Abstract
Elongation factor-2 (eEF2) catalyzes the movement of the ribosome along the mRNA. A single histidine residue in eEF2 (H715) is modified to form diphthamide. A role for eEF2 in cellular stress responses is highlighted by the fact that eEF2 is sensitive to oxidative stress and that it must be active in order to drive the synthesis of proteins that help cells to mitigate the adverse effects of oxidative stress. Many of the latter proteins are encoded by mRNAs containing a sequence called an “internal ribosomal entry site” (IRES). Under high oxidative stress conditions diphthamide-deficient cells were significantly more sensitive to cell death. These results suggest that diphthamide may play a role in protection against the degradation of eEF2. Its protection is especially important under those situations where it is necessary for the re-programming of translation from global to IRES synthesis. Indeed, we found that the expression of X-linked inhibitor of apoptosis (XIAP) and fibroblast growth factor 2 (FGF2), two proteins synthesized from mRNAs with IRES that promote cell survival are deregulated in diphthamide-deficient cells. Our findings therefore suggest that eEF2/diphthamide controls the selective translation of IRES-dependent protein targets XIAP and FGF2, critical for cell survival under conditions of oxidative stress.
Keywords: Eukaryotic elongation factor-2, Diphthamide, Lipid peroxidation, XIAP, FGF-2, internal ribosomal entry site” (IRES)
INTRODUCTION
Protein synthesis is an important, complex process that involves RNA and many regulatory proteins that are influenced by cellular metabolic status and signals from hormones, growth factors, and nutrient availability [1–3]. This regulatory machinery provides cells with the plasticity needed to respond to environmental challenges such as oxidative and energetic stress [4]. The translation process consists of three phases (initiation, elongation and termination) that each require the participation of several translation factors that transiently associate with the ribosome. Although the majority of translational control is exerted at the initiation stage [5], specific regulation of the elongation phase also occurs under certain conditions [3,6–10].
Two distinctive features of the elongation step are its high energy requirements, and the fact that it only relies on three factors. Among these factors, elongation factor 2 (eEF2), which catalyzes the ribosomal movement along the mRNA [3] is a target of several distinct regulatory signaling pathways. For instance, phosphorylation inactivates eEF2 by preventing it from binding to the ribosome [11], thereby decreasing the rate of peptide elongation and contributing to global inhibition of protein synthesis. The phosphorylation of eEF2 is regulated by nutrients and growth factors which activate mTOR (mammalian target of rapamycin) resulting in S6 kinase-mediated inhibition of eEF2 kinase, resulting in reduced phosphorylation of eEF2 and increased protein synthesis [12–14]. Phosphorylation is particularly important under conditions of an energy deficit, because translation elongation is a metabolically demanding process. For example, during fasting or exercise AMP kinase (AMPK) is activated which then phosphorylates eEF2 and thereby inhibits protein synthesis [15], thereby conserving energy for essential cellular processes that promote cell maintenance and survival. In addition to the phosphorylation status, eEF2 activity can be modified through protein-protein interactions. For example, interactions with p53 and 14-3-3 mediate eEF2 nuclear subcellular relocalization in neurons subjected to oxidative stress [16]. The stress-induced reduction in global translation is usually accompanied by the selective translation of specific proteins that are required for the cell to withstand the period of adverse conditions. This process is not dependent on the translation initiation step; instead the ribosome starts ‘reading’ the mRNA at the Internal Ribosome Entry sequence (IRES) [17,18]. Although the translation of IRES-mRNA is less dependent on initiation factors, it still requires elongation factors.
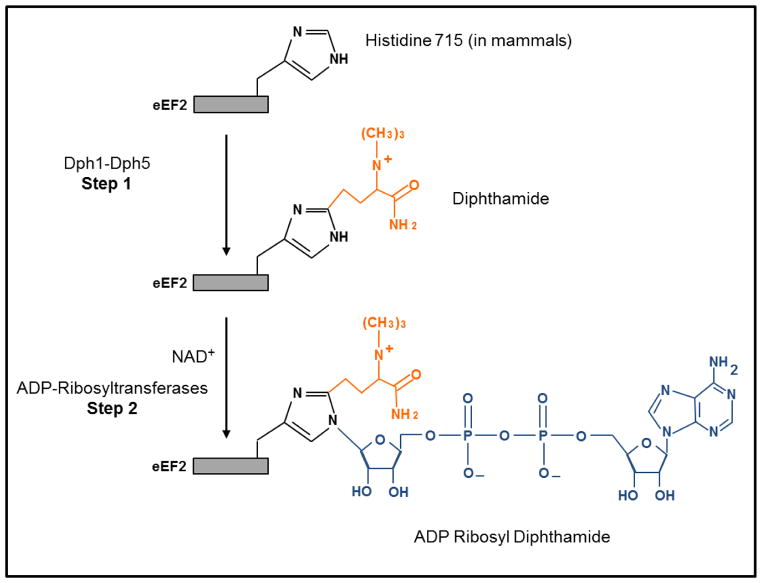
eEF2 can be ribosylated at a unique posttranslational modified derivative of histidine (H715) known as diphthamide (2-[3-carboxyamido-3-(trimethylammonio)-propyl] histidine). Diphthamide formation is mediated by the Dph enzymes of which there are five (Dph1 – Dph5) [19,20]. Diphtheria and cholera toxins catalyze the transfer of ADP-ribose from NAD+ to the diphthamide imidazole ring (Scheme 1). Neither the identity of the endogenous ADP-ribosyltransferases that ribosylate eEF2, nor the physiological roles of the diphthamide are known [19]. However, oxidative stress is known to cause significant changes in ADP-ribose/NAD metabolism and can increase eEF2 ADP-ribosylation [21].
Scheme 1.
Diphthamide synthesis (Step 1) and ADP-ribosylation (Step 2). Dph1 to Dph5 are required for the biosynthesis of diphthamide and are involved in the transfer of a 3-amino-3-carboxypropyl and a methyl group to histidine 715 of eEF-2. The diphthamide imidazole ring is the site for nicotinamide adenine dinucleotide (NAD)+ dependent ADP-ribosylation yielding ADP ribosyl diphthamide.
Because eEF2 is the target of phosphorylation, ribosylation and specific protein-protein interactions, it is likely that these regulatory mechanisms can function independently or in conjunction depending on the specific cellular metabolic circumstances. It is possible that under physiological situations, when the cell respond to nutritional and growth signals, phosphorylation of eEF2 is sufficient to switch on and off the global protein synthesis. However, under stress conditions, where ATP is needed for repair process, and eEF2 activity is affected [22–24] other post-translational modifications may play a prominent role.
In this paper we investigated the possible role played by eEF2 diphthamide in the cellular response elicited by oxidative stress. Chinese hamster ovary cells expressing or lacking the enzyme Dph3 required for the biosynthesis of diphthamide were exposed to increasing concentrations of cumene hydroperoxide (CH), which induces membrane lipid peroxidation and affects eEF2 [24,25]. Our results show that eEF-2 diphthamide is important for the synthesis of X-linked inhibitor of apoptosis (XIAP) and fibroblast growth factor 2 (FGF2), two proteins encoded by IRES-containing mRNAs and critical for cell survival under oxidative stress conditions.
MATERIALS AND METHODS
Cell cultures
Wild-type and diphthamide-deficient (Dph3 mutant) Chinese hamster ovary (CHO) cells were supplied by Stephen H. Leppla (NIAID, NIH), and were grown in α-minimal essential medium (MEM) supplemented with 8% fetal bovine serum, 25 mM HEPES, 2 mM glutamine and 50 μg/ml gentamicin.
Cell viability
Cell viability was determined using a MTS assay (Promega, Madison, WI) and a LDH activity assay (Roche Cytotoxicity detection kit, Mannheim, Germany) according to the manufacturer’s instructions.
Determination of hydroperoxides using the FOX reagent
The protocol for lipid peroxidation measurements [26] was adapted for a microplate reader. Forty micrograms of proteins were incubated with 90 μl of H2SO4 for 30 min. Following addition of 100 μl FOX reagent (0.5 mM ferrous ammonium sulfate, 0.2 mM xylenol orange and 200 mM sorbitol in 25 mM H2SO4) the mixture was incubated at room temperature for 45 min, protected from light. The formation of ferric ions was detected by measuring the resulting colored complex with xylenol orange at 540 nm.
Immunoblot analysis
Cell lines were lysed in RIPA buffer (20 mM Tris-HCl, 150mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate and 1 mM sodium orthovanadate) containing protease inhibitors. The homogenized cells were centrifuged at 12,000g for 20 min at 4°C. Protein content of the samples was estimated with Pierce BCA Protein Assay Kit (Pierce Biotechnology, Rockfold, IL). Protein samples were separated by SDS-PAGE (10% acrylamide), and transferred to a nitrocellulose membrane (Bio Rad, Hercules, CA) at 120 V for 1 h. The membranes were incubated with blocking buffer (5% dry milk in 20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.05% Tween 20) for 1 h at room temperature. Membranes were then incubated in blocking solution containing the following antibodies: eEF2 (1:5000), phospho-eEF-2 and XIAP (1: 1000) (Cell Signaling, Danvers, MA); FGF-2 (1: 500) (abcam Inc, Cambridge, MA); hnRNP (1:2000) and α-tubulin (1:1000) (Santa Cruz Biotechnology, Santa Cruz, CA), overnight at 4°C. After incubation, the membranes were washed in 20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.05% Tween 20 and incubated with peroxidase-conjugated anti-immunoglobulin secondary antibodies. The proteins were visualized using a chemiluminiscence kit from Pierce (Rockford, IL). The bands were analyzed by densitometry using Image J analysis software (NIH).
ADP-ribosylation assay
The assay was performed as described previously [27]. Briefly, 50 μg of cell lysates were incubated in ADP-ribosylation buffer (20 mM Tris-HCl, 1 mM EDTA, 50 Mm DTT; pH 7.4) with 500 ng of FP59 and 5 μM 6-Biotin-17-NAD (Trevigen, Gaithersburg, MD) for 30 min at 37°C. Samples were separated by SDS-PAGE followed by immunoblotting. The biotin-ADP-ribose-eEF-2 complexes were detected using streptavidin-IR conjugate antibody (Rockland Immunochemicals, Gilbertsville, PA) and a Typhoon 9400 scanner (GE Healthcare, Pittsburgh, PA).
Subcellular fractionation
Cells were subfractionated as described previously [28]. Cells were incubated on ice for 10 min in 800 μl of lysis buffer containing 20 mM Tris (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% IGEPAL CA-630 and protease inhibitors. Cells were centrifuged at 1,000 x g for 10 min to separate the cytoplasmic fraction of the cell extract. The nuclei were lysed by incubation on ice for 45 min with RIPA buffer containing protease inhibitors. The lysate was then centrifuged at 20,000 g for 20 min. The resulting supernatant was used as the nuclear fraction of the cell extract. The subcellular fractions were separated by SDS-PAGE and analyzed by immunoblots using antibodies against cytoplasmic and nuclear marker proteins.
Newly synthesized protein assay
Cells growing in 6-well plates were pretreated with CH, washed with warm PBS, and incubated in methionine-free D-MEM medium (Invitrogen, Grand Island, NY) for 35 min to deplete methionine reserves, after which 50 μM L-azidohomoalanine (AHA) (Invitrogen) was added for 30 min. The cells were lysed and proteins were extracted by ultrasonication in RIPA buffer containing protease inhibitors. AHA-incorporating proteins were labeled with tetramethyl rhodamine (TAMRA) using Click-iT Protein Reaction Buffer Kit (Invitrogen). The TAMRA-labeled proteins in the gel were assayed using a Typhoon 9400scanner (GE Healthcare).
Immunofluorescence
For immunostaining, cells were plated at a density of 5 × 104 cells/cm2 on 18-mm diameter round glass coverslips in 12-well plates. After treatment the cells were fixed with 4% paraformaldehyde, then permeabilized in 0.5% Triton X-100 for 10 min, rinsed with PBS, and incubated for 1 h with 10% donkey serum. The cells were incubated with the primary antibodies/antiserum overnight at 4 °C in the blocking solution. The immunocomplexes were detected using Alexa Fluor 488 (green)-conjugated or Alexa Fluor 546 (red)-conjugated secondary antibodies (1:1000; 1 h incubation at room temperature). Nuclei were counterstained with 4′, 6′-diamidino-2-phenylindole (DAPI; Vector Laboratories Inc., Burlingame, CA). Image analysis was performed using a Zeiss 510 confocal laser scanning microscope.
Statistical analysis
Data are shown as the mean ± SEMs. Statistical analysis was performed by one-way ANOVA, followed by Tukey’s test or by two-way ANOVA using GraphPadPrism Version 5.03 (Graph Pad Software). A p-value of ≤ 0.05 was considered significant.
RESULTS
Effects of CH on lipid peroxidation and cell viability
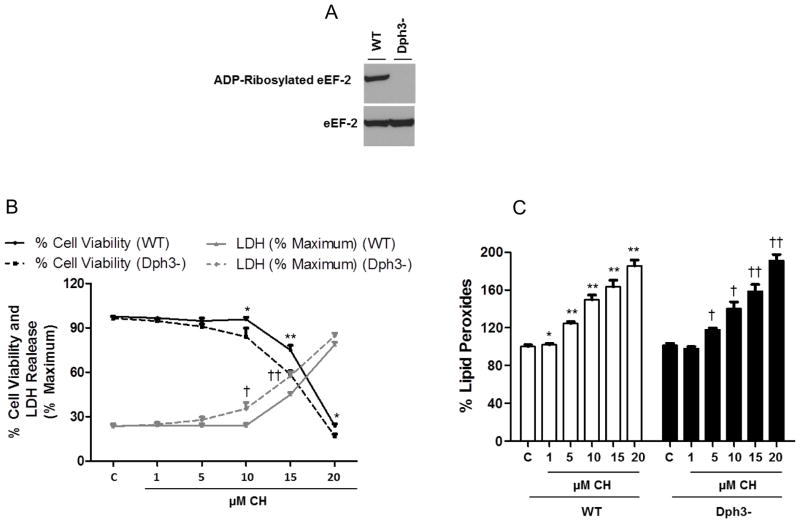
The inability of eEF-2 to be ADP-ribosylated in diphthamide-deficient (Dph3−) CHO cells was confirmed using an vitro ADP-ribosylation assay with the cell lysates. (Fig. 1A). To investigate the potential role of diphthamide on cell survival in response to oxidative stress, we analyzed the effect of increasing concentrations of CH on wild-type and Dph3− cells. Diphthamide-deficient cells were more sensitive to CH such that exposure to 10 and 15 μM of CH led to a significantly greater decrease in cell viability compared to wild-type cells (12 and 16%, respectively) (Fig. 1B). Similarly, LDH activity after 10 and 15 μM CH treatment was increased significantly more in Dph3− cells compared to wild-type cells (Fig. 1B). Lipid peroxidation increased in response to CH, in a concentration-dependent manner, with no statistically significant differences between wild-type and Dph3− cells (Fig. 1C).
Figure 1.
Diphthamide deficiency increases the vulnerability of cells to oxidative stress. A. Immunoblots for in vitro ADP-ribosylation assay with the cell lysates. The top panel shows ADP-ribosylated eEF-2 and the bottom panel shows the total eEF-2 in wild-type (WT) or diphthamide-deficient cells (Dph3−). B. Cells were incubated for 3 h in absence or presence of the indicated concentrations of CH. Cell viability was measured by the MTS assay and lactate dehydrogenase (LDH) release assay. Values are the mean and SEM of four experiments. *p< 0.05 and **p<0.01 (cell viability), †p<0.05 and ††p<0.01 (cell death); WT versus diphthamide-deficient cells. C. Lipid peroxidation levels were determined using the FOX method. Values are the mean and SEM of 5 independent experiments. *p< 0.05 and **p<0.01 versus the wild-type cell control value. †p<0.05 and ††p<0.01 versus the diphthamide- deficient cell control value.
Effects of diphthamide and CH on eEF2 subcellular localization
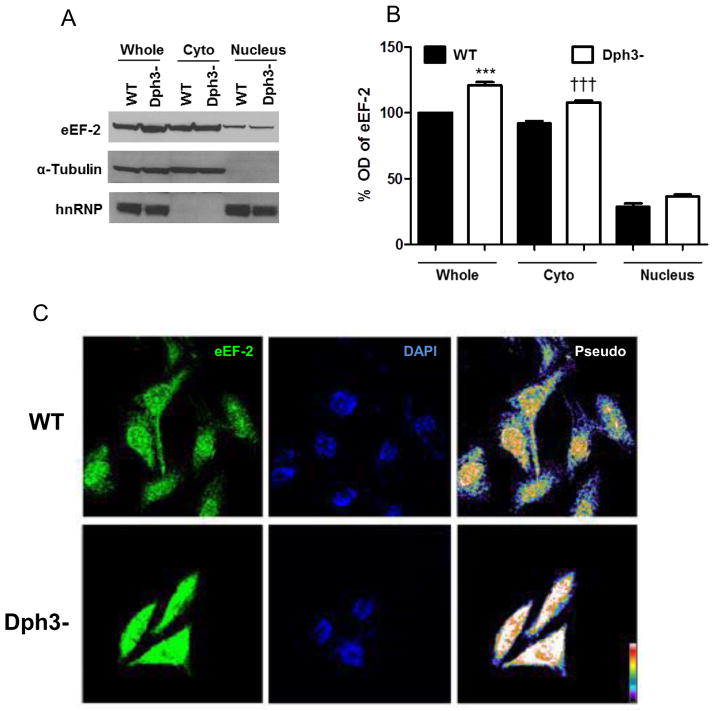
In order to determine whether diphthamide is involved in the regulation of eEF2 localization within the cell under basal and stress conditions, the levels of eEF-2 were measured in whole, cytoplasmic and nuclear fractions of wild-type and Dph3− cells by immunoblot analysis. The results showed similar levels of eEF2 in the nuclear compartment, but significantly higher levels of eEF2 in the cytoplasm and whole-cell extract of Dph3− cells (Fig 2A, B). Immunofluorescences staining confirmed that levels of eEF2 were greater in Dph3− cells compared to wild-type cells (Fig. 2C).
Figure 2.
Levels of eEF-2 are elevated in diphthamide-deficient cells. A. Subcellular fractionation and immunoblot analysis the eEF2 levels. α-tubulin and hnRNP were used as cytoplasmic and nuclear markers. B. Optical density of the eEF2 bands were normalized to a control (α-tubulin or hnRNP). Values are the mean and SEM of 4 experiments. ***p< 0.001 versus the value for whole cell extracts from wild-type cells. †††p<0.001 versus the value for cytoplasmic extracts from wild-type cells. C. Confocal localization of eEF2 in wild-type and diphthamide-deficient cells by immunofluorescence (green); nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). eEF2 integrated intensity (pixel/ μm2): WT 100 ± 5.12% n=47 cells; Dph3− 136.4 ± 8.3% n=43 cells; t test p=0.00028.
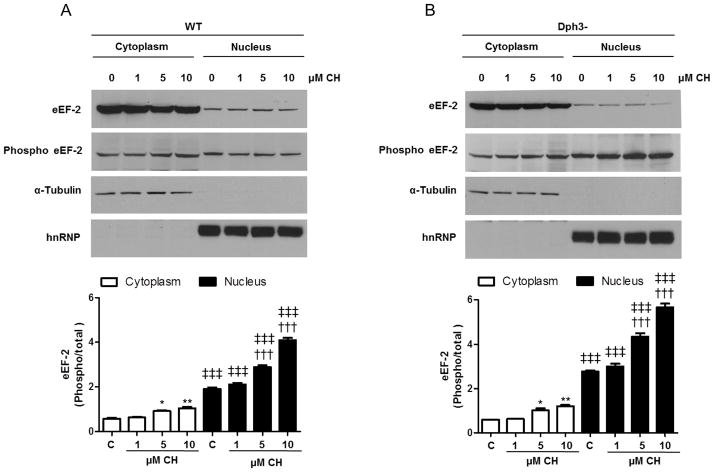
Next, we analyzed levels of total and translationally inactive (phosphorylated) eEF2 in cytoplasmic and nuclear extracts from wild-type and Dph3− cells treated with different CH concentrations. In both cell types the ratio of phosphorylated/total eEF2 was higher in nuclear extracts. Treatment with CH caused similar increases of both cytoplasmic and nuclear ratios of phosphorylated to total eEF2 (Fig. 3A, B). Interestingly, the nuclear phosphorylated/total eEF2 ratio was greater in Dph3− cells compared to wild-type cells. In wild-type cells, the ratios of phosphorylated to total eEF2 in the nuclear fraction were 3.1-fold for control, 3.3-fold for 1 μM CH, 3.1-fold for 5 μM CH and 3.9-fold for 10 μM CH compared to cytoplasm fractions. In the case of Dph3− cells, the ratios in the nuclear fraction were 4.6-fold for control, 4.7-fold for 1 μM CH, 4.3-fold for 5 μM CH and 4.7-fold for 10 μM CH.
Figure 3.
The nuclear ratio of phosphorylated to total eEF2 is higher in diphthamide-deficient cells. A and B. Subcellular fractionation and immunoblot analysis for total and phosphorylated eEF2 were performed on WT cells (A) and diphthamide-deficient cells (B), treated with CH for 3 h. α-tubulin and hnRNP were used as cytoplasmic and nuclear markers, respectively. Optical density of the eEF2 bands phosphorylated/total in cytoplasm and nucleus were normalized to a control (α-tubulin or hnRNP). Values are the mean and SEM of 4 experiments. *p<0.05 and ** p<0.01 versus the cytoplasmic control value, †††p<0.001 versus the nuclear control value. ‡‡‡ p<0.001 versus the cytoplasmic control value.
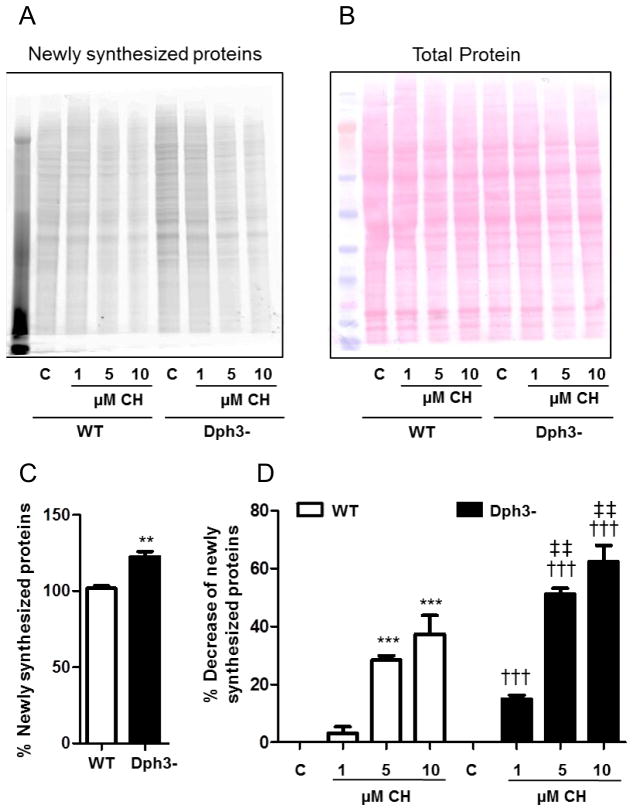
Diphthamide regulates newly synthesized in response to oxidative stress
We measured levels of newly synthesized proteins in wild-type and Dph3− cells under basal and oxidative stress conditions. Under basal conditions the rate of newly synthesized proteins was higher in diphthamide-deficient cells (22%) compared to wild- type cells (Fig. 4A, C). CH treatment produced significant reductions of newly synthesized proteins in both cell types, with a more pronounced decrease in Dph3− cells; 51% (5 μM CH) and 62% (10 μM CH) compared to Dph3− vehicle-treated cells, and 28% (5 μM CH) and 37% (10 μM CH) compared to wild-type vehicle-treated cells (Fig. 4D). The levels of total eEF-2 mirrored the changes observed in the newly synthesized proteins assays. eEF2 levels were higher in diphthamide-deficient cells compared to wild-type cells (17%) (Fig. 5A, B). However, high lipid peroxidation levels produced a significant decrease of total eEF2 levels in wild-type cells (10% for 5 μM CH and 19% for 10 μM CH) that was further exacerbated in Dph3− cells (24% for 5 μM CH and 35% for 10 μM CH) (Fig. 5C). These results suggest that diphthamide plays a role in eEF2 degradation under normal and oxidative stress conditions.
Figure 4.
Evidence that diphthamide preserves the function of eEF2 in protein synthesis under conditions of oxidative stress. A and B. To directly determine the effects of CH on newly synthesized proteins, wild-type cells (WT) and diphthamide-deficient cells (Dph3−) were pretreated with CH in methionine-free D-MEM containing L-azidohomoalanine (AHA) to label the newly synthesized proteins. After electrophoresis the gels were analyzed using a Typhoon 9400 scanner (GE Healthcare) at 545 nm excitation and 580 nm emission (A), transferred to a nitrocellulose membrane and stained with Ponceau Red (B). C. Newly synthesized proteins in WT and diphthamide-deficient cells (Dph3−) under control conditions. Values are the mean and SEM of 4 experiments. **p<0.01 versus the WT control value. D. Newly synthesized proteins in WT and diphthamide-deficient cells (Dph3−) treated with CH. Values are the mean and SEM of 4 experiments. ***p<0.001 versus the WT control value. †††p<0.001 versus the diphthamide-deficient cell control value. ‡‡ p<0.01 versus the WT control value. § p<0.05 versus the WT 1 μM CH value. Under oxidative stress, eEF2 in diphthamide deficient cells was more affected than in wild-type cells.
Figure 5.
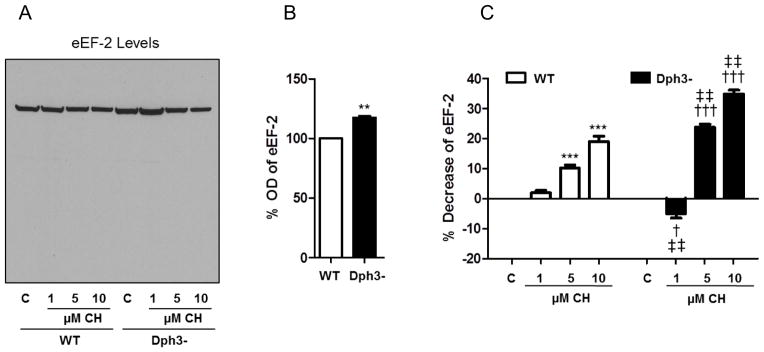
Levels of total eEF2 mirror the changes observed in the newly synthesized proteins assays. To directly determine the effects of CH on eEF2, wild-type cells (WT) and diphthamide-deficient cells (Dph3−) were pretreated with CH. A. Immunoblot analysis showing eEF-2 levels in the same nitrocellulose membrane depicted in Figure 4A-B. B. Optical density of the eEF-2 bands under basal conditions. Values are the mean and SEM of 4 experiments. *p< 0.05 and **p<0.01 vs. WT control. C. Optical density of the eEF2 bands following CH treatment. Values are the mean and SEM of 4 experiments. *p< 0.05 and ***p<0.001 versus the WT control value. †††p<0.001 versus the diphthamide-deficient cell control value. ‡‡ p<0.01 versus the WT control value. § p<0.05 versus the WT 1 μM CH value.
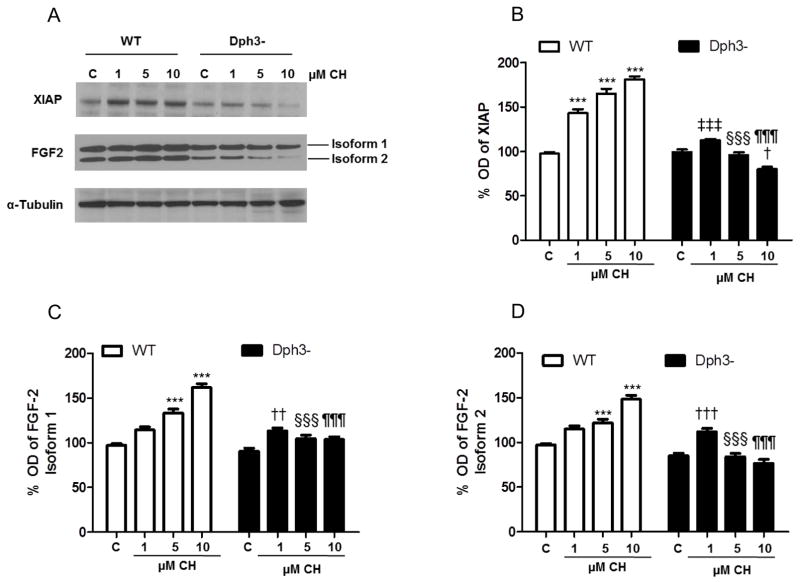
Role of Diphthamide on the synthesis of two proteins encoded by IRES-containing mRNAs (XIAP and FGF2) under oxidative stress conditions
The presence of internal ribosome entry site (IRES) elements located at the 5′-untranslated regions of specific mRNAs allows synthesis of the encoded proteins during stress conditions that cause global cap-dependent translation to be halted. We therefore analyzed the levels of the IRES-dependent protein targets XIAP and FGF2 in wild-type and Dph3− cells under basal and oxidative stress conditions (Fig. 6A). XIAP and FGF2 were chosen because they have previously been shown to promote the survival of cells subjected to oxidative and metabolic stress [29–32]. In wild-type cells CH-induced lipid peroxidation produced significant dose-dependent increases of XIAP levels compared to vehicle-treated cells [97% (C), 143% (1 μM CH), 165% (5 μM CH) and 181% (10 μM CH)]. However, no change with lower CH concentrations (1 and 5 μM), and a significant decrease in XIAP levels at the highest CH concentration (10 μM), occurred in Dph3− cells (Fig. 6B).
Figure 6.
Evidence for the involvement of eEF2/diphthamide in the control the translation of IRES-dependent protein XIAP and FGF2. A. The levels of XIAP, FGF2 were detected as described in the Materials and Methods, and the optical densities were normalized to a control (α-tubulin). B – D. Optical densities of the XIAP bands (B) and FGF2 isoforms 1 (C) and 2 (D). Values are the mean and SEM of 4 experiments. ***p<0.001 versus the WT control value. †††p<0.001 versus the diphthamide-deficient cell control value. ††p<0.01 versus the diphthamide-deficient cell control value. †p<0.05 versus the diphthamide-deficient cell control value. ‡‡‡ p<0.001 versus the WT 1 μM CH value. §§§ p<0.001 versus the WT 5 μM CH value. ¶¶¶ p<0.001 versus the WT 10 μM CH value.
Next, we measured the levels of FGF2 using an antibody that recognized two isoforms of 18 kDa (isoform 1) and 24 kDa (isoform 2) in extracts from wild-type and Dph3− cells (Fig. 6A). Similarly to what seen for XIAP, a concentration-dependent increase of FGF2 isoform 1 and 2 levels occurred in response to CH treatment in wild-type cells (Fig. 6C, D). On the other hand, exposure of Dph− cells to CH resulted in little or no change in the levels of either isoform of FGF2 (Fig. 6C, D). These results suggest that eEF2 diphthamide plays an important role in regulating the synthesis of XIAP and FGF2 under oxidative stress conditions.
DISCUSSION
Protein synthesis is a fundamental process that is critical for cellular homeostasis and responses to stress. Because of its high energy requirements it is tightly regulated, especially under stress conditions when ATP is diverted toward repair processes. In fact, global translation is reduced in response to most, if not all, types of cellular stress [4,7,10,33]. However, the stress-induced reduction in global translation is usually accompanied by the selective translation of specific proteins that are required for the cell to withstand the period of adverse conditions. This process is not dependent on the translation initiation step; instead the ribosome starts ‘reading’ the mRNA at the IRES sequence [17,18]. Although IRES-mediated protein synthesis does not require the initiation factors, it is still dependent on the elongation factors. Notably, eEF2 is very sensitive to oxidative stress [22–24], and thus its ability to participate in the IRES synthesis of proteins under adverse oxidative conditions must depend upon protective regulatory mechanisms.
A unique posttranslational derivative of histidine known as diphthamide is found only in eEF2. The biological function of diphthamide has not yet been defined. It has been suggested that diphthamide protects ribosomes against pathogenic ribosome-inactivating proteins [27]. Furthermore, diphthamide participates in the maintenance of the correct reading frame during elongation, increasing protein translational accuracy [34]. Less clear are the effects of diphthamide ribosylation on protein translation. Ribosylation induced by bacterial toxins results in the cessation of translation [19,35,36]. However other reports suggest that diphthamide is not strictly required for eEF2 elongation function [27]. Indeed, diphthamide-deficient cells are viable indicating that eEF2 must retain enough activity to drive protein synthesis at a rate compatible with cell survival, or that compensatory mechanisms are induced. Our results showing higher eEF2 cellular content (Fig. 2) and newly synthesized proteins rate (Fig. 4) in Dph3− cells support the later hypothesis. Upregulation of eEF2 has been observed in ovarian, gastric and colon cancers [37,38] and it is believed to be an adaptive mechanism of cancer cells to maintain their high protein synthesis requirements [1,6]. Modification of diphthamide synthesis genes have also been linked to cancer susceptibility. Mutations of dph1 (ovac1) gene occur in 90% of ovarian cancers, as well as other cancer cell types [39]. Given the requirement of diphthamide for translation accuracy [27], the increased rate of basal newly synthesized proteins is not necessarily advantageous for the cells under stressful conditions. In fact, our results show that under high oxidative stress conditions diphthamide-deficient cells exhibited increased eEF2 degradation (Fig. 5), and were more sensitive to cell death (Fig. 1). These findings suggest that eEF2/diphthamide is a critical determinant of the ability of cells to respond to oxidative stress in ways that promote their survival.
eEF2 is inactivated by phosphorylation resulting in reduction of global protein synthesis. However, the fact that phosphorylated eEF2 is found in the nucleus of cells and its levels increase under oxidative stress suggests that this modification may have additional biological roles. The presence of phosphorylated eEF2 in the nucleus has been associated with a lower resistance to oxidative stress [16]. Here we showed that diphthamide-deficient cells have higher levels of total eEF2 in both whole-cell extract and cytoplasm (Fig. 2). However, eEF2 levels in the nuclear fractions were similar in wild-type and diphthamide-deficient cells. When the ratio of phosphorylated/total eEF2 was analyzed, this ratio increased in cytoplasm and to a greater extent in the nucleus in response to oxidative stress. Whereas the increase in the cytoplasm was similar in both wild-type and Dph3− cells, the increase in the nucleus was more pronounced in Dph3− cells (Fig 3). These results suggest that diphthamide plays a role in the subcellular localization of eEF2 under normal and oxidative stress conditions. In addition to phosphorylation, previous findings indicated that eEF2 interacts with p53 [16,40]. However, we found that the association between eEF2 and p53 do not depends on the presence of diphthamide (Results not shown).
Our results suggest that diphthamide plays a role in the protection of cells against the degradation of eEF2 under normal and oxidative stress conditions. This protection is especially important under those situations where it is necessary to reduce global protein synthesis while selectively enabling translation of IRES-containing mRNAs that encode proteins critical for cell survival. Indeed, we found that the production of two proteins (XIAP and FGF2) from IRES-containing mRNAs [4,41] is increased in wild-type cells under oxidative stress conditions. However their expression is deregulated in diphthamide-deficient cells. This suggests that eEF2/diphthamide can play a relevant role in IRES translation under oxidative stress.
Highlights.
Diphthamide regulates global newly synthesized protein during oxidative stress
Diphthamide plays a role in the protection of cells against the degradation of eEF2
eEF2/diphthamide is important to control the translation of IRES-dependent protein XIAP and FGF2
Acknowledgments
We thank Stephen H Leppla and Christopher Bachran of the Laboratory of Bacterial Diseases, National Institute of Allergy and Infectious Diseases (NIAID), NIH for providing wild-type and diphthamide-deficient Chinese hamster ovary cells and fusion protein (FP59). We thank Myriam Gorospe and Jennifer L Martindale of the Laboratory of Molecular Biology and Immunology (LMBI), NIH for technical advice regarding protein synthesis assays. S.A.C. was supported by a Ministerio de Educacion, Cultura y Deporte of Spain postdoctoral fellowship (EX2009-0918) – A.A was supported by Ministerio de Ciencia e Innovación BFU 2010-20882. This work was supported in part by the Intramural Research Program of the National Institute on Aging
Footnotes
Conflict of Interest: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 2.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 3.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 4.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 5.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White-Gilbertson S, Kurtz DT, Voelkel-Johnson C. The role of protein synthesis in cell cycling and cancer. Mol Oncol. 2009;3:402–408. doi: 10.1016/j.molonc.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce M, Py BF, Ryazanov AG, Minden JS, Long K, Ma D, Yuan J. A pharmacoproteomic approach implicates eukaryotic elongation factor 2 kinase in ER stress-induced cell death. Cell Death Differ. 2008;15:589–599. doi: 10.1038/sj.cdd.4402296. [DOI] [PubMed] [Google Scholar]
- 8.Zelivianski S, Liang D, Chen M, Mirkin BL, Zhao RY. Suppressive effect of elongation factor 2 on apoptosis induced by HIV-1 viral protein R. Apoptosis. 2006;11:377–388. doi: 10.1007/s10495-006-4030-9. [DOI] [PubMed] [Google Scholar]
- 9.Knebel A, Haydon CE, Morrice N, Cohen P. Stress-induced regulation of eukaryotic elongation factor 2 kinase by SB 203580-sensitive and -insensitive pathways. Biochem J. 2002;367:525–532. doi: 10.1042/BJ20020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel J, McLeod LE, Vries RG, Flynn A, Wang X, Proud CG. Cellular stresses profoundly inhibit protein synthesis and modulate the states of phosphorylation of multiple translation factors. Eur J Biochem. 2002;269:3076–3085. doi: 10.1046/j.1432-1033.2002.02992.x. [DOI] [PubMed] [Google Scholar]
- 11.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 12.Iadevaia V, Wang X, Yao Z, Foster LJ, Proud CG. Evaluation of mTOR-regulated mRNA translation. Methods Mol Biol. 2012;821:171–185. doi: 10.1007/978-1-61779-430-8_10. [DOI] [PubMed] [Google Scholar]
- 13.Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, Woods A, Robinson IC, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L, Gems D, Withers DJ. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulati P, Thomas G. Nutrient sensing in the mTOR/S6K1 signalling pathway. Biochem Soc Trans. 2007;35:236–238. doi: 10.1042/BST0350236. [DOI] [PubMed] [Google Scholar]
- 15.Proud CG. The multifaceted role of mTOR in cellular stress responses. DNA Repair (Amst) 2004;3:927–934. doi: 10.1016/j.dnarep.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Arguelles S, Camandola S, Hutchison E, Cutler R, Ayala A, Mattson M. Molecular control of the amount, subcellular location and activity state of translation elongation factor 2 (eEF2) in neurons experiencing stress. Free Radic Biol Med. 2013;61:61–71. doi: 10.1016/j.freeradbiomed.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komar AA, Mazumder B, Merrick WC. A new framework for understanding IRES-mediated translation. Gene. 2012;502:75–86. doi: 10.1016/j.gene.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 19.Jørgensen R, Merrill AR, Andersen GR. The life and death of translation elongation factor 2. Biochem Soc Trans. 2006;34:1–6. doi: 10.1042/BST20060001. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bektaş M, Akçakaya H, Aroymak A, Nurten R, Bermek E. Effect of oxidative stress on in vivo ADP-ribosylation of eukaryotic elongation factor 2. Int J Biochem Cell Biol. 2005;37:91–99. doi: 10.1016/j.biocel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Argüelles S, Cano M, Machado A, Ayala A. Effect of aging and oxidative stress on elongation factor-2 in hypothalamus and hypophysis. Mech Ageing Dev. 2011;132:55–64. doi: 10.1016/j.mad.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Argüelles S, Machado A, Ayala A. Adduct formation of 4-hydroxynonenal and malondialdehyde with elongation factor-2 in vitro and in vivo. Free Radic Biol Med. 2009;47:324–330. doi: 10.1016/j.freeradbiomed.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Argüelles S, Machado A, Ayala A. ‘In vitro’ effect of lipid peroxidation metabolites on elongation factor-2. Biochim Biophys Acta. 2006;1760:445–452. doi: 10.1016/j.bbagen.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Poot M, Esterbauer H, Rabinovitch PS, Hoehn H. Disturbance of cell proliferation by two model compounds of lipid peroxidation contradicts causative role in proliferative senescence. J Cell Physiol. 1988;137:421–429. doi: 10.1002/jcp.1041370305. [DOI] [PubMed] [Google Scholar]
- 26.Jiang ZY, Woollard AC, Wolff SP. Lipid hydroperoxide measurement by oxidation of Fe2+ in the presence of xylenol orange. Comparison with the TBA assay and an iodometric method. Lipids. 1991;26:853–856. doi: 10.1007/BF02536169. [DOI] [PubMed] [Google Scholar]
- 27.Gupta PK, Liu S, Batavia MP, Leppla SH. The diphthamide modification on elongation factor-2 renders mammalian cells resistant to ricin. Cell Microbiol. 2008;10:1687–1694. doi: 10.1111/j.1462-5822.2008.01159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 29.Shan H, Ji D, Barnard AR, Lipinski DM, You Q, Lee EJ, Kamalden TA, Sun X, MacLaren RE. AAV-mediated gene transfer of human X-linked inhibitor of apoptosis protects against oxidative cell death in human RPE cells. Invest Ophthalmol Vis Sci. 2011;52:9591–9597. doi: 10.1167/iovs.10-6850. [DOI] [PubMed] [Google Scholar]
- 30.Cucchiarini M, Schetting S, Terwilliger EF, Kohn D, Madry H. rAAV-mediated overexpression of FGF-2 promotes cell proliferation, survival, and alpha-SMA expression in human meniscal lesions. Gene Ther. 2009;16:1363–1372. doi: 10.1038/gt.2009.91. [DOI] [PubMed] [Google Scholar]
- 31.Tsang AH, Lee YI, Ko HS, Savitt JM, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM, Chung KK. S-nitrosylation of XIAP compromises neuronal survival in Parkinson’s disease. Proc Natl Acad Sci U S A. 2009;106:4900–4905. doi: 10.1073/pnas.0810595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattson MP, Murrain M, Guthrie PB, Kater SB. Fibroblast growth factor and glutamate: opposing roles in the generation and degeneration of hippocampal neuroarchitecture. J Neurosci. 1989;9:3728–3740. doi: 10.1523/JNEUROSCI.09-11-03728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connolly E, Braunstein S, Formenti S, Schneider RJ. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol Cell Biol. 2006;26:3955–3965. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TK. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem. 2006;281:32639–23648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- 35.Bektaş M, Nurten R, Ergen K, Bermek E. Endogenous ADP-ribosylation for eukaryotic elongation factor 2: evidence of two different sites and reactions. Cell Biochem Funct. 2006;24:369–380. doi: 10.1002/cbf.1265. [DOI] [PubMed] [Google Scholar]
- 36.Koch-Nolte F, Reche P, Haag F, Bazan F. ADP-ribosyltransferases: plastic tools for inactivating protein and small molecular weight targets. J Biotechnol. 2001;92:81–87. doi: 10.1016/s0168-1656(01)00356-x. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura J, Aoyagi S, Nanchi I, Nakatsuka S, Hirata E, Shibata S, Fukuda M, Yamamoto Y, Fukuda I, Tatsumi N, Ueda T, Fujiki F, Nomura M, Nishida S, Shirakata T, Hosen N, Tsuboi A, Oka Y, Nezu R, Mori M, Doki Y, Aozasa K, Sugiyama H, Oji Y. Overexpression of eukaryotic elongation factor eEF2 in gastrointestinal cancers and its involvement in G2/M progression in the cell cycle. Int J Oncol. 2009;34:1181–1189. [PubMed] [Google Scholar]
- 38.Alaiya AA, Franzen B, Fujioka K, Moberger B, Schedvins K, Silfversvard C, Linder S, Auer G. Phenotypic analysis of ovarian carcinoma: polypeptide expression in benign, borderline and malignant tumors. Int J Cancer. 1997;73:678–683. doi: 10.1002/(sici)1097-0215(19971127)73:5<678::aid-ijc11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Chen CM, Behringer RR. OVCA1: tumor suppressor gene. Curr Opin Genet Dev. 2005;15:49–54. doi: 10.1016/j.gde.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Yin X, Fontoura BMA, Morimoto T, Carroll RB. Cytoplasmic complex of p53 and eEF-2. J Cell Physiol. 2003;196:474–482. doi: 10.1002/jcp.10329. [DOI] [PubMed] [Google Scholar]
- 41.Audigier S, Guiramand J, Prado-Lourenco L, Conte C, Gonzalez-Herrera IG, Cohen-Solal C, Récasens M, Prats AC. Potent activation of FGF-2 IRES-dependent mechanism of translation during brain development. RNA. 2008;14:1852–1864. doi: 10.1261/rna.790608. [DOI] [PMC free article] [PubMed] [Google Scholar]