Abstract
Variants in transmembrane protein 106 B (TMEM106B) modify the disease penetrance of frontotemporal dementia (FTD) in carriers of progranulin (GRN) mutations. We investigated whether TMEM106B is also a genetic modifier of disease in carriers of chromosome 9 open reading frame 72 (C9ORF72) expansions. We assessed the genotype of 325 C9ORF72 expansion carriers (cohort 1), 586 FTD patients lacking C9ORF72 expansions (with or without motor neuron disease [MND]; cohort 2), and a total of 1,302 controls for TMEM106B variants (rs3173615 and rs1990622) using MassArray iPLEX and Taqman genotyping assays. For our primary analysis, we focused on functional variant rs3173615, and employed a recessive genotypic model. In cohort 1, patients with C9ORF72 expansions showed a significantly reduced frequency of carriers homozygous for the minor allele as compared to controls (11.9% versus 19.1%, odds ratio (OR): 0.57, p=0.014; same direction as carriers of GRN mutations). The strongest evidence was provided by FTD patients (OR: 0.33, p=0.009) followed by FTD/MND patients (OR: 0.38, p=0.017), whereas no significant difference was observed in MND patients (OR: 0.85, p=0.55). In cohort 2, the frequency of carriers homozygous for the minor allele was not significantly reduced in patients as compared to controls (OR: 0.77, p=0.079); however, a significant reduction was observed when focusing on those patients with frontotemporal lobar degeneration and TAR DNA-binding protein 43 inclusions (FTLD-TDP; OR: 0.26, p<0.001).
Our study identifies TMEM106B as the first genetic factor modifying disease presentation in C9ORF72 expansion carriers. Homozygosity for the minor allele protects carriers from developing FTD, but not from developing MND; similar effects are seen in FTLD-TDP patients with yet unknown genetic causes. These new findings show that the protective effects of TMEM106B are not confined to carriers of GRN mutations, and might be relevant for prognostic testing, and as a promising therapeutic target for the entire spectrum of FTLD-TDP.
Keywords: C9ORF72, TMEM106B, frontotemporal dementia, motor neuron disease, amyotrophic lateral sclerosis, disease modifier
Introduction
Repeat expansions in chromosome 9 open reading frame 72 (C9ORF72) are the most common known genetic cause of frontotemporal dementia (FTD), amyotrophic lateral sclerosis (ALS), and a combination of both diseases (FTD with motor neuron disease [FTD/MND]) [7,21]. Previously, we showed that the repeat length of these expansions varies between tissues, and that repeat lengths in the cerebellum are associated with survival after disease onset [26]. Moreover, we demonstrated that additional mutations in FTD-associated genes (progranulin [GRN] and microtubule-associated protein tau [MAPT]) might also contribute to the phenotypic variability that is detected in patients with C9ORF72 repeat expansions [25].
To date, other genetic factors that could act as disease modifiers have not been reported in C9ORF72 expansion carriers. For this reason, we investigated a promising candidate: transmembrane protein 106 B (TMEM106B). Single nucleotide polymorphisms (SNPs) in TMEM106B are associated with the risk of frontotemporal lobar degeneration with TAR DNA-binding protein 43 inclusions (FTLD-TDP) [28], which is the predominant pathology in many FTD patients [18,15]. Interestingly, this association is most prominent in patients with GRN mutations; in these patients, two copies of the minor allele of TMEM106B SNPs (top SNP rs1990622) appear to reduce disease penetrance, protecting individuals from developing FTD [28,8]. Variant rs3173615, which is in linkage disequilibrium (LD) with rs1990622, dictates a change from threonine to serine at position 185 (p.T185S) [19,5,29]. This variant has been shown to regulate TMEM106B protein levels, and was proposed as the functional variant underlying the genetic association with FTLD-TDP [19].
In our present study, we assessed the frequency of TMEM106B SNPs rs3173615 and rs1990622 in a large cohort of C9ORF72 expansion carriers (n=325), in an additional cohort of patients with FTD or FTD/MND without C9ORF72 expansions (n=586), and in control subjects (n=1,302). Our results reveal that variants in TMEM106B protect against developing FTD in carriers of C9ORF72 repeat expansions, and also in patients with currently unknown causes of FTLD-TDP, emphasizing crucial similarities between C9ORF72-related FTLD-TDP, GRN-related FTLD-TDP, and other causes of FTLD-TDP.
Materials and Methods
Subjects
For cohort 1 we included all 325 carriers of C9ORF72 repeat expansions available to us. These subjects were obtained through North American/Canadian institutions: the Mayo Clinic (n=121), Coriell Research Institute (n=71), University of British Columbia, Canada (n=58), University of California, San Francisco (n=38), Robarts Research Institute (n=11), Northwestern University Feinberg School of Medicine (n=9), Drexel University College of Medicine (n=7), University of Western Ontario, Canada (n=7), and Banner Sun Health Research Institute (n=3). Subjects were clinicopathologically diagnosed with FTD (n=86), FTD/MND (n=78) or MND (n=127), with another diagnosis (n=7; e.g. Alzheimer’s disease, alcohol abuse or behavioral impairment), or they were asymptomatic at time of last evaluation (n=27; age at evaluation: 43.6±12.7). Our primary analysis focused on the 260 unrelated probands with FTD (n=69), FTD/MND (n=71) or MND (n=120), and a group of neurologically normal controls of similar age and gender provided by the Mayo Clinic (n=376). In secondary analyses, we also examined the sensitivity of our results by including the 65 remaining expansion carriers who were family members or had received another diagnosis, and by including additional controls from our major sites (n=410; Coriell Research Institute [n=264], University of British Columbia, Canada [n=17], University of California, San Francisco [n=116], and Robarts Research Institute [n=13]). All C9ORF72 expansion carriers and control subjects were screened for TMEM106B SNPs rs3173615 and rs1990622.
Cohort 2 was previously screened for TMEM106B SNP rs1990622 [8,28]; however, these subjects were investigated before the identification of repeat expansions in C9ORF72. Patients in this cohort were selected according to the following criteria: a clinical diagnosis of behavioral variant FTD, semantic dementia or progressive nonfluent aphasia, or a pathologic diagnosis of FTLD-TDP. Based on our current findings, we reanalyzed our genotype data in this cohort after exclusion of patients with C9ORF72 repeat expansions and GRN mutations, and we also genotyped all patients and controls for rs3173615. Furthermore, we extended this cohort to include all additional pathologically confirmed FTLD-TDP and FTLD/MND patients without C9ORF72 repeat expansions or GRN mutations (n=31), who were recruited at our Mayo Clinic Brain Bank since our original study in 2010. In total, cohort 2 comprised 586 patients who were ascertained at the Mayo Clinic (n=380), University of California, San Francisco (n=117), University of Western Ontario, Canada (n=31), Drexel University College of Medicine (n=21), Northwestern University Feinberg School of Medicine (n=21), University of British Columbia, Canada (n=10), and University of Texas Southwestern Medical Center (n=6). All control subjects in cohort 2 (n=765) were obtained from the Mayo Clinic (not included in cohort 1: n=516, included in cohort 1: n=133) and University of California, San Francisco (n=116) [8]. In addition, we performed a joint analysis including the controls from cohort 1 (total number of controls: n=1,302).
Characteristics of cases and controls included in both cohorts are shown in Table 1. All subjects agreed to be in the study, and biological samples were obtained after informed consent with ethical committee approval from the respective institutions.
Table 1.
Characteristics of study cohorts
| Group | N | Female gender | Age | Age at onset | FTD only | Pathological diagnosis |
|---|---|---|---|---|---|---|
| Cohort 1 – controls and C9ORF72 repeat expansion carriers (FTD, FTD/MND, and MND, or other diagnosis) | ||||||
| Controls | 376 | 173 (46.0%) | 61.2 ± 10.2 (35–90) | N/A | N/A | N/A |
| All repeat expansion carriers | 325 | 148 (45.5%) | 59.2 ± 9.9 (35–90) | 56.8 ± 9.2 (34–83) | 86 (26.5%) | 118 (36.3%) |
| FTD, FTD/MND, and MND probands | 260 | 114 (43.8%) | 59.5 ± 10.0 (35–90) | 56.9 ± 9.0 (34–83) | 69 (26.5%) | 107 (41.2%) |
| FTD probands | 69 | 28 (40.6%) | 63.1 ± 12.3 (35–90) | 58.1 ± 9.6 (34–79) | 69 (100.0%) | 40 (58.0%) |
| FTD/MND probands | 71 | 25 (35.2%) | 60.6 ± 8.5 (39–80) | 56.2 ± 9.0 (34–74) | 0 (0.0%) | 51 (71.8%) |
| MND probands | 120 | 61 (50.8%) | 56.9 ± 8.6 (37–83) | 56.5 ± 8.7 (37–83) | 0 (0.0%) | 16 (13.3%) |
| Cohort 2 – controls and FTD or FTD/MND patients without C9ORF72 repeat expansions or GRN mutations | ||||||
| Controls | 765 | 363 (47.4%) | 67.1 ± 10.0 (20–95) | N/A | N/A | N/A |
| All FTD or FTD/MND patients | 586 | 273 (46.6%) | 66.8 ± 10.4 (26–99) | 62.4 ± 9.8 (26–95) | 531 (90.6%) | 141 (24.1%) |
| All FTD patients | 531 | 251 (47.3%) | 67.1 ± 10.1 (26–99) | 62.7 ± 9.5 (26–86) | 531 (100.0%) | 101 (19.0%) |
| Pathologically diagnosed | 101 | 48 (52.5%) | 74.0 ± 11.2 (49–99) | 64.5 ± 11.1 (39–86) | 101 (100.0%) | 101 (100.0%) |
| Clinically diagnosed | 430 | 203 (47.2%) | 65.5 ± 9.1 (26–90) | 62.4 ± 9.1 (26–85) | 430 (100.0%) | 0 (0.0%) |
Continuous variables are summarized with the sample mean ± standard deviation (range). For cohort 1, the age provided is age at blood draw in controls, age at onset in clinically diagnosed patients, and age at death in pathologically diagnosed patients. For cohort 2, the age provided is age at blood draw in controls, age at diagnosis in clinically diagnosed patients, and age at death in pathologically diagnosed patients. In cohort 1, information was unavailable regarding age (n=42) and age at onset (n=57). In cohort 2, information was unavailable regarding age at onset (n=95).
Genotyping
TMEM106B SNPs rs3173615 and rs1990622 were genotyped in cohort 1 on a Sequenom MassArray iPLEX platform (San Diego, CA) and analyzed with Typer 4.0 software. In cohort 2, rs3173615 and rs1990622 were genotyped using Taqman SNP genotyping assays (C_27465458_10, C__11171598_10; Invitrogen, Carlsbad, CA) on the 7900HT Fast Real Time PCR system. Genotype calls were made using SDS v2.2 software (Applied Biosystems, Foster City, CA). In control subjects, there was no evidence of a departure from Hardy-Weinberg equilibrium for rs3173615 or rs1990622 in both cohorts (all p>0.010). The two SNPs were in almost complete LD (r2≥0.97), and given this strong LD, for simplicity we focused our analysis on functional variant rs3173615.
Immunohistochemistry
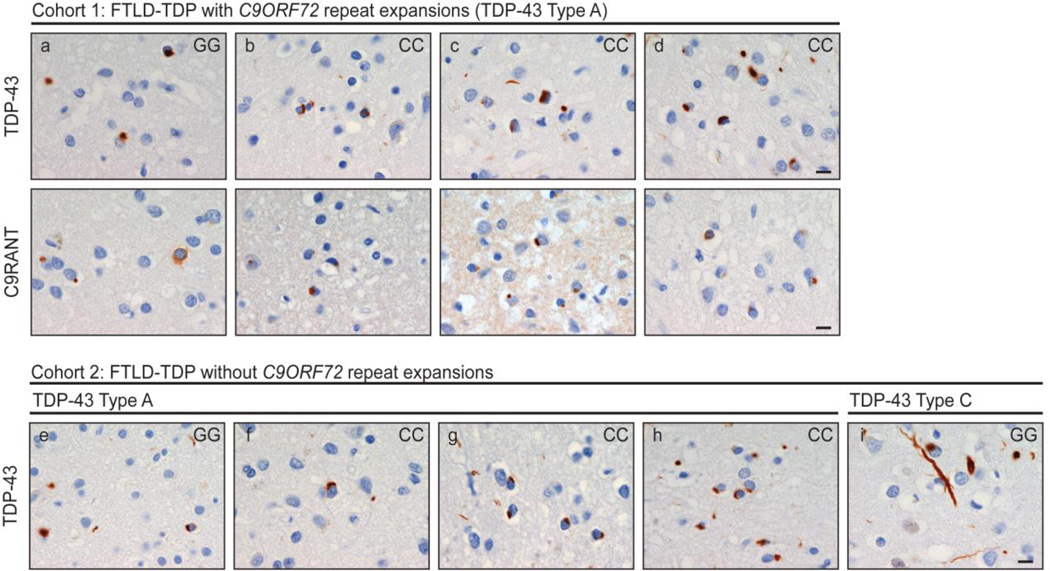
Immunohistochemistry was performed in a blinded fashion on a preliminary group of nine patients who were diagnosed with FTLD-TDP and did not show signs of MND. We included all three patients from the Mayo Clinic Brain Bank for whom fixed tissue was available and who were homozygous for the minor protective TMEM106B allele (GG genotype for rs3173615; C9ORF72 repeat expansion: n=1, without C9ORF72 repeat expansion: n=2). In addition, we also randomly selected six FTLD-TDP patients without MND who were homozygous for the major risk TMEM106B allele (CC genotype for rs3173615; C9ORF72 repeat expansion: n=3, without C9ORF72 repeat expansion: n=3), to allow comparisons of TDP-43 burden between patients homozygous for the minor or major allele.
We stained 5-µm-thick sections from the frontal cortex for TDP-43 (pS409/410, 1:5,000, mouse monoclonal, Cosmobio Co., Tokyo, Japan), and repeat-associated non-ATG translation peptides (C9RANT, Rb5823, 1:5,000, Mayo Clinic) [1], the latter has recently been shown to be distinctive for C9ORF72 expansions [1,17]. Sections were cut from formalin-fixed paraffin-embedded blocks, deparaffinized in xylene, rehydrated in a graded series of ethanol, and washed in distilled water. To process stains, we used DAKO Autostainer Plus (DAKO, Carpinteria, CA) and DAKO EnVision+ System–horseradish peroxidase (diaminobenzidine). Immunostains were subsequently counterstained with Lerner’s hematoxylin, dehydrated, and coverslipped. Stained slides were viewed using an Olympus BX40 microscope (Olympus Corporation of the Americas, Center Valley, PA). TDP-43 pathologic subtype was assigned in accordance with the harmonized criteria (Type A-C) [14]. Neuronal lesion counts were performed in a blinded fashion on layer 2 of the midfrontal gyrus. Counts were performed in six 10x microscopic fields randomly selected over the gyrus, and averaged for a composite pathologic burden score.
Statistical Analysis
Separately for both cohorts, comparisons of TMEM106B SNPs with controls were made using logistic regression models; odds ratios (ORs) and 95% confidence intervals (CIs) were estimated. Given our previous studies related to TMEM106B in carriers of GRN mutations [19,8], we hypothesized that TMEM106B SNPs would affect disease penetrance in C9ORF72 expansion carriers under a recessive genotypic model (presence versus absence of two copies of the minor allele). For this reason, we utilized this model in our primary analysis, though in secondary analyses we also examined other genotypic models (additive, dominant, and co-dominant).
In cohort 1, models were adjusted for gender, and the primary analysis involved unrelated probands with FTD, FTD/MND, MND, and controls. Comparisons with controls were made for all patients, and also separately for FTD, FTD/MND, and MND subgroups. Sensitivity of results to the inclusion of related family members and individuals with other diagnoses, and to the inclusion of additional controls were considered in secondary analyses. For cohort 2, models were adjusted for gender and age (age at blood draw in controls, age at diagnosis in clinically diagnosed patients, age at death in pathologically diagnosed patients). We compared controls to all patients, and additionally, we made comparisons separately for the FTD subgroup and according to the method of diagnosis (clinical or pathological).
To evaluate associations of TMEM106B SNPs with age at onset, we also used linear regression models adjusted for gender and disease subgroup for cohort 1 (using only FTD, FTD/MND, and MND probands); our primary analysis involved a recessive genotypic model, while other genotypic models (additive, dominant, and co-dominant) were examined in secondary analyses.
In order to adjust for multiple testing in our main disease-association analysis using recessive genotypic models, we utilized a Holm step-down adjustment [11] separately for both cohorts. After this adjustment, p≤0.025 (cohort 1) and p≤0.0125 (cohort 2) were considered statistically significant. All statistical analyses were performed with R Statistical Software (version 2.14.0; R Foundation for Statistical Computing, Vienna, Austria).
Results
Cohort 1
We first compared the frequency of carriers homozygous for the minor rs3173615 TMEM106B allele (corresponding to homozygous p.185S carriers [GG genotype]) between our FTD, FTD/MND or MND probands with C9ORF72 expansions and our control subjects (Table 1). Homozygosity for the minor allele was detected in 11.9% of the 260 cases as compared to 19.1% of the 376 controls (OR: 0.57, p=0.014; Table 2). When including all 65 family members and individuals with other diagnoses who harbored C9ORF72 expansions, the frequency of carriers homozygous for the minor allele remained lower in individuals with C9ORF72 expansions than in controls (11.4% versus 19.1%).
Table 2.
Associations of TMEM106B rs3173615 with disease
| TMEM106B rs3173615 genotype information | Comparison with controls under a recessive model |
||||||
|---|---|---|---|---|---|---|---|
| Group | N | MAF | CC | CG | GG | OR (95% CI) | P-value |
| Cohort 1 – controls and C9ORF72 repeat expansion carrier probands (FTD, FTD/MND, MND) | |||||||
| Controls | 376 | 43.2% | 123 (32.7%) | 181 (48.1%) | 72 (19.1%) | 1.00 (reference) | N/A |
| FTD, FTD/MND, and MND patients | 260 | 38.8% | 89 (34.2%) | 140 (53.8%) | 31 (11.9%) | 0.57 (0.36 – 0.90) | 0.014 |
| FTD patients | 69 | 35.5% | 25 (36.2%) | 39 (56.5%) | 5 (7.2%) | 0.33 (0.13 – 0.85) | 0.009 |
| FTD/MND patients | 71 | 42.3% | 17 (23.9%) | 48 (67.6%) | 6 (8.5%) | 0.38 (0.16 – 0.92) | 0.017 |
| MND patients | 120 | 38.8% | 47 (39.2%) | 53 (44.2%) | 20 (16.7%) | 0.85 (0.49 – 1.46) | 0.55 |
| Cohort 2 – controls and FTD or FTD/MND patients without C9ORF72 repeat expansions or GRN mutations | |||||||
| Controls | 765 | 41.0% | 280 (36.6%) | 342 (44.7%) | 143 (18.7%) | 1.00 (reference) | N/A |
| FTD and FTD/MND patients | 586 | 39.3% | 213 (36.3%) | 285 (48.6%) | 88 (15.0%) | 0.77 (0.58 – 1.03) | 0.079 |
| FTD patients | 531 | 39.8% | 187 (35.2%) | 265 (49.9%) | 79 (14.9%) | 0.76 (0.56 – 1.03) | 0.071 |
| Pathologically diagnosed | 101 | 30.6% | 45 (44.6%) | 50 (49.5%) | 6 (5.9%) | 0.26 (0.11 – 0.61) | <0.001 |
| Clinically diagnosed | 430 | 42.0% | 142 (33.0%) | 215 (50.0%) | 73 (17.0%) | 0.91 (0.67 – 1.25) | 0.56 |
FTD=frontotemporal dementia; MND=motor neuron disease; OR=odds ratio; CI=confidence interval; MAF=minor allele frequency. ORs, 95% CIs, and p-values result from logistic regression models where rs3173615 was considered under a recessive model. For cohort 1, models were adjusted for gender. For cohort 2, models were adjusted for age (age at blood draw in controls, age at diagnosis in clinically diagnosed patients, and age at death in pathologically diagnosed patients) and gender. After applying a Holm step-down adjustment for multiple testing, p≤ 0.025 are considered as statistically significant in cohort 1, and p≤ 0.0125 are considered as statistically significant in cohort 2.
Subsequently, we determined whether the lower frequency of homozygous carriers was present in all disease subgroups (Table 2). Interestingly, the lowest frequency was encountered in FTD patients (7.2%, OR: 0.33, p=0.009) followed by FTD/MND patients (8.5%, OR: 0.38, p=0.017), whereas no significant difference was detected in MND patients (16.7%, OR: 0.85, p=0.55). These findings were consistent when including family members (FTD: 7.0%, FTD/MND: 9.0%, and MND: 16.7%).
A similar trend was seen for all aforementioned differences with controls, when including 410 extra controls from other sites to form a combined control group of 786 individuals (Online Resource Table 1), and when additionally adjusting the logistic regression models for age in the subset of individuals with that information available (data not shown). We also considered other genotypic models (additive, dominant, and co-dominant), but the differences stayed most significant under the recessive genotypic model (Online Resource Table 2).
Furthermore, we investigated whether homozygosity for the minor allele might influence age at onset. In the 240 FTD, FTD/MND or MND probands with age at onset information available, the mean age at onset was 1.26 years lower in patients homozygous for the minor allele in comparison to other patients, when utilizing a recessive genotypic model (95% CI: 4.78 years lower to 2.25 years higher; Table 3); it is important to realize, however, that this did not approach significance (p=0.48). The lack of association between rs3173615 and age at onset was also apparent when examining other genotypic models (additive, dominant, and co-dominant; Table 3). All association analyses regarding disease and age at onset were comparable for rs1990622 (data not shown).
Table 3.
Associations of TMEM106B rs3173615 with age at onset for probands in cohort 1
| Association between rs3173615 and age at onset | ||
|---|---|---|
| Model | Regression coefficient (95% CI) | P-value |
| Recessive (GG vs. CC or CG) | −1.26 (−4.78, 2.25) | 0.48 |
| Additive (effect of each additional G allele) | −0.24 (−2.04, 1.55) | 0.79 |
| Dominant (CG or GG vs. CC) | 0.16 (−2.30, 2.63) | 0.90 |
| Co-dominant | ||
| CG vs. CC | 0.45 (−2.12, 3.02) | 0.73 |
| GG vs. CC | −0.99 (−4.84, 2.85) | 0.61 |
Regression coefficients, 95% CIs, and p-values result from linear regression models adjusted for age and disease group. Regression coefficients are interpreted as the change in mean age at onset corresponding to presence of the GG genotype for rs3173615 (recessive model), to each additional G allele for rs3173615 (additive model), and to presence of the CG or GG genotype for rs3173615 (dominant model). For the co-dominant model, regression coefficients are interpreted as the change in mean age at onset in comparison to the CC genotype for rs3173615.
Cohort 2
Before the identification of repeat expansions in C9ORF72, we assessed a large cohort of FTD patients, FTD/MND patients, and control subjects for TMEM106B SNPs [28,8]. For our present study, we excluded all carriers of C9ORF72 expansions and GRN mutations from this cohort, and reanalyzed our data. In addition, we extended our cohort with 31 newly ascertained FTLD-TDP patients. In this cohort of 586 patients and 765 controls, we showed that the frequency of carriers homozygous for the minor rs3173615 TMEM106B allele (GG genotype) did not differ significantly between groups, when using a recessive genotypic model (15.0% versus 18.7%, OR: 0.77, p=0.079; Table 2). Because our findings in cohort 1 were most pronounced in FTD patients, we subsequently focused our analysis on patients with FTD (without signs of MND); however, in this group we also did not detect a significant difference between patients and controls (14.9% versus 18.7%, OR: 0.76, p=0.071). Whilst this group included both clinically diagnosed patients (n=430) and pathologically diagnosed patients (n=101), we next investigated these subgroups separately. Although no significant differences were observed when comparing clinically diagnosed patients and controls (17.0% versus 18.7%, OR: 0.91, p=0.56; reported diagnosis of behavioral variant FTD: 14.3%; remaining FTD patients: 18.1%), the frequency of carriers homozygous for the minor allele was significantly lower in patients who received a diagnosis of FTLD-TDP than in controls (5.9% versus 18.7%, OR: 0.26, p<0.001), even though patients with C9ORF72 expansions and GRN mutations had already been excluded.
Comparable results were obtained after inclusion of all additional controls from cohort 1 to form a combined control group of 1,302 subjects (Online Resource Table 1). Differences between patients and controls were also examined under other genotypic models (additive, dominant, and co-dominant); however, differences were not as strong as those observed under a recessive genotypic model (Online Resource Table 2). All results were similar when evaluating associations with TMEM106B rs1990622 (data not shown).
Immunohistochemistry
All nine patients for whom immunohistochemistry was performed displayed TDP-43 pathology (Table 4, Fig. 1). Only one patient had pathologic lesions consistent with TDP-43 Type C (predominant long dystrophic neuritis; patient i), and therefore, this patient was excluded from further analyses. The remaining eight patients showed pathologic lesions consistent with TDP-43 Type A (neuronal cytoplasmic and intranuclear inclusions and small dystrophic neuritis; patients a–h). When performing a very preliminary comparison in TDP-43 Type A burden within each of the cohorts, the lowest pathologic burden was consistently present in homozygous carriers of the minor, protective, TMEM106B allele. Of note, all patients with C9ORF72 repeat expansions demonstrated C9RANT pathology, characterized by neuronal cytoplasmic inclusions, as opposed to patients without these expansions.
Table 4.
TDP-43 burden of patients with TDP-43 Type A pathology
| Group | C9ORF72 expansion | Genotype | Age at Death | Gender | TDP-43 Count | |
|---|---|---|---|---|---|---|
| Cohort 1 | a | Yes | GG | 90 | F | 7.7 |
| b | Yes | CC | 83 | F | 10.5 | |
| c | Yes | CC | 74 | M | 11.0 | |
| d | Yes | CC | 71 | M | 20.5 | |
| Cohort 2 | e | No | GG | 90 | F | 4.5 |
| f | No | CC | 84 | M | 6.0 | |
| g | No | CC | 86 | M | 16.7 | |
| h | No | CC | 77 | M | 24.2 |
Subjects are homozygous carriers of the minor TMEM106B allele (GG), or of the major TMEM106B allele (CC) in rs3173615. TDP-43 counts were performed in six 10× microscopic fields randomly selected over the gyrus, and averaged for a composite pathologic burden score. Identifiers correspond to Fig. 1.
Fig. 1. Immunohistochemistry of patients homozygous for minor or major TMEM106B allele.
TAR DNA-binding protein 43 (TDP-43; cohort 1 and cohort 2) and dipeptide-repeat protein (C9RANT; cohort 1) pathology in the midfrontal gyrus (n=9; 3 homozygous carriers of the minor allele [GG], and 6 homozygous carriers of the major allele [CC] in rs3173615). The case identifier (Table 4) is shown in the upper left corner, and the genotype is shown in the upper right corner. [measure bar = 10 µm]
Discussion
Our results identify rs3173615 (corresponding to TMEM106B p.T185S) as an important genetic modifying factor in C9ORF72 expansion carriers. We focused our analysis on a recessive genotypic model, and this model revealed that individuals who carry a C9ORF72 repeat expansion and harbor two copies of the minor TMEM106B allele (GG genotype) are less likely to develop disease symptoms (OR: 0.57), in particular FTD (OR: 0.33). Although these findings suggest that the protective effects of TMEM106B variants are not limited to previously reported individuals with GRN mutations, these protective effects seem more pronounced in carriers of GRN mutations, who demonstrated a frequency of homozygous carriers as low as 2% [19,30,8], corresponding to an OR of approximately 0.12 [8].
Repeat expansions in C9ORF72 and mutations in GRN are common causes of FTD: they account for up to 50% of familial FTD patients [27,16,2,6,9]. Importantly, in our second patient cohort in which all carriers of C9ORF72 expansions and GRN mutations were excluded, the frequency of two copies of the minor TMEM106B allele in FTLD-TDP patients remained significantly lower as compared to the more heterogeneous group of clinically diagnosed FTD patients or controls. These findings suggest that the protective effects of TMEM106B variants are not confined to C9ORF72-related FTLD-TDP or GRN-related FTLD-TDP, but that they can also be encountered in the remaining FTLD-TDP patients with a presently unknown cause.
Interestingly, other well-known types of FTD, such as FTLD-tau caused by MAPT mutations, do not appear to show a low frequency of carriers homozygous for minor TMEM106B alleles (18%) [8]. Moreover, we found no significant differences in TMEM106B variants between MND patients with C9ORF72 repeat expansions and controls, which is in line with a report that assessed these variants in a cohort of ALS patients and controls [30]. Importantly, while our analyses focused on the protective effect of the minor allele of TMEM106B SNPs, our findings are compatible with previous studies which identified the major allele of TMEM106B SNPs as potential risk-alleles for the development of FTD [28,29,8,23,30,5]. In fact, given our new findings, it is also important to emphasize that differences in the composition of cohorts (e.g. percentage of patients with C9ORF72 repeat expansions, GRN mutations, other causes of FTLD-TDP, or the presence of MND), may have greatly impacted the results of these previously published TMEM106B association studies, and likely explain the reported conflicting results related to TMEM106B risk in FTD [28,29,8,23,30,5].
TMEM106B is a 274 amino acid glycosylated type-II transmembrane protein that localizes in late endosomes and lysosomes, and co-localizes with the progranulin protein (PGRN) [3,4,19,12]. Elevated levels of TMEM106B have been shown to result in abnormal lysosomal morphology, and are found to delay the degradation of endocytic cargoes [3]. Although the effects of TMEM106B overexpression on PGRN levels seem small, PGRN levels in plasma appear to correlate with TMEM106B variants [3,19,4,8,5]. In vitro experiments that assessed the risk isoform (T185) and protective isoform (S185) of TMEM106B, showed that the protective isoform is consistently expressed at lower levels (as low as half) [19]. It has also been revealed that this probably results from a more rapid protein degradation (nearly 4-fold), potentially caused by an abnormal glycosylation of S185 [19]. Taken together, these findings suggest that lysosomal pathways may play a critical role in the pathogenesis of FTD; for instance, they may affect the degradation and subsequent aggregation of TDP-43, and thereby, they could influence the likelihood of developing FTD.
Because we did detect homozygous carriers of the minor TMEM106B allele in a small percentage of FTLD-TDP patients, we postulate that another as yet unknown factor (or combination of factors) has such a strong effect in these patients that the protective effects of the minor allele are lessened. Importantly, our highly preliminary immunohistochemical findings may support the hypothesis that TMEM106B variants affect TDP-43: patients with the protective allele who did develop FTLD-TDP seemed to show less TDP-43 burden than patients with the risk allele. Since homozygous carriers of protective alleles are rare in FTD patients, however, we were only able to investigate a very limited number of samples, and consequently, additional cases should be assessed to confirm these suggestive findings.
Based on our present results, and the aforementioned studies, we speculate that a cascade of lysosomal dysfunction might contribute to C9ORF72-related FTD, and possibly, to other types of FTD. In fact, it has already been suggested that FTD-associated genes valosin containing protein (VCP) and charged multivesicular body protein 2b (CHMP2B), which account for a small subgroup of FTD patients, are involved in accumulation of autophagosomes, affect endolysosomal sorting, regulate the maturation and size of early endosomes, impair endosome-lysosome fusion, and delay the degradation process of cargo proteins [20,24,10,13,22], and thus, lysosomal dysfunction could represent a shared mechanism that underlies many types of FTD.
Our study has several limitations. Despite the relatively large sample size, for example, the possibility of a type II error (i.e. false-negative association) should be considered. Moreover, due to the low frequency of two copies of the minor TMEM106B allele, especially in FTD patients, detailed analysis of clinical and pathological characteristics was obviously hampered. This could have also affected differences in frequencies detected when utilizing a recessive genotypic model, and may have contributed to the lack of association with age at onset. Furthermore, although we excluded individuals with known familial relationships from our primary analysis, and C9ORF72 and TMEM106B are located on different chromosomes, we cannot exclude the possibility that some subjects may be distantly related and share loci. Nevertheless, our study was able to thoroughly assess TMEM106B variants in a unique cohort of both clinically and pathologically diagnosed C9ORF72 expansion carriers, in an additional large cohort of FTD patients and controls, and in a disease spectrum ranging from FTD to MND.
To summarize, our findings reveal a new link between C9ORF72-related FTLD-TDP, GRN-related FTLD-TDP, and other causes of FTLD-TDP: a decreased frequency of carriers homozygous for the minor G-allele of TMEM106B rs3173615, suggesting a protective effect of this particular genotype. In C9ORF72 mutation carriers, TMEM106B protects specifically against the development of FTD, but not MND; providing the first genetic modifier factor involved in disease presentation in this important genetic subgroup. We hypothesize that TMEM106B variants might protect C9ORF72 expansion carriers from developing FTD, potentially by affecting lysosomal pathways. These findings provide novel opportunities for the development of prognostic tests, and for new treatment strategies aiming at C9ORF72 expansion carriers, and the larger subgroup of patients within the disease spectrum of FTD with TDP-43 pathology.
Supplementary Material
Acknowledgements
This project was supported by NIH grants R01 NS080882, R01 NS065782, R01 AG026251, P01 AG017586, P50 NS072187, P50 AG016574, P30 AG013854, P30 AG012300, P30 AG019610, U01 AG006786, the ALS Therapy Alliance, and the Consortium for Frontotemporal Dementia Research. Data collection at University of British Columbia is supported by CIHR grant #179009. Dr. Van Blitterswijk is supported by the Milton Safenowitz Post-Doctoral Fellowship for ALS research from the ALS Association.
Footnotes
Conflicts of Interest
Mrs. DeJesus-Hernandez and Dr. Rademakers hold a patent on methods to screen for the hexanucleotide repeat expansion in the C9ORF72 gene; the other authors declare that they have no conflict of interest.
References
- 1.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 3.Brady OA, Zheng Y, Murphy K, Huang M, Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet. 2013;22(4):685–695. doi: 10.1093/hmg/dds475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, Busch JI, Akle S, Grossman M, Van Deerlin V, Trojanowski JQ, Lee VM. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32(33):11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N, Bertelsen S, Mayo K, Norton JB, Morris JC, Goate A. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68(5):581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, Kumar-Singh S, Van Broeckhoven C. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 7.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, Crook J, Finger E, Hantanpaa KJ, Karydas AM, Sengdy P, Gonzalez J, Seeley WW, Johnson N, Beach TG, Mesulam M, Forloni G, Kertesz A, Knopman DS, Uitti R, White CL, 3rd, Caselli R, Lippa C, Bigio EH, Wszolek ZK, Binetti G, Mackenzie IR, Miller BL, Boeve BF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Geschwind DH, Rademakers R. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76(5):467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, Crook R, Melquist S, Kuntz K, Petersen R, Josephs K, Pickering-Brown SM, Graff-Radford N, Uitti R, Dickson D, Wszolek Z, Gonzalez J, Beach TG, Bigio E, Johnson N, Weintraub S, Mesulam M, White CL, 3rd, Woodruff B, Caselli R, Hsiung GY, Feldman H, Knopman D, Hutton M, Rademakers R. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15(20):2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 10.Han JH, Ryu HH, Jun MH, Jang DJ, Lee JA. The functional analysis of the CHMP2B missense mutation associated with neurodegenerative diseases in the endo-lysosomal pathway. Biochem Biophys Res Commun. 2012;421(3):544–549. doi: 10.1016/j.bbrc.2012.04.041. [DOI] [PubMed] [Google Scholar]
- 11.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 12.Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D, Capell A, Haass C. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem. 2012;287(23):19355–19365. doi: 10.1074/jbc.M112.365098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17(18):1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 14.Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, Perry RH, Trojanowski JQ, Mann DM, Lee VM. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122(1):111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, Ince PG, Kamphorst W, Revesz T, Rozemuller AJ, Kumar-Singh S, Akiyama H, Baborie A, Spina S, Dickson DW, Trojanowski JQ, Mann DM. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117(1):15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majounie E, Abramzon Y, Renton AE, Keller MF, Traynor BJ, Singleton AB. Large C9orf72 repeat expansions are not a common cause of Parkinson's disease. Neurobiol Aging. 2012;33(10):2527. doi: 10.1016/j.neurobiolaging.2012.05.007. e2521–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 18.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB, 3rd, Castanedes-Casey M, Rousseau L, Benussi L, Binetti G, Ghidoni R, Hsiung GY, Mackenzie IR, Finger E, Boeve BF, Ertekin-Taner N, Graff-Radford NR, Dickson DW, Rademakers R. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem. 2013;126(6):781–791. doi: 10.1111/jnc.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramanathan HN, Ye Y. The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. 2012;22(2):346–359. doi: 10.1038/cr.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritz D, Vuk M, Kirchner P, Bug M, Schutz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, Gstaiger M, Aebersold R, Weihl CC, Meyer H. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol. 2011;13(9):1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rollinson S, Mead S, Snowden J, Richardson A, Rohrer J, Halliwell N, Usher S, Neary D, Mann D, Hardy J, Pickering-Brown S. Frontotemporal lobar degeneration genome wide association study replication confirms a risk locus shared with amyotrophic lateral sclerosis. Neurobiol Aging. 2011;32(4):758. doi: 10.1016/j.neurobiolaging.2010.12.005. e751–757. [DOI] [PubMed] [Google Scholar]
- 24.Urwin H, Authier A, Nielsen JE, Metcalf D, Powell C, Froud K, Malcolm DS, Holm I, Johannsen P, Brown J, Fisher EM, van der Zee J, Bruyland M, Van Broeckhoven C, Collinge J, Brandner S, Futter C, Isaacs AM. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19(11):2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Blitterswijk M, Baker MC, Dejesus-Hernandez M, Ghidoni R, Benussi L, Finger E, Hsiung GY, Kelley BJ, Murray ME, Rutherford NJ, Brown PE, Ravenscroft T, Mullen B, Ash PE, Bieniek KF, Hatanpaa KJ, Karydas A, McCarty Wood E, Coppola G, Bigio EH, Lippa C, Strong MJ, Beach TG, Knopman DS, Huey ED, Mesulam M, Bird T, White CL, 3rd, Kertesz A, Geschwind DH, Van Deerlin VM, Petersen RC, Binetti G, Miller BL, Petrucelli L, Wszolek ZK, Boylan KB, Graff-Radford NR, Mackenzie IR, Boeve BF, Dickson DW, Rademakers R. C9ORF72 repeat expansions in cases with previously identified pathogenic mutations. Neurology. 2013 doi: 10.1212/WNL.0b013e3182a8250c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Blitterswijk M, Dejesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO, Serrano G, Beach TG, Josephs KA, Knopman DS, Petersen RC, Boeve BF, Graff-Radford NR, Boylan KB, Petrucelli L, Dickson DW, Rademakers R. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 2013;12(10):978–988. doi: 10.1016/S1474-4422(13)70210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Blitterswijk M, DeJesus-Hernandez M, Rademakers R. How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr Opin Neurol. 2012;25(6):689–700. doi: 10.1097/WCO.0b013e32835a3efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DM, Pickering-Brown SM, Seelaar H, Heutink P, van Swieten JC, Murrell JR, Ghetti B, Spina S, Grafman J, Hodges J, Spillantini MG, Gilman S, Lieberman AP, Kaye JA, Woltjer RL, Bigio EH, Mesulam M, Al-Sarraj S, Troakes C, Rosenberg RN, White CL, 3rd, Ferrer I, Llado A, Neumann M, Kretzschmar HA, Hulette CM, Welsh-Bohmer KA, Miller BL, Alzualde A, Lopez de Munain A, McKee AC, Gearing M, Levey AI, Lah JJ, Hardy J, Rohrer JD, Lashley T, Mackenzie IR, Feldman HH, Hamilton RL, Dekosky ST, van der Zee J, Kumar-Singh S, Van Broeckhoven C, Mayeux R, Vonsattel JP, Troncoso JC, Kril JJ, Kwok JB, Halliday GM, Bird TD, Ince PG, Shaw PJ, Cairns NJ, Morris JC, McLean CA, DeCarli C, Ellis WG, Freeman SH, Frosch MP, Growdon JH, Perl DP, Sano M, Bennett DA, Schneider JA, Beach TG, Reiman EM, Woodruff BK, Cummings J, Vinters HV, Miller CA, Chui HC, Alafuzoff I, Hartikainen P, Seilhean D, Galasko D, Masliah E, Cotman CW, Tunon MT, Martinez MC, Munoz DG, Carroll SL, Marson D, Riederer PF, Bogdanovic N, Schellenberg GD, Hakonarson H, Trojanowski JQ, Lee VM. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42(3):234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Zee J, Van Langenhove T, Kleinberger G, Sleegers K, Engelborghs S, Vandenberghe R, Santens P, Van den Broeck M, Joris G, Brys J, Mattheijssens M, Peeters K, Cras P, De Deyn PP, Cruts M, Van Broeckhoven C. TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain. 2011;134(Pt 3):808–815. doi: 10.1093/brain/awr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vass R, Ashbridge E, Geser F, Hu WT, Grossman M, Clay-Falcone D, Elman L, McCluskey L, Lee VM, Van Deerlin VM, Trojanowski JQ, Chen-Plotkin AS. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121(3):373–380. doi: 10.1007/s00401-010-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.