Abstract
To investigate circulation of mimiviruses in the Amazon Region of Brazil, we surveyed 513 serum samples from domestic and wild mammals. Neutralizing antibodies were detected in 15 sample pools, and mimivirus DNA was detected in 9 pools of serum from capuchin monkeys and in 16 pools of serum from cattle.
Keywords: Amazon, mimivirus, megavirus, megavirales, monkey serum, bovine serum, vertebrates, Brazil, viruses
The group of nucleocytoplasmic large DNA viruses includes viruses that are able to infect different hosts, such as animals, green algae, and unicellular eukaryotes (1). Several members of this group are widely distributed in various environments, actively circulate in nature, and are responsible for outbreaks of medical importance (2,3). Mimiviridae, the newest family in this group, has been researched as a putative pneumonia agent and found in different biomes worldwide (3,5–9). The ubiquity of freeliving amebas and their parasitism by mimiviruses enhances the prospect that diverse environments could shelter these giant viruses (8–10). Mimiviruses can induce infection in a murine model, have had antibodies detected in patients with pneumonia, and can replicate in murine and human phagocytes (11,12). Moreover, although some authors suggest that mimivirus is a not frequent pneumonia agent (4), mimivirus has been isolated from a human with pneumonia (3).
The biomes in Brazil, particularly in the Amazon region, provide the diversity, species richness, and ecologic relationships ideal for identifying circulation of mimiviruses. Preliminary studies found Acanthamoeba polyphaga mimivirus (APMV) genomes in samples of bovine serum from Germany (13,14), indicating that the analysis of samples from vertebrates could be a way to explore and understand the circulation of this group of viruses in nature. We describe the detection of mimivirus antibodies and DNA in 2 mammalian species in the Amazon region of Brazil.
The Study
We selected 321 serum samples collected from wild monkeys from the Amazon region of Brazil during 2001–2002: 91 from black howler monkeys (Alouatta caraya) and 230 from capuchin monkeys (Cebus apella). Samples were collected in an overflow area of a fauna rescue program during the construction of a hydroelectric dam in Tocantins State (Figure 1, Appendix). The monkeys had no previous contact with humans. After blood collection, the animals were released into areas selected by environmental conservation programs. We also collected serum samples from cattle (Bos taurus): 147 samples from Pará and Maranhão States in the Amazon region and 45 from Bahia and Espírito Santo States in the Caatinga and Mata Atlântica biomes.
Figure 1.
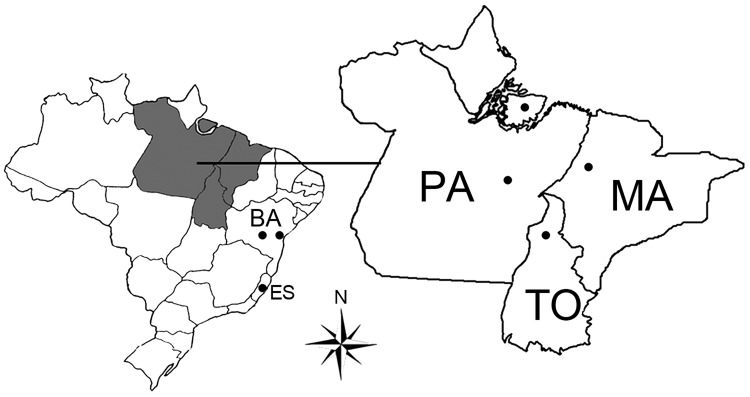
States in Brazil where serum samples were collected for study of mimivirus in mammals. Dots indicate collection sites. ES, Espírito Santos; BA, Bahia; PA, Pará; TO, Tocantins; MA, Maranhão.
All samples underwent serologic and molecular testing for mimivirus (Figure 2). Because total serum volumes were low, the specimens were grouped into pools of 2–5 serum samples (20 μL for each sample) from animals belonging to the same species that were from the same collection area. A total of 210 pools were compiled (Table). Pools were tested by real-time PCR targeting the conserved helicase viral gene (primers 5′-ACCTGATCCACATCCCATAACTAA-3′ and 5′-GCCTCATCAACAAATGGTTTCT-3′). DNA extractions were performed by using phenol-chloroform-isoamyl alcohol, and DNA quality and concentration were checked by using a nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). PCRs were performed by using the One Step SYBr Green Master Mix (Applied Biosystems, Foster City, CA, USA), and real-time PCR quality and sensitivity parameters were adjusted, including efficiency (102.6%) and R2 (0.992). APMV (kindly provided by Didier Raoult, Marseille, France) was used as a positive control. The serum pools were manipulated in a laminar flow cabinet, separate from any virus samples, to avoid cross-contamination.
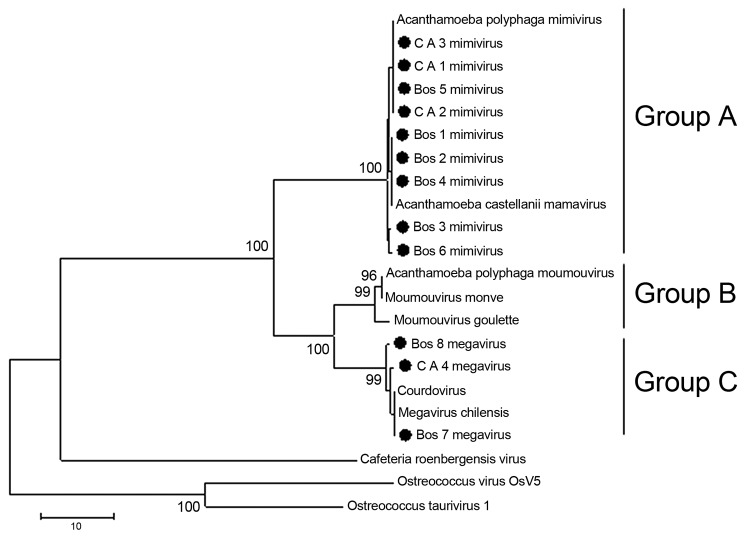
Figure 2.
Consensus bootstrap phylogenetic neighbor-joining tree of helicase gene from nucleocytoplasmic large DNA viruses showing alignment of mimivirus and megavirus isolates obtained from Cebus apella (CA) and bovids (Bos) in Brazil. Tree was constructed by using MEGA version 4.1 (www.megasoftware.net) on the basis of the nucleotide sequences with 1,000 bootstrap replicates. Bootstrap values >90% are shown. Nucleotide sequences were obtained from GenBank. Scale bar indicates rate of evolution.
Table. Sources and test results for serum samples from wild and domestic animals analyzed for presence of mimivirus, Brazil*.
| Species | States where samples were collected | Total no. samples | Total no. pools | Real-time PCR, helicase gene |
VN >90%, no. (%) pools | |
|---|---|---|---|---|---|---|
| No. (%) negative pools | No. (%) positive pools | |||||
| Black howler monkeys | Tocantins | 91 | 21 | 21 (100.0) | 0 | 0 |
| Capuchin monkeys |
Tocantins |
230 |
106 |
97 (91.5) |
9 (8.5) |
5 (4.72) |
| Cattle | Pará and Maranhão | 147 | 63 | 47 (74.6) | 16 (25.4) | 10 (15.9) |
|
|
Espírito Santo and Bahia |
45 |
20 |
20 (100.0) |
0 |
0 |
| Total | 513 | 210 | 185 (88.1) | 25 (11.9) | 15 (7.14) | |
*VN, virus neutralization test.
Of the 210 pools, 25 (11.9%) were positive for APMV (viral loads 1.4 × 103 to 2.3 × 106 copies/mL); 9 (4.3%) pools were capuchin monkey serum and 16 (7.6%) were bovine serum, all from the Amazon region. Mimivirus DNA was not detected in serum from black howler monkeys or cattle from Bahia and Espírito Santo States (Table). Using external primers 5′-ACCTGATCCACATCCCATAACTAAA-3′ and 5′-ATGGCGAACAATATTAAAACTAAAA-3′, we amplified a larger fragment of the helicase gene (390 bp) from all 25 positive samples; 12 positive serum pools, 4 from capuchin monkeys and 8 from cattle, were chosen for helicase gene sequencing and analysis. The helicase fragments were directly sequenced in both orientations and in triplicate (MegaBACE sequencer; GE Healthcare, Buckinghamshire, UK). The sequences were aligned with previously published sequences from GenBank by using ClustalW (www.clustal.org) and manually aligned by using MEGA software version 4.1 (www.megasoftware.net). Modeltest software (www.molecularevolution.org/software/phylogenetics/modeltest) was used determine which model of evolution was most appropriate for our analysis.
Optimal alignment of the predicted highly conserved helicase amino acid sequences showed several amino acid substitutions in the mimivirus amplicons we derived compared with other available sequences (Figure 3). Nine of the 12 sequenced amplicons showed high identity among themselves and with the APMV sequence (GenBank accession no. HQ336222). The other 3 amplicons showed a high identity with Megavirus chilensis (GenBank accession no. JN258408), a giant virus isolated in 2011 from seawater off the coast of Chile (15). A neighbor-joining phylogenetic tree constructed on the basis of the helicase gene revealed that all the amplicons we derived clustered with mimivirus isolates; however, according to the sequences alignment analysis, 3 of them clustered directly with the Megavirus chilensis group (Figure 2). The sequences have been deposited in GenBank.
Figure 3.
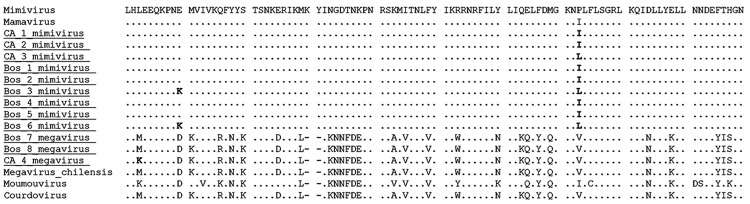
Amino acid inferred sequence of a fragment of the nucleocytoplasmic large DNA virus helicase gene (130 aa were inferred from the obtained 390-bp sample). Samples obtained in this study are underlined; boldface indicates polymorphic.
Concomitantly with molecular analysis, the pools of samples were submitted to a virus neutralization (VN) test to detect mimivirus neutralizing antibodies. VN was used rather than ELISA because the secondary antibodies required for an ELISA for monkey species were unavailable. Before being arranged in pools, the serum samples were inactivated separately by heating at 56°C for 30 min. Inactivated samples were diluted 1:20, mixed with 107 APMV particles to a final volume of 400 μL (peptone yeast glucose medium), then incubated at 37°C for 1 h to optimize virus–antibody interaction. This solution was added to 2 × 105 Acanthamoeba castellanii cells (ATCC 30234). To improve virus–ameba contact, the adsorption step was performed while rotating for 60 min. The samples were then centrifuged at 400 × g for 5 min, the supernatants were discarded, and the amebas were cultivated at 32°C for 8 h in PYG medium. Afterward, the samples were titrated in A. castellannii cells by using the 50% tissue culture infective dose method. Antimimivirus serum produced in Balb/c mice was used as VN positive control, and bovine serum collected during previous studies by our group was used as VN negative control. The percentage of reduction was calculated, and the cutoff for positive serum was defined as 90% of the reduction in comparison with the negative control. VN results showed that 15 of the 25 PCR-positive pools contained neutralizing antibodies against mimivirus, 5 from C. apella monkeys and 10 from cattle (Table).
Conclusions
We found evidence of mimivirus circulation in wild and domestic animals in the Amazon region of Brazil. Several agents of emerging infectious diseases in humans have reservoirs in wild and domestic animals, which act as a regular source for these agents. Anthropogenic disturbance of the Amazon ecosystem and increases in agricultural and livestock areas result in more contact between wildlife and rural human populations (2). Therefore, although mimivirus-associated pneumonia has not been studied in human patients in Brazil, surveillance of wild and domestic animals can help predict outbreaks and lead to establishment of control measures.
Although mimiviruses are known to be present in water and soil environments, new studies are necessary to determine if these viruses are a part of a vertebrate’s normal microbiota and act as opportunistic pathogens for pneumonia and to clarify whether viruses that are associated with pneumonia have any special genetic and physiologic features. Ecologic and public health studies should be designed to evaluate the risk for infection by mimiviruses during wildlife conservation efforts and to determine whether surveillance systems can predict outbreaks by monitoring mimivirus infections in wild and domestic animals.
Acknowledgments
We thank João Rodrigues dos Santos, Gisele Cirilo, and their colleagues for excellent technical support.
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, the Fundação de Amparo à Pesquisa do Estado de Minas Gerais, the Ministério da Agricultura, Pecuária e Abastecimento, and the Pro-Reitoria de Pesquisa da Universidade Federal de Minas Gerais. F.P.D. was supported by a fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior. E.G.K., C.A.B., G.S.T., and P.C.P.F. are researchers of the Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Biography
Mr Dornas is a pharmacist and a PhD student at the Laboratório de Vírus, Departamento de Microbiologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil. His research interests focus on diagnosing, monitoring, controlling, and preventing emerging infectious diseases.
Footnotes
Suggested citation for this article: Dornas FP, Rodrigues FP, Boratto PVM, Silva LCF, Ferreira PCP, Bonjardim CA, et al. Mimivirus circulation among wild and domestic mammals, Amazon Region, Brazil. Emerg Infect Dis [Internet]. 2014 Mar [date cited]. http://dx.doi.org/10.3201/eid2003.131050
References
- 1.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 2009;6:223 . 10.1186/1743-422X-6-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahão JS, Silva-Fernandes AT, Lima LS, Campos RK, Guedes MI, Cota MM, et al. Vaccinia virus infection in monkeys, Brazilian Amazon. Emerg Infect Dis. 2010;16:976–9. 10.3201/eid1606.091187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M, et al. First isolation of mimivirus in a patient with pneumonia. Clin Infect Dis. 2013;57:e127–34. 10.1093/cid/cit354 [DOI] [PubMed] [Google Scholar]
- 4.Vanspauwen MJ, Schnabel RM, Bruggeman CA, Drent M, van Mook WN, Bergmans DC, et al. Mimivirus is not a frequent cause of ventilator-associated pneumonia in critically ill patients. J Med Virol. 2013;85:1836–41. 10.1002/jmv.23655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colson P, Fancello L, Gimenez G, Armougom F, Desnues C, Fournous G, et al. Evidence of the megavirome in humans. J Clin Virol. 2013;57:191–200. 10.1016/j.jcv.2013.03.018 [DOI] [PubMed] [Google Scholar]
- 6.Boughalmi M, Saadi H, Pagnier I, Colson P, Fournous G, Raoult D, et al. High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Environ Microbiol. 2013;15:2000–7. 10.1111/1462-2920.12068 [DOI] [PubMed] [Google Scholar]
- 7.Colson P, Gimenez G, Boyer M, Fournous G, Raoult D. The giant Cafeteria roenbergensis virus that infects a widespread marine phagocytic protist is a new member of the fourth domain of Life. PLoS ONE. 2011;6:e18935. 10.1371/journal.pone.0018935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, et al. A giant virus in amoebae. Science. 2003;299:2033. 10.1126/science.1081867 [DOI] [PubMed] [Google Scholar]
- 9.Fouque E, Trouilhé MC, Thomas V, Hartemann P, Rodier MH, Héchard Y. Cellular, biochemical, and molecular changes during encystment of free-living amoebae. Eukaryot Cell. 2012;11:382–7. 10.1128/EC.05301-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–33. 10.1128/CMR.17.2.413-433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raoult D, La Scola B, Birtles R. The discovery and characterization of mimivirus, the largest known virus and putative pneumonia agent. Clin Infect Dis. 2007;45:95–102. 10.1086/518608 [DOI] [PubMed] [Google Scholar]
- 12.Ghigo E, Kartenbeck J, Lien P, Pelkmans L, Capo C, Mege JL, et al. Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 2008;4:e1000087. 10.1371/journal.ppat.1000087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger P, Papazian L, Drancourt M, La Scola B, Auffray JP, Raoult D. Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg Infect Dis. 2006;12:248–55. 10.3201/eid1202.050434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, et al. Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis. 2012;18:469–72. 10.3201/eid1803.111905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Distant mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci U S A. 2011;108:17486–91. 10.1073/pnas.1110889108 [DOI] [PMC free article] [PubMed] [Google Scholar]