ABSTRACT
Most viruses that replicate in the cytoplasm of host cells form neo-organelles that serve as sites of viral genome replication and particle assembly. These highly specialized structures concentrate viral replication proteins and nucleic acids, prevent the activation of cell-intrinsic defenses, and coordinate the release of progeny particles. Despite the importance of inclusion complexes in viral replication, there are key gaps in the knowledge of how these organelles form and mediate their functions. Reoviruses are nonenveloped, double-stranded RNA (dsRNA) viruses that serve as tractable experimental models for studies of dsRNA virus replication and pathogenesis. Following reovirus entry into cells, replication occurs in large cytoplasmic structures termed inclusions that fill with progeny virions. Reovirus inclusions are nucleated by viral nonstructural proteins, which in turn recruit viral structural proteins for genome replication and particle assembly. Components of reovirus inclusions are poorly understood, but these structures are generally thought to be devoid of membranes. We used transmission electron microscopy and three-dimensional image reconstructions to visualize reovirus inclusions in infected cells. These studies revealed that reovirus inclusions form within a membranous network. Viral inclusions contain filled and empty viral particles and microtubules and appose mitochondria and rough endoplasmic reticulum (RER). Immunofluorescence confocal microscopy analysis demonstrated that markers of the ER and ER-Golgi intermediate compartment (ERGIC) codistribute with inclusions during infection, as does dsRNA. dsRNA colocalizes with the viral protein σNS and an ERGIC marker inside inclusions. These findings suggest that cell membranes within reovirus inclusions form a scaffold to coordinate viral replication and assembly.
IMPORTANCE
Viruses alter the architecture of host cells to form an intracellular environment conducive to viral replication. This step in viral infection requires the concerted action of viral and host components and is potentially vulnerable to pharmacological intervention. Reoviruses form large cytoplasmic replication sites called inclusions, which have been described as membrane-free structures. Despite the importance of inclusions in the reovirus replication cycle, little is known about their formation and composition. We used light and electron microscopy to demonstrate that reovirus inclusions are membrane-containing structures and that the endoplasmic reticulum (ER) and the ER-Golgi intermediate compartment interact closely with these viral organelles. These findings enhance our understanding of the cellular machinery usurped by viruses to form inclusion organelles and complete an infectious cycle. This information, in turn, may foster the development of antiviral drugs that impede this essential viral replication step.
INTRODUCTION
The replication and assembly of many viruses occur in specialized intracellular compartments known as virus factories, viral inclusions, or viroplasms. These neo-organelles formed during viral infection concentrate viral replication proteins and nucleic acids, prevent the activation of cell-intrinsic defenses, and coordinate the release of progeny particles (1–3). Many RNA viruses build factories by remodeling host membranes and creating new interorganelle contacts (4). Interestingly, membrane rearrangements are induced by both enveloped and nonenveloped viruses, suggesting that viral replication requires the physical support of cell membranes, even for those viruses that do not incorporate membranes into progeny particles (5). The growing interest in understanding how virus factories form, coupled with technical advances in genomics, proteomics, and cell imaging, has advanced our knowledge of the biogenesis and architecture of these unique structures. However, for many viruses, it is not known how these structures form and mediate their functions.
Mammalian orthoreoviruses (reoviruses) are nonenveloped, double-shelled viruses that contain a genome of 10 double-stranded RNA (dsRNA) segments (6). Following the entry of reovirus into cells, the outer capsid is removed within the endocytic compartment (7–9), which allows the release of transcriptionally active core particles into the cytoplasm (10–13). These particles initiate a primary round of transcription to produce full-length, message-sense, single-stranded RNAs (ssRNAs) corresponding to each viral gene segment (14, 15). Reovirus ssRNAs can be translated and also serve as templates for minus-strand synthesis to generate nascent genomic dsRNA within replicase particles (16). These particles initiate a secondary round of transcription that fuels most viral protein synthesis (17). Particle assembly is completed by the addition of outer-capsid proteins onto nascent cores.
Viral RNA assortment, genome replication, secondary transcription, and particle assembly occur within specialized virus-derived neo-organelles, termed inclusions, which form in the host cell cytoplasm (14, 16, 18–22). Inclusions can be detected by confocal microscopy as early as 4 h postinfection (hpi) and contain viral proteins and dsRNA, as well as particles at various stages of morphogenesis (23–26). Mature viral progeny are arranged in paracrystalline arrays at late times postinfection prior to viral release (18, 21, 27). It is not thought that reovirus inclusions require membrane for their formation or function.
Most cell lines are readily infected by numerous reovirus strains (6). However, Madin-Darby canine kidney (MDCK) cells are susceptible to infection by reovirus strain type 1 Lang (T1) but not strain type 3 Dearing (T3) (28, 29). The primary determinant of this replication difference is the viral M1 gene (29), which encodes μ2, a multifunctional replication protein. The critical μ2-dependent function in the replication cycle in MDCK cells occurs at a postentry step in the viral life cycle, following primary transcription and translation but prior to dsRNA synthesis (29). Further characterization of T3 replication in MDCK cells revealed that particle assembly is defective (30). These studies indicate that events within the inclusion involving the interaction of viral proteins and cell-type-specific factors promote the development of inclusions with the capacity to generate fully assembled infectious virions. Thus, the formation of reovirus inclusions is a determinant of virus cell tropism.
In this study, we used transmission electron microscopy (TEM), three-dimensional (3D) image reconstructions, and immunofluorescence confocal microscopy to study the composition and organization of reovirus neo-organelles. Surprisingly, we found that reovirus inclusions formed in HeLa cells are membranous webs in which viral particles are embedded. Using organelle markers, we found that the endoplasmic reticulum (ER) and ER-Golgi intermediate compartment (ERGIC) are remodeled during infection and codistribute with inclusions. Membrane recruitment also occurs during reovirus infection of MDCK cells, suggesting a function for cellular membranes in reovirus replication in different cell types. These results suggest that reovirus inclusions are complex structures that recruit cell membranes to house functions required for viral replication.
RESULTS
Reovirus inclusions are surrounded by membranes and mitochondria.
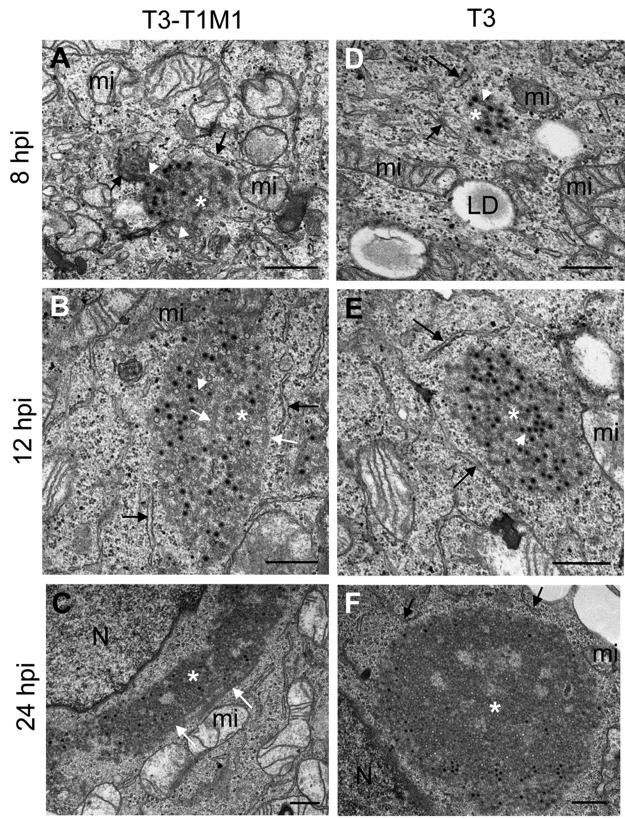
To define the organization of reovirus inclusions, we performed ultrastructural studies of infected cells. HeLa cells were infected with reovirus strain T3-T1M1 or T3, each of which establishes productive infection of this cell line (data not shown), and imaged by TEM. We observed that inclusions formed by either strain were partially surrounded by ER membranes (Fig. 1). Mitochondria and rough ER (RER) cisternae surrounded and attached to inclusions at 8 and 12 hpi (Fig. 1A and B and D and E). Quantification of imaging data from 53 randomly selected inclusions showed that RER elements associated with all of them. Smooth membranes were frequently observed to be connected with the interior of inclusions (white arrowheads in Fig. 1A). Thick, coated microtubules were detected inside and adjacent to the inclusions formed by both strains at 12 and 24 hpi but were not observed at 8 hpi (Fig. 1B and C and 2). At 24 hpi, inclusions developed into large structures that occupied extensive areas of the cytosol (Fig. 1C and F and 2A and C) with mitochondria and RER cisternae remaining closely apposed. No significant changes in mitochondrial morphology or integrity were observed during infection. Most of the viral particles within inclusions at early times (8 and 12 hpi) exhibited mature morphology, as demonstrated by dark staining inside the viral particle surrounded by lighter staining (Fig. 1A and B and D and E). However, at 24 hpi, empty particles were more numerous (Fig. 2B and D).
FIG 1 .
Ultrastructure of reovirus inclusions in HeLa cells. HeLa cells were infected with reovirus strain T3-T1M1 (A to C) or T3 (D to F) and fixed at 8, 12, or 24 hpi. Ultrathin (~60- to 70-nm) sections were imaged by TEM. Inclusions are marked with white asterisks, RER cisternae are marked with black arrows, smooth membranes inside inclusions are marked with white arrowheads, and coated microtubules are marked with white arrows. T3-T1M1 inclusions at 8 (A), 12 (B), and 24 (C) hpi and T3 inclusion at 8 (D), 12 (E), and 24 (F) hpi are shown. LD, lipid droplet; mi, mitochondria; N, nucleus. Scale bars: 0.5 µm.
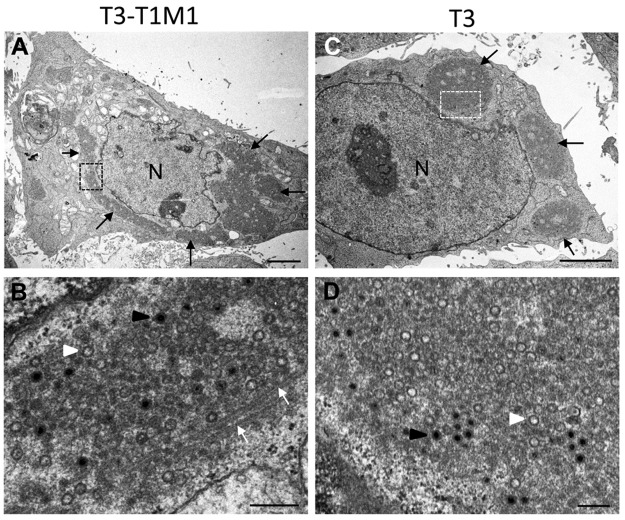
FIG 2 .
TEM of HeLa cells infected with reovirus strain T3-T1M1 or T3 at 24 hpi. Low (A) and high (B) magnifications of cells infected with T3-T1M1 are shown. (A) Perinuclear inclusion (black arrows). (B) Enlargement of highlighted area in panel A. The inclusion contains filled viral particles (black arrowhead), empty viral particles (white arrowhead), and coated microtubules (white arrows). (C, D) Low (C) and high (D) magnifications of cells infected with T3. In panel C, perinuclear inclusions are marked with black arrows. Panel D is an enlargement of the boxed area in panel C. The inclusion contains filled (black arrowhead) and empty (white arrowhead) particles. Scale bars: 3 µm in panels A and C, 0.25 µm in panels B and D.
Imaging of cells at higher magnification showed details of the membranes and microtubules contained within reovirus inclusions (Fig. 3). RER elements were observed attached at discrete points to the inclusion periphery, but characteristic RER cisternae were rarely observed inside these structures. However, the smooth membranes associated with cytosolic viral particles and seen inside inclusions appeared to be connected to RER (Fig. 3A and B). Occasionally, RER elements on the periphery of inclusions were joined with cubic membrane sheets (Fig. 3C) reminiscent of the membranous platforms assembled by other viruses (5, 31). Coated microtubules were found to fill the inclusions at 24 hpi (Fig. 3D). Both filled and empty viral particles appeared to attach to coated microtubules but not to uncoated microtubules (Fig. 3E).
FIG 3 .
TEM of viral inclusions in T3-T1M1-infected HeLa cells. Inclusions are marked with white asterisks. (A) Viral particles (arrows) in small inclusions attached to membranes in a region containing RER and the Golgi compartment at 8 hpi. (B) Smooth membranes (white arrowheads) inside an inclusion surrounded by RER at 8 hpi. (C) Inclusion at 12 hpi surrounded by mitochondria and ER membranes. Some RER elements close to the inclusion (arrow on the right) appear to be connected to a sheet of cubic membranes (black asterisk). (D) Inclusion at 24 hpi filled with coated microtubules (white arrows). (E) Higher magnification of a different inclusion with viral particles attached to coated (~60 nm, white arrows) but not uncoated (~30 nm, black arrows) microtubules. N, nucleus; C, centriole. Scale bars: 0.25 µm.
Cell membranes are associated with inclusions during nonproductive T3 infection in MDCK cells.
In contrast to HeLa cells, reovirus strains T3 and T3-T1M1 differ in the capacity to complete a replication cycle in MDCK cells (28, 29). Although T3 can infect MDCK cells and form inclusion-like structures, yields of viral progeny in these cells are markedly less than those produced by T3-T1M1 (29, 30). To determine whether membranes are associated with the inclusions formed by reovirus in another cell type and to gain insight into the elements required for the biogenesis of functional inclusions, we imaged MDCK cells infected with either T3-T1M1 (permissive) or T3 (nonpermissive) reovirus (see Fig. S1 in the supplemental material). Inclusions formed in MDCK cells infected with T3-T1M1 at 8, 12, and 24 hpi were similar to those observed in HeLa cells. In contrast, inclusion formation was delayed in T3-infected MDCK cells, consistent with previous observations (29; data not shown). Accordingly, inclusions in T3-infected MDCK cells were scarce at 8 and 12 hpi but numerous and large at 24 hpi (data not shown). ER membranes and mitochondria were associated with inclusions formed in cells infected with either virus strain (see Fig. S1A, D, and E). Vacuoles containing fibers and a few viral particles were observed in inclusions assembled by T3 reovirus (see Fig. S1E). However, microtubules were observed only in the inclusions assembled by T3-T1M1 (see Fig. S1A and B). Inclusions formed in T3-T1M1-infected cells contained numerous mature virions and few empty particles (see Fig. S1C), whereas in cells infected by T3, the inclusions contained numerous empty particles and few mature virions (see Fig. S1F). These data corroborate previous TEM studies of reovirus infection of MDCK cells (29, 30). In addition, T3 inclusions were filled with many smaller particles, which appeared to be subassemblies of macromolecules containing nucleic acid, as shown by a density darker than that in empty particles. These particles were irregular in size and did not have a shell (see Fig. S1F). These findings suggest that reovirus recruits membranes to the inclusions formed in HeLa and MDCK cells.
Reovirus inclusions associate with ERGIC and ER elements.
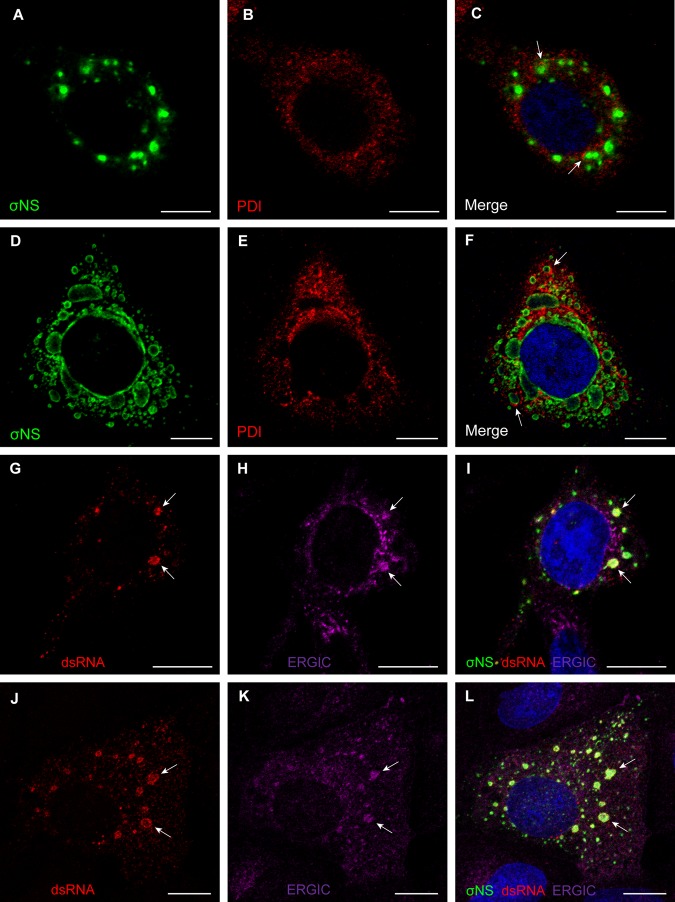
To elucidate the cellular source of the inclusion-associated membranes, we stained T3-T1M1-infected HeLa and MDCK cells with different organelle markers (Fig. 4; see Fig. S2 in the supplemental material). Viral inclusions were marked with an antibody specific for the σNS protein, which is an essential reovirus inclusion component (23, 25, 32). ER marker protein disulfide isomerase (PDI) distributed with inclusions in both HeLa cells (see Fig. S2A to C) and MDCK cells (data not shown). Golgi compartment markers giantin and wheat germ agglutinin (WGA) were not associated with inclusions during infection, and the staining patterns of each did not differ between infected and uninfected cells (see Fig. S2D to I). These findings make it unlikely that the Golgi compartment serves as a source of membranes associated with reovirus inclusions. Higher-magnification images showed ER marker PDI in close apposition to viral inclusions at early and late times after infection (Fig. 4A to F), whereas the PDI staining was mostly perinuclear in uninfected cells (see Fig. S2A to C). Type I transmembrane protein ERGIC-53, which has been used in previous studies to mark the ERGIC (33), localized within inclusion structures in both HeLa and MDCK cells (Fig. 4G to L), as does the KDEL receptor (see Fig. S2J to L), which marks the ERGIC and Golgi compartment. Collectively, these observations suggest that the ERGIC, a membranous system that functionally links the ER and the Golgi compartment, is a source of inclusion-associated membranes, consistent with our TEM studies.
FIG 4 .
Reovirus inclusions codistribute with ER and ERGIC elements. (A to F) HeLa cells were infected with T3-T1M1 for 12 h (A to C) or 24 h (D to F). Cells were fixed; permeabilized; stained for σNS (green), PDI (red), or nuclei (blue); and visualized by confocal microscopy. Arrows indicate viral inclusions associated with RER elements on the periphery. (G to L) HeLa cells (G to I) and MDCK cells (J to L) were infected with T3-T1M1 for 12 h; fixed; permeabilized; stained for dsRNA (red), ERGIC-53 (magenta), σNS (green), or nuclei (blue); and visualized by confocal microscopy. Arrows indicate viral inclusions that contain dsRNA, the ERGIC, and σNS. Scale bars: 10 µm.
To confirm that inclusions localizing with ERGIC-53 support viral replication, T3-T1M1-infected HeLa and MDCK cells were stained with a dsRNA-specific antibody (Fig. 4G to L). Staining for dsRNA codistributed with inclusion marker σNS and ERGIC-53 inside the inclusions. These data suggest that membrane-filled inclusions are sites of reovirus RNA replication.
Serial sections and 3D reconstructions reveal that reovirus inclusions are membranous webs that contain microtubules.
To understand the global architecture of inclusions and the contribution of cell membranes and specific organelles to the construction of these structures, we generated 3D reconstructions of reovirus inclusions. Oriented serial sections of reovirus-infected cells were imaged by TEM and processed for 3D reconstruction (see Fig. S3 and 4 in the supplemental material). The resulting 3D reconstructions showed that inclusions in HeLa cells are membranous webs surrounded by mitochondria (Fig. 5; see Fig. S3A and B and Fig. S5 in the supplemental material). Smooth membranes of the inclusions were observed to contact mitochondria in a variety of orientations (arrows in Fig. 5; see Movie S1 in the supplemental material). RER cisternae also were observed to contact the inclusion membranes and adjacent mitochondria (see Fig. S5 and Movie S2). Microtubules and viral particles were embedded in the inclusion membranous webs (Fig. 5; see Fig. S5A, and B). As detected by 3D TEM, viral particles appeared to be attached to microtubules (see Fig. S5C and D). These features were observed in all of the 3D reconstructions prepared from serial sections of reovirus-infected cells (data not shown).
FIG 5 .

3D model of reovirus inclusions in HeLa cells. HeLa cells were infected with T3-T1M1 and fixed at 12 hpi. The inclusion was visualized by TEM of serial sections, 3D reconstruction, and image processing. (A) Mitochondria (red) surround a network of membranes (yellow). RER (brown) and nuclear envelope (blue) are adjacent to the inclusion. Filled viral particles (black), empty viral particles (white), and microtubules (green) are integrated into the inclusion membrane network. (B) Rotation of the same reconstruction showing contacts between the inclusion membranes and mitochondria (arrows). The volume has been superimposed onto the 2D image of one of the sections in the series.
To analyze the morphology of inclusions formed by reovirus strains that vary in the capacity to complete an infectious cycle, TEM images were processed to generate 3D reconstructions of inclusions formed in MDCK cells infected with strain T3-T1M1 or T3 (Fig. 6). Numerous mitochondria and RER cisternae were observed to surround the inclusions formed in T3-T1M1-infected cells (Fig. 6A). RER membranes appeared to contact the smooth membranes of the inclusions at 24 hpi, which are filled with mature virions, empty particles, and microtubules (Fig. 6A; see Movie S3 in the supplemental material). However, inclusions formed in T3-infected cells do not contain microtubules. In addition, most of the viral particles inside the membranous web in T3-infected cells were empty, and fewer filled particles were observed (Fig. 6B; see Movie S4). RER and mitochondria surrounded the inclusions formed by T3 reovirus. Inclusions assembled during T3-T1M1 infection were frequently observed close to the plasma membrane at 24 hpi. Image reconstructions of those inclusions showed viral particles adjacent to the plasma membrane and attached to smooth membranes, RER cisternae, and microtubules (Fig. 6C to E; see Movie S5). Together, our data indicate that reovirus inclusions are associated with ER and ERGIC membranes and mitochondria, highlighting these cellular organelles as important constituents of viral replication factories.
FIG 6 .
3D model of reovirus inclusions in MDCK cells. MDCK cells were infected with T3-T1M1 (A and C to E) or T3 (B) and fixed at 24 hpi. Membranes (yellow), mitochondria (red), RER (light brown), microtubules (green), filled viral particles (black), and empty viral particles (white) are shown. (A) Image reconstruction of an inclusion formed in T3-T1M1-infected cells. RER membranes surround and incorporate into the inclusion membrane network that contains numerous mature virions, fewer empty particles, and microtubules. (B) Image reconstruction of an inclusion formed in T3-infected cells. Most of the viral particles inside the membranous web are empty. No microtubules were observed inside or around the inclusion. Mitochondria, RER, and lipid droplets (large white structures) surround the inclusion, which is in contact with the nuclear envelope (blue). (C) Image reconstruction of an inclusion close to the plasma membrane (brown) in a cell infected with T3-T1M1. The plasma membrane from another cell is colored blue. The inclusion contains membranes (yellow) and filled (black) and empty (white) viral particles. Peripheral RER elements, microtubules, and viral particles are in contact with the cytosolic face of the plasma membrane. Enlargements of this region are shown in panels D and E. Viral particles attached to RER, microtubules, and the plasma membrane (arrows) are visible.
DISCUSSION
In this study, we demonstrated that inclusions formed by reovirus are associated with cell membranes. The participation of cell membranes within inclusions is a new finding for reoviruses, which previously had been thought to form inclusion complexes devoid of membranes. However, other Reoviridae viruses require membranes to replicate. Hyposoter exiguae reovirus (a reovirus of the parasitoid wasp H. exiguae) is released from infected cells exclusively by budding (34). Orbiviruses are variously described as being membranophilic (35), pseudoenveloped (36), or transiently enveloped (37). Rotavirus morphogenesis requires penetration of the ER to acquire outer-shell proteins VP4 and VP7 (38, 39). Therefore, the use of cell membranes to promote replication, assembly, or release may be a general feature of dsRNA viruses.
Previous studies of the ultrastructure of reovirus inclusions clearly demonstrate the presence of microtubules, but membranes in association with inclusions have not been described (18, 30). The discordance between our results and those obtained previously might be attributable to differences in the time postinfection chosen for analysis and the strategies used for TEM. In conventional TEM, samples must be thin, <100 nm, for electrons to be able to traverse them and generate a projection image. These ultrathin sections of eukaryotic cells are single planes of much larger volumes. Moreover, when analysis is restricted to random sections of cells, scarce or nonrandomly distributed elements can be missed. Oriented serial sections and 3D reconstructions avoid these limitations and allow imaging of whole cells and a comprehensive analysis of intracellular contacts (5, 40, 41). This analysis was instrumental in our finding that reovirus inclusions are embedded in membrane.
Our 3D image reconstructions have revealed the complex organization of reovirus inclusions and the participation of cell membranes (most likely derived from the ERGIC), mitochondria, and microtubules in inclusion structure. Confocal microscopy studies confirmed an association of the inclusions with the ER and ERGIC and suggest that the Golgi complex does not contribute to inclusion formation. Viral particles appear to attach to coated microtubules and membranes inside the inclusion and remain attached when reaching the plasma membrane. Microtubule coats are likely to contain the viral μ2 protein, which associates with microtubules (42), as well as other viral and perhaps cellular proteins. When a complete analysis is performed with serial sections and 3D reconstructions, all of the inclusions identified in both HeLa and MDCK cells are associated with mitochondria, ER elements, and membranous webs. We think that the cell membranes within inclusions serve as a physical scaffold for inclusion formation and organize the reovirus nonstructural and structural proteins required for viral genome replication and progeny particle assembly. In this regard, RER cisternae make numerous contacts with reovirus inclusions but do not surround them completely. The association of RER with reovirus inclusions is reminiscent of the structures of rubella virus replication organelles (43). In rubella virus factories, RER cisternae provide a framework for newly synthesized viral proteins to incorporate into viral replication complexes (43).
Mitochondria are recruited to factories assembled by many viruses (5). In keeping with this general feature of viral replication, our 3D image reconstructions showed numerous contacts between reovirus inclusion membranes and mitochondria. Moreover, new interorganelle contacts have been visualized within reovirus factories. For example, attachment of mitochondria to ERGIC, the Golgi compartment, or lysosomes is not observed in uninfected mammalian cells (5). Mitochondria may be used as an energy source to power viral replication and also supply host factors required for viral genome synthesis and particle assembly, as reported for rubella virus and tombusviruses (5, 43). In reovirus-infected cells, the transfer of energy and host factors likewise might be mediated by the physical contacts between mitochondria and the inclusion membranous webs. RER also might serve as the source of viral and cellular proteins required for inclusion activities.
The inclusions assembled in MDCK cells infected with strain T3, which enters these cells but does not complete an infectious cycle (28, 29), contain membrane but lack microtubules. This finding suggests that microtubules are required for the functional organization of inclusions and assembly of progeny particles. Moreover, this finding provides a potential explanation for the genetic linkage between the replication differences displayed by reovirus strains in MDCK cells and the μ2-encoding M1 gene (28, 29). The μ2 protein interacts with microtubules and dictates inclusion morphology (42). Concordantly, microtubules are observed in inclusions assembled by both T3 and T3-T1M1 in HeLa cells, which support the replication of both strains (data not shown). Microtubule function might be required to transport essential components to inclusions, organize inclusion content, or compartmentalize inclusion activities. Microtubules also might be required to transport progeny particles to the plasma membrane for subsequent release. It is not known whether microtubules incorporate into small inclusions and enhance inclusion coalescence and growth, as reported for some other viruses (44, 45). The delay in inclusion formation during T3 infection of MDCK cells also could point to a defect in the recruitment or remodeling of cell membranes. We observed membranes in association with inclusions from the initial stages of inclusion assembly in HeLa cells infected with either T3 or T3-T1M1, suggesting that membranes could potentiate steps required for inclusion biogenesis.
An important question left unanswered by our study is the mechanism by which progeny viral particles migrate from inclusions to the cell periphery and exit infected cells. Reovirus exits some cells by lysis (28, 46, 47). However, egress of virus from polarized endothelial cells (48) and epithelial cells (49) is noncytolytic. The mechanism by which nonenveloped viral particles are released from infected cells without inducing cell lysis is not known. ER membranes could be used by reovirus to traffic to the cell periphery through some type of vesicular transport pathway that bypasses the Golgi apparatus, as suggested by our 3D reconstructions (Fig. 6). Unconventional vesicular trafficking has been suggested for rotavirus egress, which also bypasses the Golgi apparatus to reach the cell periphery (50). The rotavirus spike protein VP4 is particularly important for this process. Despite not having transmembrane domains, VP4 interacts with lipids and is enriched in lipid rafts (51). VP4 is essential for rotavirus release, as it interacts with actin filaments in infected cells (52). A homologous reovirus protein responsible for interaction with lipids is not known. All reovirus proteins lack transmembrane domains, and posttranslational modifications are not completely defined, although the μ1 protein contains a myristoyl moiety (53). Lipidation of reovirus proteins, specifically at late times after infection, might be required for membrane recruitment and particle transport to the cell periphery.
New technologies now offer access to the analysis of viruses in cells in unprecedented detail. The complexity of the interaction networks established in these contexts is changing our concept of viruses from that of inert chemicals to living entities capable of performing a wide variety of intracellular functions. Reovirus inclusions appear to assemble as membranous replication organelles that are usually associated with mitochondria. However, it is not known how the cellular organelles are recruited, how macromolecular transport operates inside the inclusions to connect viral replication and morphogenesis, and how nonenveloped viruses like reovirus exit the membranous scaffolds and leave the cell. With the information provided by this study, we now can initiate functional and mechanistic studies to identify the signals that regulate the biogenesis and activities of viral inclusions. This work will enhance our knowledge of the cell biology of virus replication and provide potential new therapeutic targets to ameliorate diseases caused by pathogenic viruses.
MATERIALS AND METHODS
Cells and viruses.
Spinner-adapted murine L929 fibroblast cells were grown in either suspension or monolayer cultures as previously described (54). HeLa CCL2 cells and MDCK cells were grown in Dulbecco’s modified Eagle’s medium containing 4.5 g/liter glucose, l-glutamine, and sodium pyruvate (Mediatech) supplemented to contain 10% fetal bovine serum, 100 U/ml penicillin G (Gibco), 100 µg/ml streptomycin (Gibco), and 0.25 µg/ml amphotericin B (Sigma). Reovirus strains T3 and T3-T1M1 were recovered by using plasmid copies of gene segment cDNAs as previously described (29). Virus was purified by cesium gradient centrifugation as previously described (55). Viral titers were determined by plaque assay with L929 cells (56).
TEM.
Virus was allowed to adsorb to monolayers of HeLa and MDCK cells at a multiplicity of infection (MOI) of 20 PFU/cell. Following incubation at 37°C for 8, 12, or 24 h, cells were fixed at room temperature for 1 h with a mixture of 4% paraformaldehyde and 1% glutaraldehyde in phosphate-buffered saline (pH 7.4), postfixed with 1% osmium tetroxide, dehydrated in increasing concentrations of acetone, and processed for embedding in epoxy resin EML-812 (TAAB Laboratories) as previously described (57, 58). Osmium tetroxide is a lipid-staining agent used in TEM to provide image contrast. Osmium(VIII) oxide binds phospholipid head groups, thus providing contrast and allowing ultrastructural identification of membranes, which are apparent as dark, flexible lines with a thickness of ~5 nm (59, 60). Ultrathin (~60- to 70-nm) sections were collected on uncoated 300-mesh copper grids (TAAB Laboratories), stained with uranyl acetate and lead citrate, and imaged by TEM. Images were acquired with a JEOL JEM 1011 electron microscope operating at 100 kV.
Confocal microscopy.
Virus at an MOI of 20 PFU/cell was allowed to adsorb to HeLa CCL2 cells cultivated on untreated glass coverslips and MDCK cells cultivated on poly-l-lysine (Sigma)-treated glass coverslips in 24-well plates. Following incubation at 37°C for 12 or 24 h, cells were fixed with ice-cold methanol (−20°C), permeabilized with 1% Triton X-100, and stained with σNS-specific antibody (25) and ERGIC-53-specific antibody H-245 (Santa Cruz Biotechnology) or KDEL receptor-specific antibody FL-212 (Santa Cruz Biotechnology) for ERGIC staining, anti-giantin antibody ab24586 (Abcam) or WGA conjugated with Alexa Fluor 555 (Invitrogen) to mark the Golgi compartment, PDI-specific antibody DL-11 (Sigma) to mark the ER, or K2 antibody (40) to mark dsRNA. ToPro3 or 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) was used to stain nuclei. Alexa Fluor-conjugated antibodies (Invitrogen) were used as secondary antibodies. Images were acquired with Zeiss LSM 510 Meta and 710 Meta inverted confocal microscopes.
3D image reconstructions.
Consecutive ultrathin (~60- to 70-nm) sections were collected on Formvar-coated copper slot grids (TAAB Laboratories), stained, and imaged by TEM. A total of eight series of 15 were selected and processed for 3D reconstruction as previously described (40, 61) (see Fig. S4 in the supplemental material). Photographs of reovirus inclusions were taken at a nominal magnification of ×10,000 or ×12,000. Plates were digitized as 8-bit images with a 3.39-nm final pixel size and a 600-dpi resolution with an Epson Perfection Photo 3170 scanner. Digital images were aligned by selected tracers between two consecutive sections with the free editor for serial section microscopy Reconstruct (62) (http://synapses.clm.utexas.edu/tools/index.stm). Segmentation and 3D visualization were performed with Amira (http://amira.zib.de). Movies were prepared from the 3D reconstructions with the Camera Rotate and Movie Maker applications of the Amira software suite.
SUPPLEMENTAL MATERIAL
Movie of the reconstruction in Fig. 5. The inclusion membranes (yellow) clearly contact mitochondria (red). Download
Movie of the reconstruction in Fig. S5A. RER cisternae (light brown) surround and contact the inclusion membranes (yellow) and mitochondria (red). Microtubules (green), filled viral particles (black), and empty viral particles (white) are visible within the inclusion membrane network. The nuclear envelope is colored blue. Download
Movie of the reconstruction in Fig. 6A. RER membranes (brown) surround and incorporate into the inclusion membrane network (yellow) that contains numerous mature virions (black), fewer empty particles (white), and microtubules (green). Download
Movie of the reconstruction in Fig. 6B. Mitochondria (red), RER (brown), and lipid droplets (large white structures) surround and contact the inclusion membranes (yellow), which are attached to the nuclear envelope (blue). The inclusion membrane network contains many empty particles (white) and fewer mature virions (black). Download
Movie of the reconstruction in Fig. 6C. The inclusion contains membranes (yellow) and filled (black) and empty (white) viral particles. Peripheral RER elements (light brown), microtubules (green), and viral particles (black) are in contact with the cytosolic face of the plasma membrane (dark brown). The plasma membrane from another cell is colored blue. Download
Ultrastructure of reovirus inclusions in MDCK cells. MDCK cells were infected with T3-T1M1 (A to C) or T3 (D to F) and fixed at 24 hpi. Ultrathin (~60- to 70-nm) sections were imaged by TEM. (A) T3-T1M1 inclusion surrounded by RER cisternae (black arrows). (B) Enlargement of the highlighted region in panel A showing coated microtubules (white arrows). (C) Enlargement of the central region in panel B. Filled (black arrowhead) and empty (white arrowhead) viral particles are apparent. (D) Large inclusion near the nucleus of a cell infected with T3. ER membranes (black arrows) are shown. (E) Highlighted region in panel D showing membranes (black arrows) inside the inclusion. A vacuole (V), which contains fibers and a few viral particles, appears attached to the inclusion periphery. (F) Enlargement of the central area in panel E. The inclusion contains a few filled particles (black arrowhead), numerous empty particles (white arrowhead), and many smaller particles (yellow arrowheads). LD, lipid droplet; mi, mitochondria; N, nucleus. Scale bars: 1.5 µm in panels A and D; 0.25 µm in panels B, C, E, and F. Download
Reovirus inclusions codistribute with the ER and ERGIC and do not associate with the Golgi compartment. HeLa cells were infected with T3-T1M1 for 24 h. Cells were fixed, permeabilized, stained, and visualized by confocal microscopy. (A to C) Cells were stained for σNS (green), PDI (red), or nuclei (blue). (D to F) Cells were stained for σNS (green), giantin (red), or nuclei (blue). (G to I) Cells were stained for σNS (green), the Golgi compartment with WGA (red), or nuclei (blue). (J to L) Cells were stained for σNS (green), KDEL receptor (red), or nuclei (blue). Asterisks mark noninfected cells. Scale bars: 10 µm. Download
Organelles and membranes in reovirus inclusions. (A) Ultrathin sections of mock-infected HeLa cells display a nonuniform distribution of organelles (left). Ultrathin sections of HeLa cells infected with T3 for 24 h show viral inclusions associated with mitochondria and RER (right). (B) Quantification of mitochondria associated with 53 randomly selected inclusions. (C) TEM images of 9 of the 15 ultrathin sections from the series used to generate the 3D reconstruction shown in Fig. 5 (raw data). Only the central area within the inclusion is shown. Smooth membranes inside the inclusion are indicated by yellow arrows. G, Golgi compartment; mi, mitochondria; N, nucleus. Scale bars: 0.5 µm in panel A; 0.25 µm in panel B. Download
Schematic of the generation of 3D reconstructions from serial sections. After the collection of serial sections (ultramicrotomy) and imaging by TEM, several computational steps were used to render the final model. The 3D reconstruction at the bottom is a different view of the inclusion shown in Fig. 6B. Download
3D model of reovirus inclusions in HeLa cells. HeLa cells were infected with T3-T1M1 and fixed at 12 hpi. The inclusions were visualized by TEM of serial sections, 3D reconstruction, and image processing. (A) The volume shows two inclusions (yellow) surrounded by mitochondria (red) and RER (light brown) near the nuclear envelope (blue). Filled particles (black), empty particles (white), and microtubules (green) are apparent within the inclusion membrane network. (B) Same volume as in panel A after rotation. The mitochondrion-associated density has been omitted for better visualization of contacts between the inclusion and surrounding RER elements. (C) Enlargement of the volume corresponding to the inclusion on the left. The membrane-associated density has been omitted for better visualization of contacts (arrows) between filled (black) and empty (white) particles with microtubules (green). (D) Same volume as in panel A after rotation. The volume has been superimposed onto the 2D image (inverted contrast) of one of the sections in the series. Download
ACKNOWLEDGMENTS
We thank members of the Risco and Dermody laboratories for many useful discussions and Jim Chappell and Jennifer Konopka for critical review of the manuscript. Confocal microscopy experiments were conducted in the Vanderbilt Cell Imaging Shared Resource.
This research was supported by an FPI Program fellowship (I.F.C.) and research grants BIO2009-07255 and BIO2012-33314 from the Spanish Ministry of Economy and Competitiveness (CR), Public Health Service awards T32 GM007347 (C.M.L.), F31 NS074596 (C.M.L.), T32 HL007751 (B.A.M.), F32 AI080108 (B.A.M.), and R01 AI032539 (T.S.D.); the Vanderbilt International Scholar Program (P.F.Z.); and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards P30 CA68485 for the Vanderbilt-Ingram Cancer Center and P60 DK20593 for the Vanderbilt Diabetes Research and Training Center.
Footnotes
Citation Fernández de Castro I, Zamora PF, Ooms L, Fernández JJ, Lai CM-H, Mainou BA, Dermody TS, Risco C. 2014. Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes. mBio 5(1):e00931-13. doi:10.1128/mBio.00931-13.
REFERENCES
- 1. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374. 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2:32–48. 10.5501/wjv.v2.i2.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 87:10612–10627. 10.1128/JVI.01370-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147–172. 10.1042/BC20040058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Castro IF, Volonté L, Risco C. 2013. Virus factories: biogenesis and structural design. Cell. Microbiol. 15:24–34. 10.1111/cmi.12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dermody TS, Parker JS, Sherry B. 2013. Orthoreoviruses, p 1304–1346 In Knipe DM, Howley PM. (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7. Sturzenbecker LJ, Nibert M, Furlong D, Fields BN. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baer GS, Ebert DH, Chung CJ, Erickson AH, Dermody TS. 1999. Mutant cells selected during persistent reovirus infection do not express mature cathepsin L and do not support reovirus disassembly. J. Virol. 73:9532–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ebert DH, Deussing J, Peters C, Dermody TS. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609–24617. 10.1074/jbc.M201107200 [DOI] [PubMed] [Google Scholar]
- 10. Borsa J, Morash BD, Sargent MD, Copps TP, Lievaart PA, Szekely JG. 1979. Two modes of entry of reovirus particles into L cells. J. Gen. Virol. 45:161–170. 10.1099/0022-1317-45-1-161 [DOI] [PubMed] [Google Scholar]
- 11. Odegard AL, Chandran K, Zhang X, Parker JS, Baker TS, Nibert ML. 2004. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78:8732–8745. 10.1128/JVI.78.16.8732-8745.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agosto MA, Ivanovic T, Nibert ML. 2006. Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. U. S. A. 103:16496–16501. 10.1073/pnas.0605835103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ivanovic T, Agosto MA, Zhang L, Chandran K, Harrison SC, Nibert ML. 2008. Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. EMBO J. 27:1289–1298. 10.1038/emboj.2008.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watanabe Y, Millward S, Graham AF. 1968. Regulation of transcription of the reovirus genome. J. Mol. Biol. 36:107–123. 10.1016/0022-2836(68)90223-4 [DOI] [PubMed] [Google Scholar]
- 15. Shatkin AJ, LaFiandra AJ. 1972. Transcription by infectious subviral particles of reovirus. J. Virol. 10:698–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Antczak JB, Joklik WK. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760–776. 10.1016/0042-6822(92)90478-8 [DOI] [PubMed] [Google Scholar]
- 17. Skup D, Millward S. 1980. Reovirus-induced modification of cap dependent translation in infected L cells. Proc. Natl. Acad. Sci. U. S. A. 77:152–156. 10.1073/pnas.77.1.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dales S, Omatos PJ. 1965. The uptake and development of reovirus in strain L cells followed with labelled viral ribonucleic acid and ferritin-antibody conjugates. Virology 25:193–211. 10.1016/0042-6822(65)90199-6 [DOI] [PubMed] [Google Scholar]
- 19. Morgan EM, Zweerink HJ. 1974. Reovirus morphogenesis: core-like particles in cells infected at 39 degrees with wild-type reovirus and temperature-sensitive mutants of groups B and G. Virology 59:556–565. 10.1016/0042-6822(74)90465-6 [DOI] [PubMed] [Google Scholar]
- 20. Morgan EM, Zweerink HJ. 1975. Characterization of transcriptase and replicase particles isolated from reovirus infected cells. Virology 68:455–466. 10.1016/0042-6822(75)90286-X [DOI] [PubMed] [Google Scholar]
- 21. Rhim JS, Jordan LE, Mayor HD. 1962. Cytochemical, fluorescent-antibody and electron microscopic studies on the growth of reovirus (echo 10) in tissue culture. Virology 17:342–355. 10.1016/0042-6822(62)90125-3 [DOI] [PubMed] [Google Scholar]
- 22. Zweerink HJ, Morgan EM, Skyler JS. 1976. Reovirus morphogenesis: characterization of subviral particles in infected cells. Virology 73:442–453. 10.1016/0042-6822(76)90405-0 [DOI] [PubMed] [Google Scholar]
- 23. Becker MM, Goral MI, Hazelton PR, Baer GS, Rodgers SE, Brown EG, Coombs KM, Dermody TS. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459–1475. 10.1128/JVI.75.3.1459-1475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Broering TJ, Parker JS, Joyce PL, Kim J, Nibert ML. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285–8297. 10.1128/JVI.76.16.8285-8297.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Becker MM, Peters TR, Dermody TS. 2003. Reovirus ςNS and μNS proteins form cytoplasmic inclusion structures in the absence of viral infection. J. Virol. 77:5948–5963. 10.1128/JVI.77.10.5948-5963.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Broering TJ, Kim J, Miller CL, Piggott CD, Dinoso JB, Nibert ML, Parker JS. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882–1892. 10.1128/JVI.78.4.1882-1892.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dales S. 1963. Association between the spindle apparatus and reovirus. Proc. Natl. Acad. Sci. U. S. A. 50:268–275. 10.1073/pnas.50.2.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodgers SE, Barton ES, Oberhaus SM, Pike B, Gibson CA, Tyler KL, Dermody TS. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 71:2540–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ooms LS, Kobayashi T, Dermody TS, Chappell JD. 2010. A post-entry step in the mammalian orthoreovirus replication cycle is a determinant of cell tropism. J. Biol. Chem. 285:41604–41613. 10.1074/jbc.M110.176255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ooms LS, Jerome WG, Dermody TS, Chappell JD. 2012. Reovirus replication protein M2 influences cell tropism by promoting particle assembly within viral inclusions. J. Virol. 86:10979–10987. 10.1128/JVI.01172-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deng Y, Almsherqi ZA, Ng MM, Kohlwein SD. 2010. Do viruses subvert cholesterol homeostasis to induce host cubic membranes? Trends Cell Biol. 20:371–379. 10.1016/j.tcb.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller CL, Broering TJ, Parker JS, Arnold MM, Nibert ML. 2003. Reovirus sigmaNS protein localizes to inclusions through an association requiring the μNS amino terminus. J. Virol. 77:4566–4576. 10.1128/JVI.77.8.4566-4576.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hauri HP, Kappeler F, Andersson H, Appenzeller C. 2000. ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 113:587–596 [DOI] [PubMed] [Google Scholar]
- 34. Stoltz D, Makkay A. 2000. Co-replication of a reovirus and a polydnavirus in the ichneumonid parasitoid Hyposoter exiguae. Virology 278:266–275. 10.1006/viro.2000.0652 [DOI] [PubMed] [Google Scholar]
- 35. Foster N, Alders M. 1979. Bluetongue virus: a membraned structure, p 48–49 In Proc. Annu. Meet. Microsc. Soc. Am. Microscopy Society of America, Reston, VA [Google Scholar]
- 36. Els HJ, Verwoerd DW. 1969. Morphology of bluetongue virus. Virology 38:213–219. 10.1016/0042-6822(69)90362-6 [DOI] [PubMed] [Google Scholar]
- 37. Hyatt AD, Zhao Y, Roy P. 1993. Release of bluetongue virus-like particles from insect cells is mediated by BTV nonstructural protein NS3/NS3A. Virology 193:592–603. 10.1006/viro.1993.1167 [DOI] [PubMed] [Google Scholar]
- 38. Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401 In Knipe DM, Howley PM. (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 39. Poruchynsky MS, Maass DR, Atkinson PH. 1991. Calcium depletion blocks the maturation of rotavirus by altering the oligomerization of virus-encoded proteins in the ER. J. Cell Biol. 114:651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fontana J, López-Montero N, Elliott RM, Fernández JJ, Risco C. 2008. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell. Microbiol. 10:2012–2028. 10.1111/j.1462-5822.2008.01184.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Risco C, Fernández de Castro I. 2013. Virus morphogenesis in the cell: methods and observations. Subcell. Biochem. 68:417–440. 10.1007/978-94-007-6552-8_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parker JS, Broering TJ, Kim J, Higgins DE, Nibert ML. 2002. Reovirus core protein M2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483–4496. 10.1128/JVI.76.9.4483-4496.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fontana J, López-Iglesias C, Tzeng WP, Frey TK, Fernández JJ, Risco C. 2010. Three-dimensional structure of rubella virus factories. Virology 405:579–591. 10.1016/j.virol.2010.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eichwald C, Arnoldi F, Laimbacher AS, Schraner EM, Fraefel C, Wild P, Burrone OR, Ackermann M. 2012. Rotavirus viroplasm fusion and perinuclear localization are dynamic processes requiring stabilized microtubules. PLoS One 7:e47947. 10.1371/journal.pone.0047947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Howard AR, Moss B. 2012. Formation of orthopoxvirus cytoplasmic A-type inclusion bodies and embedding of virions are dynamic processes requiring microtubules. J. Virol. 86:5905–5914. 10.1128/JVI.06997-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tyler KL, Squier MK, Rodgers SE, Schneider BE, Oberhaus SM, Grdina TA, Cohen JJ, Dermody TS. 1995. Differences in the capacity of reovirus strains to induce apoptosis are determined by the viral attachment protein sigma1. J. Virol. 69:6972–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Connolly JL, Barton ES, Dermody TS. 2001. Reovirus binding to cell surface sialic acid potentiates virus-induced apoptosis. J. Virol. 75:4029–4039. 10.1128/JVI.75.9.4029-4039.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lai CM, Mainou BA, Kim KS, Dermody TS. 2013. Directional release of reovirus from the apical surface of polarized endothelial cells. mBio 4:e00049–e00013. 10.1128/mBio.00049-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Excoffon KJ, Guglielmi KM, Wetzel JD, Gansemer ND, Campbell JA, Dermody TS, Zabner J. 2008. Reovirus preferentially infects the basolateral surface and is released from the apical surface of polarized human respiratory epithelial cells. J. Infect. Dis. 197:1189–1197. 10.1086/529515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jourdan N, Maurice M, DeLautier D, Quero AM, Servin AL, Trugnan G. 1997. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J. Virol. 71:8268–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cuadras MA, Bordier BB, Zambrano JL, Ludert JE, Greenberg HB. 2006. Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway. J. Virol. 80:3935–3946. 10.1128/JVI.80.8.3935-3946.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gardet A, Breton M, Trugnan G, Chwetzoff S. 2007. Role for actin in the polarized release of rotavirus. J. Virol. 81:4892–4894. 10.1128/JVI.02698-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nibert ML, Schiff LA, Fields BN. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boehme KW, Guglielmi KM, Dermody TS. 2009. Reovirus nonstructural protein sigma1s is required for establishment of viremia and systemic dissemination. Proc. Natl. Acad. Sci. U. S. A. 106:19986–19991. 10.1073/pnas.0907412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Furlong DB, Nibert ML, Fields BN. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Virgin HW, IV, Bassel-Duby R, Fields BN, Tyler KL. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 62:4594–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Risco C, Rodríguez JR, López-Iglesias C, Carrascosa JL, Esteban M, Rodríguez D. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839–1855. 10.1128/JVI.76.4.1839-1855.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fontana J, Tzeng WP, Calderita G, Fraile-Ramos A, Frey TK, Risco C. 2007. Novel replication complex architecture in rubella replicon-transfected cells. Cell. Microbiol. 9:875–890. 10.1111/j.1462-5822.2006.00837.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bozzola JJ, Russell LD. 1999. Specimen preparation for transmission electron microscopy, p 21–31 In Bozzola JJ, Russell LD. (ed), Electron microscopy: principles and techniques for biologists, 2nd ed. Jones and Bartlett Publishers, Sudbury, MA [Google Scholar]
- 60. Hayat MA. 2000. Principles and techniques of electron microscopy: biological applications, 4th ed. Cambridge University Press, Cambridge, United Kingdom: [Google Scholar]
- 61. Sanz-Sánchez L, Risco C. 2013. Multilamellar structures and filament bundles are found on the cell surface during bunyavirus egress. PLoS One 8:e65526. 10.1371/journal.pone.0065526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fiala JC. 2005. Reconstruct: a free editor for serial section microscopy. J. Microsc. 218:52–61. 10.1111/j.1365-2818.2005.01466.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie of the reconstruction in Fig. 5. The inclusion membranes (yellow) clearly contact mitochondria (red). Download
Movie of the reconstruction in Fig. S5A. RER cisternae (light brown) surround and contact the inclusion membranes (yellow) and mitochondria (red). Microtubules (green), filled viral particles (black), and empty viral particles (white) are visible within the inclusion membrane network. The nuclear envelope is colored blue. Download
Movie of the reconstruction in Fig. 6A. RER membranes (brown) surround and incorporate into the inclusion membrane network (yellow) that contains numerous mature virions (black), fewer empty particles (white), and microtubules (green). Download
Movie of the reconstruction in Fig. 6B. Mitochondria (red), RER (brown), and lipid droplets (large white structures) surround and contact the inclusion membranes (yellow), which are attached to the nuclear envelope (blue). The inclusion membrane network contains many empty particles (white) and fewer mature virions (black). Download
Movie of the reconstruction in Fig. 6C. The inclusion contains membranes (yellow) and filled (black) and empty (white) viral particles. Peripheral RER elements (light brown), microtubules (green), and viral particles (black) are in contact with the cytosolic face of the plasma membrane (dark brown). The plasma membrane from another cell is colored blue. Download
Ultrastructure of reovirus inclusions in MDCK cells. MDCK cells were infected with T3-T1M1 (A to C) or T3 (D to F) and fixed at 24 hpi. Ultrathin (~60- to 70-nm) sections were imaged by TEM. (A) T3-T1M1 inclusion surrounded by RER cisternae (black arrows). (B) Enlargement of the highlighted region in panel A showing coated microtubules (white arrows). (C) Enlargement of the central region in panel B. Filled (black arrowhead) and empty (white arrowhead) viral particles are apparent. (D) Large inclusion near the nucleus of a cell infected with T3. ER membranes (black arrows) are shown. (E) Highlighted region in panel D showing membranes (black arrows) inside the inclusion. A vacuole (V), which contains fibers and a few viral particles, appears attached to the inclusion periphery. (F) Enlargement of the central area in panel E. The inclusion contains a few filled particles (black arrowhead), numerous empty particles (white arrowhead), and many smaller particles (yellow arrowheads). LD, lipid droplet; mi, mitochondria; N, nucleus. Scale bars: 1.5 µm in panels A and D; 0.25 µm in panels B, C, E, and F. Download
Reovirus inclusions codistribute with the ER and ERGIC and do not associate with the Golgi compartment. HeLa cells were infected with T3-T1M1 for 24 h. Cells were fixed, permeabilized, stained, and visualized by confocal microscopy. (A to C) Cells were stained for σNS (green), PDI (red), or nuclei (blue). (D to F) Cells were stained for σNS (green), giantin (red), or nuclei (blue). (G to I) Cells were stained for σNS (green), the Golgi compartment with WGA (red), or nuclei (blue). (J to L) Cells were stained for σNS (green), KDEL receptor (red), or nuclei (blue). Asterisks mark noninfected cells. Scale bars: 10 µm. Download
Organelles and membranes in reovirus inclusions. (A) Ultrathin sections of mock-infected HeLa cells display a nonuniform distribution of organelles (left). Ultrathin sections of HeLa cells infected with T3 for 24 h show viral inclusions associated with mitochondria and RER (right). (B) Quantification of mitochondria associated with 53 randomly selected inclusions. (C) TEM images of 9 of the 15 ultrathin sections from the series used to generate the 3D reconstruction shown in Fig. 5 (raw data). Only the central area within the inclusion is shown. Smooth membranes inside the inclusion are indicated by yellow arrows. G, Golgi compartment; mi, mitochondria; N, nucleus. Scale bars: 0.5 µm in panel A; 0.25 µm in panel B. Download
Schematic of the generation of 3D reconstructions from serial sections. After the collection of serial sections (ultramicrotomy) and imaging by TEM, several computational steps were used to render the final model. The 3D reconstruction at the bottom is a different view of the inclusion shown in Fig. 6B. Download
3D model of reovirus inclusions in HeLa cells. HeLa cells were infected with T3-T1M1 and fixed at 12 hpi. The inclusions were visualized by TEM of serial sections, 3D reconstruction, and image processing. (A) The volume shows two inclusions (yellow) surrounded by mitochondria (red) and RER (light brown) near the nuclear envelope (blue). Filled particles (black), empty particles (white), and microtubules (green) are apparent within the inclusion membrane network. (B) Same volume as in panel A after rotation. The mitochondrion-associated density has been omitted for better visualization of contacts between the inclusion and surrounding RER elements. (C) Enlargement of the volume corresponding to the inclusion on the left. The membrane-associated density has been omitted for better visualization of contacts (arrows) between filled (black) and empty (white) particles with microtubules (green). (D) Same volume as in panel A after rotation. The volume has been superimposed onto the 2D image (inverted contrast) of one of the sections in the series. Download