Abstract
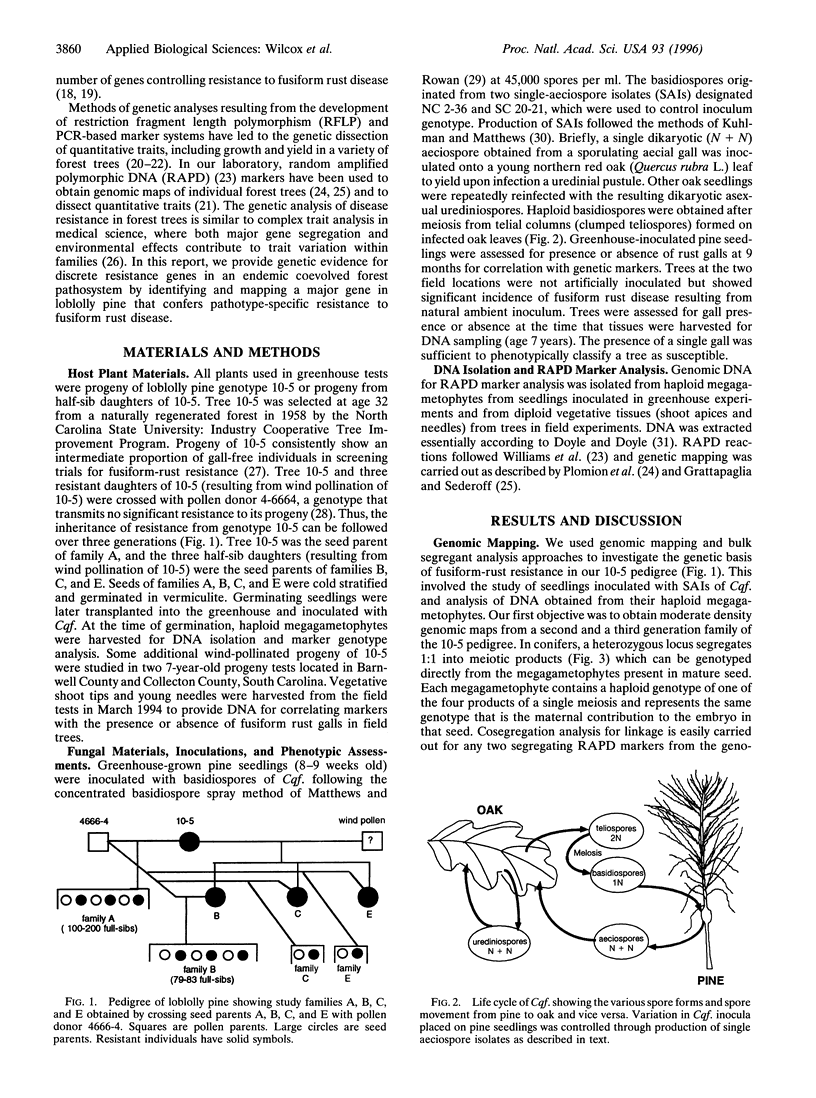
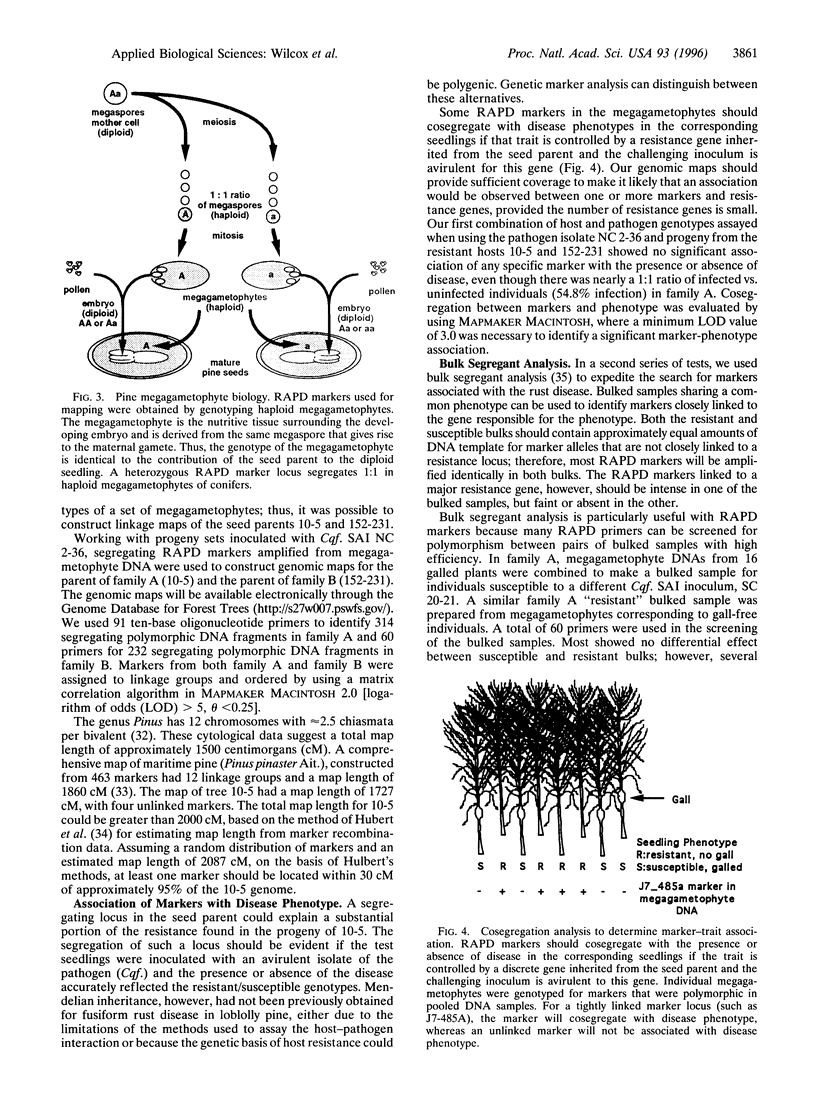
Genomic mapping has been used to identify a region of the host genome that determines resistance to fusiform rust disease in loblolly pine where no discrete, simply inherited resistance factors had been previously found by conventional genetic analysis over four decades. A resistance locus, behaving as a single dominant gene, was mapped by association with genetic markers, even though the disease phenotype deviated from the expected Mendelian ratio. The complexity of forest pathosystems and the limitations of genetic analysis, based solely on phenotype, had led to an assumption that effective long-term disease resistance in trees should be polygenic. However, our data show that effective long-term resistance can be obtained from a single qualitative resistance gene, despite the presence of virulence in the pathogen population. Therefore, disease resistance in this endemic coevolved forest pathosystem is not exclusively polygenic. Genomic mapping now provides a powerful tool for characterizing the genetic basis of host pathogen interactions in forest trees and other undomesticated, organisms, where conventional genetic analysis often is limited or not feasible.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradshaw H. D., Jr, Stettler R. F. Molecular genetics of growth and development in populus. IV. Mapping QTLs with large effects on growth, form, and phenology traits in a forest tree. Genetics. 1995 Feb;139(2):963–973. doi: 10.1093/genetics/139.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devey M. E., Delfino-Mix A., Kinloch B. B., Jr, Neale D. B. Random amplified polymorphic DNA markers tightly linked to a gene for resistance to white pine blister rust in sugar pine. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):2066–2070. doi: 10.1073/pnas.92.6.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia D., Sederoff R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994 Aug;137(4):1121–1137. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groover A., Devey M., Fiddler T., Lee J., Megraw R., Mitchel-Olds T., Sherman B., Vujcic S., Williams C., Neale D. Identification of quantitative trait loci influencing wood specific gravity in an outbred pedigree of loblolly pine. Genetics. 1994 Dec;138(4):1293–1300. doi: 10.1093/genetics/138.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. D., Axelrod R., Tanese R. Sexual reproduction as an adaptation to resist parasites (a review). Proc Natl Acad Sci U S A. 1990 May;87(9):3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert S. H., Ilott T. W., Legg E. J., Lincoln S. E., Lander E. S., Michelmore R. W. Genetic analysis of the fungus, Bremia lactucae, using restriction fragment length polymorphisms. Genetics. 1988 Dec;120(4):947–958. doi: 10.1093/genetics/120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinloch B. B., Jr, Parks G. K., Fowler C. W. White pine blister rust: simply inherited resistance in sugar pine. Science. 1970 Jan 9;167(3915):193–195. doi: 10.1126/science.167.3915.193. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Schork N. J. Genetic dissection of complex traits. Science. 1994 Sep 30;265(5181):2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
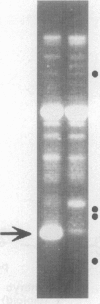
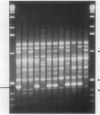
- Michelmore R. W., Paran I., Kesseli R. V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]