mycobacterium tuberculosisd-alanine:d-alanine ligase by the antibiotic d-cycloserine. FEBS J. 280, 1150–1166. [DOI] [PubMed] [Google Scholar]
- Mullins L. S.; Zawadzke L. E.; Walsh C. T.; Raushel F. M. (1990) Kinetic evidence for the formation of d-alanyl phosphate in the mechanism of d-alanyl-d-alanine ligase. J. Biol. Chem. 265, 8993–8998. [PubMed] [Google Scholar]
- Neuhaus F. C.; Lynch J. L. (1964) The Enzymatic Synthesis of d-Alanyl-d-Alanine. 3. On the Inhibition of d-Alanyl-d-Alanine Synthetase by the Antibiotic d-Cycloserine. Biochemistry 3, 471–480. [DOI] [PubMed] [Google Scholar]
- Morrison J. F.; Walsh C. T. (1988) The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 61, 201–301. [DOI] [PubMed] [Google Scholar]
- Lassen F. O.; Stammer C. H. (1971) Cycloserine Dimer Hydrolysis and Its Equilibration with Cycloserine. J. Org. Chem. 36, 2631–2634. [DOI] [PubMed] [Google Scholar]
- Bugg T. D.; Dutka-Malen S.; Arthur M.; Courvalin P.; Walsh C. T. (1991) Identification of vancomycin resistance protein VanA as a d-alanine:d-alanine ligase of altered substrate specificity. Biochemistry 30, 2017–2021. [DOI] [PubMed] [Google Scholar]
- Liu S.; Chang J. S.; Herberg J. T.; Horng M. M.; Tomich P. K.; Lin A. H.; Marotti K. R. (2006) Allosteric inhibition of Staphylococcus aureusd-alanine:d-alanine ligase revealed by crystallographic studies. Proc. Natl. Acad. Sci. U.S.A. 103, 15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M.; Kawahara Y.; Ichikawa A.; Matoba Y.; Matsuo H.; Lee D. G.; Kumagai T.; Sugiyama M. (2004) Self-protection mechanism in d-cycloserine-producing Streptomyces lavendulae. Gene cloning, characterization, and kinetics of its alanine racemase and d-alanyl-d-alanine ligase, which are target enzymes of d-cycloserine. J. Biol. Chem. 279, 46143–46152. [DOI] [PubMed] [Google Scholar]
- Zawadzke L. E.; Bugg T. D.; Walsh C. T. (1991) Existence of two d-alanine:d-alanine ligases in Escherichia coli: Cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry 30, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Copeland R. A.; Pompliano D. L.; Meek T. D. (2006) Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discovery 5, 730–739. [DOI] [PubMed] [Google Scholar]
- Lu H.; England K.; am Ende C.; Truglio J. J.; Luckner S.; Reddy B. G.; Marlenee N. L.; Knudson S. E.; Knudson D. L.; Bowen R. A.; Kisker C.; Slayden R. A.; Tonge P. J. (2009) Slow-onset inhibition of the FabI enoyl reductase from Francisella tularensis: Residence time and in vivo activity. ACS Chem. Biol. 4, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney D. C. (2004) Biochemical mechanisms of drug action: What does it take for success?. Nat. Rev. Drug Discovery 3, 801–808. [DOI] [PubMed] [Google Scholar]
- Bruning J. B.; Murillo A. C.; Chacon O.; Barletta R. G.; Sacchettini J. C. (2011) Structure of the Mycobacterium tuberculosisd-alanine:d-alanine ligase, a target of the antituberculosis drug d-cycloserine. Antimicrob. Agents Chemother. 55, 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C.; Moews P. C.; Walsh C. T.; Knox J. R. (1994) Vancomycin resistance: Structure of d-alanine:d-alanine ligase at 2.3 Å resolution. Science 266, 439–443. [DOI] [PubMed] [Google Scholar]
- Neuhaus F. C. (1962) The enzymatic synthesis of d-alanyl-d-alanine. II. Kinetic studies on d-alanyl-d-alanine synthetase. J. Biol. Chem. 237, 3128–3135. [PubMed] [Google Scholar]
- Cook P. F., and Cleland W. W. (2007) Enzyme Kinetics and Mechanism, pp 121–123, Garland Science, New York. [Google Scholar]
- Carlson H. A.; Briggs J. M.; McCammon J. A. (1999) Calculation of the pKa values for the ligands and side chains of Escherichia colid-alanine:d-alanine ligase. J. Med. Chem. 42, 109–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.