Abstract
S-glutathionylation (SSG) is an important regulatory posttranslational modification on protein cysteine (Cys) thiols, yet the role of specific cysteine residues as targets of modification is poorly understood. We report a novel quantitative mass spectrometry (MS)-based proteomic method for site-specific identification and quantification of S-glutathionylation across different conditions. Briefly, this approach consists of initial blocking of free thiols by alkylation, selective reduction of glutathionylated thiols and covalent capture of reduced thiols using thiol affinity resins, followed by on-resin tryptic digestion and isobaric labeling with iTRAQ (isobaric tags for relative and absolute quantitation) for MS-based identification and quantification. The overall approach was initially validated by application to RAW 264.7 mouse macrophages treated with different doses of diamide to induce glutathionylation. A total of 1071 Cys-sites from 690 proteins were identified in response to diamide treatment, with ~90% of the sites displaying >2-fold increases in SSG-modification compared to controls. This approach was extended to identify potential SSG- modified Cys-sites in response to H2O2, an endogenous oxidant produced by activated macrophages and many pathophysiological stimuli. The results revealed 364 Cys-sites from 265 proteins that were sensitive to S-glutathionylation in response to H2O2 treatment, thus providing a database of proteins and Cys-sites susceptible to this modification under oxidative stress. Functional analysis revealed that the most significantly enriched molecular function categories for proteins sensitive to SSG modifications were free radical scavenging and cell death/survival. Overall the results demonstrate that our approach is effective for site-specific identification and quantification of SSG-modified proteins. The analytical strategy also provides a unique approach to determining the major pathways and cellular processes most susceptible to S-glutathionylation under stress conditions.
Keywords: S-glutathionylation, redox regulation, resin assisted enrichment, macrophage, proteomics, hydrogen peroxide, protein thiols
Introduction
The importance of reactive oxygen species (ROS) and reactive nitrogen species (RNS) as second messengers in signal transduction has recently gained recognition [1, 2]. Reversible posttranslational modifications of protein cysteine thiols represent a major form of cellular regulation mediated by ROS and RNS in redox signaling [1, 3–5]. The formation of mixed disulfides between protein cysteine thiols and cellular glutathione (GSH), known as protein S-glutathionylation (SSG), is one of the most prevalent forms of reversible posttranslational modifications of protein thiols. Emerging evidence supports the significance of protein-SSG in regulating a variety of cellular processes from bacteria to mammals, including human pathologies under oxidative and nitrosative stress [6–9].
Protein-SSG can be induced by ROS or RNS under physiological or pathological conditions. Although not exactly resolved, a number of potential mechanisms that either occur spontaneously or are catalyzed by enzymes such as glutaredoxins (Grx) have been recognized for the formation of protein-SSG [1, 8, 10], including: 1) protein thiols react with glutathione disulfide (GSSG) via thiol-disulfide exchange reaction; 2) protein thiol or GSH reacts with the corresponding oxidized thiol derivatives (e.g., S-nitroso, sulfenic acid, thiyl radical, etc). Conversely, the protein SSG-modification can be reversed by means of reactions catalyzed by the thiol-disulfide oxidoreductases glutaredoxins (Grx), and potentially other enzymes [10].
A growing number of proteins have been identified as regulated by SSG covering a wide spectrum of cellular signaling pathways [6, 11]. Examples of reported SSG-modified proteins include enzymes with active-site thiols such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH) [12] and caspase-3 [13], signaling proteins such as protein kinase A [14] and protein kinase C [15], transcription factors c-Jun and NF-κB [16–18], ion channels and calcium dependent proteins such as sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA) [19, 20], and apoptotic death receptor protein Fas (CD95) [21]. Nevertheless, our knowledge of the relevance of S-glutathionylation in physiological and pathological processes is still limited due to the lack of effective approaches for the identification and quantification of protein-SSGs and their specific modification sites. One early established method utilizes in vivo metabolic labeling of GSH with [35S] cysteine coupled with SDS-PAGE separation and autoradiography for the detection of modified thiols [22–24]. Another conventional method is based on western blot coupled with anti-GSH antibodies [25] or biotinylated glutathione S-transferase and anti-biotin antibodies [26]. However, these methods have limited specificity and sensitivity and are unable to distinguish individual S-glutathionylated (SSG) sites within a target protein which may have different functional consequences.
Mass spectrometry (MS)-based proteomics coupled with affinity or chemical enrichment strategies can overcome these limitations and enable large-scale identification of specific sites subject to modifications. Recent approaches for identification of glutathionylated proteins have been reported which incorporate a biotin tag via an exogenous glutathionylation reagent [27–29] or via a modified biotin-switch technique involving selective reduction and immediate alkylation of protein-SSG sites [30] followed by avidin-biotin-based enrichment. These former methods involve the reaction of cysteine thiols with biotinylated GSSG or similar reagents to form protein-SSGs, which might not reflect the true endogenous level of SSG modifications. The effectiveness of the modified biotin switch technique was also not demonstrated for identification of specific sites of modification. Moreover, there are currently no effective approaches for quantitative measurement of the dynamic changes of S-glutathionylation at a broad proteome scale. It will contribute greatly to an increased understanding of the biological role of S-glutathionylation if a more sensitive detection method for site-specific identification and quantification of SSG modified proteins were available.
Herein we report a quantitative MS-based proteomic method for profiling protein-SSGs and their specific modification sites by adapting a recently developed resin-assisted enrichment method used for S-nitrosylation [31, 32] with on-resin isobaric labeling with iTRAQ (isobaric tags for relative and absolute quantitation) reagents. A number of previous studies have reported that the resin-assisted covalent enrichment offers a simpler, more efficient means of capturing cysteine-containing peptides [33] and other PTMs such as S-nitrosylation [32, 34]. The resin-assisted enrichment minimizes the degree of non-specific binding that is often encountered with non-covalent avidin-biotin enrichment, thus providing an overall better specificity and sensitivity [31–33]. This approach was initially validated and applied to RAW 264.7 macrophage cells treated with diamide and H2O2 to identify potential cysteine redox switches that are sensitive to S-glutathionylation. Macrophage cells are selected as a model due to the potential significance of redox regulation in oxidative stress response and inflammation [16]. The capacity of macrophages to generate substantial amounts of ROS is an important property of their activation by foreign particulates and pathogens. Although macrophages must deal with high oxidative stress levels, surprisingly little is known about the specific macrophage proteins susceptible to SSG modification and the potential signaling pathways impacted. We identified 364 SSG-modified Cys-sites from 265 proteins in macrophages that were sensitive to S-glutathionylation in response to H2O2 treatment. These SSG-modified proteins cover a range of enzymes involved in ROS metabolism, stress response signaling, and apoptosis pathways.
Materials and Methods
Materials
E. coli Glutaredoxin (Grx 3) [C14S/C65Y] was from IMCO Corporation Ltd AB (Stockholm, Sweden). Glutathione Reductase (GR) was from Roche Diagnostics Corporation (Indianapolis, IN). NADPH tetrasodium Salt (β-NADPH·4Na, β-Nicotinamide adenine dinucleotide phosphate (reduced form)·4Na), BCA protein assay reagents, silver stain kit, spin columns, cell culture RPMI-1640 media and reagents (penicillin, streptomycin, L-glutamine), and hydrogen peroxide were purchased from Thermo Fisher Scientific (Rockford, IL). Sequencing grade modified porcine trypsin was from Promega (Madison, WI). iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) reagents were from AB SCIEX (Foster City, CA). The SeeBlue Plus2 protein standard was from Invitrogen (Carlsbad, CA). Thiol-affinity resin (Thiopropyl Sepharose 6B) was from GE Healthcare (Uppsala, Sweden). Tris/glycine/SDS (TGS) buffer, Laemmli sample loading buffer, and Tris-HCl precast gel with 4–20% linear gradient were all from Bio-Rad Laboratories (Hercules, CA). Unless otherwise noted, all other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Cell culture, diamide and hydrogen peroxide treatments, and protein extraction
Murine RAW 264.7 macrophages (TIB-71) from American Type Culture Collection (ATCC) (Manassas, VA, USA) were cultured and maintained in 100 mm culture plates with RPMI-1640 media containing 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. Cells were cultured at 37 °C with 5% CO2. Prior to treatment, cells were seeded into 100 mm culture plates and grown until 80% confluent. Original growth media was removed and replaced with media containing either hydrogen peroxide or diamide at the desired concentration for 30 min. After treatment, cells were rinsed twice with cold RPMI-1640 media containing no supplements and harvested in lysis buffer (50 mM HEPES, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, pH 7.7) containing freshly prepared 50 mM N-ethylmaleimide (NEM). Cell lysates were centrifuged at 14,000 rpm at 4° C, for 10 min and soluble protein fraction was retained. Protein concentration was determined using the BCA assay.
De-nitrosylation and alkylation of free thiols
To block free protein thiols, ~ 1 mg of the above lysates were resuspended to a final protein concentration of 0.5 μg/μL in alkylation buffer consisting of 250 mM HEPES (pH 7.7), 20 mM NEM and 2% SDS. 1 mM sodium ascorbate (NaASC) and 1μM CuCl were added to the alkylation buffer to reduce S-nitrosylated cysteines back to free cysteines. Both free cysteines and S-nitrosylated cysteines (reduced to free cysteines) were alkylated. The alkylation reaction was carried out in 4 mL Amicon Ultra 30K molecular weight cut-off (MWCO) filter units (EMD Millipore, Billerica, MA) in a thermomixer (Fisher Scientific, Pittsburgh, PA) in the dark at 55 °C and 850 rmp for 30 min. Excess reagents were removed by buffer exchange with 8 M urea 3 times and once with water. The protein concentration of the concentrated samples was measured by the BCA assay.
Selective reduction of protein-SSG
To selectively reduce glutathionylated proteins, ~480 μg of the above alkylated samples were diluted to a final concentration of 1 μg/μL in 25 mM HEPES (pH 7.7) followed by the addition of 2.5 μg/mL Grx3, 0.5 mM GSH, 1 mM NADPH, and 4 U/mL of GR in 1.5 mL Eppendorf centrifuge tubes. Samples were incubated at 37 °C for 10 min, and transferred to 0.5 mL Amicon Ultra 10K filters on ice. Excess reagents were removed by buffer exchange with 8 M urea three times, resulting in a final volume of 30–40 μL. Protein concentrations of the de-glutathionylated samples were measured by the BCA assay.
Resin-assisted enrichment, on-resin digestion, iTRAQ labeling, and peptide elution
Following selective reduction, the formerly glutathionylated proteins (350 μg) were diluted to ~120 μL with 25mM HEPES buffer with 0.2% SDS and loaded to Handee Mini-Spin columns with 35 mg of pre-washed Thiopropyl Sepharose 6B resin. The de-glutathionylated proteins containing free thiols were covalently captured by the resin through the formation of mixed disulfides as described previously [31, 33]. Enrichment was carried out in a thermomixer at room temperature with shaking at 850 rpm for 2 h. Nonspecifically bound proteins were removed by washing the resin five times with the following solutions: (1) 8M urea, (2) 80% acetonitrile (ACN) with 0.1% trifluoroacetic acid (TFA), and (3) 25 mM HEPES (pH 7.7).
On-resin protein digestion was performed at 37° C with shaking at 850 rpm for 3 h in ~120 μL of digestion buffer containing 25 mM HEPES (pH 7.7), 0.1% SDS, 7 μg trypsin, and 1 mM CaCl2. Non-specifically bound peptides were again removed by washing with the following solutions: 1) 2 M NaCl (5X), 2) 80% ACN with 0.1% TFA (5X), 3) 25 mM HEPES (pH 7.7) (3X), and 4) 50mM triethylammonium bicarbonate (TEAB) buffer (pH 8.5) (2X). On-resin isobaric labeling with iTRAQ reagents was performed as previously described [31]. Briefly, 140 μL of ethanol was added to the manufacturer-provided iTRAQ reagent vials. Thirty μL of dissolution buffer and 75 μL of the iTRAQ reagent solutions were added to the spin columns. The labeling reaction was carried out at room temperature for 1 h. The reaction was stopped by adding 8 μL of 5% NH2OH·HCl in 200 mM TEAB buffer for 15 min. The excess iTRAQ reagents were removed by washing five times each with 1) 80% ACN with 0.1% TFA and 2) 25 mM ammonium bicarbonate.
Labeled cysteine containing peptides (Cys-peptides) were eluted by incubation with 20 mM dithiothreitol (DTT) in 100 μL of 25 mM ammonium bicarbonate twice at room temperature for 30 min and10 min, respectively. Any potential remaining peptides were further eluted by incubation with 100 μL of 80% ACN/0.1% TFA for 10 min. All eluted Cys-peptide samples were combined, and concentrated in a Thermo Scientific Savant SpeedVac® concentrator and adjusted to a final volume of 30 μL with water. The eluted Cys-peptide samples from each iTRAQ labeling channel were combined with equal amounts of each labeled sample and cleaned up with Omix C18 (10 μL) tips (Agilent Technologies, Walnut Creek, CA) according to the manufacturer protocol. The final eluted peptides were concentrated down to ~15 μL and DTT was added to reach 20 mM final concentration to prevent the oxidation of cysteines prior to LC-MS/MS analysis.
SDS-polyacrylamide gel electrophoresis (PAGE) and Western Blot
For protein-level SDS-PAGE, glutathionylated proteins were directly eluted from the resin using 20 mM DTT as stated above. To perform SDS-PAGE analysis, sample volumes were adjusted so that equal amounts of eluted proteins or final eluted peptides were diluted in Laemmli sample loading buffer containing 5 mM TCEP and heated at 95°C for 5 min. Samples were separated electrophoretically on either a 4–12% (for proteins) or a 4–20% (for peptides) Tris-HCl polyacrylamide gel in Tris-glycine/SDS (TGS) running buffer. Separated proteins/peptides were then subjected to silver staining according to the manufacturer’s instructions.
For Western Blot analysis, equal amounts of isolated proteins were diluted in LDS sample buffer and separated electrophoretically under non-reducing conditions on a 4–12% Bis-Tris polyacrylamide gel in MES SDS running buffer (Invitrogen, Carlsbad, CA). Separated proteins were then subjected to electrophoretic transfer to a polyvinylidene difluoride (PVDF) membrane (Invitrogen, Carlsbad, CA). Membranes were blocked for 1 h at room temperature in a 5% solution of membrane blocking agent (GE Healthcare, Pittsburgh, PA) in TBS-T (Tris buffered saline with 1% Tween-20). To measure the total protein-SSG level, a primary mouse monoclonal anti-glutathione antibody, IgG2a, (Virogen, Watertown, MA) was added to membranes at a 1:1000 dilution and membranes were incubated overnight at 4°C with gentle rocking. Membranes were washed in TBS-T before being incubated with the peroxidase-conjugated goat anti-mouse IgG (Sigma-Aldrich) secondary antibody at 1:2000 in TBS-T for 1 hr at room temperature. The membranes were washed in TBS-T prior to development with SuperSignal West Femto ECL (enhanced chemiluminescent substrate) (Thermo Scientific, Rockford, IL) according to the manufacturer’s instructions.
To measure the level of SSG modification of specific proteins, equal amounts of total proteins were first subjected to SSG enrichment using thiol-affinity resin and eluted at the protein level. Equal volumes of enriched proteins were separated on a 4–12% Bis-Tris polyacrylamide gel and transferred to a PVDF membrane. Membranes intended for probing with mouse anti-GAPDH primary antibody (Life Technologies, Grand Island, NY) and rabbit anti-thioredoxin (TXN) primary antibody (Abcam, Cambridge, MA) were blocked for 1 hr at room temperature in a 1% w/v solution of dry milk (LabTechnologies, Livingston, NJ)) in PBS. Membranes intended for probing with rabbit anti-Annexin A1(ANXA1) primary antibody (Cell Signaling, Danvers, MA) and mouse anti- peroxiredoxin 3(PRDX3) primary antibody (Abcam, Cambridge, MA) were blocked for 1 hr at room temperature in a 5% w/v solution of dry milk (LabTechnologies, Livingston, NJ)) in TBS-T. GAPDH and TRX antibodies were diluted 1:1000 in a 1% w/v solution of dry milk in PBS, while ANXA1 and PRDX3 antibodies were diluted 1:1000 in a 5% w/v solution of bovine serum albumin (BSA) in TBS-T. Membranes were then incubated with the intended primary antibody overnight at 4°C with gentle rocking. Membranes were washed as previously described and incubated with the appropriate peroxidase-conjugated secondary antibody for 1 h at room temperature; either goat anti-rabbit IgG (Jackson Laboratories, Bar Harbor, Maine) diluted 1:4000 in a 1% w/v solution of dry milk in PBS, or goat anti-mouse IgG (Thermo Fisher Scientific, Rockford, IL) diluted 1:4000 in a 5% w/v solution of dry milk in TBS-T. Membranes were washed as previously described and developed using SuperSignal West Femto ECL according to the manufacturer’s instructions.
LC-MS/MS analyses
All peptide samples were analyzed by a Waters nanoAquity UPLC® system (Waters Corporation, Milford, MA) with a reversed-phase capillary HPLC column manufactured in-house using 3-μm Jupiter C18 stationary phase packed into a 70-cm length of 360 μm o.d. × 75 μm i.d. fused silica capillary tubing. The column was equilibrated with 99 % mobile phase A (0.1% formic acid in water) and 1% mobile phase B (0.1% formic acid in acetonitrile) conditions prior to sample injection onto a 5 μL sample loop. The gradient ramps from mobile phase A to 95% of mobile phase B over 3 h with a flow rate of 300 nL/min. MS analysis was performed using a LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) coupled with a custom electrospray ionization (ESI) interface as previously described [31]. Full MS spectra were collected at a resolution of 60K over the range of m/z 400–2000 with an automated gain control (AGC) value of 1 × 106 followed by data-dependent orbitrap HCD MS/MS for the top six abundant parent ions with an AGC target value of 5 × 104 and a normalized collision energy setting of 45%. Precursor ion activation was performed with an isolation width of 2.5 Da. The ion transfer tube temperature and spray voltage were 350 °C and 2.2 kV respectively. A dynamic exclusion time of 60 sec was used.
Data Analysis
MS/MS data analyses were conducted as previously described [31]. Briefly, MS/MS spectra were identified by SEQUEST (version 27, revision 12) search against the Uniprot mouse protein database (released on May 05, 2010). The key search parameters used were: 50 ppm tolerance for precursor ion masses, 0.05 Da for fragment ion masses, a maximum of 2 missed tryptic cleavages, dynamic oxidation of methionine (+15.9949 Da), dynamic NEM modification of cysteine (+125.0477 Da), and static iTRAQ modification of lysine and N-termini (+144.1021 Da). To calculate the false discovery rate (FDR), the decoy-database searching methodology [35, 36] was used for the SEQUEST search and MS Generating-Function (MSGF) scores were generated for each identified spectrum by computing rigorous p-values (spectral probabilities) [37]. To achieve <1% FDR for peptide identifications, the following criteria were applied: (1) for those with mass measurement error within 5 ppm, fully tryptic peptides with MSGF score <1E-8 and partially tryptic peptides with MSGF score <1E-10; (2) for those with mass measurement error >5 ppm (presumably due to picking the wrong monoisotopic peak), only fully tryptic peptides with a MSGF score <1E-10. Functional and protein interaction network analysis were performed using Ingenuity Pathway Analysis (IPA) (www.ingenuity.com).
Results
Overall analytical strategy and initial method optimization
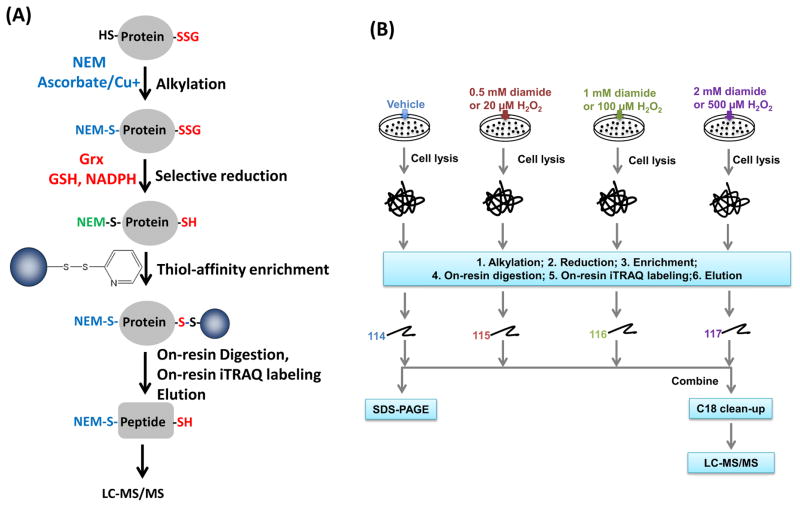
To enable identification and quantification of SSG at specific modified Cys-sites, we adapted a recently developed approach used for quantitative site-specific reactivity profiling of S-nitrosylation [31]. As shown in Fig. 1A, free thiols were initially blocked by alkylation with NEM. Formerly S-nitrosylated cysteines were also reduced by selective reduction using ascorbate and subsequently blocked by NEM. SSG-modified proteins were selectively reduced by a reduction cocktail containing the Grx enzyme, GSH, and NADPH. The newly formed free thiols were then specifically captured by Thiopropyl Sepharose 6B resin. The enriched proteins were subjected to on-resin trypsin digestion, followed by the removal of non-Cys-peptides. Resin bound Cys-peptides can be further labeled with amine-reactive isobaric labeling reagents such as iTRAQ, tandem mass tags (TMT) [38], or deuterium isobaric amine-reactive tags [39] to facilitate peptide quantification. In this work, iTRAQ reagents were used for labeling. The iTRAQ labeled Cys-peptides were subjected to LC-MS/MS analyses for site-specific identification and quantification of the SSG modified cysteines. Fig. 1B illustrates the details of quantification strategy across multiple conditions for cells treated with either diamide or hydrogen peroxide (H2O2), for induction of S-glutathionylation.
Fig. 1.
(A) Strategy for enriching and site-specific identification of SSG-modified Cys-peptides. Ascorbate coupled with CuCl was used for selective reduction of SNO. NEM was used to alkylate free thiols. The cocktail of Grx, GSH, GR, and NADPH was used for selectively reducing protein-SSG. iTRAQ labeling of enriched Cys-peptides was carried out on-resin, followed by DTT elution. (B) Experimental strategy for multiplex quantification of SSG-modified peptides. Cells were treated by different doses of exogenous stimuli. After on-resin iTRAQ labeling, the eluted peptides are subjected to either SDS-PAGE or combined for LC-MS/MS analyses.
To assess the effectiveness of the overall approach, RAW 264.7 murine macrophage cells were initially treated with different doses of diamide (0, 0.5, 1, and 2 mM), a chemical oxidizing agent, for 30 min to induce S-glutathionylation of proteins. The dose-dependent induction of S-glutathionylation by diamide [40] was confirmed by Western Blot using anti-GSH antibody (Fig. 2A). Based on this observation, 1 mM diamide induced a clear level of S-glutathionylation, which was the condition chosen for further optimization of selective reduction of S-glutathionylation.
Fig. 2.
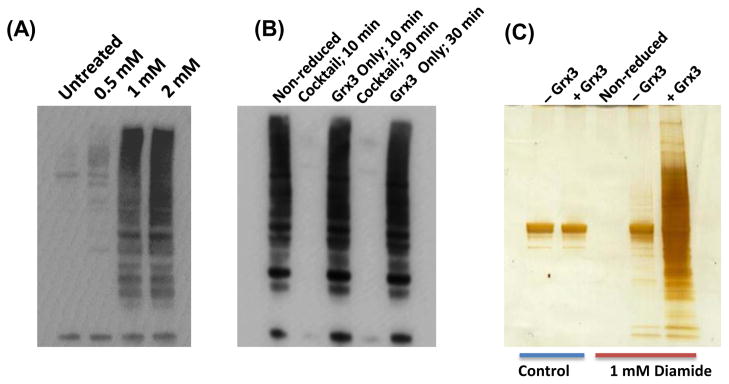
Optimization of reduction conditions. (A) Anti-SSG Western blot image of cell lysates from macrophages treated with different concentrations of diamide. (B) Anti-SSG Western blot images of non-reduced and reduced cell lysates from diamide treated macrophages. All cells were treated with 1 mM diamide followed by enrichment of cellular proteins using Grx reduction cocktail or using Grx only. (C) SDS-PAGE of enriched proteins. Samples were reduced by the complete reduction cocktail or with Grx3 omitted followed by resin-assisted enrichment and protein-level elution. The reduction cocktail contains 2.5 μg/mL Grx3, 0.5 mM GSH, 1 mM NADPH, and 4 U/mL of GR.
To evaluate the reduction efficiency, cell lysates were subjected to alkylation with NEM to block the free thiols, and the SSG-modified Cys-sites were selectively reduced by Grx3 [C14S/C65Y], a mutated form of E. coli Grx3 [41], in the presence of GSH, NADPH, and GR. As shown in Fig. 2B, Grx alone did not reduce the SSG-modified proteins significantly for either a 10 or 30 min incubation based on the anti-GSH Western Blot; however, the reduction cocktail of Grx3 in the presence of GSH, NADPH, and GR led to complete reduction within the 10 min incubation. The 10 minute incubation time using the reduction cocktail was chosen as the optimal reduction condition and used for all subsequent experiments.
To examine the specificity of SSG reduction, samples were reduced with the complete reduction cocktail containing Grx3 and with Grx3 omitted from the reduction cocktail. After reduction, proteins were enriched on the resin and eluted at the protein level for SDS-PAGE analysis. Without Grx3, a relatively low level of background signal was observed for both the untreated and diamide treated samples (Fig. 2C, Lane 1 and 4), suggesting a low degree of non-specific reduction of disulfides and SSG by GSH in the absence of Grx3. Moreover, the level of background signals from non-specific reduction was markedly lower compared to that reduced by the cocktail containing Grx3 for the diamide treated sample. These results suggest that the Grx3 reduction cocktail offers a relative good reduction specificity of protein-SSGs with only a minimal level of non-specific reduction of protein disulfides and SSGs.
MS-Quantification of diamide-induced S-glutathionylation
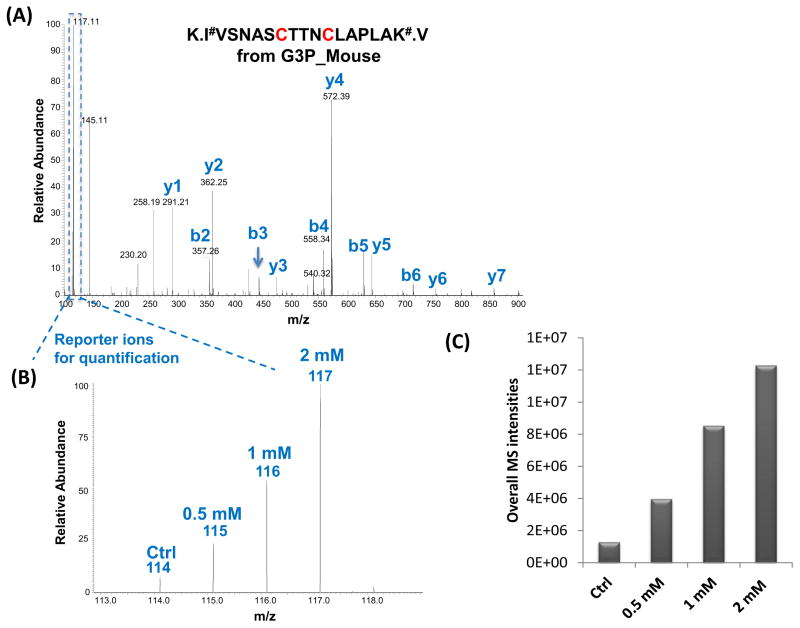
To identify potential S-glutathionylated sites, RAW 264.7 cells were treated with three different doses of diamide. Cellular lysates were selectively reduced, enriched, and labeled with 4-plex iTRAQ reagents followed by LC-MS/MS analysis. Specific cysteine sites of glutathionylation were identified based on MS/MS spectra as illustrated in Fig. 3, where both Cys 150 and 154 of GAPDH, a well-known glutathionylated protein [12], were identified as being SSG-modified. The reporter ion intensities enabled quantification of the glutathionylated cysteine sites in response to diamide treatment (Fig. 3B). A total of 1531 unique peptides were identified with a FDR ~0.4% and 94% of the identified peptides were free cysteine-containing peptides (Supplementary Table 1), revealing the high enrichment specificity and sensitivity offered by this approach. These peptides correspond to 1071 potential SSG-modified sites, covering 690 proteins. Of all the Cys-sites, ~90% sites showed at least two-fold increase in response to 2 mM diamide treatment when compared to the control samples, according to the observed reporter ion intensity. Fig. 3C shows the overall MS reporter ion intensities in response to different doses of diamide, reflecting the overall level of glutathionylated peptides within the cells. Those peptides that showed at least a five-fold increase in intensity induced by 2 mM diamide were further listed in Supplementary Table 2, in which 689 Cys-sites were identified. The observation of specific sites with a greater increase in signal intensity might reflect increased susceptibility of these sites to S-glutathionylation by diamide treatment.
Fig. 3.
(A) An MS/MS spectrum of a Cys-peptide from GAPDH; (B) Zoom-in spectrum of reporter-ion region showing the SSG abundance increased in response to diamide treatments; (C) Overall MS reporter ion intensities from summing all identified SSG-peptides from control and diamide-treated samples.
MS-quantification of H2O2-induced S-glutathionylation
Next we applied the above established quantification method for investigation of the Cys-sites and pathways affected by S-glutathionylation in response to H2O2, a much milder oxidant compared to diamide. Hydrogen peroxide serves as a signaling messenger, playing an important role in host defense and oxidative stress, particularly in higher organisms [42]. RAW 264.7 cells were treated with 0, 20, 100, and 500 μM H2O2 for 30 min, using a similar experimental design as shown in Fig. 1B. Fig. 4A shows that S-glutathionylation was clearly induced by 100 and 500 μM H2O2 treatment based on the SDS-PAGE analysis of the enriched Cys-peptides.
Fig. 4.
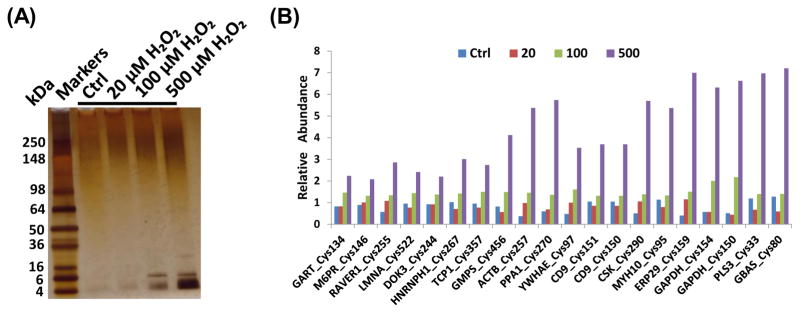
(A) Silver-staining image of SDS-PAGE of enriched Cys-peptides; (B) Selected Cys-sites showing increased levels of SSG modifications in response to H2O2 treatments. The treatment concentrations of H2O2 were in μM.
The LC-MS/MS experiments led to the identification of a total of 1191 unique peptides, corresponding to 1077 Cys-sites from 647 proteins, with a FDR of ~0.3%. Of all the identified peptides, 99% contained cysteine residues (Supplementary Table 3), again demonstrating the high degree of specificity and sensitivity of this approach. Based on the reporter ion intensities, ~55% of Cys-sites displayed a greater than 1.5-fold increase in response to H2O2 treatment, which is consistent with the fact that H2O2 represents a much milder stimulant compared to diamide. To further identify Cys-sites that are prone to H2O2-induced S-glutathionylation, we applied the following criteria: 1) the reporter ion intensity for 500-μM-H2O2-treated condition > 500, 2) the intensity increase for both 500-μM-H2O2 over control and 500-μM H2O2 over 20-μM H2O2 >1.5-fold. As a result, 364 Cys-sites and 384 Cys-peptides from 265 proteins were confidently identified as being susceptible to S-glutathionylation by H2O2 treatment (Supplemental Table 4). The reproducibility of the quantification method was also assessed based on two independent 4-plex experiments where a median coefficient of variation (CV) values of 17% for the fold-changes between 500 μM and 100 μM H2O2 treatments was observed (Supplemental Table 5). Fig. 4B further illustrates the levels of H2O2-induced S-glutathionylation at specific Cys-sites of selected proteins; these Cys-sites are most sensitive to SSG modification. For most sites, little or no modification was observed for samples treated with 20 μM H2O2, while dose-dependent increases of SSG were observed for 100 μM and 500 μM H2O2 treatments.
Western blot confirmation of selected SSG-modified proteins
To perform an orthogonal verification of the MS-observed increased S-glutathionylation on selected proteins, Western blot experiments were performed for GAPDH, TXN, PRDX3, and ANXA1. GAPDH, TXN, and PRDX3 were previously reported to be regulated by S-glutathionylation [12] [22, 43]. While ANXA1 is a novel SSG-modified protein identified in this work, the regulation by S-glutathionylation of ANXA2, another protein in the annexin family, has been previously reported [44]. For these experiments, total SSG-modified proteins were first enriched by the resin-assisted method and the eluted proteins were separated by SDS-PAGE and probed with antibodies specific to each of the selected proteins. Fig. 5A shows the levels of SSG-modified proteins in response to both H2O2 and diamide treatment as measured by western blot. As shown, each of the proteins exhibit an increase in the total amount of SSG-modification in response to both 0.5 and 1.0 mM diamide treatments, while only GAPDH displayed a clear increase in S-glutathionylation in response to 500 μM H2O2 treatment. The results confirm the MS-based identification of these protein modifications and support the fact that diamide is a much more potent oxidant than H2O2. Fig. 5B further illustrates the observed relative abundance levels of SSG-modification on specific Cys-sites in these proteins according to different doses of H2O2 exposure. In contrast to the limited sensitivity of Western blot analysis, the MS results showed a significant increase in SSG levels for each of these sites. The results also illustrate the greater susceptibility of GAPDH to S-glutathionylation in response to H2O2 treatment. The overall results demonstrate that our MS-based quantification method is more sensitive than Western blot for measuring levels of SSG modification.
Fig. 5.
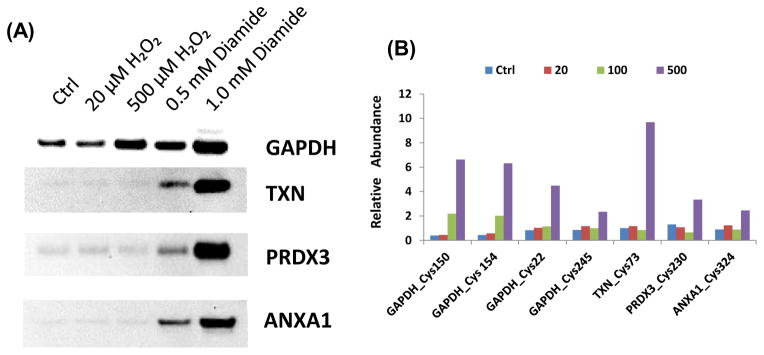
(A) Western blots of selected SSG-modified proteins. SSG-modified proteins were first enriched by thiol-affinity resin. The eluted proteins were subjected to Western blotting using specific antibodies against individual proteins. (B) Increased levels of SSG-modifications on individual Cys-sites from these proteins in response to H2O2 treatments. The treatment concentrations of H2O2 were in μM.
Functional analyses of S-glutathionylation target proteins responsive to H2O2 treatment
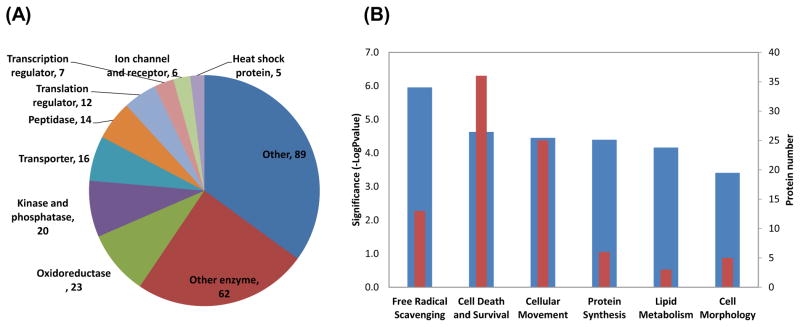
To gain an overall functional picture of SSG-modifications induced by H2O2 treatment, functional analyses of these modified proteins were performed using Ingenuity Pathway Analysis (IPA) tools. Fig. 6A shows the distribution of different types of SSG-modified proteins based on protein function. Among the annotated proteins, the majority were enzymes including oxidoreductases, kinases, phosphatases, and peptidases. Transcriptional and translational regulators, ion channel proteins, membrane receptor proteins, and heat shock proteins were also observed to be modified by SSG, which is in agreement with previous reports of these proteins being regulated by S-glutathionylation [6, 11]. Table 1 lists selected proteins based on their molecular function along with their relative SSG levels on specific Cys-sites in response to H2O2 treatment. It is noteworthy that many of the observed modification sites involve active-sites of enzymes (GAPDH, GATM, UCHL3, and CASP1), consistent with the hypothesis that these enzymes are sensitive to redox regulation through cysteine switch like mechanisms. In addition, previously reported sites of S-glutathionylation or S-nitrosylation were also identified in our analyses, providing additional confidence in our results [12, 14, 20, 22, 45, 46].
Fig. 6.
(A) Distribution of molecular types of identified sensitive SSG-modified proteins from H2O2 treatments; (B) Top significant molecular function categories. Significance levels are indicated by −log Pvalue (blue bar). The red bar indicates the number of proteins in each category.
Table 1.
Selected proteins observed with increased S-glutathionylation levels in response to H2O2 treatments.
| Function | Accession | Description | Symbol | Cys-sites | Relative abundances of SSG*
|
|||
|---|---|---|---|---|---|---|---|---|
| Ctrl | 20 μM | 100 μM | 500 μM | |||||
| Oxidoreductase | P50544 | Very long-chain specific acyl-CoA dehydrogenase | ACADVL | 238§ | 1.0 | 1.3 | 0.7 | 2.5 |
| Q569X5 | G3P-dehydrogenase | GAPDH | 154# | 0.4 | 0.6 | 2.0 | 6.3 | |
| 245 | 0.9 | 1.2 | 1.0 | 2.3 | ||||
| 150# | 0.4 | 0.4 | 2.2 | 6.6 | ||||
| 22 | 0.8 | 1.0 | 1.1 | 4.5 | ||||
| Q9CR21 | Acyl carrier protein, mitochondrial | NDUFAB1 | 140 | 0.9 | 1.1 | 1.0 | 11.0 | |
| P20108 | Thioredoxin-dependent peroxide reductase, mitochondrial | PRDX3 | 230 | 1.3 | 1.1 | 0.6 | 3.3 | |
| P08228 | Superoxide dismutase [Cu-Zn] | SOD1 | 7@ | 0.8 | 1.0 | 1.3 | 1.7 | |
| P10639 | Thioredoxin | TXN | 73@ | 1.0 | 1.2 | 0.8 | 9.7 | |
| Kinases | O09110 | Dual specificity mitogen-activated protein kinase kinase 3 | MAP2K3 | 29 | 1.0 | 1.1 | 0.8 | 4.3 |
| P63085 | Mitogen-activated protein kinase 1 | MAPK1 | 159 | 1.0 | 1.0 | 1.0 | 3.5 | |
| P28867 | Protein kinase C delta type | PRKCD | 507@ | 1.0 | 1.1 | 0.9 | 2.0 | |
| 127 | 1.1 | 0.8 | 1.1 | 15.9 | ||||
| Other enzymes | P20108 | Glycine amidinotransferase, mitochondrial | GATM | 407# | 1.0 | 1.1 | 0.9 | 1.6 |
| Q6PAV2 | Probable E3 ubiquitin-protein ligase HERC4 | HERC4 | 1025# | 0.5 | 1.2 | 1.3 | 3.3 | |
| Q1HFZ0 | tRNA-methyltransferase NSUN2 | NSUN2 | 321# | 0.0 | 1.6 | 1.4 | 7.6 | |
| Q7TQI3 | Ubiquitin thioesterase | OTUB1 | 91# | 0.8 | 1.0 | 1.1 | 2.8 | |
| P61080 | Ubiquitin-conjugating enzyme | UBE2D1 | 85# | 0.8 | 1.0 | 1.2 | 1.9 | |
| Q9JKB1 | Ubiquitin carboxyl-terminal hydrolase isozyme L3 | UCHL3 | 95# | 0.0 | 0.9 | 2.1 | 18.5 | |
| Ion channels | Q8R429 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | ATP2A1 | 349@ | 0.9 | 0.9 | 1.2 | 1.8 |
| Heat shock proteins | P07901 | Heat shock protein HSP 90-alpha | HSP90AA1 | 598§ | 1.0 | 0.7 | 1.2 | 2.1 |
| P11499 | Heat shock protein HSP 90-beta | HSP90AB1 | 590§ | 1.0 | 1.1 | 0.9 | 1.9 | |
| HSP90AB1 | 366 | 1.1 | 0.9 | 1.1 | 2.5 | |||
| Cytoskeletal proteins | Q6ZWM3 | Actin, cytoplasmic 1 | ACTB | 257@ | 0.6 | 1.0 | 1.5 | 5.4 |
| ACTB | 272@ | 0.8 | 1.4 | 0.7 | 4.1 | |||
| P10107 | Annexin A1 | ANXA1 | 324 | 0.9 | 1.2 | 0.9 | 2.5 | |
| Apoptosis | P29452 | Caspase-1 | CASP1 | 284# | 1.3 | 0.9 | 0.8 | 12.4 |
Known active-sites of the enzymes.
Previously reported sites of S-nitrosylation.
Cells were treated with different concentrations of H2O2 and the quantified SSG levels for each Cys-site were normalized against the average level across the first three conditions.
Fig. 6B further illustrates the top categories of molecular functions of enriched SSG-modified proteins in response to H2O2 treatment. Interestingly, the top molecular functions that are identified as being regulated by S-glutathionylation are free radical scavenging, cell death and survival, and cellular movement. These functional categories are in agreement with the well-known knowledge of oxidative stress/cell death induced by exposure to H2O2 treatments [47]. A number of proteins involved in free radical scavenging or the metabolism of ROS are well-known oxidoreductases such as PRDX3, SOD1, and LDHA. The observation of increased SSG levels in these free radical scavenging proteins along with heat shock proteins and well-known targets of ROS such as TXN, GAPDH, and CASP1 suggest that S-glutathionylation is an important underlying mechanism for regulating oxidative stress response signaling and cell death and survival [48].
Discussion
S-glutathionylation is an important post-translational modification that plays a significant role in redox signaling and regulation of a variety of cellular functions and processes involved in disease pathology [6–8, 48, 49]. While many protein targets of S-glutathionylation have been identified [1, 6, 11], it has been challenging to determine the specific sites at which these modification occur, and their relative abundance. In this work, we developed and demonstrated a quantitative proteomic approach for proteome-wide identification and quantification of site-specific S-glutathionylation. The effectiveness of this approach was demonstrated by applying the method for enrichment of SSG modified proteins from mouse macrophages treated with exogenous oxidants. In total, we identified more than 1000 Cys-sites from ~650 proteins which are susceptible to glutathionylation. It is worthy to note that the present proteomic approach offers two advantages; site-specific identification of SSG-modifications and the ability to quantify relative abundance of SSG-modified sites. With the global perspective of this approach, this study identified many novel SSG-modified proteins as well as a number of proteins previously reported to be regulated by glutathionylation [1, 6, 11]. Our work also extends beyond previous approaches and provides reactivity information regarding the individual cysteine residues susceptible to S-glutathionylation. Due to the increased sensitivity of the method, the reactivity of individual cysteine residues to SSG modification is measurable by employing a range of doses for each exogenous oxidant (Fig. 4B).
Potential limitations of this approach can be attributed to the indirect nature of the method as well as the specificity of the reduction step by the Grx enzyme cocktail, which is important for confidence of final identification of SSG-modified sites. Previous studies have shown that Grx reduces protein-SSG to protein-thiol through a monothiol mechanism [50, 51] and becomes S-glutathionylated itself at the active-site C22 [52, 53], which is then regenerated to free thiol by glutathione reductase (GR), GSH, and NADPH. We chose E. coli Grx3 [C14S/C65Y], which is mutated at the active-sites responsible for disulfide reduction, to increase the specificity of the SSG reduction step [41]. The utility of this enzyme for selective reduction of protein glutathionylation has been demonstrated previously [30, 54, 55]. Our results provide further support of the specificity of this enzyme based on the observation of very low basal levels of enriched proteins in untreated samples (Fig. 2C). We also note that there are low levels of artifacts resulting from non-specific reduction of protein disulfides and SSGs by GSH alone in the absence of Grx3 (Fig. 2C). However, we anticipate that Cys-sites from reduced stable disulfide bonds would be detected by the presence of similar reporter ion intensities (Fig. 3B) without a significant increase in intensity in response to exogenous stimuli [56]. Only those Cys-sites which were sensitive to exogenous stimuli were included as potential SSG sites (Supplemental Table 2 and 4). Moreover, the reduction condition with Grx3 omitted from the reduction cocktail can serve as a negative control to identify levels of non-specific reduction and differentiate potential artifacts from SSG signals.
When RAW cells were treated with H2O2, 364 Cys-sites from 265 proteins were identified as potential sites of endogenous S-glutathionylation or sites susceptible to SSG modification. The SSG-modified proteins we identified cover a variety of molecular functions (Fig. 6) and many of these proteins have been previously reported to be regulated by S-glutathionylation [1, 6, 11]. However, it should be noted that a 500 μM concentration of H2O2 treatment to cells is much higher than the physiological cellular concentrations of H2O2 [42]. Therefore, it is unclear whether these identified SSG modifications have physiological relevance to redox regulation. Future work needs to be focused on the detection of in vivo S-glutathionylation under physiological conditions (e.g., macrophages stimulated by lipopolysaccharide).
Despite this general uncertainty about physiological relevance, the site-specific and quantitative data in response to different doses of H2O2 provides important information regarding the potential regulatory sites for many previous reported redox-regulated proteins [47, 57]. In particular, a number of the observed Cys-sites that were most sensitive to S-glutathionylation in the present work are also known to be active-sites of the enzymes in which they were identified, supporting the regulatory role of S-glutathionylation in protein functions. For example, thioredoxin (TXN), one of the most important enzymes in redox regulation, was reported to be S-glutathionylated at Cys73, which leads to inactivation of the enzyme [22]. Our observation of Cys73 as an SSG-sensitive site further supports the idea of TXN playing a regulatory role involved in crosstalk between the glutathione and thioredoxin systems [22]. GAPDH is also a well-studied enzyme of redox regulation that plays a role in multiple cellular functions such as survival and apoptosis [58, 59]. The regulation of GAPDH by S-glutathionylation has been well characterized [12] while the exact sites of SSG modifications were still not confirmed [45]. Our quantitative reactivity data confirm that both Cys150 (the known active-site) and Cys154 were sensitive to S-glutathionylation in response to H2O2 treatment while Cys245 and Cys22 were also susceptible to modification. Another example is superoxide dismutase (SOD1), whose posttranslational modifications were suggested to play an important role in SOD1 aggregation in the familial form of amyotrophic lateral sclerosis [60]. Our study confirmed that Cys7, a suggested SSG-modification site [45], was sensitive to SSG modification and was supportive of the possible functional regulation of SOD1 by S-glutathionylation. Caspase-1 (CASP1), a thiol protease, is another example of an enzyme known to play an essential role in the initiation of inflammation and cell apoptosis. While a previous study reported the regulation of its activity by S-glutathionylation at Cys397 and Cys362 as well as potential oxidation at active-site Cys284 [61], our observation of Cys284 as the only site sensitive to SSG-modification suggests that S-glutathionylation of the active site of CASP1 is likely a mechanism for modulating its activity under oxidative stress.
Besides these proteins previously reported for S-glutathionylation, the proteome-wide identifications also provided a valuable database of macrophage proteins and specific cysteine residues susceptible to SSG modification, including many proteins not previously known about their susceptibility to SSG. This knowledge base will be useful for studying the function of specific proteins and the role of S-glutathionylation in cellular regulation.
Conclusions
We have developed an effective approach for enrichment, site-specific identification, and multiplex quantification of SSG-modifications. Our results from macrophages treated with diamide and H2O2 demonstrated the overall sensitivity of this resin-assisted enrichment approach for broad quantification of SSG-modified Cys-sites. We identified more than 1000 SSG-modified sites from both treatment conditions as well as 364 SSG-sites from 265 proteins in response to H2O2 treatment alone. Many of the SSG-modified proteins were enzymes containing active-site thiols and as well as proteins involved in free radical scavenging, oxidative stress signaling, and cell death and survival. The results provided further support of the significance of S-glutathionylation as a regulatory mechanism in oxidative stress and inflammation. Furthermore, the data provided identification of specific SSG-modified sites and enabled quantification of their relative reactivity to S-glutathionylation for both known and novel proteins.
Supplementary Material
Highlights.
Novel proteomics approach for site-specific quantification of S-glutathionylation
>1000 Cys-sites identified in RAW macrophages treated with diamide or H2O2
364 Cys-sites from 265 proteins identified as sensitive to H2O2 treatments
Potential significance of SSG in oxidative stress signaling pathways revealed
Acknowledgments
This work was supported by NIH Director’s New Innovator Award Program DP2OD006668, a DOE Early Career Research Award, P41 GM103493, the PNNL LDRD program, and the DOE Office of Biological and Environmental Research Genome Sciences Program under the Pan-omics project. This work was performed in the Environmental Molecular Science Laboratory, a DOE/BER national scientific user facility at PNNL in Richland, Washington. PNNL is operated by Battelle for the DOE under contract DE-AC05-76RLO-1830.
Abbreviations
- SSG
S-glutathionylation or S-glutathionylated, ROS, reactive oxygen species
- RNS
reactive nitrogen species
- GSH
glutathione
- GSSG
glutathione disulfide
- Grx
glutaredoxin
- GR
glutathione reductase
- iTRAQ
isobaric tags for relative and absolute quantitation
- NEM
N-ethylmaleimide
- BST
biotin switch technique
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bachi A, Dalle-Donne I, Scaloni A. Redox proteomics: chemical principles, methodological approaches and biological/biomedical promises. Chem Rev. 2013;113:596–698. doi: 10.1021/cr300073p. [DOI] [PubMed] [Google Scholar]
- 2.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Held JM, Gibson BW. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Molecular & cellular proteomics : MCP. 2012;11:R111–013037. doi: 10.1074/mcp.R111.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastore A, Piemonte F. S-Glutathionylation signaling in cell biology: progress and prospects. Eur J Pharm Sci. 2012;46:279–292. doi: 10.1016/j.ejps.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shelton MD, Mieyal JJ. Regulation by reversible S-glutathionylation: molecular targets implicated in inflammatory diseases. Mol Cells. 2008;25:332–346. [PMC free article] [PubMed] [Google Scholar]
- 10.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. S-glutathionylation in protein redox regulation. Free Radic Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Mohr S, Hallak H, de Boitte A, Lapetina EG, Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. The Journal of biological chemistry. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 13.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase 3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circul Res. 2007;100:213–219. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 14.Humphries KM, Juliano C, Taylor SS. Regulation of cAMP-dependent protein kinase activity by glutathionylation. The Journal of biological chemistry. 2002;277:43505–43511. doi: 10.1074/jbc.M207088200. [DOI] [PubMed] [Google Scholar]
- 15.Ward NE, Stewart JR, Ioannides CG, O’Brian CA. Oxidant-induced S-glutathiolation inactivates protein kinase C-alpha (PKC-alpha): a potential mechanism of PKC isozyme regulation. Biochemistry. 2000;39:10319–10329. doi: 10.1021/bi000781g. [DOI] [PubMed] [Google Scholar]
- 16.Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, Wouters EF, Janssen-Heininger YM. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pineda-Molina E, Klatt P, Vazquez J, Marina A, Garcia de Lacoba M, Perez-Sala D, Lamas S. Glutathionylation of the p50 subunit of NF-kappaB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry. 2001;40:14134–14142. doi: 10.1021/bi011459o. [DOI] [PubMed] [Google Scholar]
- 18.Klatt P, Molina EP, De Lacoba MG, Padilla CA, Martinez-Galesteo E, Barcena JA, Lamas S. Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1999;13:1481–1490. doi: 10.1096/fasebj.13.12.1481. [DOI] [PubMed] [Google Scholar]
- 19.Lancel S, Zhang J, Evangelista A, Trucillo MP, Tong X, Siwik DA, Cohen RA, Colucci WS. Nitroxyl activates SERCA in cardiac myocytes via glutathiolation of cysteine 674. Circul Res. 2009;104:720–723. doi: 10.1161/CIRCRESAHA.108.188441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, Cohen RA. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nature medicine. 2004;10:1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 21.Anathy V, Aesif SW, Guala AS, Havermans M, Reynaert NL, Ho YS, Budd RC, Janssen-Heininger YM. Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of Fas. J Cell Biol. 2009;184:241–252. doi: 10.1083/jcb.200807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casagrande S, Bonetto V, Fratelli M, Gianazza E, Eberini I, Massignan T, Salmona M, Chang G, Holmgren A, Ghezzi P. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fratelli M, Demol H, Puype M, Casagrande S, Villa P, Eberini I, Vandekerckhove J, Gianazza E, Ghezzi P. Identification of proteins undergoing glutathionylation in oxidatively stressed hepatocytes and hepatoma cells. Proteomics. 2003;3:1154–1161. doi: 10.1002/pmic.200300436. [DOI] [PubMed] [Google Scholar]
- 24.Gao XH, Bedhomme M, Veyel D, Zaffagnini M, Lemaire SD. Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol Plant. 2009;2:218–235. doi: 10.1093/mp/ssn072. [DOI] [PubMed] [Google Scholar]
- 25.Fratelli M, Gianazza E, Ghezzi P. Redox proteomics: identification and functional role of glutathionylated proteins. Expert Rev Proteomics. 2004;1:365–376. doi: 10.1586/14789450.1.3.365. [DOI] [PubMed] [Google Scholar]
- 26.Cheng G, Ikeda Y, Iuchi Y, Fujii J. Detection of S-glutathionylated proteins by glutathione S-transferase overlay. Arch Biochem Biophys. 2005;435:42–49. doi: 10.1016/j.abb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Brennan JP, Miller JI, Fuller W, Wait R, Begum S, Dunn MJ, Eaton P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol Cell Proteomics. 2006;5:215–225. doi: 10.1074/mcp.M500212-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Zaffagnini M, Bedhomme M, Groni H, Marchand CH, Puppo C, Gontero B, Cassier-Chauvat C, Decottignies P, Lemaire SD. Glutathionylation in the photosynthetic model organism Chlamydomonas reinhardtii: a proteomic survey. Mol Cell Proteomics. 2012;11:M111–014142. doi: 10.1074/mcp.M111.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang BY, Chou CC, Hsieh FT, Gao S, Lin JC, Lin SH, Chen TC, Khoo KH, Lin CH. In vivo tagging and characterization of S-glutathionylated proteins by a chemoenzymatic method. Angew Chem Int Ed Engl. 2012;51:5871–5875. doi: 10.1002/anie.201200321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, Cotgreave IA. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 31.Su D, Shukla AK, Chen B, Kim JS, Nakayasu E, Qu Y, Aryal U, Weitz K, Clauss TR, Monroe ME, Camp DG, II, Bigelow DJ, Smith RD, Kulkarni RN, Qian WJ. Quantitative site-specific reactivity profiling of S-nitrosylation in mouse skeletal muscle using cysteinyl peptide enrichment coupled with mass spectrometry. Free Radic Biol Med. 2013;57:68–78. doi: 10.1016/j.freeradbiomed.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T, Qian WJ, Strittmatter EF, Camp DG, Anderson GA, Thrall BD, Smith RD. High throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 34.Liu M, Hou J, Huang L, Huang X, Heibeck TH, Zhao R, Pasa-Tolic L, Smith RD, Li Y, Fu K, Zhang Z, Hinrichs SH, Ding SJ. Site-specific proteomics approach for study protein S-nitrosylation. Analytical Chemistry. 2010;82:7160–7168. doi: 10.1021/ac100569d. [DOI] [PubMed] [Google Scholar]
- 35.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 36.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-Based Evaluation of Peptide and Protein Identifications from Tandem Mass Spectrometry and SEQUEST Analysis: The Human Proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical Chemistry. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Wang Y, Li S. Deuterium isobaric amine-reactive tags for quantitative proteomics. Analytical Chemistry. 2010;82:7588–7595. doi: 10.1021/ac101306x. [DOI] [PubMed] [Google Scholar]
- 40.Gilge JL, Fisher M, Chai YC. The effect of oxidant and the non-oxidant alteration of cellular thiol concentration on the formation of protein mixed-disulfides in HEK 293 cells. PLoS One. 2008;3:e4015. doi: 10.1371/journal.pone.0004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordstrand K, slund F, Holmgren A, Otting G, Berndt KD. NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism. J Mol Biol. 1999;286:541–552. doi: 10.1006/jmbi.1998.2444. [DOI] [PubMed] [Google Scholar]
- 42.Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxidants & redox signaling. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 43.Chae HZ, Oubrahim H, Park JW, Rhee SG, Chock PB. Protein glutathionylation in the regulation of peroxiredoxins: a family of thiol-specific peroxidases that function as antioxidants, molecular chaperones, and signal modulators. Antioxidants & redox signaling. 2012;16:506–523. doi: 10.1089/ars.2011.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caplan JF, Filipenko NR, Fitzpatrick SL, Waisman DM. Regulation of annexin A2 by reversible glutathionylation. The Journal of biological chemistry. 2004;279:7740–7750. doi: 10.1074/jbc.M313049200. [DOI] [PubMed] [Google Scholar]
- 45.Tao L, English AM. Protein S-glutathiolation triggered by decomposed S-nitrosoglutathione. Biochemistry. 2004;43:4028–4038. doi: 10.1021/bi035924o. [DOI] [PubMed] [Google Scholar]
- 46.Chen FC, Ogut O. Decline of contractility during ischemia-reperfusion injury: actin glutathionylation and its effect on allosteric interaction with tropomyosin. Am J Physiol Cell Physiol. 2006;290:C719–727. doi: 10.1152/ajpcell.00419.2005. [DOI] [PubMed] [Google Scholar]
- 47.Forman HJ, Torres M. Redox signaling in macrophages. Mol Aspects Med. 2001;22:189–216. doi: 10.1016/s0098-2997(01)00010-3. [DOI] [PubMed] [Google Scholar]
- 48.Anathy V, Roberson EC, Guala AS, Godburn KE, Budd RC, Janssen-Heininger YM. Redox-based regulation of apoptosis: S-glutathionylation as a regulatory mechanism to control cell death. Antioxidants & redox signaling. 2012;16:496–505. doi: 10.1089/ars.2011.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mieyal JJ, Chock PB. Posttranslational modification of cysteine in redox signaling and oxidative stress: Focus on s-glutathionylation. Antioxid Redox Signal. 2012;16:471–475. doi: 10.1089/ars.2011.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghezzi P, Di Simplicio P. Glutathionylation pathways in drug response. Curr Opin Pharmacol. 2007;7:398–403. doi: 10.1016/j.coph.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Zaffagnini M, Bedhomme M, Lemaire SD, Trost P. The emerging roles of protein glutathionylation in chloroplasts. Plant Sci. 2012;185–186:86–96. doi: 10.1016/j.plantsci.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Jao S, Nanduri S, Starke DW, Mieyal JJ, Qin J. Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity. Biochemistry. 1998;37:17145–17156. doi: 10.1021/bi9806504. [DOI] [PubMed] [Google Scholar]
- 53.Reynaert NL, Ckless K, Guala AS, Wouters EF, van der Vliet A, Janssen-Heininger YM. In situ detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochim Biophys Acta. 2006;1760:380–387. doi: 10.1016/j.bbagen.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Hamnell-Pamment Y, Lind C, Palmberg C, Bergman T, Cotgreave IA. Determination of site-specificity of S-glutathionylated cellular proteins. Biochem Biophys Res Commun. 2005;332:362–369. doi: 10.1016/j.bbrc.2005.04.130. [DOI] [PubMed] [Google Scholar]
- 55.Zhang C, Rodriguez C, Circu ML, Aw TY, Feng J. S-Glutathionyl quantification in the attomole range using glutaredoxin-3-catalyzed cysteine derivatization and capillary gel electrophoresis with laser-induced fluorescence detection. Anal Bioanal Chem. 2011;401:2165–2175. doi: 10.1007/s00216-011-5311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray CI, Uhrigshardt H, O’Meally RN, Cole RN, Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Molecular & cellular proteomics : MCP. 2012;11:M111–013441. doi: 10.1074/mcp.M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavakoli S, Asmis R. Reactive oxygen species and thiol redox signaling in the macrophage biology of atherosclerosis. Antioxidants & redox signaling. 2012;17:1785–1795. doi: 10.1089/ars.2012.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholls C, Li H, Liu JP. GAPDH: a common enzyme with uncommon functions. Clinical and experimental pharmacology & physiology. 2012;39:674–679. doi: 10.1111/j.1440-1681.2011.05599.x. [DOI] [PubMed] [Google Scholar]
- 59.Butterfield DA, Hardas SS, Lange ML. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer’s disease: many pathways to neurodegeneration. Journal of Alzheimer’s disease : JAD. 2010;20:369–393. doi: 10.3233/JAD-2010-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furukawa Y, Kaneko K, Yamanaka K, O’Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. The Journal of biological chemistry. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nature immunology. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.