Background: ASC is the common adaptor of caspase-1 activation in several inflammasomes.
Results: A novel interaction between PML and ASC is identified. PML-deficient macrophages display enhanced levels of IL-1β secretion and higher levels of ASC in the cytosol.
Conclusion: PML retains ASC in the nucleus limiting inflammasome activation.
Significance: Understanding the regulation of inflammasome components will aid in the development of therapeutics to alleviate IL-1β-mediated diseases.
Keywords: Cancer, Cytokine, Inflammation, Innate Immunity, Nod-like Receptors (NLR), Inflammasome
Abstract
The apoptosis-associated speck-like protein containing a caspase-activating recruitment domain (ASC) is an essential component of several inflammasomes, multiprotein complexes that regulate caspase-1 activation and inflammation. We report here an interaction between promyelocytic leukemia protein (PML) and ASC. We observed enhanced formation of ASC dimers in PML-deficient macrophages. These macrophages also display enhanced levels of ASC in the cytosol. Furthermore, IL-1β production was markedly enhanced in these macrophages in response to both NLRP3 and AIM2 inflammasome activation and following bone marrow-derived macrophage infection with herpes simplex virus-1 (HSV-1) and Salmonella typhimurium. Collectively, our data indicate that PML limits ASC function, retaining ASC in the nucleus.
Introduction
Inflammasomes are large cytoplasmic multiprotein complexes that act to regulate the activation of caspase-1, the enzyme that processes the pro-forms of the cytokines IL-1β and IL-18 into their active forms (1). Six pathogen recognition receptors are known to drive inflammasome formation. These are the cytoplasmic nucleotide-binding oligomerization domain-like receptors, NLRP1, NLRP3, NLRP6, and NLRC4, and certain proteins of the pyrin and HIN domain (PYHIN) family termed AIM2-like receptors such as AIM2 and IFI16 (2). The recruitment of pro-caspase-1 to the inflammasome is facilitated by the adaptor protein, apoptosis associated speck-like protein (ASC)3 containing a CARD (also known as PYCARD or TMS1) (3, 4). ASC is a 22-kDa protein containing an N-terminal pyrin domain and a C-terminal CARD (3, 4). During NLRP3 and AIM2 inflammasome activation, homotypic interactions are formed between the pyrin domain of ASC and those of NLRP3 or AIM2 (5, 6). Subsequently, the CARD domain of ASC recruits pro-caspase-1 leading to its auto-cleavage and activation (5, 7). Deletion of the ASC gene in mice abolishes caspase-1 activation and secretion of IL-1β in response to Salmonella typhimurium (8), lipopolysaccharide (LPS) (9), and influenza virus (10), highlighting its central role in inflammasome function.
Overexpression systems have been used to study and visualize the formation of an ASC aggregate or “speck” indicative of inflammasome assembly (3, 11). Splice variants of ASC are also known to exist and have been shown to compete with full-length ASC for association with NLRP3, hampering the processing of IL-1β (12, 13). ASC has been shown to be primarily in the nucleus of resting monocytes, and upon pathogen infection, it rapidly redistributes to the cytosol (14). ASC has also been shown to redistribute to the cytoplasm along with the p20 subunit of caspase-1 following infection of endothelial cells with Kaposi sarcoma-associated herpesvirus (15). Furthermore, ASC redistribution has been detected in THP-1 cells stimulated with microbial lipopeptides (16). Nothing is known about the mechanisms controlling ASC trafficking from the nucleus to the cytosol.
Here, we report an interaction between ASC and the nuclear body component promyelocytic leukemia protein (PML). PML acts as a scaffold protein for subnuclear structures termed PML nuclear bodies. It is a member of the tripartite motif protein family and contains a characteristic tripartite domain termed RBCC consisting of a RING (R), one or two B-boxes (B), and a predicted coiled-coil region at its N terminus. The PML transcript undergoes alternative splicing to generate several isoforms, all of which differ solely in their C-terminal region (17, 18). In this regard, a nuclear export sequence is present and retained in the C-terminal portion of PMLI. This allows PMLI to shuttle between the nucleus and the cytoplasm (19). Here, we report that the interaction between PML and ASC is involved in the retention of ASC in the nucleus. We have found that PML is a negative regulator of ASC, attenuating inflammasome activation by retaining ASC in the nucleus.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
pBabe-GFP-PMLI was a kind gift from Prof. Avri Ben-Ze'ev, Department of Molecular Cell Biology, Weizmann Institute of Science, Israel. 12-Myristate 13-acetate, puromycin, poly(deoxyadenylic-thymidylic) acid sodium salt (poly(dA-dT)), and adenosine 5′-triphosphate disodium salt hydrate (ATP) were purchased from Sigma. The following antibodies were used: rabbit anti-ASC (AL177) (AdipoGen), mouse anti-β-actin, mouse anti-GFP, or mouse anti-GAPDH (Sigma) and lamin B (AbCam), mouse monoclonal PML antibody (PG-M3), GFP antibody, and protein A/G-plus-agarose beads were purchased from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Jackson ImmunoResearch. Ultrapure rough LPS (from Escherichia coli, serotype EH100) was purchased from Alexis. Lipofectamine 2000TM was from Invitrogen. StrataCleanTM resin was from Agilent Technologies. TNFα and IL-1β ELISA DuoSet® kits were purchased from R&D Systems. For viral infection, herpes simplex virus (HSV-1) (a gift from I. Julkunen) was used. S. typhimurium UK-1 strain was obtained from Dr. Sinead Corr (Trinity College Dublin, Ireland).
Cell Culture
The human embryonic kidney T cell (HEK-293T) and the human acute monocytic leukemia (THP-1) cell lines were purchased from the European Cell Culture Collection. HEK293 cells and THP-1 parental cells were maintained in DMEM and RPMI, respectively, supplemented with 10% (v/v) FCS and 1% (v/v) penicillin/streptomycin solution. Bone marrow from wild type and PML−/− 129Sv mice was differentiated for 7 days in DMEM supplemented with 10% (v/v) FCS and 1% (v/v) penicillin/streptomycin solution and M-CSF (20% (v/v) L929 mouse fibroblast supernatant). For experiments, differentiated BMDM were seeded at 5 × 105 cells/ml.
Mass Spectrometry
Cell-free extracts were prepared from THP-1 cells as described previously (20). Samples were diluted in immunoprecipitation buffer (20 mm HEPES-KOH, pH 7.5, 50 mm NaCl, 0.3% CHAPS, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, I mm dithiothreitol, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 250 μm phenylmethylsulfonyl fluoride) and precleared with protein A/G-agarose beads (Santa Cruz Biotechnology) at 4 °C for 1 h. Precleared cell-free extracts were incubated with fresh protein A/G-agarose beads (Santa Cruz Biotechnology) and anti-ASC antibody (Adipogen) overnight at 4 °C. Captured complexes were washed three times in immunoprecipitation buffer and eluted into two-dimensional gel sample buffer (8 m urea, 4% CHAPS, 0.05% SDS, 100 mm dithiothreitol, 0.03% bromphenol blue, 0.2% ampholytes). Proteins were separated using two-dimensional gel electrophoresis and silver-stained, and in-gel protein digestion and protein identification by MALDI-TOF using a Voyager DE-PRO mass spectrometer were carried out as described previously (20).
Co-immunoprecipitation Assay
HEK-293T cells were seeded at 2 × 105 cells/ml in 10-cm dishes. 24 h later, cells were transfected with a total of 8 μg of the indicated plasmids using GeneJuice® and cultured for a further 48 h before harvesting. 50 μl of each sample was removed as whole cell lysate. Co-immunoprecipitations (co-IPs) were performed with GFP-TRAP beads (Chromotek) according to the manufacturer's instructions. Cell lysates were incubated with the beads at 4 °C for 1–2 h. Following this, the lysate and beads were centrifuged at 2,200 × g for 3 min at 4 °C; the supernatant was removed, and the beads were washed three times in 1 ml of wash buffer. Immune complexes were eluted by the addition of 50 μl of SDS/Laemmli buffer and boiling the samples. Co-IPs were analyzed by SDS-PAGE and Western blotting.
PML Truncations
The PML truncations PML(1–166), PML(1–223), PML(1–286), and PML(1–491) were generated by PCR amplification of inserts with primers incorporating restriction sites for BamHI and EcoRI (primers shown in Table 1). Resulting BamHI and EcoRI fragments were ligated into pBabe-puro. The PML truncations PML(1–166), PML(1–223), PML(1–286), and PML(1–491) were generated by PCR amplification of inserts with primers incorporating restriction sites for BamHI and EcoRI. Resulting BamHI and EcoRI fragments were ligated into pBabe-puro. PML truncation primers used are as follows: sense (all), CTGAGGATCCATGGTGAGCAAGGGCGAGGAGCTG, and antisense, 1–166, ATTAGAATTCCTATGCTAGGGGCCGGGCCTCGTGCTTGAGGAA; 1–223, ATTAGAATTCCTACTGTGGCTGCTGTCAAGGAGCGCGCACGAGCA; 1–286, ATTAGAATTCTACGTGAGCTACCACCTGGCGCACGCGCTC; and 1–491, ATTAGAATTCCTAGACTCCATCTTGATGACCTTCCTGGGGCACTGGGTCTG.
TABLE 1.
Proteins associating with ASC as identified by mass spectrometry from cell-free extracts of THP-1 cells
| Protein | Mr | MS FIT SCORE | |
|---|---|---|---|
| Q9NX36 | J domain-containing protein C21orf55 | 45.8 | 483 |
| O43586 | PSTPIP1 | 47.5 | 453 |
| Q8NB25 | Uncharacterized protein C6orf60 | 119 | 23080 |
| P29590 | PML | 97.5 | 12915 |
| O15327 | Type II inositol 3,4-biphosphate-4-phosphate | 104.7 | 5169 |
| Q9Y2U8 | Inner nuclear membrane protein Man1 | 99.9 | 4665 |
| Q9WJR | HER V-K-19q12 povirus ancestral PoI protein | 108 | 4564 |
| Q9UPN4 | 5-Azactidine-induced protein 1 | 122 | 4533 |
| Q8NEN0 | Armadillo repeat-containing protein 2 | 96 | 3817 |
| O00159 | Myosin | 118 | 3814 |
| P40189 | IL-6 receptor subunit β precursor | 103 | 3361 |
| P33991 | DNA replication licensing factor MCM4 | 96 | 3345 |
| Q9NRE2 | Teashirt homolog 2 | 115 | 2263 |
| P57740 | Nuclear pore complex protein Nup107 | 106 | 2172 |
| Q8N475 | Follistatin-related protein 5 precursor | 95 | 2171 |
| Q9BXJ9 | NMDA receptor-regulated protein 1 | 101 | 1711 |
| Q8IY82 | Coiled-coil domain-containing protein 135 | 135 | 1663 |
| P49916 | DNA ligase 3 | 102 | 1583 |
| Q13873 | Bone morphogenetic protein receptor type-2 precursor | 115 | 1435 |
| P98073 | Enteropeptidase precursor | 112 | 1381 |
| P67936 | Tropomyosin α-4 chain | 28.5 | 1115 |
Enzyme-linked Immunosorbent Assay
For cytokine measurements, BMDM were seeded at 5 × 105 cell/ml in a 12-well plate and stimulated in triplicate. Supernatants were removed and analyzed for IL-1β and TNF-α, IL-6 (R&D Systems) or IL-18 (eBiosciences) using enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturers' instructions.
Viral and Bacterial Infection
BMDM cells were seeded at 5 × 105 cells/ml in 12-well plates and left to rest overnight. Cells were then left unstimulated or stimulated with LPS (100 ng/ml) for 3 h and then mock-infected or infected with HSV-1 (m.o.i. 10) for 22 h. For infection with S. typhimurium, BMDM were stimulated with 10 ng/ml LPS for 3 h followed by infection with S. typhimurium (m.o.i. 20) for 2 h. Cells were stimulated in triplicate. Supernatants were removed and analyzed by enzyme-linked immunosorbent assay for IL-1β, TNF-α, and IL-6 or lactate dehydrogenase as described.
Cytotoxicity Assay
A CytoTox96® nonradioactive cytotoxicity assay kit was used according to the manufacturer's instructions (Promega) to determine lactate dehydrogenase release from cells.
Western Blotting
Cell lysates were prepared by direct lysis in 5× Laemmli sample buffer. The protein content of supernatants was concentrated using StrataCleanTM resin according to the manufacturer's instructions. Protein samples were resolved on 12 or 15% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes using a semi-dry transfer system (Cleaver Scientific). Membranes were blocked in 5% (w/v) dried milk in TBS-T (50 mm Tris/HCl, pH 7.6, 150 mm NaCl, and 0.1% (v/v) Tween 20) for 1 h at room temperature (RT). Membranes were incubated with antibodies against ASC (AL177), β-actin, lamin B, GFP, IL-1β, or GAPDH at a 1:1000 dilution in 5% (w/v) dried milk in TBS-T overnight at 4 °C. Membranes were probed for PML using mouse anti-PML (PG-M3) at a 1:500 dilution in 5% (w/v) BSA in TBS. Membranes were then incubated with the appropriate HRP-conjugated secondary antibody diluted 1:2000 in 5% (w/v) dried milk in TBS-T for an hour before being developed by enhanced chemiluminescence (ECL) according to the manufacturer's instructions (Cell Signaling Technology, Inc.). Some membranes were stripped using RestoreTM PLUS Western blot stripping buffer according to the manufacturer's instructions (Thermo Fisher Scientific Inc.) before being reprobed.
Quantitative Real Time RT-PCR
Total RNA samples were isolated using RNeasy mini kit (Qiagen, MD) according to the manufacturer's instructions. For quantitative real time PCR, cDNA was transcribed using the High Capacity cDNA reverse transcription kit (Applied Biosystems). Primers for human PMLI were purchased from MWG, and probes for human and mouse PML (encompassing exons common to all PML isoforms) were purchased from Applied Biosystems.
Stable Cell Line Generation
The shRNAmir clones were purchased from Open Biosystems (V3LHS_304380 and V3LHS_400341). 293T cells were seeded at 2 × 105 cells/ml in 10-cm dishes. Plasmids encoding shRNA (4 μg) or nonsilencing control (4 μg) and viral proteins pSPAX2 (3 μg) and pMD2 (1 μg) were transfected into 293T cells using GeneJuice according to the manufacturer's protocol. 48 h post-transfection, viruses containing supernatants were harvested and replaced with fresh media. Collected supernatants were centrifuged, filtrated with 0.2-μm filters, and stored at 4 °C. The harvesting step was repeated after 24 h, and supernatants were mixed together. THP-1 target cells were plated at 2 × 105 cells/ml in 10-cm dishes. After 24 h, cells were replated with 50% RPMI culture medium with 10% (v/v) FCS and 1% (v/v) penicillin/streptomycin solution and 50% harvested virus-containing supernatants. 4 μg of Polybrene (Sigma) was added into the cells. Transduced cells were cultured for 48 h. Puromycin selection (3 μg/ml) was initiated after another 48 h. THP1-stable PML knockdown cells were maintained in RPMI. All media contained 10% (v/v) FCS, 1% (v/v) penicillin/streptomycin solution, and 3 μg/ml puromycin. Knockdown efficiency was assessed by real time PCR as described.
ASC Oligomerization Assay
2 × 106 BMDM cells were seeded per well in a 6-well plate. The following day, cells were stimulated with 100 ng/ml LPS for 3 h followed by the addition of 5 mm ATP for 1 h or transfection of 1 μg/ml poly(dA-dT) for 5 h. Supernatants were removed, and cells were rinsed in ice-cold PBS. 500 μl of ice-cold buffer (20 mm HEPES-KOH, pH 7.5, 150 mm KCl, 1% Nonidet P-40, 0.1 mm PMSF, 1 μg/ml leupeptin, 11.5 μg/ml aprotinin, and 1 mm sodium orthovanadate) was added. Cells were removed using a cell scraper and transferred to microcentrifuge tubes. Cells were lysed by shearing 10× through a 21-gauge needle. 50 μl of lysate was removed for Western blot analysis. The lysates were centrifuged at 330 × g for 10 min at 4 °C. The pellets were washed twice in 1 ml of ice-cold PBS (centrifuged 330 × g for 3 min at 4 °C) and resuspended in 500 μl of PBS. 2 mm disuccinimidyl suberate (from a fresh 100 mm stock prepared from disuccinimidyl suberate equilibrated to RT and made up in dry DMSO) was added to the resuspended pellets, which were incubated at RT for 30 min with rotation. The samples were then centrifuged at 330 × g for 10 min at 4 °C. The supernatant was removed, and cross-linked pellets were resuspended in 40 μl of Laemmli sample buffer. The samples were boiled for 5 min at 99 °C and analyzed by Western blotting.
Subcellular Fractionation
Subcellular fractionations were performed using the nuclear extraction kit as per the manufacturer's instructions (Active Motif). Briefly, 8 × 106 cells were isolated by scraping off the dish and washing in ice-cold PBS. Cells were resuspended in hypotonic buffer and incubated for 15 min on ice before addition of 0.5% final Nonidet P-40. The use of Nonidet P-40, which is ionic, ensures a high stringency lysis of cells and the nuclear compartment. Homogenates were briefly centrifuged, and the supernatant containing the cytoplasmic fraction was stored at −80 °C. Pellets were resuspended in complete lysis buffer and incubated on ice for 30 min while rocking. Lysates were centrifuged for 10 min at 14,000 × g at 4 °C. Supernatants containing the nuclear fraction were stored at −80 °C.
RESULTS
PML Interacts with ASC in the Nucleus
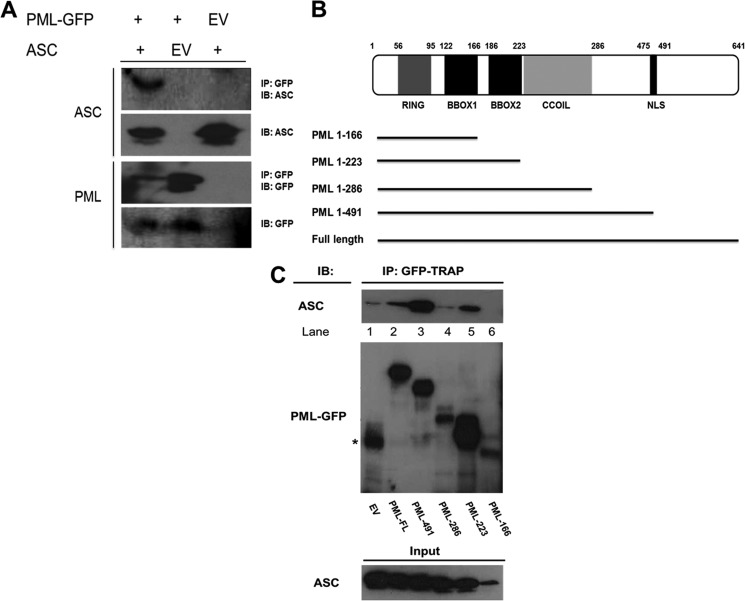
To identify proteins that interact with ASC, we isolated ASC by immunoprecipitation from whole cell lysates prepared from THP-1 cells. Proteins in the complex were identified by MALDI-TOF mass spectrometry. Peak lists were submitted to databases MASCOT and MS-FIT. A highly significant MS-FIT score was obtained for PML (Table 1). We confirmed this interaction by co-IP. Co-IPs were performed in HEK293T cells transiently overexpressing ASC and PMLI-GFP. ASC immunoprecipitated with PMLI-GFP and not with EV-GFP (Fig. 1A, top row, compare first and second lanes). These results confirmed the interaction between PML and ASC identified by mass spectrometry.
FIGURE 1.
PML interacts with ASC. A, HEK293T cells were transfected with plasmids encoding empty vector green fluorescent protein (EV-GFP), PMLI-GFP, and/or ASC as indicated. 48 h post-transfection, cells were lysed, and 50 μl of whole cell lysate was kept for analysis. The remainder was immunoprecipitated (IP) with GFP-TRAPTM beads for 1–2 h at 4 °C. Whole cell lysates and immunoprecipitated samples were analyzed by Western blotting using anti-GFP and anti-ASC (AL177) antibodies. GFP pulldown resulted in the precipitation of PML-GFP (1st and 2nd lanes, 3rd panel) as expected and also ASC (1st lane, top panel). B, PML truncations were generated to identify the domains involved in the interaction with ASC. C, HEK293T cells were transfected with plasmids encoding ASC and EV-GFP or PML truncations as indicated. Proteins were precipitated as in A and analyzed by Western blotting. GFP pulldown resulted in the precipitation of ASC with PML(1–223), PML(1–491), and PML-FL. C, top panel, * denotes nonspecific band. Results presented are representative of four (A) and three (C) independent experiments. IB, immunoblot.
Next, we generated truncated mutants of PML-GFP to identify the regions of PML that interact with ASC. A schematic of the truncations is shown in Fig. 1B. Expression levels of the different mutants varied as can be seen in Fig. 1C, middle panel. The full-length form (Fig. 1C, lane 2) and PML-491 were equally expressed. PML-286 (Fig. 1C, lane 4) and PML-166 (lane 6) had low expression, whereas PML-223 was highly expressed (lane 5). PML-491 showed a stronger interaction with ASC than the full-length form (Fig. 1C, top panel, compare lane 3 with lane 2), possibly indicating an inhibitory region between amino acids 491 and 641. PML-223 could still interact with ASC (Fig. 1C, lane 5) indicating that the interaction could be occurring with the RING, BBOX1, or BBOX2 domains.
Effect of PML on Inflammasome-induced ASC Oligomerization
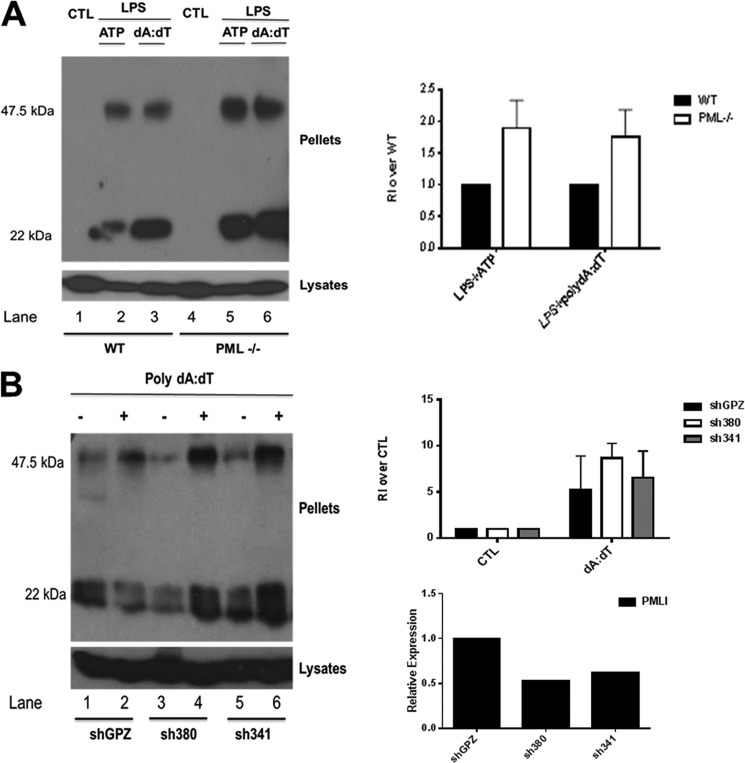
We next examined the effect of PML deficiency on ASC oligomerization. The formation of ASC dimers and higher order oligomers can be detected in cross-linked cell pellets analyzed by Western blotting. As shown in Fig. 2A, activation of the NLRP3 inflammasome with LPS and ATP led to ASC dimer formation (lane 2), an effect that was enhanced in PML-deficient macrophages (lane 5). This was also the case with activation of the AIM2 inflammasome (Fig. 2A, compare lane 3 for wild type with lane 6 for PML-deficient macrophages). Densitometric analysis of oligomerization is shown in Fig. 2A (right panel).
FIGURE 2.
Loss of PML enhances NLRP3- and AIM2-stimulated ASC oligomerization. A, left panel, primary BMDM from 129Sv wild type (WT) and PML-deficient mice were left unstimulated or stimulated with 100 ng/ml LPS for 3 h with or without the addition of 5 mm ATP for 1 h or transfected with 1 μg/ml poly(dA-dT) for 5 h as indicated. Cell pellets were washed and resuspended in PBS and then cross-linked by incubation with 20 mm DSS for 30 min. Cross-linked pellets and cell lysates were analyzed by Western blotting with anti-ASC antibody (AL177). Right panel, densitometry of oligomerization experiments is presented. Each dimer band was normalized to whole cell lysate ASC content, and the relative intensity (RI) of the dimer band (47.5 kDa) over the whole cell lysate (set at 1) was calculated. The data shown are representative of three independent experiments. CTL, control. B, left panel, all three THP-1 cell lines were differentiated with 12-myristate 13-acetate for 16 h. Following this, cells were stimulated with 2 μg/ml poly(dA-dT) for 5 h. Cell lysates were centrifuged at 330 × g for 10 min at 4 °C. Cell pellets were again analyzed for ASC by Western blotting as in A. Right panel, top row, densitometry of oligomerization in THP-1 cells lines with stable knockdown of PML. Each band was normalized to its corresponding whole cell lysate ASC content, and the relative intensity (RI) of the dimer band (47.5 kDa) over the whole cell lysate (set at 1) was calculated. The data shown are representative of two independent experiments. Right panel, bottom row, mRNA knockdown of PMLI in THP-1 cell lines generated using two shRNA lentiviral vectors (sh380 and sh341). Nonsilencing lentiviral vector pGIPZ was used for generation of nontargeting control cell line, shGPZ.
We confirmed this effect in two PMLI-deficient stable THP-1 cell lines generated by lentiviral infection of short hairpin RNA, sh380 and sh341. A greater amount of dimer was detected in both sh380 and sh341 cell lines stimulated with poly(dA-dT) compared with the nonsilencing control, shGPZ (Fig. 2B, left panel, compare 4th and 6th lanes with 2nd lane). The relative knockdown of PMLI mRNA is shown in Fig. 2B (right panel, bottom) along with densitometric analysis of oligomerization Fig. 2B (right panel, top).
Subcellular Localization of ASC in Wild Type Versus PML-deficient BMDM
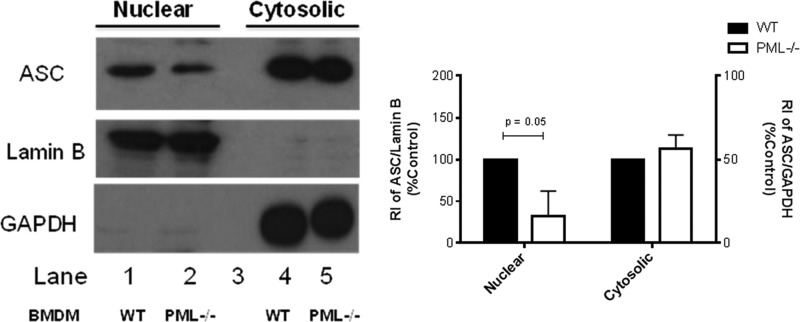
We next considered that if PML retains ASC in the nucleus, then resting PML-deficient cells would have a different subcellular pattern of ASC compared with wild types. Subcellular fractionation was performed on PML-deficient and wild type BMDM. Levels of ASC in the nuclear fraction of PML-deficient cells was markedly lower than that detected in WT controls (Fig. 3, left panel, top row, compare lane 2 with lane 1). Antibodies to lamin B and GAPDH were used as nuclear and cytoplasmic loading controls, respectively. Densitometric analysis of fractionation experiments is presented (Fig. 3, right panel). This result indicates that the inhibitory effect of PML on ASC dimerization following NLRP3 or AIM2 activation (which occurs in the cytosol) may in part be due to retaining ASC in the nucleus.
FIGURE 3.
PML-deficient BMDM display reduced levels of ASC in the nucleus compared with WT. 8 × 106 unstimulated BMDM from PML−/− and wild type mice were fractionated into nuclear and cytosolic compartments. Antibodies targeting lamin B and GAPDH were used as nuclear and cytosolic loading controls, respectively (left panel, middle and bottom rows). Levels of ASC in both nuclear and cytosolic fractions were analyzed by Western blot (top panel). Results are representative of three independent experiments. Densitometric analysis of ASC levels in the nucleus and cytosol of PML-deficient cells are presented relative to lamin B and GAPDH content and normalized to wild type controls (right panel).
Enhanced IL-1β and IL-18 Secretion in PML-deficient Bone Marrow-derived Macrophage
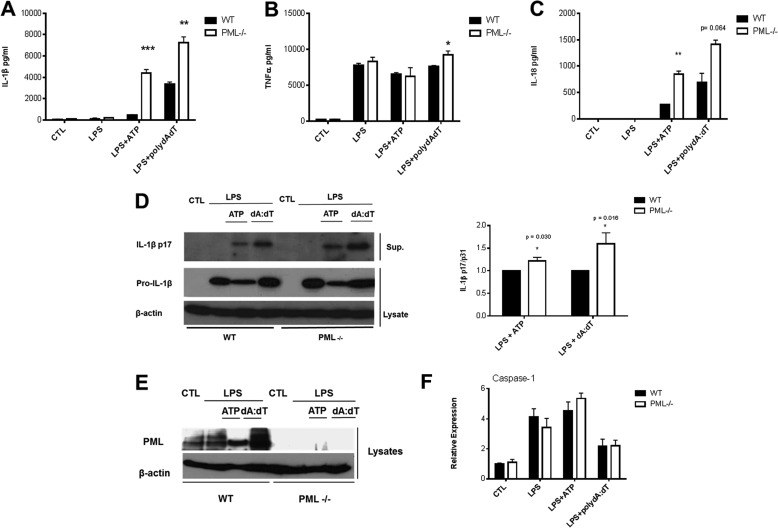
We next tested if the interaction between ASC and PML might have a functional consequence for IL-1β and IL-18 production. As shown in Fig. 4, A and C, the amount of IL-1β and IL-18, respectively, produced by BMDM stimulated with lipopolysaccharide (LPS) and the NLRP3 activator ATP was markedly enhanced in PML-deficient cells compared with wild type (WT) controls. Similarly, elevated levels of IL-1β and IL-18 were detected in PML-deficient cells stimulated with LPS and the AIM2 activator poly(dA-dT). This effect on IL-1β and IL-18 secretion was specific as no significant effect was observed on TNFα production in response to NLRP3 activation, and only a very marginal enhancement in TNFα production in response to AIM2 activation was observed (Fig. 4B). This effect on IL-1β was further confirmed by Western blotting for the mature subunit of IL-1β-p17 (Fig. 4D, left panel). PML deficiency was confirmed by Western blot (Fig. 4E), with poly(dA-dT) causing an apparent induction of PML in WT macrophages (lane 4). We were unable to obtain consistent blots for the p10 subunit of caspase-1. However, comparable levels of caspase-1 mRNA was detected in WT and PML−/− cells following activation of the inflammasome with LPS + ATP and LPS + poly(dA-dT) (Fig. 4F). Comparable levels of caspase-1 were also observed following infection with S. typhimurium (data not shown).
FIGURE 4.
PML-deficient BMDM display enhanced levels of IL-1β and IL-18 in response to NLRP3 and AIM2 inflammasome activation. Primary BMDM from 129Sv wild type (WT) and PML−/− mice were left unstimulated or stimulated with 100 ng/ml LPS for 3 h alone with or without the addition of 5 mm ATP for 1 h or transfected with 1 μg/ml poly(dA-dT) for 5 h as indicated. Supernatants were analyzed by ELISA for IL-1β (A), TNFα (B), and IL-18 (C) or by Western blotting for pro-IL-1β and the mature subunit IL-1β p17 (D). Densitometric analysis of IL-1β p17 levels in WT versus PML-deficient cells is also presented (right panel). BMDMs were also assessed for PML deficiency by Western blot (E). A representative blot from one of three independent experiments is shown (D and E). Relative mRNA expression of caspase-1 in WT and PML−/− primary BMDMs in response to specific NLRP3 and AIM2 activation is also shown (F). RNA was isolated and the quantity of caspase-1 analyzed by real time PCR. A–C, results are expressed as mean ± S.D. for triplicate determinations. Results are representative of three individual experiments, ***, p < 0.001; **, p < 0.01; *, p < 0.05 PML−/− versus WT controls (CTL).
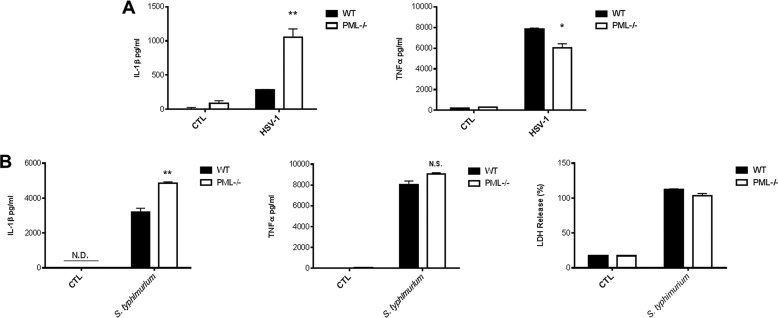
Finally, we tested the ability of a virus and bacterium to induce IL-1β in PML-deficient BMDM. Herpes simplex virus-1 (HSV-1) is a strong inducer of IL-1β in macrophages with in vitro experiments highlighting a role for the NLRP3 inflammasome (24, 25). As can be seen in Fig. 5A (left panel), induction of IL-1β by HSV-1 was markedly enhanced in PML-deficient BMDM with no enhancement in TNFα production. TNFα production was slightly lower in the PML-deficient BMDM (Fig. 5A, right panel). This indicated that PML would also limit inflammasome activation in response to a virus.
FIGURE 5.
PML deficiency enhances levels of IL-1β in response to HSV-1 and S. typhimurium and has no effect on caspase-1-dependent pyroptosis. A, for infection with HSV-1, BMDMs were left unstimulated and mock-infected or stimulated with LPS (100 ng/ml) for 3 h and infected with HSV-1 (m.o.i. 10) for 22 h. Supernatants were analyzed by ELISA for IL-1β and TNFα. B, for S. typhimurium infection, cells were stimulated with 10 ng/ml LPS for 3 h followed by infection (m.o.i. 20) for 2 h. Supernatants were analyzed for IL-1β and TNFα by ELISA (left and middle panel) and lactate dehydrogenase cytotoxicity (right panel). Results are expressed as mean ± S.D. for triplicate determinations. N.D., not detected; N.S., not significant. Results are representative of three individual experiments, **, p < 0.01; *, p < 0.05 PML−/− versus WT controls (CTL)
Furthermore, an ASC-independent caspase-1 complex has been found to induce pyroptotic cell death in response to S. typhimurium (26). To confirm that the interaction between PML and ASC has a specific effect on the inflammasome complex and not the “pyroptosome,” we examined the effect of S. typhimurium on pyroptotic cell death in PML−/− BMDM. As expected, there was no difference in ASC-independent caspase-1-dependent pyroptotic cell death in PML−/− cells compared with WT controls (Fig. 5B, right panel). There was, however, significantly enhanced secretion of IL-1β (p < 0.01) (Fig. 5B, left panel) in PML−/− cells in response to S. typhimurium challenge in accordance with our earlier findings (Figs. 4A and 5A). There was no significant effect on TNFα in response to S. typhimurium (Fig. 5B, middle panel).
DISCUSSION
The redistribution of ASC from the nucleus to the cytosol during inflammasome activation is well documented (14–16). Although the exact molecular mechanism governing this process remains elusive, in this study we identify a role for PML in the localization of ASC to the nucleus. ASC was shown to interact with PML, and PML deficiency led to decreased localization of ASC in the nucleus. Furthermore, several experimental approaches in this study highlighted the enhanced activation of the inflammasome upon stimulation in PML-deficient macrophages. In PML-deficient cells, there was enhanced formation of ASC dimers in response to NLRP3 and AIM2 activation. This was also observed in THP-1 cells in which PMLI was stably knocked down. Importantly, enhanced levels of IL-1β were detected upon activating NLRP3 and AIM2 inflammasomes in PML-deficient cells. Therefore, we conclude that PML plays a role in the localization of ASC to the nucleus limiting its capacity to activate the inflammasome.
PML may retain ASC in the nucleus simply to prevent exacerbated activation of inflammasomes. That which would trigger the dissociation of ASC from PML in the nucleus was not explored here. However, several viruses have been shown to disrupt PML-nuclear bodies with a subsequent mislocalization of PML to the cytosol (27), and this could be a mechanism by which viruses enhance inflammasome activation. Furthermore, PML protein expression is most notably reduced or abolished in several cancers, including prostate cancer, adenocarcinoma, non-Hodgkin lymphoma (28), and gastric cancer (29). The loss of PML in these cancers may contribute to the inflammatory potential of cells via altered localization of ASC to the cytosol. This could have a pro-tumor effect because IL-1β has been shown to have a role in the development of tumors in the aforementioned cancers (30–32).
It is also possible that PML might play a role in the recently characterized nuclear inflammasome. The PYHIN protein IFI16 was found to facilitate the activation of a nuclear inflammasome via the recruitment of ASC and caspase-1 in response to Kaposi sarcoma-associated herpesvirus infection (15). PML could also limit the function of this inflammasome. It will also be of interest to examine individual PML isoforms as regulators of ASC. Alternative splicing in the C terminus of PML generates isoforms I–VII (19). Specifically, PMLVII, also known as cytoplasmic PML (cPML/PMLVII) (33), could influence ASC in the cytosol.
During the preparation of this manuscript, Lo et al. (34), published data demonstrating that PML can promote NLRP3 inflammasome activation suggesting that instead of limiting the inflammasome PML might promote its function. The study did not examine an interaction between PML and ASC. The reason for this difference is not clear but could be related to the different genetic background of the mice (C57BL/6 versus 129Sv) or the different functions of the multiple isoforms of PML. As Lo et al. (34) mentioned, PML appears to inhibit NF-κB, a transcription factor required for induction of pro-IL-1β, so why it would then have a positive role in NLRP3 activation is unclear. More work is clearly needed, but overall both studies confirm an interaction between PML and the NLRP3 inflammasome and highlight the complexity already associated with the many functions of PML.
In summary, our study has identified PML as a novel regulator of ASC localization acting to limit its function. This could have consequences for the role of ASC in cancer and inflammation.
This work was supported by the Victorian Government's Operational Infrastructure Support Program along with Science Foundation Ireland.
- ASC
- apoptosis-associated speck-like protein
- PML
- promyelocytic leukemia protein
- CARD
- caspase-activating recruitment domain
- BMDM
- bone marrow-derived macrophage
- m.o.i.
- multiplicity of infection
- co-IP
- co-immunoprecipitation.
REFERENCES
- 1. Rock K. L., Latz E., Ontiveros F., Kono H. (2010) The sterile inflammatory response. Annu. Rev. Immunol. 28, 321–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dowling J. K., O'Neill L. A. (2012) Biochemical regulation of the inflammasome. Crit. Rev. Biochem. Mol. Biol. 47, 424–443 [DOI] [PubMed] [Google Scholar]
- 3. Masumoto J., Taniguchi S., Ayukawa K., Sarvotham H., Kishino T., Niikawa N., Hidaka E., Katsuyama T., Higuchi T., Sagara J. (1999) ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 274, 33835–33838 [DOI] [PubMed] [Google Scholar]
- 4. Conway K. E., McConnell B. B., Bowring C. E., Donald C. D., Warren S. T., Vertino P. M. (2000) TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 60, 6236–6242 [PubMed] [Google Scholar]
- 5. Srinivasula S. M., Poyet J. L., Razmara M., Datta P., Zhang Z., Alnemri E. S. (2002) The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 277, 21119–21122 [DOI] [PubMed] [Google Scholar]
- 6. Arlehamn C. S., Evans T. J. (2011) Pseudomonas aeruginosa pilin activates the inflammasome. Cell Microbiol. 13, 388–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) NALP3 Forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 8. Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., Dixit V. M. (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto M., Yaginuma K., Tsutsui H., Sagara J., Guan X., Seki E., Yasuda K., Yamamoto M., Akira S., Nakanishi K., Noda T., Taniguchi S. (2004) ASC is essential for LPS-induced activation of procaspase-1 independently of TLR-associated signal adaptor molecules. Genes Cells 9, 1055–1067 [DOI] [PubMed] [Google Scholar]
- 10. Ichinohe T., Lee H. K., Ogura Y., Flavell R., Iwasaki A. (2009) Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206, 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernandes-Alnemri T., Wu J., Yu J. W., Datta P., Miller B., Jankowski W., Rosenberg S., Zhang J., Alnemri E. S. (2007) The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14, 1590–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsushita K., Takeoka M., Sagara J., Itano N., Kurose Y., Nakamura A., Taniguchi S. (2009) A splice variant of ASC regulates IL-1beta release and aggregates differently from intact ASC. Mediators Inflamm. 2009, 287387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bryan N. B., Dorfleutner A., Kramer S. J., Yun C., Rojanasakul Y., Stehlik C. (2010) Differential splicing of the apoptosis-associated speck like protein containing a caspase recruitment domain (ASC) regulates inflammasomes. J. Inflammmation 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bryan N. B., Dorfleutner A., Rojanasakul Y., Stehlik C. (2009) Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J. Immunol. 182, 3173–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karur N., Veettil M. V., Sharma-Walia N., Bottero V., Sadagopan S., Otageri P., Chandran B. (2011) IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khare S., Dorfleutner A., Bryan N. B., Yun C., Radian A. D., de Almeida L., Rojanasakul Y., Stehlik C. (2012) An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fagioli M., Alcalay M., Pandolfi P. P., Venturini L., Mencarelli A., Simeone A., Acampora D., Grignani F., Pelicci P. G. (1992) Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene 7, 1083–1091 [PubMed] [Google Scholar]
- 18. Jensen K., Shiels C., Freemont P. S. (2001) PML protein isoforms and the RBCC/TRIM motif. Oncogene 20, 7223–7233 [DOI] [PubMed] [Google Scholar]
- 19. Condemine W., Takahashi Y., Zhu J., Puvion-Dutilleul F., Guegan S., Janin A., de Thé H. (2006) Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 66, 6192–6198 [DOI] [PubMed] [Google Scholar]
- 20. Creagh E. M., Brumatti G., Sheridan C., Duriez P. J., Taylor R. C., Cullen S. P., Adrain C., Martin S. J. (2009) Bicaudal is a conserved substrate for Drosophila and mammalian caspases and is essential for cell survival. PLoS One 4, e5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deleted in proof.
- 22. Deleted in proof.
- 23. Deleted in proof.
- 24. Rathinam V. A., Jiang Z., Waggoner S. N., Sharma S., Cole L. E., Waggoner L., Vanaja S. K., Monks B. G., Ganesan S., Latz E., Hornung V., Vogel S. N., Szomolanyi-Tsuda E., Fitzgerald K. A. (2010) The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muruve D. A., Pétrilli V., Zaiss A. K., White L. R., Clark S. A., Ross P. J., Parks R. J., Tschopp J. (2008) The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452, 103–107 [DOI] [PubMed] [Google Scholar]
- 26. Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V. M., Monack D. M. (2010) Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207, 1745–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geoffroy M. C., Chelbi-Alix M. K. (2011) Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 31, 145–158 [DOI] [PubMed] [Google Scholar]
- 28. Gurrieri C., Capodieci P., Bernardi R., Scaglioni P. P., Nafa K., Rush L. J., Verbel D. A., Cordon-Cardo C., Pandolfi P. P. (2004) Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J. Natl. Cancer Inst. 96, 269–279 [DOI] [PubMed] [Google Scholar]
- 29. Kim H. J., Song D. E., Lim S. Y., Lee S. H., Kang J. L., Lee S. J., Benveniste E. N., Choi Y. H. (2011) Loss of the promyelocytic leukemia protein in gastric cancer: implications for IP-10 expression and tumor-infiltrating lymphocytes. PLoS One 6, e26264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. El-Omar E. M., Carrington M., Chow W. H., McColl K. E., Bream J. H., Young H. A., Herrera J., Lissowska J., Yuan C. C., Rothman N., Lanyon G., Martin M., Fraumeni J. F., Jr., Rabkin C. S. (2000) Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 404, 398–402 [DOI] [PubMed] [Google Scholar]
- 31. O'Riordan J. M., Abdel-latif M. M., Ravi N., McNamara D., Byrne P. J., McDonald G. S., Keeling P. W., Kelleher D., Reynolds J. V. (2005) Proinflammatory cytokine and nuclear factor κ-B expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am. J. Gastroenterol. 100, 1257–1264 [DOI] [PubMed] [Google Scholar]
- 32. Zhu P., Baek S. H., Bourk E. M., Ohgi K. A., Garcia-Bassets I., Sanjo H., Akira S., Kotol P. F., Glass C. K., Rosenfeld M. G., Rose D. W. (2006) Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell 124, 615–629 [DOI] [PubMed] [Google Scholar]
- 33. Salomoni P., Bellodi C. (2007) New insights into the cytoplasmic function of PML. Histol. Histopathol. 22, 937–946 [DOI] [PubMed] [Google Scholar]
- 34. Lo Y. H., Huang Y. W., Wu Y. H., Tsai C. S., Lin Y. C., Mo S. T., Kuo W. C., Chuang Y. T., Jiang S. T., Shih H. M., Lai M. Z. (2013) Selective inhibition of the NLRP3 inflammasome by targeting to promyelocytic leukemia protein in mouse and human. Blood 121, 3185–3194 [DOI] [PubMed] [Google Scholar]