Abstract
Rhodopsin is a G-protein-coupled receptor, in which retinal chromophore acts as inverse-agonist or agonist depending on its configuration and protonation state. Photostimulation of rhodopsin results in a pH-dependent equilibrium between the active state (Meta-II) and its inactive precursor (Meta-I). Here, we monitored conformational changes of rhodopsin using a fluorescent probe Alexa594 at the cytoplasmic surface, which shows fluorescence increase upon the generation of active state, by single-molecule measurements. The fluorescence intensity of a single photoactivated rhodopsin molecule alternated between two states. Interestingly, such a fluorescence alternation was also observed for ligand-free rhodopsin (opsin), but not for dark-state rhodopsin. In addition, the pH-dependences of Meta-I/Meta-II equilibrium estimated by fluorescence measurements deviated notably from estimates based on absorption spectra, indicating that both Meta-I and Meta-II are mixtures of two conformers. Our observations indicate that rhodopsin molecules intrinsically adopt both active and inactive conformations, and the ligand retinal shifts the conformational equilibrium. These findings provide dynamical insights into the activation mechanisms of G-protein-coupled receptors.
Introduction
Rhodopsin, the photoreceptor protein of retinal rod cells, is a member of G-protein-coupled receptors (GPCRs) (1). GPCRs are ubiquitous membrane proteins with a seven transmembrane structure, which respond to a variety of stimuli. Unlike most GPCRs, which are activated by diffusible ligands, rhodopsin has a covalently bound retinal as an intrinsic ligand, which also functions as a light-absorbing chromophore. Light absorption causes the isomerization of the inverse agonist 11-cis-retinal into the agonist all-trans-retinal (2), resulting in the formation of the active state of rhodopsin (3). Despite the uniqueness of rhodopsin, recent progress in x-ray crystallography has revealed that the agonist-induced conformational change of rhodopsin, which involves an outward movement of helix VI, is similar to that of other GPCRs (4–7). Therefore, understanding the mechanisms of conformational changes that lead to the activation of rhodopsin could provide a new, to our knowledge, paradigm for the signaling mechanism of GPCRs.
The snapshots of resting and activated structures of GPCRs presented by x-ray crystallographic studies have provided valuable insights into the activation mechanisms of GPCRs. However, the analysis focusing on the conformational dynamics would be essential to better understand the activation mechanism of rhodopsin (8–11). In this work, we explored the conformational dynamics of individual receptor molecules by single-molecule analysis.
Single-molecule analysis using total internal reflection fluorescence microscopy (TIRFM) has been successfully applied to analyses of intramolecular conformational changes (12,13) as well as intermolecular interactions (14–16). Unlike bulk measurements, single-molecule analysis of individual molecules provides direct information about the kinetics of forward and backward reactions in an equilibrium mixture. Here we used single-molecule detection to directly monitor the conformational fluctuations between active and inactive states of rhodopsin.
Activation of vertebrate rhodopsin proceeds through the formation of an equilibrium mixture of the active state Meta-II and its inactive precursor Meta-I, which have characteristic absorption spectra due to the protonation state of the retinal chromophore (17). Because the retinal chromophore also acts as the ligand, the protonation state of the retinal is commonly used as proxy to monitor the formation of the active state. However, crystal structures of the active state of rhodopsin and other GPCRs indicate that large conformational changes occur at the cytoplasmic surface away from the ligand-binding site. In agreement with this notion, a previous study reported that Alexa594 immobilized at Cys-316 on cytoplasmic helix VIII of bovine rhodopsin is sensitive to the generation of G protein activating state (Meta-II) (18).
In this work, we examined the conformational dynamics of opsin (the ligand-free rhodopsin), dark-state rhodopsin (resting state containing 11-cis-retinal), as well as photoactivated rhodopsin (equilibrium mixture of Meta-I and Meta-II) by single-molecule fluorescence measurements of Alexa594 bound to Cys-316. Our single-molecule analyses strongly suggest that a rhodopsin molecule intrinsically adopts a conformational equilibrium between active and inactive conformations, which is regulated by the presence/absence, configuration, or the protonation state of the retinal chromophore.
Materials and Methods
Sample preparation
Bovine rod outer segments (ROS) were isolated from bovine retinas, and rhodopsin in ROS was labeled with Alexa594 as described previously (18). Rhodopsin labeled with Alexa594 (Rh/Alexa594) was solubilized by OG/asolectin buffer (60 mM n-octyl-β-D-glucoside, 1 mg/mL asolectin, 50 mM HEPES, 140 mM NaCl, pH 7.5) and purified by concanavalin A-Sepharose. The purity of rhodopsin achieved by our protocol was assessed by the optical purity index (Abs280/Abs500) of the unlabeled rhodopsin sample purified by this method, and it was typically 1.88 (Fig. S1 b in the Supporting Material). The molar ratio of rhodopsin and Alexa594 in the purified Rh/Alexa594 sample was 1:0.98 based on the visible absorption spectra (Fig. S1 b).
Purified Rh/Alexa594 sample, in which the molar ratio of asolectin and Rh/Alexa594 was 1300:1, was dialyzed against 500 volumes of dialysis buffer containing 140 mM NaCl and 50 mM MES (pH 6.0), 50 mM MOPS (pH 7.0), 50 mM HEPES (pH 8.0), or 50 mM CHES (pH 9.0) at 4°C in the dark for 36 h with six buffer exchanges.
Opsin was prepared by photobleaching of Rh/Alexa594 in the presence of 50 mM NH2OH. For TIRFM imaging of the dark-state, photoisomerization of the chromophore by excitation laser was prevented by using a retinal analog in which C11=C12 double bond is locked in cis configuration by a 7-membered ring (7m-Ret) (19,20). Rh/Alexa594 regenerated with this retinal analog (7m-Rh/Alexa594) was also purified and incorporated into asolectin liposomes. To suppress the light-induced conformational change, Rh/Alexa594 was incorporated into 1,2-dimyristoyl-phosphatidylcholine (DMPC) liposomes, in which conformational change is arrested at Meta-I stage (21). For control experiments, Rh/Alexa594 in asolectin liposome was denatured by incubating it at 90°C for 3 min. Suspensions of liposomes were centrifuged (20,400 × g, 20 min) and the unprecipitated liposomes in the supernatant, which contained a small number of pigment or opsin molecules, were subjected to the single-molecule analysis. Bovine rod G protein (transducin), was extracted from irradiated bovine rod outer segment membranes by adding GTP, and purified by DEAE Toyopearl 650S (Tosoh, Tokyo, Japan) column chromatography (22).
Spectrophotometry
Absorption spectra were recorded using a Shimadzu (Shimadzu, Kyoto, Japan) UV2450 spectrophotometer. Fluorescence spectra were recorded with a Shimadzu RF-5300PC spectrofluorometer. To maintain the sample temperature at 6°C, an optical cell holder was connected to a Neslab RTE-7 temperature controller. The sample was irradiated with light from a 1 kW tungsten halogen lamp (Rikagaku Seiki, Tokyo, Japan) that had been passed through a glass cut-off filter (Y52, Toshiba, Tokyo, Japan). Alexa594 bound to rhodopsin was excited at 605 nm. Time course of fluorescence changes of Alexa594 were recorded at 630 nm.
G protein activation assay
GDP/GTPγS exchange by G protein transducin was examined by a radionucleotide filter-binding assay (22). All procedures were carried out at 0°C. Reaction mixtures (∼125 nM pigment, 600 nM G protein, 50 mM MOPS, 140 mM NaCl, 2.25 mM MgCl2, 1 mM DTT, 2 μM GDP, pH 7.0) were kept in the dark or irradiated with light generated by a 1 kW tungsten halogen lamp and Y52 filter for 30 s. The GDP/GTPγS exchange reaction was then initiated by the addition of [35S]GTPγS (final concentration, 1 μM). After incubation for the selected time in the dark, an aliquot (20 μL) of the reaction mixture was mixed with 200 μL of stop solution (20 mM Tris/HCl, 100 mM NaCl, 25 mM MgCl2, 1 μM GTPγS, and 2 μM GDP, pH 7.4), and it was immediately filtered through a nitrocellulose membrane to trap [35S]GTPγS bound to G protein. The amount of bound [35S]GTPγS was quantitated by a liquid scintillation counter (Tri-Carb 2910 TR; PerkinElmer, Waltham, MA).
Single-molecule imaging by total internal reflection fluorescence microscopy
Fluorescence of Alexa594 was observed using an inverted microscope (TE 2000, Nikon, Tokyo, Japan) with a 60× oil-immersion objective (ApoTIRF 60× 1.49 NA, Nikon). Fluorescence of Alexa594 was generated by 559 nm laser (WS-0559-050, 50 mW, NTT Electronics, Kanagawa, Japan). Energy of the laser was reduced by a neutral density filter (ND50). The images were acquired using an EMCCD camera (C9100-134, ImagEM, Hamamatsu Photonics, Hamamatsu, Japan) with a 345× EM gain. The image of 512 × 512 pixels were recorded with a time resolution of 20 frame/s and space resolution of 108 nm/pixel (2.5× relay lens) or 67 nm/pixel (4.0× relay lens). To prevent photobleaching of Alexa594 fluorophore, 2-mercaptoethanol and Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) were added to the sample at the final concentrations of 5% (w/v) and ∼1 mM, respectively. The sample dish was maintained at 6°C with an air-cooling device (Peltier-4, Taiei Denki, Tokyo, Japan). All sample manipulations were performed under dim red light.
Data analyses
Time course of fluorescence intensity was obtained from TIRFM images by using ImageJ. Fluorescence intensity steps were detected by a laboratory-written analytical program based on a hidden Markov model (23,24) developed by LabVIEW (National Instruments, Austin, TX). All statistical and kinetic analyses were performed with IGOR Pro 6 (WaveMetrics, Lake Oswego, OR).
Results
Preparation of asolectin liposomes containing rhodopsin labeled by Alexa594
To monitor the light-induced conformational change of rhodopsin, rhodopsin was labeled by Alexa594 at Cys-316 on cytoplasmic helix VIII (Rh/Alexa594) (18). The optical purity index (Abs280/Abs500) of unlabeled rhodopsin sample prepared by our method was typically 1.88, and the molar ratio of the rhodopsin and Alexa594 in Rh/Alexa594 sample was estimated to be 1:0.98 based on the visible absorption spectra (Fig. S1 b). Bovine rhodopsin has two solvent-exposed Cys residues (Cys-140 and Cys-316), which are potentially labeled by Alexa594 (18). However, the binding efficiency for Cys-140 is substantially lower than that for Cys-316. Taken together, Alexa594 in our preparation is virtually homogeneous and the effect of contaminant was not taken into consideration.
Alexa594 immobilized at Cys-316 shows fluorescence increase upon the generation of the G protein activating state (Meta-II) (18). We prepared asolectin liposomes containing Rh/Alexa594 to disperse molecules for single-molecule analysis. Electron microscopy of these liposomes showed that our preparation was uniform and relatively small (diameter = ∼70 nm, Fig. S1 a). Of importance, asolectin liposomes reproduced the membrane environment of native ROS membranes, as evidenced by the pH-dependent equilibrium of Meta-I/Meta-II, which is a sensitive indicator of the membrane environment (17,25). Fig. S1 c clearly shows that the pH-dependent Meta-I/Meta-II equilibrium observed in ROS membranes was preserved in asolectin liposomes, with Meta-II formation favored at lower pH having a pKa of 6.5 (25,26). We also compared the fluorescence changes observed during photoactivation of Rh/Alexa594 in asolectin liposomes and ROS membranes. As shown in Figs. S1, d and e, the fluorescence increase upon light irradiation of Rh/Alexa594 and its pH dependency were very similar between asolectin liposomes and ROS membranes. These findings indicate that Rh/Alexa594 incorporated into asolectin liposomes was suitable for studying the conformational change of rhodopsin by single-molecule analysis. The increase in the amount of Meta-II in the Meta-I/Meta-II equilibrium correlates well with the increase in Alexa594 fluorescence, indicating that Meta-II is mainly in a high fluorescence state, whereas dark-state and Meta-I are mainly in a low fluorescence state (see below). On the other hand, fluorescence increase was reduced in DMPC liposomes, where the conformational change of photoactivated rhodopsin is arrested at Meta-I stage (Fig. S1 f). At all pH, the fluorescence intensity was reduced in the presence of hydroxylamine, which converts Meta-I and Meta-II into retinal oxime and opsin (Fig. S1 d, broken lines), indicating that opsin is mainly in a low fluorescence state (see below).
Fluorescent spots of Rh/Alexa594 detected by single-molecule imaging
Fig. 1 a shows a typical TIRFM image of Rh/Alexa594 in asolectin liposomes. The typical time courses of fluorescence changes for fluorescent spots are shown in Fig. 1, b and c, and Movie S1. The interconversion between the high fluorescence state (Fhigh) and low fluorescence state (Flow) is immediately apparent. Fluorescence intensity steps were detected by a laboratory-written program based on a hidden Markov model (black lines in Fig. 1, b and c). Fluorescence changes due to blinking of the fluorophore, migration of liposome, instrumental noise, etc., were excluded manually (Fig. S2).
Figure 1.
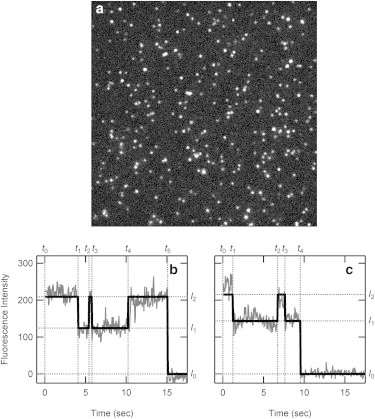
Typical fluorescent spots of Rh/Alexa594 observed by TIRFM. (a) A typical TIRFM image. The images were obtained at 20 frames/s. The background was subtracted by a Rolling Ball Background Subtraction in ImageJ. (b and c) Analysis of fluorescence intensity of the fluorescent spot. The fluorescence intensity was estimated by averaging 12 × 12 pixels square containing a fluorescent spot. The average intensity of five 12 × 12 pixels squares in which no spot existed was subtracted as the background. Fluorescence intensity steps were detected by using a laboratory-written program based on a hidden Markov model (black line). See also Movie S1.
Fluorescence intensity of single Rh/Alexa594 molecule
It has been reported that rhodopsin has a significant tendency to form dimers in proteoliposomes (27,28). Therefore, we examined the fluorescence intensity of a single Alexa594 molecule bound to rhodopsin to discriminate between monomers and oligomers. The histogram of the maximum fluorescence intensity (I2 in Fig. 1, b and c), was constructed from the time course of fluorescent spots (Fig. 2 a). As each spot may contain one, two, or more Rh/Alexa594 molecules, the histogram was fitted with a sum of three Gaussian functions having stepwise mean values as follows:
| (1) |
where I is a maximum fluorescence intensity of each spot, μ is the mean intensity for a single molecule, Ai (i = 1, 2, 3) is the frequency at iμ, and iσ2 is the variance of each distribution. Using Eq. 1, the histogram of the maximum fluorescence intensity was fitted with mean intensity μ = 154 for a single molecule. Furthermore, the last fluorescence decrease, corresponding to the photobleaching of the last Alexa594 molecule in each fluorescent spot was examined. Since the photobleaching of Alexa594 in Fhigh (Fig. 1 b) and Flow (Fig. 1 c) occurred, the histograms of the last fluorescence decrease were separately constructed (Fig. 2, b and c, respectively). In addition, the histogram for the fluorescent spots, which were photobleached before the fluorescence alternation was constructed, where we could not determine which state of Alexa594 photobleached (Fig. 2 d). These histograms showed the single Gaussian distributions with μ = 160, 126, and 145 (Fig. 2, b–d, respectively). The good agreement between μ of maximum intensity (Fig. 2 a) and the photobleaching of Fhigh (Fig. 2 b) confirmed that most of the spots in asolectin liposomes originated from single Rh/Alexa594 molecules. Because μ varied between the measurements, we chose the spots having the maximum intensity (I2) of μ ± half width at half-maximum for further analysis.
Figure 2.
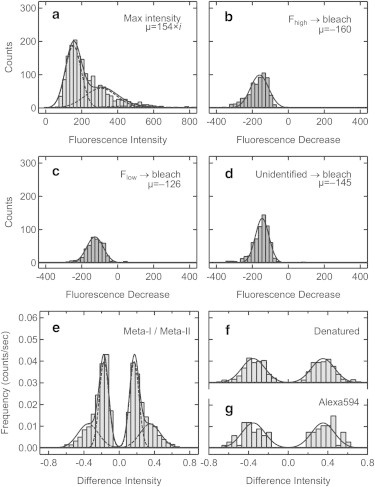
Histograms of fluorescence intensity of spots and difference intensity observed by TIRFM. (a) Histograms of maximum intensity in asolectin liposomes at pH 6 (n = 1850) fitted with three Gaussian functions shown in Eq. 1 (μ = 154, σ = 45, A1 = 186, A2 = 59, A3 = 6). (b–d) Histograms of the fluorescence decrease upon photobleaching of Alexa594 in Fhigh (b), Flow (c), and unidentified state because of photobleaching before the fluorescence alternation (d). They were fitted with the single Gaussian functions [n = 676, μ = −160, σ = 51, A = 99 for (b), n = 471, μ = −126, σ = 46, A = 78 for (c), and n = 703, μ = −145, σ = 38, A = 134 for (d)]. (e–g) Difference intensity histograms for photoactivated rhodopsin (n = 924) (e), denatured rhodopsin (n = 505) (f), and free Alexa594 molecules in solution (n = 129) (g). The difference intensity histogram in (e) was well fitted with two mirror-imaged pairs of Gaussian functions (Eq. 2: μ1 = ±0.304, σ1 = 0.111, A1 = 0.014, and μ2 = ±0.167, σ2 = 0.049, A2 = 0.037), whereas those in (f) and (g) were fitted with a single mirror-imaged pair of Gaussian functions [μ = ±0.344, σ = 0.112, A = 0.011 (f), and μ = ±0.377, σ = 0.127, A = 0.011 (g)].
Fluorescence changes of single photoactivated rhodopsin molecules
The histograms of the photobleaching of Fhigh and Flow showed that μ of Flow (−126) was ∼80% of that of Fhigh (−160) (Fig. 2, b and c). This is consistent with the ratio of the fluorescence increases for formation of Meta-II (pH 6) and Meta-I (pH 9) in bulk fluorescence measurement (Fig. S1 d), showing the consistency between the bulk measurement and the single-molecule measurement. The histograms of difference fluorescence intensity (e.g., I1-I2 for t1 and t3, and I2-I1 for t2 and t4 in Fig. 1 b) were constructed, in which the difference fluorescence intensities were normalized by the maximum fluorescence intensity of each spot (Fig. 2, e–g). Thus, the abscissa of these histograms shows the difference intensities relative to the maximum intensity of each spot. The difference intensity histogram of photoactivated rhodopsin (Fig. 2 e) was fitted with the sum of two mirror-imaged pairs of Gaussian functions as follows:
| (2) |
where ΔI is the relative difference intensity, μi is the mean difference, Ai is the frequency at μi, and σi is the standard deviation of the ith distribution. The results showed that the difference intensity histogram of photoactivated rhodopsin was well fitted with Eq. 2 (μ1 = 0.304 and μ2 = 0.167) (Fig. 2 e).
To examine whether or not these fluorescence changes were derived from conformational changes of rhodopsin, denatured rhodopsin and free Alexa594 molecules in solution were subjected to the same TIRFM imaging and analyses (Fig. 2, f and g). The difference intensity histogram of denatured Rh/Alexa594 was fitted with a single mirror-imaged pair of small Gaussian curves (μ = ±0.344) (Fig. 2 f). A similar pair of small Gaussian peaks were observed in the histogram of free Alexa594 molecules (μ = ±0.377) (Fig. 2 g). These components (noise component) likely originate from the instrumental noise and/or intrinsic fluorescence fluctuation of Alexa594, and represent the false-detection rate of conformational change in our experimental setup and analytical software. Therefore, the smaller peaks of photoactivated rhodopsin (μ1 = ±0.304) correspond to the noise components, whereas greater ones (μ2 = ±0.167) were derived from the conformational changes (Fig. 2 e). It should be noted that the frequency at ΔI = ∼0 is almost 0 because small fluorescence changes were hardly detected by our program based on a hidden Markov model.
Although the hidden Markov model provided the most likely number of states (∼3.5 states including bleached state in average), we assumed two conformational states (Fhigh and Flow) because the fluorescence changes from the conformational changes were expressed by a single mirror-imaged pair of Gaussian curves at μ2 = ±0.167.
Duration time after fluorescence increase or decrease in photoactivated rhodopsin
The previous results showed that the Gaussian distributions of fluorescence changes at μ = ±0.167 for photoactivated rhodopsin (Fig. 2 e) were due to conformational changes. To kinetically characterize the conformational equilibrium, we next extracted changes with difference intensity (ΔI) of μ ± half width at half-maximum (0.167 ± 0.058). We estimated the duration time of each conformation of photoactivated rhodopsin (e.g., t1-t0, t3-t2, and t5-t4 for Fhigh, t2-t1 and t4-t3 for Flow in Fig. 1 b), and constructed a probability histogram of duration times (d) of Flow and Fhigh with a bin-width of 100 ms (Fig. 3, a–d). Assuming an exponential decay of Flow and Fhigh with rate constants of klow and khigh, respectively, duration time histograms for Flow and Fhigh can be expressed with a single-exponential function with klow and khigh as follows:
| (3) |
where Plow(d) and Phigh(d) are the probabilities that Flow and Fhigh are converted at d and Clow and Chigh are amplification constants for Flow and Fhigh, respectively. Because the decay of Fhigh is caused by the conversion from Fhigh to Flow or photobleaching of Alexa594, khigh is expressed as follows:
| (4) |
where khigh→low and kbleach are the rate constants for the conversion from Fhigh to Flow and photobleaching of Alexa594, respectively. Similarly, klow is expressed as follows:
| (5) |
Duration time histograms were fitted with Eq. 3 to obtain klow and khigh. Using kbleach obtained by the single-exponential fitting of the histogram of the fluorescence retention time (e.g., t5-t0 in Fig. 1 b), khigh→low and klow→high were calculated (Table S1).
Figure 3.
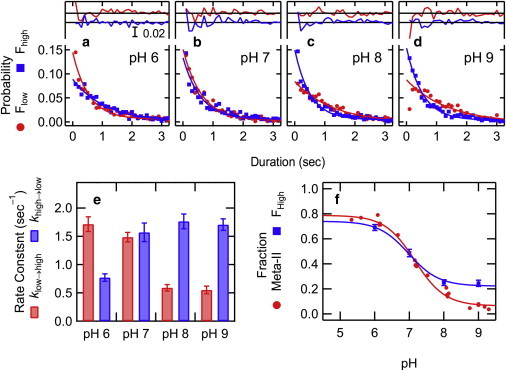
Duration times for Fhigh and Flow in photoactivated rhodopsin. Histograms of duration time after fluorescence increase (Fhigh, blue squares) and decrease (Flow, red circles) at pH 6 (a), pH 7 (b), pH 8 (c), and pH 9 (d). Plots were fitted with a single-exponential function, and the residuals of the fit are shown above. Rate constants for conformational changes (klow→high or khigh→low) were obtained according to Eqs. 4 and 5 (Table S1). (e) pH dependency of klow→high (red) and khigh→low (blue). (f) pH dependency of the ratio of Fhigh (blue square) and Meta-II (red circle). The ratio of Meta-II was determined by UV-visible absorption measurements. Data were fitted with a Henderson-Hasselbalch equation . pKa was 7.16 for Meta-I/Meta-II (red), and 6.98 for Flow/Fhigh (blue). To see this figure in color, go online.
The pH-dependency of khigh→low and klow→high are shown in Fig. 3 e. Using these values, the pH-dependency of the ratio of Fhigh was calculated (Table S1), which is consistent with the result of bulk fluorescence measurement. The profile of the pH-dependency is similar to that of the ratio of Meta-I and Meta-II estimated by ultraviolet (UV)-visible absorption spectroscopy, indicating that Flow and Fhigh essentially correspond to Meta-I and Meta-II, respectively. However, they showed a small but notable deviation at pH <6 and considerable differences at pH >8 (Fig. 3 f). UV-visible absorption measurement monitors the protonation state of the Schiff base linkage, whereas Alexa594 at Cys-316 is sensitive to the conformation around the cytoplasmic surface. Because Meta-I is favored at higher pH, a greater fraction of Fhigh than that of Meta-II at pH >8 suggests that Meta-I is in a mixture of Fhigh and Flow. These results suggest that, Meta-I, which has a protonated Schiff base linkage, can partially adopt an active (open) conformation. Similarly, the small deviation between Flow/Fhigh and Meta-I/Meta-II at pH <6 suggests that Meta-II, in which the Schiff base linkage is deprotonated, partially adopts an inactive (close) conformation.
Previous analyses have proposed an activation sequence consisting of an equilibrium between Meta-I and multiple states of Meta-II (Meta-IIa, Meta-IIb and Meta-IIbH+) (29,30). Although Meta-IIa, Meta-IIb, and Meta-IIbH+ all have a deprotonated retinal Schiff base, the conformational changes for Meta-IIa formation are significantly smaller than those for Meta-IIb and Meta-IIbH+ (31). Thus, Flow and Fhigh of Meta-II may correspond to Meta-IIa and Meta-IIb, respectively. Under our experimental condition (6°C), however, Meta-II is predominantly in Meta-IIbH+ form (30). Our results therefore suggested that the protein moiety of Meta-IIbH+ is intrinsically interconverted between Fhigh and Flow conformations.
Correlation between frequencies of conformational changes and G protein activation efficiencies
Bulk fluorescence measurements showed that fluorescence intensities of dark-state rhodopsin and opsin are significantly smaller than that of photoactivated rhodopsin (Fig. S1 d). However, opsin (32–35) or constitutively active mutants of rhodopsin (36–38) have low G protein activation efficiency, suggesting that dark-state rhodopsin and opsin could generate the G protein activating state with low frequency. Therefore, we analyzed the fluorescence changes of dark-state rhodopsin and opsin in single molecules, to assess the possible conformational equilibrium.
Because the excitation laser for TIRFM caused the photolysis of rhodopsin, the C11=C12 double bond of the chromophore was fixed in the 11-cis configuration by the 7-membered ring retinal (7m-Ret) (19,20) for the measurements of the dark-state rhodopsin (7m-Rh) (Fig. 4 a, inset). It is possible that the structural constraint of ring retinal influences the conformation or dynamics of rhodopsin. However, 7-membered ring is relatively flexible, and the circular dichroism of the chromophore of 7m-Rh is similar to that of native rhodopsin (20). In addition, the absorption spectrum of 7m-Rh is also similar to native rhodopsin. These findings suggest that the 7m-Ret, which would function as an inverse-agonist, is accommodated to the chromophore binding pocket similar to the native 11-cis-retinal. The time courses of fluorescence intensity for 7m-Rh and opsin were obtained by TIRFM images and analyzed in the same manner as that for photoactivated rhodopsin to generate the respective intensity change histograms (see Fig. 4, a and d). In addition, the photoactivated rhodopsin in DMPC liposome, in which conformational change is arrested at the Meta-I stage (Fig. S1 f, inset) (21), was also analyzed (Fig. 4 b). The bulk fluorescence measurement showed that the fluorescence increase upon the formation of Meta-I in DMPC liposomes was ∼5% (Fig. S1 f). This value is significantly smaller than that of Meta-I in asolectin liposomes (∼10%, Fig. S1 d, pH 9), indicating that the conformational equilibrium of Meta-I in DMPC liposomes (Meta-IDMPC) is biased to Flow as compared to that in asolectin liposomes.
Figure 4.
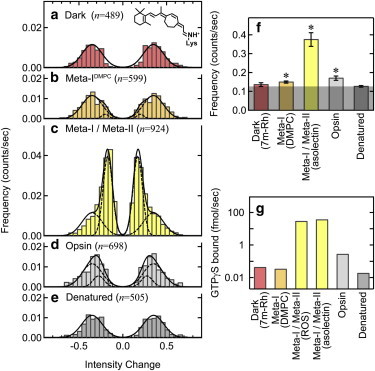
Correlation between frequencies of conformational changes and G protein activation efficiencies. Intensity change histograms for dark-state (7m-Rh) (a), Meta-I in DMPC liposome (b), Meta-I/Meta-II mixture in asolectin liposomes (c), opsin (d), and denatured rhodopsin (e). These histograms were fitted with two mirror-imaged pairs of Gaussian curves (Eq. 2), where the parameters for noise components were global (μ1 = 0.346, σ1 = 0.113, A1 = 0.012). Additional components were observed for Meta-IDMPC (μ2 = 0.201, σ2 = 0.041, A2 = 0.002), Meta-I/Meta-II (μ2 = 0.171, σ2 = 0.054, A2 = 0.040), and opsin (μ2 = 0.279, σ2 = 0.072, A2 = 0.005). (f) The frequency of fluorescence changes obtained by the summations of difference intensity histogram. Error bar indicates the standard deviation of two to five independent data sets. Frequency for the noise component is shown by shade. The asterisk indicates that the difference from denatured rhodopsin is significant (p < 0.05; Student’s t-test, two-tailed). (g) G protein activation efficiencies estimated by GTPγS binding assay. To see this figure in color, go online.
Fluorescence changes between Fhigh and Flow occurred with a significantly lower frequency in the dark-state, Meta-IDMPC, opsin, or denatured rhodopsin than in photoactivated rhodopsin (Meta-I/Meta-II mixture) (Fig. 4, a–e). These difference intensity histograms were global fitted with the sum of two mirror-imaged pairs of Gaussian functions (Eq. 2), where the parameters for the noise component (μ1, σ1, and A1) were set as global variables. The excellent result of the global fitting indicates that the quantity of the noise component was equal among the histograms in Fig. 4. The histogram of dark-state (7m-Rh) and denatured rhodopsin agreed with the noise component, which represents the frequency of false detection of conformational changes (Fig. 4, a and e). On the other hand, an additional Gaussian distribution was required to reproduce the histograms for opsin and Meta-IDMPC, indicating that conformational changes occur in these states. However, the intensity of the Gaussian distribution for conformational change of Meta-IDMPC and opsin (Fig. 4, b and d) was significantly smaller than that of photoactivated rhodopsin (Fig. 4 c). The percentages of the occurrence of fluorescence changes derived from conformational changes of Meta-IDMPC and opsin were estimated to be 6% and 27% of total occurrence of fluorescence changes, respectively. Due to this low occurrence of conformational changes, the retention times of Flow and Fhigh of opsin and Meta-IDMPC could not be reliably estimated. However, when we compared the frequencies of fluorescence changes by calculating the summations of these histograms (Fig. 4 f), we found that the frequencies of fluorescence changes for Meta-IDMPC and opsin were significantly higher than that of the dark-state or denatured rhodopsin. These results indicate that not only photoactivated rhodopsin but also Meta-IDMPC and opsin exhibited conformational interconversions between Flow and Fhigh.
Unlike Meta-I, Meta-II, and opsin, 7m-Rh did not show fluorescence changes due to conformational changes. It is important to note that frequency of fluorescence change in each state (Fig. 4 f) was consistent with the fluorescence increase observed in bulk measurements. Radionucleotide filter-binding assay showed that G protein activation efficiency of opsin was 1/100 of that of photoactivated rhodopsin in asolectin liposomes, whereas those of 7m-Rh and Meta-IDMPC were negligible (Fig. 4 g). This tendency is consistent with the frequency of fluorescence changes as well. These findings suggested that the active state (Fhigh) is accumulated by the increase of klow→high rather than the decrease of khigh→low (see Discussion).
Discussion
In this study, we observed the conformational changes of single rhodopsin molecules using Alexa594 as a fluorescent probe. We first monitored the conformational dynamics in single photoactivated rhodopsin molecules. The fluorescence intensity alternated between Fhigh and Flow, which are likely to correspond to active and inactive conformations, respectively. Such fluorescence alternation was also observed in the denatured rhodopsin and free Alexa594 molecule, implying that the fluorescence intensity steps detected by our analysis included stochastic noise and/or intrinsic fluorescence fluctuation of Alexa594 as well as conformational changes. However, the excellent result of global fitting of histograms of fluorescence changes with two mirror-imaged pairs of Gaussian distributions, in which the parameters for the noise component were global, demonstrated that the quantity of noise component was equal in all samples and the frequency of fluorescence changes derived from the conformational changes was quantitatively evaluated (Fig. 4).
When the fluorescence spots having ΔI within 0.167 ± 0.058 were chosen in photoactivated rhodopsin in asolectin liposome (Fig. 2 e), the occurrence of fluorescence changes derived from conformational changes was significantly greater than that from noise components (Fig. 2, f and g). Thus, the dynamics of the equilibrium between Fhigh and Flow was examined in detail for photoactivated rhodopsin. The difference between the pH-dependent profile of absorption (Meta-I/Meta-II, Fig. 3 f, red) and fluorescence (Flow/Fhigh, Fig. 3 f, blue) measurements indicates that Meta-I as well as Meta-II are not in a uniform conformation, but they are in an equilibrium between active and inactive conformations. Using the acidic and alkaline values obtained by fitting the absorption and fluorescence data with the Henderson-Hasselbalch equation and assuming that the pH-dependency of the ratio of Fhigh to Flow within Meta-I and Meta-II are negligible, the fractions of Fhigh in Meta-I and Meta-II were calculated to be 0.18 and 0.89, respectively. Therefore, Gibbs free energy difference (ΔG) for the formation of the active state is +3.6 kJ/mol when the ligand is a protonated all-trans-retinal (Meta-I), whereas ΔG is −4.9 kJ/mol when the ligand is a deprotonated all-trans-retinal (Meta-II) (Fig. 5), implying that the disruption of the ionic lock by the deprotonation of the chromophore decreases ΔG by 8.5 kJ/mol. Note that the conformation of Meta-IDMPC is biased to Flow and ΔG would be greater than that for Meta-I in asolectin liposomes.
Figure 5.
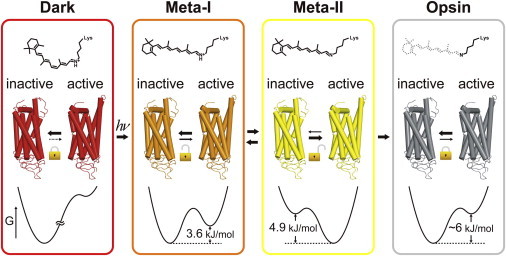
Activation mechanism of rhodopsin based on ligand-induced population shift. Meta-I, Meta-II, opsin, and possibly dark-state rhodopsin are in the equilibria between active and inactive conformations. Agonist all-trans retinal shifts the equilibrium toward active conformation by depressing ΔG, whereas inverse agonist 11-cis retinal shifts the equilibrium toward inactive conformation by elevating ΔG. Note that ΔG for Meta-IDMPC would be greater than that for Meta-I in asolectin liposomes. To see this figure in color, go online.
We have shown that the conformational equilibrium within individual photoactivated rhodopsin molecules can be observed by single-molecule analysis. Using this method, we then measured the possible conformational equilibrium in the dark-state, opsin, and Meta-IDMPC, which are thought to be single species. We found that although no fluorescence changes other than noise component were observed for the dark-state rhodopsin (containing 7m-Ret), Meta-IDMPC and opsin exhibited clear fluorescence changes, albeit with smaller frequency than photoactivated rhodopsin (Fig. 4 f). In addition, Meta-I and Meta-II in asolectin liposome are also in the equilibrium between Flow and Fhigh, as shown by the deviation of pH-dependence between Flow/Fhigh and Meta-I/Meta-II (Fig. 3 f), indicating that rhodopsin molecule intrinsically adopts both active and inactive conformation. Interestingly, the frequencies of conformational changes correlated with the fluorescence increase observed in bulk measurements and the G protein activation efficiencies (Fig. 4 g). G protein activity should linearly correlate with the fraction of Fhigh in the mixture of Flow and Fhigh,
whereas frequency of fluorescence changes is given by the sum of frequency from Flow to Fhigh
and that from Fhigh to Flow
These equations imply that the fraction of Fhigh is increased by the increase of klow→high or the decrease of khigh→low, whereas the increases of klow→high and khigh→low both result in the increase of frequency of fluorescence changes. Therefore, it is suggested that the active state (Fhigh) is accumulated by the increase of klow→high rather than the decrease of khigh→low. Rhodopsin is locked in the inactive conformation mainly by an ionic lock between Arg-135 in the conserved ERY motif (Helix 3) and Glu-247 (Helix 6). It is proposed that this ionic lock is coupled with the ionic bond between protonated Schiff base of the chromophore and its counterion Glu-113 (Helix 3) (39,40), which is replaced by the ionic bond between Glu-113 and Lys-296 in opsin. Our current findings of the presence of Fhigh and Flow within one species suggest that the ionic lock and chromophore are not strictly coupled. Because Alexa594 is likely to be sensitive to the disruption of the ionic lock, the rate of the disruption of the ionic lock would be regulated by the presence/absence, configuration, or the protonation state of the chromophore.
Fourier transform infrared spectroscopy experiments have suggested that opsin adopts a Meta-II-like conformation at acidic pH (pKa = 4.1) (41), however the amount of active conformation would be quite low in our experimental condition (pH 6–9) (41). In addition, crystal structure of opsin was similar to that of Meta-II (42), although the structure of C-terminal ∼20 amino acid residues was not resolved. These findings suggest that the conformation of Fhigh of opsin is similar to Fhigh of the photoactivated rhodopsin. However, the difference in the fluorescence intensities between Fhigh and Flow (mean difference intensity, μ) for opsin (μ = ±0.279) was greater than that for photoactivated rhodopsin (μ = ±0.171) (Fig. 4, d and c). This difference suggests that the difference in local conformations between Fhigh of Meta-II and that of opsin around Cys-316 may be one of the reasons why the relation between the frequency and the G protein activation efficiency was not linear. The bulk fluorescence measurements demonstrated that the fluorescence increase upon formation of opsin is ∼60% of that of Meta-I (Fig. S1 d, pH 9.0). If two-state equilibrium of opsin, similar to that of Meta-I, is assumed and the difference in μ is taken into consideration, ΔG of opsin is calculated to be ∼6 kJ/mol (Fig. 5).
Unlike Meta-I, Meta-II, and opsin, fluorescence alternation due to the conformational changes was not observable for dark-state rhodopsin (7m-Rh). This suggests that conformational conversion to Fhigh is suppressed by binding to 11-cis-retinal, which functions as an inverse agonist. It is known that the extremely low G protein activation efficiency of dark state rhodopsin accounts for the low dark noise of rod cells so as to function as a single photon detector. Low G protein activation efficiency of the dark state rhodopsin is explained by the low rate of thermal isomerization of the chromophore, but it would be also caused by the suppression of the generation of Fhigh.
Our current results show that the conformational dynamics between Flow and Fhigh is essential for the activation and inactivation of rhodopsin. The retinal chromophore changes its potency from inverse-agonist to agonist through photoisomerization. In agreement with the potency of the ligand, the ΔG between active and inactive conformations is changed and the equilibrium is shifted (Fig. 5). The ligand-induced population shift during the activation process of rhodopsin substantiates the conceptual two-state model, which can explain characteristic biological responses of GPCRs to several pharmacologically distinguishable ligands (43–46).
The present single-molecule study successfully monitored the dynamics within individual rhodopsin molecules and quantitatively analyzed these conformational equilibria. Accumulated evidence about crystal structures of GPCRs revealed that the helical arrangements of active and inactive conformations in rhodopsin are similar to those in other GPCRs. The present single-molecule analysis using a fluorophore-labeled receptor molecule is applicable to the evaluation of conformational fluctuation of active and inactive states in a variety of GPCRs. Further combination of these two techniques would accelerate extending the theoretical framework of activation to better explain drug behavior.
Acknowledgments
We are grateful to Dr. Koji Nakanishi at Columbia University for provision of the 11-cis-locked 7-membered ring retinal. We thank Dr. Yoshinori Fujiyoshi at Nagoya University for technical support and helpful discussion. We also thank Dr. Takesi Matsuyama at Kyoto University for critical reading of our manuscript and invaluable comments.
This work was supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) [Grants-in-Aid for Scientific Research to T.Y. (25440167), Y.Shichida. (24121716, 25251036), and Y.I. (23370070), and Grants for Excellent Graduate Schools].
Supporting Material
References
- 1.Shichida Y., Imai H. Visual pigment: G-protein-coupled receptor for light signals. Cell. Mol. Life Sci. 1998;54:1299–1315. doi: 10.1007/s000180050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kandori H., Shichida Y., Yoshizawa T. Photoisomerization in rhodopsin. Biochemistry (Mosc.) 2001;66:1197–1209. doi: 10.1023/a:1013123016803. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann K.P. Effect of GTP on the rhodopsin-G-protein complex by transient formation of extra metarhodopsin II. Biochim. Biophys. Acta. 1985;810:278–281. doi: 10.1016/0005-2728(85)90143-4. [DOI] [PubMed] [Google Scholar]
- 4.Choe H.W., Kim Y.J., Ernst O.P. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 5.Standfuss J., Edwards P.C., Schertler G.F. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen S.G., Choi H.J., Kobilka B.K. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen S.G., DeVree B.T., Kobilka B.K. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Struts A.V., Salgado G.F., Brown M.F. Retinal dynamics underlie its switch from inverse agonist to agonist during rhodopsin activation. Nat. Struct. Mol. Biol. 2011;18:392–394. doi: 10.1038/nsmb.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angel T.E., Gupta S., Chance M.R. Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc. Natl. Acad. Sci. USA. 2009;106:14367–14372. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orban T., Jastrzebska B., Palczewski K. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012;20:826–840. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye S., Huber T., Sakmar T.P. FTIR analysis of GPCR activation using azido probes. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozuka J., Yokota H., Yanagida T. Dynamic polymorphism of single actin molecules in the actin filament. Nat. Chem. Biol. 2006;2:83–86. doi: 10.1038/nchembio763. [DOI] [PubMed] [Google Scholar]
- 13.Arai Y., Iwane A.H., Yanagida T. Dynamic polymorphism of Ras observed by single molecule FRET is the basis for molecular recognition. Biochem. Biophys. Res. Commun. 2006;343:809–815. doi: 10.1016/j.bbrc.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 14.Teramura Y., Ichinose J., Sako Y. Single-molecule analysis of epidermal growth factor binding on the surface of living cells. EMBO J. 2006;25:4215–4222. doi: 10.1038/sj.emboj.7601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiroshima M., Saeki Y., Sako Y. Dynamically varying interactions between heregulin and ErbB proteins detected by single-molecule analysis in living cells. Proc. Natl. Acad. Sci. USA. 2012;109:13984–13989. doi: 10.1073/pnas.1200464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibino K., Shibata T., Sako Y. Activation kinetics of RAF protein in the ternary complex of RAF, RAS-GTP, and kinase on the plasma membrane of living cells: single-molecule imaging analysis. J. Biol. Chem. 2011;286:36460–36468. doi: 10.1074/jbc.M111.262675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews R.G., Hubbard R., Wald G. Tautomeric forms of metarhodopsin. J. Gen. Physiol. 1963;47:215–240. doi: 10.1085/jgp.47.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamoto Y., Kataoka M., Palczewski K. Light-induced conformational changes of rhodopsin probed by fluorescent alexa594 immobilized on the cytoplasmic surface. Biochemistry. 2000;39:15225–15233. doi: 10.1021/bi0018685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao B., Tsuda M., Nakanishi K. Flash photolysis and low temperature photochemistry of bovine rhodopsin with a fixed 11-ene. Biophys. J. 1981;35:543–546. doi: 10.1016/S0006-3495(81)84809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akita H., Tanis S.P., Nakanishi K. Non-bleachable rhodopsins retaining the full natural chromophore. J. Am. Chem. Soc. 1980;102:6370–6372. [Google Scholar]
- 21.Baldwin P.A., Hubbell W.L. Effects of lipid environment on the light-induced conformational changes of rhodopsin. 1. Absence of metarhodopsin II production in dimyristoylphosphatidylcholine recombinant membranes. Biochemistry. 1985;24:2624–2632. doi: 10.1021/bi00332a006. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita T., Terakita A., Shichida Y. Distinct roles of the second and third cytoplasmic loops of bovine rhodopsin in G protein activation. J. Biol. Chem. 2000;275:34272–34279. doi: 10.1074/jbc.M002954200. [DOI] [PubMed] [Google Scholar]
- 23.Rabiner L.R. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- 24.Bronson J.E., Fei J., Wiggins C.H. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkes J.H., Liebman P.A. Temperature and pH dependence of the metarhodopsin I-metarhodopsin II kinetics and equilibria in bovine rod disk membrane suspensions. Biochemistry. 1984;23:5054–5061. doi: 10.1021/bi00316a035. [DOI] [PubMed] [Google Scholar]
- 26.Sato K., Morizumi T., Shichida Y. Direct observation of the pH-dependent equilibrium between metarhodopsins I and II and the pH-independent interaction of metarhodopsin II with transducin C-terminal peptide. Biochemistry. 2010;49:736–741. doi: 10.1021/bi9018412. [DOI] [PubMed] [Google Scholar]
- 27.Mansoor S.E., Palczewski K., Farrens D.L. Rhodopsin self-associates in asolectin liposomes. Proc. Natl. Acad. Sci. USA. 2006;103:3060–3065. doi: 10.1073/pnas.0511010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fotiadis D., Jastrzebska B., Engel A. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr. Opin. Struct. Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Knierim B., Hofmann K.P., Hubbell W.L. Sequence of late molecular events in the activation of rhodopsin. Proc. Natl. Acad. Sci. USA. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahalingam M., Martínez-Mayorga K., Vogel R. Two protonation switches control rhodopsin activation in membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17795–17800. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaitseva E., Brown M.F., Vogel R. Sequential rearrangement of interhelical networks upon rhodopsin activation in membranes: the Meta IIa conformational substate. J. Am. Chem. Soc. 2010;132:4815–4821. doi: 10.1021/ja910317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surya A., Foster K.W., Knox B.E. Transducin activation by the bovine opsin apoprotein. J. Biol. Chem. 1995;270:5024–5031. doi: 10.1074/jbc.270.10.5024. [DOI] [PubMed] [Google Scholar]
- 33.Jäger S., Palczewski K., Hofmann K.P. Opsin/all-trans-retinal complex activates transducin by different mechanisms than photolyzed rhodopsin. Biochemistry. 1996;35:2901–2908. doi: 10.1021/bi9524068. [DOI] [PubMed] [Google Scholar]
- 34.Han M., Lou J., Smith S.O. Partial agonist activity of 11-cis-retinal in rhodopsin mutants. J. Biol. Chem. 1997;272:23081–23085. doi: 10.1074/jbc.272.37.23081. [DOI] [PubMed] [Google Scholar]
- 35.Surya A., Knox B.E. Modulation of opsin apoprotein activity by retinal. Dark activity of rhodopsin formed at low temperature. J. Biol. Chem. 1997;272:21745–21750. doi: 10.1074/jbc.272.35.21745. [DOI] [PubMed] [Google Scholar]
- 36.Robinson P.R., Cohen G.B., Oprian D.D. Constitutively active mutants of rhodopsin. Neuron. 1992;9:719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- 37.Cohen G.B., Yang T., Oprian D.D. Constitutive activation of opsin: influence of charge at position 134 and size at position 296. Biochemistry. 1993;32:6111–6115. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- 38.Han M., Smith S.O., Sakmar T.P. Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry. 1998;37:8253–8261. doi: 10.1021/bi980147r. [DOI] [PubMed] [Google Scholar]
- 39.Vogel R., Sakmar T.P., Siebert F. Coupling of protonation switches during rhodopsin activation. Photochem. Photobiol. 2007;83:286–292. doi: 10.1562/2006-06-19-IR-937. [DOI] [PubMed] [Google Scholar]
- 40.Vogel R., Mahalingam M., Sakmar T.P. Functional role of the “ionic lock”—an interhelical hydrogen-bond network in family A heptahelical receptors. J. Mol. Biol. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Vogel R., Siebert F. Conformations of the active and inactive states of opsin. J. Biol. Chem. 2001;276:38487–38493. doi: 10.1074/jbc.M105423200. [DOI] [PubMed] [Google Scholar]
- 42.Park J.H., Scheerer P., Ernst O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 43.Lefkowitz R.J., Cotecchia S., Costa T. Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol. Sci. 1993;14:303–307. doi: 10.1016/0165-6147(93)90048-O. [DOI] [PubMed] [Google Scholar]
- 44.Samama P., Cotecchia S., Lefkowitz R.J. A mutation-induced activated state of the β2-adrenergic receptor. Extending the ternary complex model. J. Biol. Chem. 1993;268:4625–4636. [PubMed] [Google Scholar]
- 45.Robertson M.J., Dougall I.G., Leff P. Agonist-antagonist interactions at angiotensin receptors: application of a two-state receptor model. Trends Pharmacol. Sci. 1994;15:364–369. doi: 10.1016/0165-6147(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 46.Leff P. The two-state model of receptor activation. Trends Pharmacol. Sci. 1995;16:89–97. doi: 10.1016/s0165-6147(00)88989-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


