Abstract
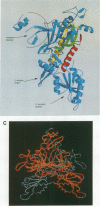
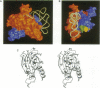
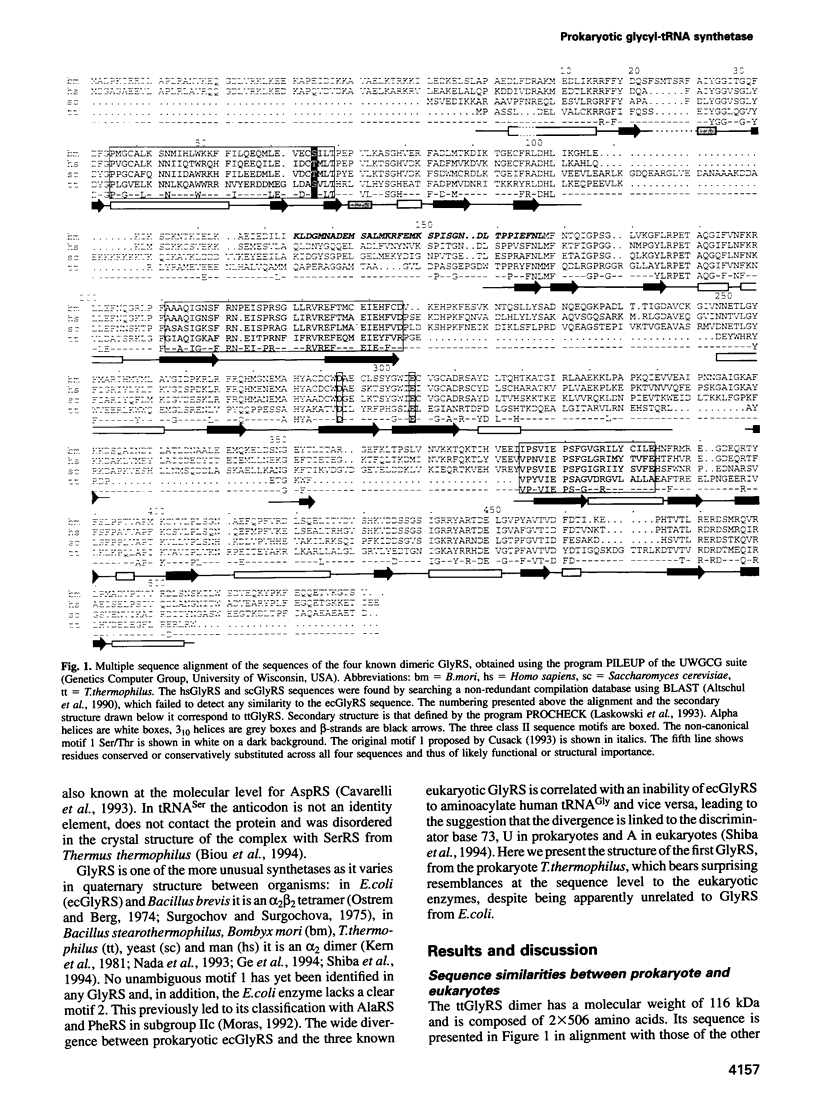
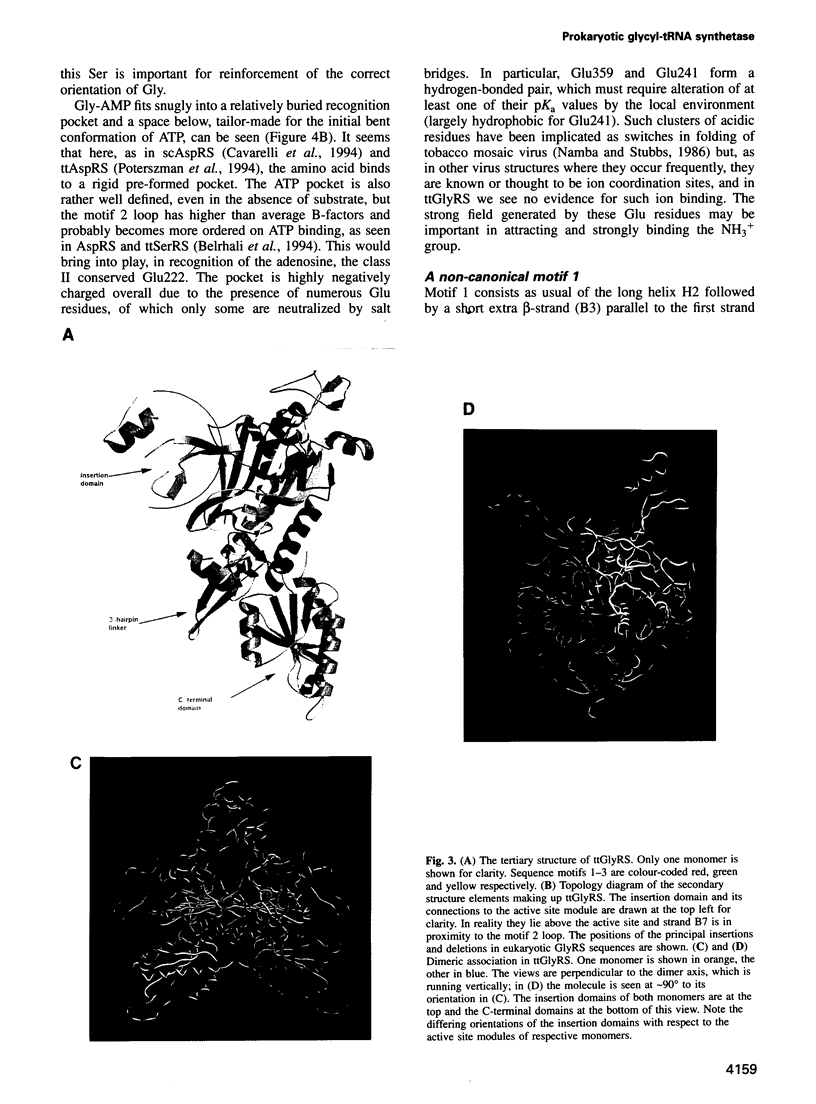
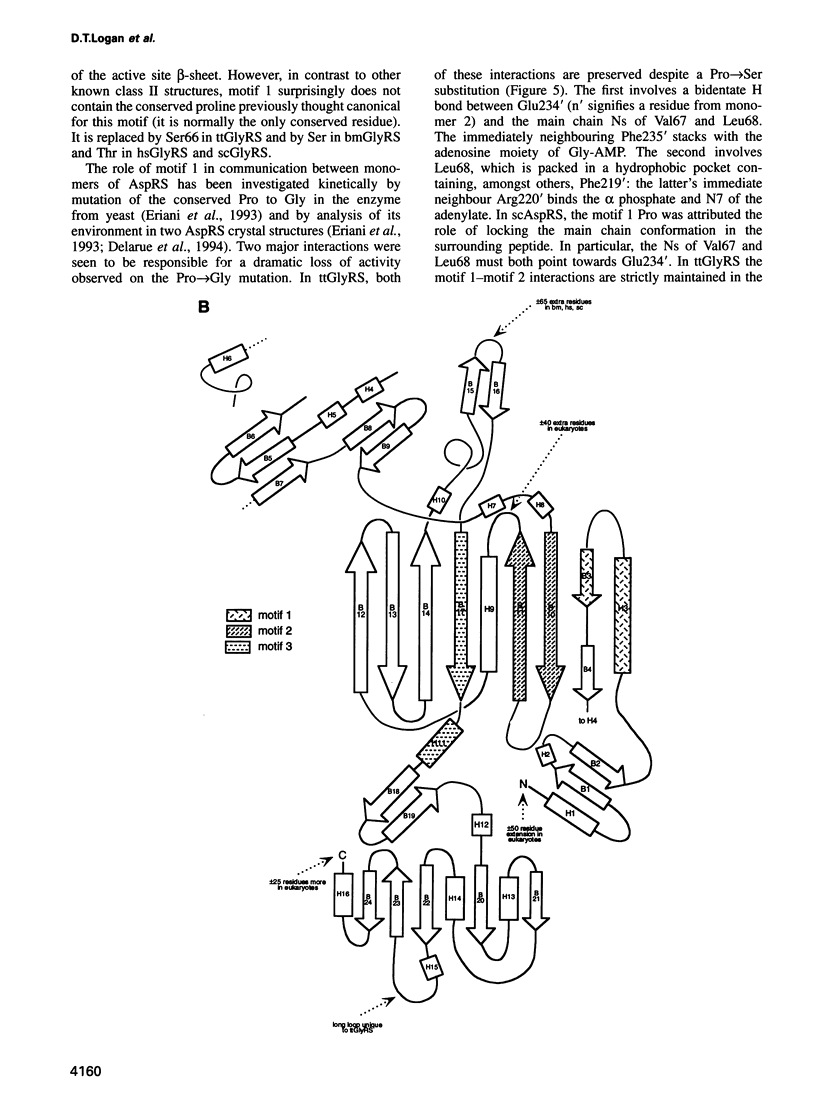
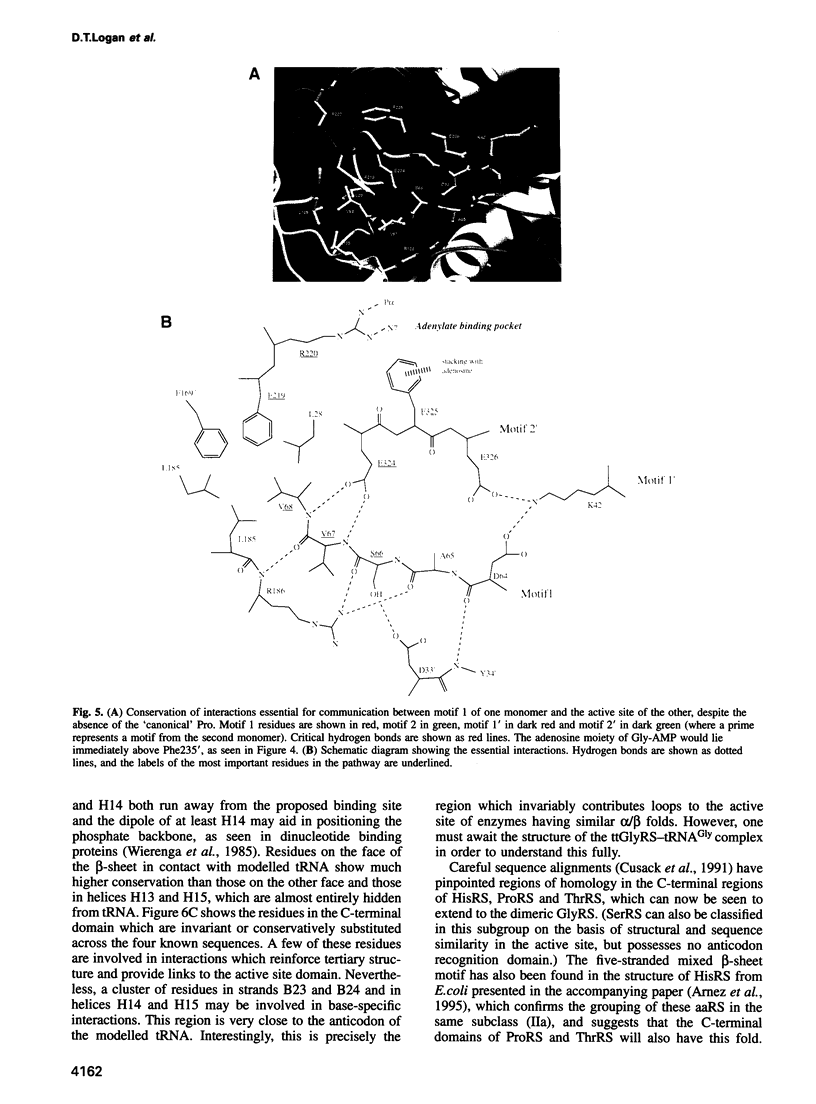
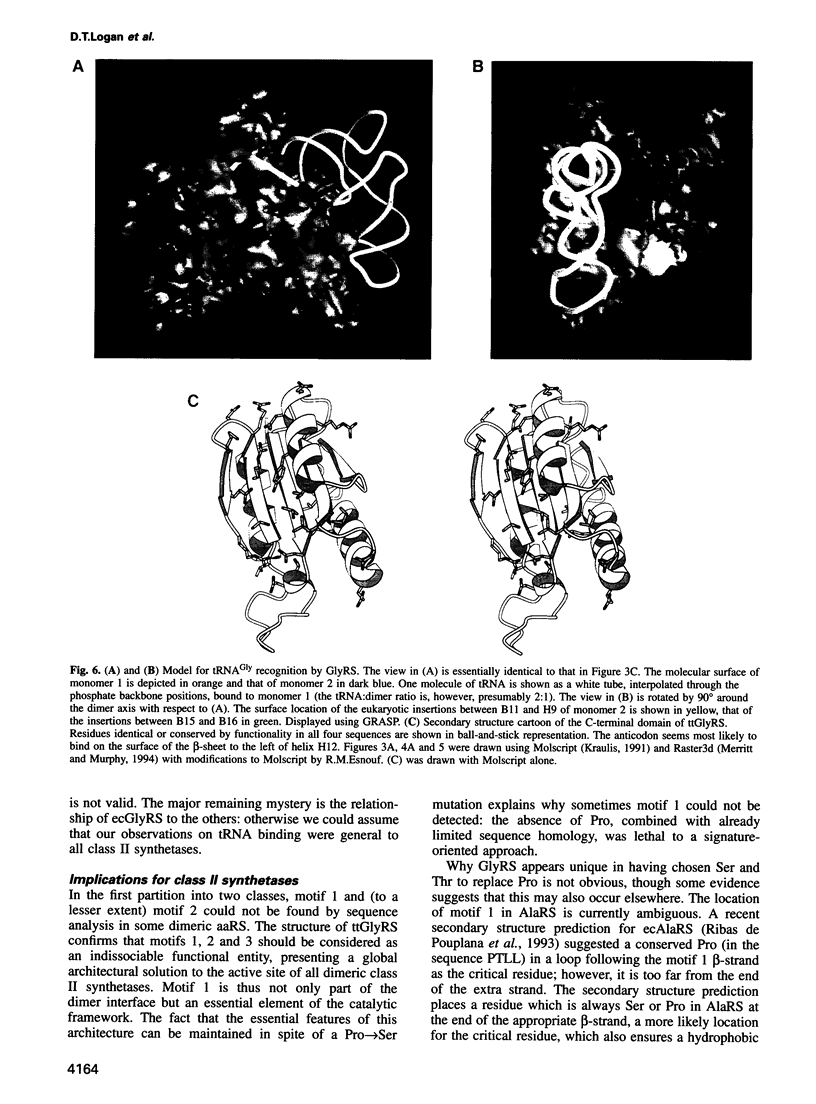
The sequence and crystal structure at 2.75 A resolution of the homodimeric glycyl-tRNA synthetase from Thermus thermophilus, the first representative of the last unknown class II synthetase subgroup, have been determined. The three class II synthetase sequence motifs are present but the structure was essential for identification of motif 1, which does not possess the proline previously believed to be an essential class II invariant. Nevertheless, crucial contacts with the active site of the other monomer involving motif 1 are conserved and a more comprehensive description of class II now becomes possible. Each monomer consists of an active site strongly resembling that of the aspartyl and seryl enzymes, a C-terminal anticodon recognition domain of 100 residues and a third domain unusually inserted between motifs 1 and 2 almost certainly interacting with the acceptor arm of tRNA(Gly). The C-terminal domain has a novel five-stranded parallel-antiparallel beta-sheet structure with three surrounding helices. The active site residues most probably responsible for substrate recognition, in particular in the Gly binding pocket, can be identified by inference from aspartyl-tRNA synthetase due to the conserved nature of the class II active site.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnez J. G., Harris D. C., Mitschler A., Rees B., Francklyn C. S., Moras D. Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. EMBO J. 1995 Sep 1;14(17):4143–4155. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artymiuk P. J., Rice D. W., Poirrette A. R., Willet P. A tale of two synthetases. Nat Struct Biol. 1994 Nov;1(11):758–760. doi: 10.1038/nsb1194-758. [DOI] [PubMed] [Google Scholar]
- Belrhali H., Yaremchuk A., Tukalo M., Berthet-Colominas C., Rasmussen B., Bösecke P., Diat O., Cusack S. The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure. 1995 Apr 15;3(4):341–352. doi: 10.1016/s0969-2126(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Belrhali H., Yaremchuk A., Tukalo M., Larsen K., Berthet-Colominas C., Leberman R., Beijer B., Sproat B., Als-Nielsen J., Grübel G. Crystal structures at 2.5 angstrom resolution of seryl-tRNA synthetase complexed with two analogs of seryl adenylate. Science. 1994 Mar 11;263(5152):1432–1436. doi: 10.1126/science.8128224. [DOI] [PubMed] [Google Scholar]
- Biou V., Yaremchuk A., Tukalo M., Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994 Mar 11;263(5152):1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Eriani G., Rees B., Ruff M., Boeglin M., Mitschler A., Martin F., Gangloff J., Thierry J. C., Moras D. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994 Jan 15;13(2):327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Cura V., Krishnaswamy S., Podjarny A. D. Heavy-atom refinement against solvent-flattened phases. Acta Crystallogr A. 1992 Sep 1;48(Pt 5):756–764. doi: 10.1107/s0108767392003416. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Cusack S., Härtlein M., Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991 Jul 11;19(13):3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S. Sequence, structure and evolutionary relationships between class 2 aminoacyl-tRNA synthetases: an update. Biochimie. 1993;75(12):1077–1081. doi: 10.1016/0300-9084(93)90006-e. [DOI] [PubMed] [Google Scholar]
- Delarue M., Moras D. The aminoacyl-tRNA synthetase family: modules at work. Bioessays. 1993 Oct;15(10):675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poterszman A., Nikonov S., Garber M., Moras D., Thierry J. C. Crystal structure of a prokaryotic aspartyl tRNA-synthetase. EMBO J. 1994 Jul 15;13(14):3219–3229. doi: 10.1002/j.1460-2075.1994.tb06623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublié S., Bricogne G., Gilmore C., Carter C. W., Jr Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995 Jan 15;3(1):17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Cavarelli J., Martin F., Dirheimer G., Moras D., Gangloff J. Role of dimerization in yeast aspartyl-tRNA synthetase and importance of the class II invariant proline. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10816–10820. doi: 10.1073/pnas.90.22.10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Shi J. P., Schimmel P. Overlapping nucleotide determinants for specific aminoacylation of RNA microhelices. Science. 1992 Feb 28;255(5048):1121–1125. doi: 10.1126/science.1546312. [DOI] [PubMed] [Google Scholar]
- Fraser T. H., Rich A. Amino acids are not all initially attached to the same position on transfer RNA molecules. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3044–3048. doi: 10.1073/pnas.72.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. L., Tzagoloff A. Structure and evolution of a group of related aminoacyl-tRNA synthetases. J Mol Biol. 1991 Apr 5;218(3):557–568. doi: 10.1016/0022-2836(91)90701-7. [DOI] [PubMed] [Google Scholar]
- Ge Q., Trieu E. P., Targoff I. N. Primary structure and functional expression of human Glycyl-tRNA synthetase, an autoantigen in myositis. J Biol Chem. 1994 Nov 18;269(46):28790–28797. [PubMed] [Google Scholar]
- Imada K., Sato M., Tanaka N., Katsube Y., Matsuura Y., Oshima T. Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution. J Mol Biol. 1991 Dec 5;222(3):725–738. doi: 10.1016/0022-2836(91)90508-4. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kern D., Giegé R., Ebel J. P. Glycyl-tRNA synthetase from baker's yeast. Interconversion between active and inactive forms of the enzyme. Biochemistry. 1981 Jan 6;20(1):122–131. doi: 10.1021/bi00504a021. [DOI] [PubMed] [Google Scholar]
- Logan D. T., Cura V., Touzel J. P., Kern D., Moras D. Crystallisation of the glycyl-tRNA synthetase from Thermus thermophilus and initial crystallographic data. J Mol Biol. 1994 Sep 2;241(5):732–735. doi: 10.1006/jmbi.1994.1547. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Rapid determination of nucleotides that define tRNA(Gly) acceptor identity. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6147–6151. doi: 10.1073/pnas.88.14.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E. A., Murphy M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994 Nov 1;50(Pt 6):869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Moras D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem Sci. 1992 Apr;17(4):159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- Mosyak L., Safro M. Phenylalanyl-tRNA synthetase from Thermus thermophilus has four antiparallel folds of which only two are catalytically functional. Biochimie. 1993;75(12):1091–1098. doi: 10.1016/0300-9084(93)90008-g. [DOI] [PubMed] [Google Scholar]
- Murzin A. G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993 Mar;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S., Chang P. K., Dignam J. D. Primary structure of the gene for glycyl-tRNA synthetase from Bombyx mori. J Biol Chem. 1993 Apr 15;268(11):7660–7667. [PubMed] [Google Scholar]
- Namba K., Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science. 1986 Mar 21;231(4744):1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Onesti S., Miller A. D., Brick P. The crystal structure of the lysyl-tRNA synthetase (LysU) from Escherichia coli. Structure. 1995 Feb 15;3(2):163–176. doi: 10.1016/s0969-2126(01)00147-2. [DOI] [PubMed] [Google Scholar]
- Ostrem D. L., Berg P. Glycyl transfer ribonucleic acid synthetase from Escherichia coli: purification, properties, and substrate binding. Biochemistry. 1974 Mar 26;13(7):1338–1348. doi: 10.1021/bi00704a006. [DOI] [PubMed] [Google Scholar]
- Poterszman A., Delarue M., Thierry J. C., Moras D. Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase. J Mol Biol. 1994 Nov 25;244(2):158–167. doi: 10.1006/jmbi.1994.1716. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana L., Buechter D. D., Davis M. W., Schimmel P. Idiographic representation of conserved domain of a class II tRNA synthetase of unknown structure. Protein Sci. 1993 Dec;2(12):2259–2262. doi: 10.1002/pro.5560021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz M., Fourme R., Broutin I., Prangé T. The catalytic site of serine proteinases as a specific binding cavity for xenon. Structure. 1995 Mar 15;3(3):309–316. doi: 10.1016/s0969-2126(01)00161-7. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Shiba K., Schimmel P., Motegi H., Noda T. Human glycyl-tRNA synthetase. Wide divergence of primary structure from bacterial counterpart and species-specific aminoacylation. J Biol Chem. 1994 Nov 25;269(47):30049–30055. [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Francklyn C. Cytosine 73 is a discriminator nucleotide in vivo for histidyl-tRNA in Escherichia coli. J Biol Chem. 1994 Apr 1;269(13):10022–10027. [PubMed] [Google Scholar]