Background: LRP1 is an endocytic receptor of ADAMTS-5 (aggrecanase 2) in cartilage.
Results: ADAMTS-4 (aggrecanase 1) is also internalized via LRP1 but at a slower rate than ADAMTS-5.
Conclusion: LRP1 differently regulates the extracellular activities of the two key aggrecanases in cartilage.
Significance: LRP1 is a major traffic controller of the two aggrecanases.
Keywords: Chondrocytes, Endocytosis, Extracellular Matrix, Osteoarthritis, Trafficking, Aggrecanase
Abstract
Degradation of the cartilage proteoglycan aggrecan is an early event in the development of osteoarthritis, and a disintegrin and metalloproteinase with thrombospondin motifs-4 (ADAMTS-4) and ADAMTS-5 are considered to be the major aggrecan-degrading enzymes. We have recently found that ADAMTS-5 is rapidly endocytosed via low density lipoprotein receptor-related protein 1 (LRP1) and degraded by chondrocytes. Here we report that this regulatory mechanism also applies to ADAMTS-4, although its rate of endocytosis is slower than that of ADAMTS-5. Domain deletion mutagenesis of ADAMTS-4 identified that the cysteine-rich and spacer domains are responsible for binding to LRP1, whereas the thrombospondin 1 and spacer domains are responsible in ADAMTS-5. The estimated t½ value of ADAMTS-4 endocytosis was about 220 min, whereas that of ADAMTS-5 was 100 min. The difference in half-lives between the two enzymes is explained by the 13-fold lower affinity of ADAMTS-4 for LRP1 compared with that of ADAMTS-5. Studies using soluble ligand binding clusters of LRP1 showed that ADAMTS-4 binds to clusters II and IV with similar KD,app values of 98 and 73 nm, respectively, whereas ADAMTS-5 binds to cluster II, III, and IV with KD,app values of 3.5, 41, and 9 nm, respectively. Thus, ADAMTS-5 competitively inhibits ADAMTS-4 endocytosis but not vice versa. This study highlights that the affinity between a ligand and LRP1 dictates the rate of internalization and suggests that LRP1 is a major traffic controller of the two aggrecanases, especially under inflammatory conditions, where the protein levels of ADAMTS-4 increase, but those of ADAMTS-5 do not.
Introduction
Aggrecan is a major extracellular matrix component of articular cartilage, and its degradation is an early event in the development of osteoarthritis (OA)6 (1–3). The major proteinases responsible for aggrecan degradation are matrix metalloproteinases (MMPs) and aggrecanases that are members of a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family (4). Aggrecanases are defined by their ability to cleave the Glu373-Ala374 bond of the aggrecan core protein (5, 6). Elevated aggrecanase-generated aggrecan fragments have been found in synovial fluids of patients with OA and inflammatory joint disease (7, 8). These fragments were also detected in normal synovial fluid and serum of animals (6), suggesting that aggrecanases function in both physiological and pathological catabolism of aggrecan. Among the ADAMTS that have aggrecanolytic activity, ADAMTS-4 and ADAMTS-5 have been considered as the major aggrecanases involved in cartilage matrix turnover because of their effective aggrecan-degrading activity in vitro (9–11). Although mouse gene ablation studies have indicated ADAMTS-5 is the key aggrecanase for the development of arthritis in mice (12–14), both ADAMTS-4 and ADAMTS-5 are considered to play roles in human OA (15, 16). ADAMTS-5 is about 30× more active than ADAMTS-4 on aggrecan (11), but we have recently found that it is rapidly endocytosed by chondrocytes via the scavenger receptor low density lipoprotein receptor-related protein 1 (LRP1). This suggests that the post-translational regulation is a major regulatory mechanism of the extracellular levels of ADAMTS-5, and this regulation is impaired in OA cartilage due to the reduction in LRP1 levels largely caused by shedding from the cell membrane (17). This in part explains an increased extracellular activity of ADAMTS-5 and aggrecan degradation leading to slowly progressing OA in which little significant increase in ADAMTS-5 mRNA was observed (15, 18, 19). In contrast, the expression of ADAMTS-4 mRNA and its protein levels correlate with the progression of OA in humans (15).
LRP1 is a multifunctional endocytic type 1 transmembrane receptor consisting of a 515-kDa α-chain containing the extracellular ligand binding domains and a non-covalently associated 85-kDa β-chain containing a transmembrane domain and a short cytoplasmic tail (20). LRP1 internalizes >40 ligands from the pericellular environment, including lipoproteins, extracellular matrix (ECM) proteins, growth factors, cell surface receptors, proteinases, and proteinase-proteinase inhibitor complexes (21–23). LRP1 is widely expressed (24, 25), and its expression is particularly high in articular chondrocytes and macrophages.7 The ablation of the LRP1 gene in mice is embryonically lethal (26), but tissue specific deletion of the LRP1 gene has demonstrated that it protects the vasculature and controls β-amyloid precursor protein trafficking, lipid metabolism in adipocytes, and macrophage biology (27). In cartilage, LRP1 can endocytose MMP-13 (28, 29) and tissue inhibitor of metalloproteinases (TIMP-3), which inhibits collagenases and aggrecanases (30, 31). LRP1 interacts with frizzled-1 and down-regulates the canonical Wnt-β-catenin signaling pathway (32). It also represses the hypertrophy of chondrocytes during endochondral ossification by removing connective tissue growth factor (33, 34). LRP1 is, therefore, an important regulator of skeletal development and maintenance of cartilage homeostasis.
In this study we have re-addressed whether ADAMTS-4, which has a similar homologous domain composition to ADAMTS-5, is endocytosed by chondrocytes by either the same mechanism or by different pathways. The initial discovery of ADAMTS-5 endocytosis stemmed from our observation that aggrecanase activity is reduced when ADAMTS-5 was incubated with live porcine cartilage compared with when it was incubated with freeze-thawed (dead) cartilage. This led us to discover that ADAMTS-5 is endocytosed via LRP1 by viable chondrocytes. In those studies we observed no significant differences in aggrecan degradation between live and dead cartilage when ADAMTS-4, MMP-1, or MMP-13 was added (17), although MMP-13 has been reported to be endocytosed by chondrocytes via LRP1 (28, 29). However, the concentrations of ADAMTS-4 and MMP-13 used in those studies were 10-fold higher than that of ADAMTS-5, as ADAMTS-5 is the most active aggrecanase (10, 11). Furthermore, we noticed that the basal level of aggrecan degradation in live cartilage was slightly but significantly higher than that of dead cartilage. Subtraction of these values from those in which ADAMTS-4 was added revealed a significant reduction in aggrecanolytic activity detected with live cartilage compared with that with dead cartilage, and this difference is more prominent at lower concentrations of ADAMTS-4. The present study demonstrates bona fide LRP1-mediated endocytosis of ADAMTS-4. We also show the similarities and differences in LRP1 interaction between ADAMTS-4 and ADAMTS-5. Our endocytic competition studies between the two aggrecanases provide further insights into their role in normal turnover and pathological degradation of aggrecan.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
The sources of materials used were as follows: dimethylmethylene blue and the anti-FLAG M2 mouse monoclonal antibody from Sigma; BC-3 mouse monoclonal antibody that recognizes the N-terminal 374ARGSV aggrecan core protein fragments generated by aggrecanase, the anti-early endosome antigen 1 (EEA1) rabbit polyclonal antibody, and the anti-LRP1 mouse monoclonal antibodies 5A6 and 8G1 from Abcam (Cambridge, UK) and from Calbiochem; a hydroxamate-based MMP inhibitor CT-1746 from Celltech (Slough, UK); the anti-actin antibody from Santa Cruz Biotechnology (Santa Cruz, CA); the anti-tubulin antibody from Cell Signaling (Danvers, MA); the anti-Myc tag antibody from Merck Millipore (Darmstadt, Germany); purified human full-length LRP1 from BioMac (Leipzig, Germany). The anti-human ADAMTS-4 catalytic domain rabbit polyclonal antibody was raised in rabbit and characterized (9). Recombinant human ADAMTS-4, ADAMTS-5, their domain deletion mutants, MMP-1, MMP-13, and RAP were prepared as described previously (9, 10, 35, 36). Recombinant human IL-1α was kindly provided by Prof. J. Saklatvala (Kennedy Institute of Rheumatology, Oxford, UK). All other reagents used were of the highest analytical grade available.
Human and Porcine Articular Cartilage Culture
Human articular cartilage was obtained from patients undergoing amputations at the Royal National Orthopaedic Hospital (Stanmore, UK) following informed consent and approval by the Riverside Research Ethics Committee. Healthy cartilage was obtained from the knee after amputation due to soft tissue sarcoma and osteosarcoma with no involvement of the cartilage. Tissues were obtained from 5 patients (all male; aged 8–41 years, mean age 21.2 years).
Dissected human articular cartilage (∼20 mm3: 25 mg wet volume/weight) or porcine articular cartilage (∼6 mm3: 10 mg wet volume/weight) from the metacarpophalangeal joints of 3–9-month-old pigs was placed in one well of a round-bottom 96-well plate and allowed to rest for 24 h in 200 μl of Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum before use. The medium was then replaced, and the cartilage was rested for a further 24 h in 200 μl of DMEM at 37 °C (for fresh live cartilage) before aggrecan degradation and endocytosis assays. For freeze-thawed cartilage experiments, cartilage pieces were frozen at −80 °C and thawed in the same medium to render the chondrocytes non-viable. The medium was replaced with fresh medium before subsequent assays.
Analysis of Aggrecan Degradation
Each piece of cartilage was incubated in 100 μl of DMEM with or without IL-1α (10 ng/ml) or various concentrations of MMPs and ADAMTS. Three pieces of cartilage were subjected to each treatment. After incubation for various periods of time, the conditioned media were harvested and glycosaminoglycan (GAG) released into the medium was measured using the dimethylmethylene blue assay (37). Aggrecan fragments in the medium were deglycosylated as described previously (9) and analyzed by Western blotting using an anti-ARGSV aggrecan neoepitope antibody (BC-3). Immunoreactive bands were quantified using NIH ImageJ, and results are presented as relative intensities.
Isolation of Chondrocytes and Cell Culture
Chondrocytes were isolated as described previously (31). Primary porcine cells and both primary and passaged human cells were used in the experiments. For the endocytosis assay, cells were plated at a density of 2.5 × 105 cells/well (24-well plate) in DMEM containing 10% FCS.
Analysis of ADAMTS-4 Clearance
Cartilage explants or cells were incubated in 100 or 400 μl of DMEM with or without 500 nm RAP. After incubation for 30 min, media were replaced with DMEM containing 10 nm ADAMTS-4 or its domain deletion mutants with or without 500 nm RAP. Four pieces of cartilage were subjected to each treatment. After incubation for various periods of time, media were collected, and the protein was precipitated with TCA and dissolved in 50 μl of 1× SDS-sample buffer (50 mm Tris-HCl (pH 6.8), 5% 2-mercaptoethanol, 2% SDS, and 10% glycerol). The cartilage explants and cells were washed with DMEM and lysed in 50 μl of 2× SDS sample buffer. All samples were analyzed by SDS-PAGE and Western blotting using an anti-ADAMTS-4 catalytic domain antibody. Immunoreactive bands were quantified using NIH ImageJ, and the amount of ADAMTS-4 remaining in the medium at each time point was calculated as a percentage of the amount of ADAMTS-4 at 0 h. The amount of ADAMTS-4 incubated alone in the medium at each time point was also calculated as described above, and each value was normalized by subtracting the amount of auto-degraded ADAMTS-4 at each time point.
Immunocytochemical Localization of Endocytosed ADAMTS-4
Cultured cells on 4-well Lab-Tek chamber slides (Nunc, Roskilde, Denmark) were incubated in DMEM with 20 nm ADAMTS-4 in the absence or presence of 500 nm RAP for 4 h. Cells were washed with DMEM, fixed with 3% paraformaldehyde in Tris-buffered saline (TBS; 10 min, room temperature), and permeabilized with TBS containing 10 mm CaCl2 (TNC) and 0.1% Triton X-100 (15 min, room temperature). Each sample was incubated with anti-FLAG M2 mouse monoclonal antibody and anti-EEA1 rabbit polyclonal antibody (3 h, room temperature). Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 568-conjugated anti-rabbit IgG (Molecular Probes, Eugene, OR) were used to visualize the antigen signals (1 h, room temperature). Actin was stained with actin stain 670 phalloidin (Cell Signaling), and nuclei were stained with DAPI. Samples were viewed using a Nikon Eclipse TE2000-U confocal laser scanning microscope. The data were collated using Volocity software (Improvision, Coventry, UK).
siRNA Knockdown of LRP1 in Human Articular Chondrocytes
siRNA oligonucleotides for LRP1 (On-TargetPlus SMARTpool siRNA), and nontargeting oligonucleotide were purchased from Thermo Scientific Dharmacon (Lafayette, CO). Human articular chondrocytes were plated at a density of 1.5 × 106 cells/dish (6-cm dish) in DMEM containing 10% FCS and incubated until 50% confluent. Lipofectamine 2000 (Invitrogen) was used to transfect cells with siRNA at a final concentration of 10 nm in Opti-MEM I. At 4 h after transfection, the Opti-MEM was removed and replaced with DMEM containing 10% FCS.
Quantitative Reverse Transcriptase-PCR
Quantitative reverse transcriptase-PCR was carried out as described previously (36). Briefly, cDNA was generated using a reverse-transcription kit (Applied Biosystems, Foster City, CA) and random primers from RNA were extracted and prepared using the RNeasy mini kit (Qiagen, Valencia, CA) following the manufacturer's guidelines. cDNA was then used for real time PCR assays using TaqMan technology. The ΔΔ threshold cycle (ΔΔCt) method of relative quantitation was used to calculate relative mRNA levels for each transcript examined. The 60 S acidic ribosomal protein P0 (RPLP0) gene was used to normalize the data. Pre-developed primer/probe sets for LRP1 and RPLP0 were purchased from Applied Biosystems.
ELISA for Binding of ADAMTS-4 and ADAMTS-5 to LRP1 or Soluble LRP1 Fragments
Human LRP1 or soluble LRP1 fragments (5 or 25 nm, respectively, in 100 μl of TNC) was coated overnight at 4 °C onto microtiter plates (Corning). Wells were blocked with 3% BSA in TNC (1 h; 37 °C) and washed in TNC containing 0.05% (v/v) Brij-35 after this and each subsequent step. Wells were then incubated with various concentrations of ADAMTS-4, its domain deletion mutants, or domain deletion mutants of ADAMTS-5 in blocking solution containing 100 μm CT-1746 for 3 h at room temperature. Bound enzymes were detected using anti-FLAG M2 mouse monoclonal antibody (1 h; room temperature) and then with an anti-mouse secondary antibody coupled to horseradish peroxidase (Abcam; 1 h; room temperature). Hydrolysis of tetramethylbenzidine substrate (KPL, Gaithersburg, MA) was measured at 450 nm using a BioTek EL-808 absorbance microplate reader (BioTek, Winooski, VT). Each value was normalized by subtracting the amount of enzyme bound to control wells that were not coated with LRP1 or soluble LRP1 fragments.
Expression and Purification of Soluble LRP1 Fragments
The expression vector for the selected portions (ligand binding domain clusters I-IV) of LRP1 (see Fig. 7A) has been prepared previously (38). Secreted fragments of LRP1 containing individual ligand binding domain cluster were transiently expressed in HEK293 cells using TransIT-2020 transfection reagent (Mirus, Madison, WI) according to the manufacture's protocol. Cells growing in 150-mm dishes (∼70% confluence) were transfected in serum-containing DMEM with 30 μg of pSecTagB (Invitrogen) carrying cDNA for various LRP1 fragments. After 6 h of transfection, the medium was replaced with serum-free DMEM, and cells were further incubated for 72 h. The resultant conditioned medium was directly applied to a nickel-nitrilotriacetic acid affinity column (Qiagen) that had previously been equilibrated with 50 mm HEPES (pH 7.5) buffer containing 150 mm NaCl and 5 mm CaCl2. The column was washed with 10 column volumes of 50 mm HEPES (pH 7.5) buffer containing 1 m NaCl, 5 mm CaCl2, and 20 mm imidazole. It was then eluted by 500 mm imidazole in HEPES (pH 7.5) buffer containing 150 mm NaCl and 5 mm CaCl2. For analysis of N-linked glycosylation of soluble LRP1 fragments, the preparations were treated with peptide N-glycosidase F (New England Biolabs, Ipswich, MA) according to the manufacturer's protocol.
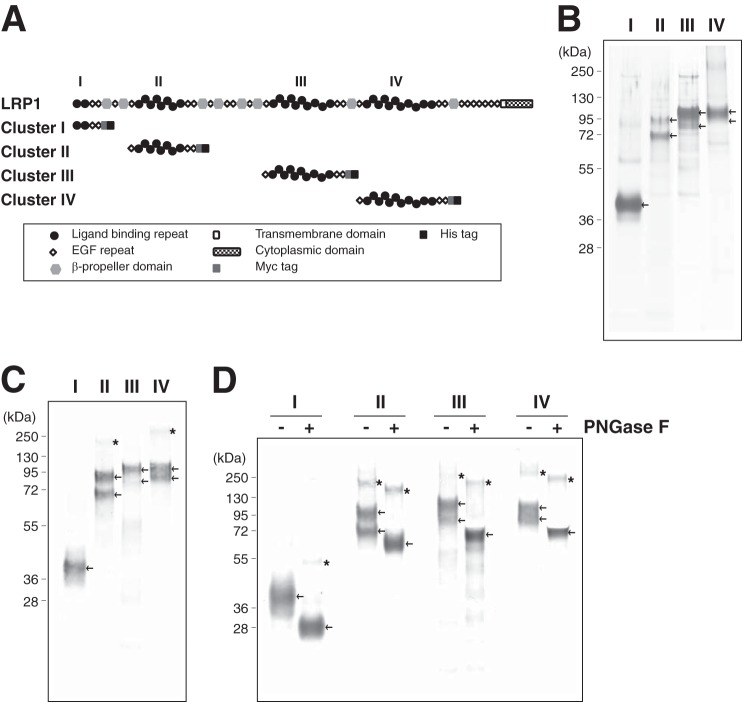
FIGURE 7.
Expression and purification of soluble recombinant LRP1 fragments. A, modular domain organization of LRP1 and its soluble receptor fragments used in this study. The four clusters of ligand-binding clusters are numbered I-IV. The symbols for the various domains are indicated in the inset. B, SDS-PAGE of LRP1 fragments containing each ligand binding cluster purified from conditioned medium of HEK293 cells transfected with the designated LRP1 clusters. Proteins were separated on a 4–12% gel under reducing conditions and stained with silver. C, purified proteins were further analyzed by Western blotting using an anti-Myc antibody. D, Western blotting of each clusters before (−) and after (+) peptide N-glycosidase F (PNGase F) treatment. Proteins were probed by anti-Myc antibody. The recombinant LRP1 fragments are indicated by the arrows, and asterisks indicate possible dimer or oligomer of the fragments.
RESULTS
Aggrecanase Activity of ADAMTS-4 Is Reduced in Live Cartilage Compared with Dead Cartilage
We first evaluated the ability of ADAMTS-4 to cleave aggrecan in cartilage by adding recombinant ADAMTS-4 lacking the C-terminal spacer (Sp) domain to live or freeze-thawed (dead) porcine cartilage explants in culture and measuring GAG release after 24 h. Dead cartilage was used to eliminate the involvement of metabolically active live chondrocytes (36). Live cartilage responded to IL-1 and released GAG, but this was negligible with dead cartilage, indicating that the latter is metabolically inactive (Fig. 1A). At 10–50 nm ADAMTS-4, the amount of GAG released from live cartilage was significantly lower than that released from dead cartilage; the ratio of GAG release from dead cartilage compared with live cartilage was 4.7, 3.3, and 1.6 with 10, 20, and 50 nm ADAMTS-4, respectively. There was no significant difference in GAG release when 100 nm ADAMTS-4 was added (Fig. 1B). Western blot analysis of the products generated by 20 nm ADAMTS-4 with an anti-ARGSV neoepitope antibody indicated that aggrecan was cleaved at the Glu373-Ala374 bond, characteristic of aggrecanase activity (Fig. 1C).
FIGURE 1.
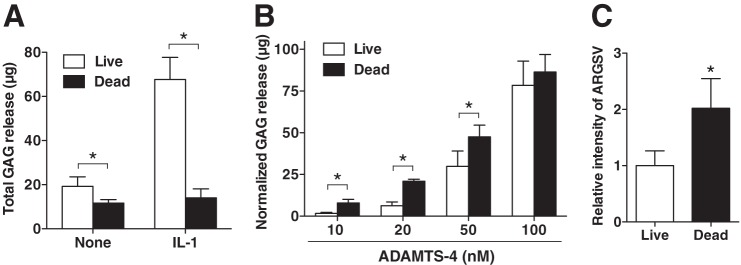
Aggrecanase activity of ADAMTS-4 is reduced in live compared with dead cartilage. A, live and freeze-thawed (Dead) porcine cartilage explants (n = 3) were incubated without or with 10 ng/ml IL-1α for 24 h. GAG released into the medium was measured by the dimethylmethylene blue assay. B, live and dead porcine cartilage were incubated with various concentrations of ADAMTS-4 for 24 h. GAG released into the medium was measured as in A. C, densitometric analysis of immunoreactive aggrecan fragments detected in the medium after 24 h of culture (from B, 20 nm) by Western blotting using an anti-ARGSV neoepitope antibody. The amount of aggrecan fragment in the medium of live cartilage explants incubated with 20 nm ADAMTS-4 was taken as 1. Bars represent the means ± S.D. *, p < 0.05; unpaired t test.
ADAMTS-4 Is Internalized and Degraded by Articular Cartilage Chondrocytes via LRP
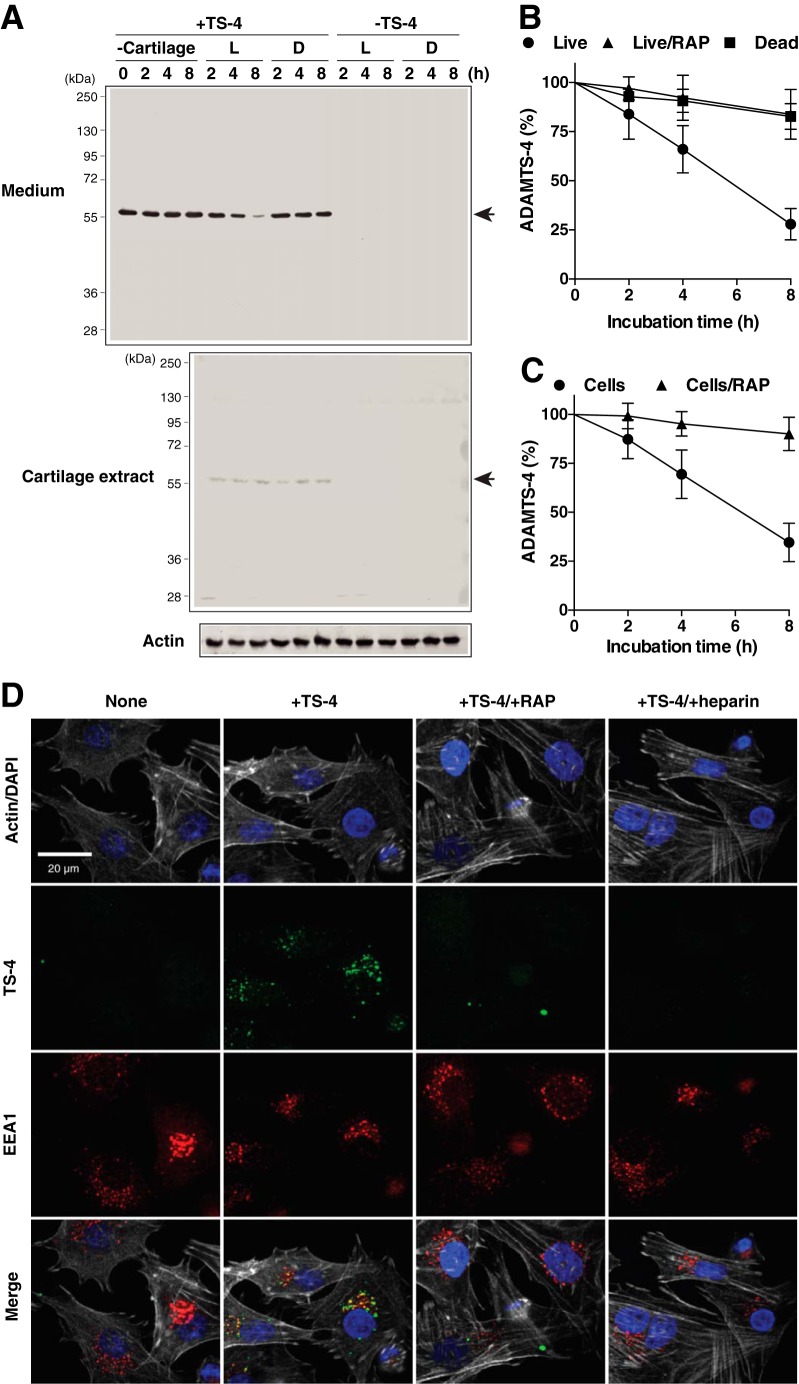
When 10 nm ADAMTS-4 was added to live cartilage in culture, ∼75% of the enzyme was depleted from the medium after 8 h without any detectable degradation fragments as shown by Western blot analysis (Fig. 2A and B). In contrast, most of ADAMTS-4 remained in the medium when exposed to dead cartilage. A small portion of ADAMTS-4 was also detected in extracts from both live and dead cartilage after 2–8 h, suggesting its binding to cartilage ECM. We could not detect endogenous ADAMTS-4, as its level was too low to detect by this method. The addition of RAP, an antagonist of ligand binding to LRP receptors, to this system almost completely inhibited ADAMTS-4 depletion from the medium of live cartilage (Fig. 2B), suggesting that ADAMTS-4 is endocytosed by a member of the LRP family. Isolated chondrocytes endocytosed ADAMTS-4 with similar kinetics as live cartilage (half-life of ∼6 h, and their endocytosis was also inhibited by RAP) (Fig. 2C). The internalization of ADAMTS-4 was examined by immunofluorescent confocal microscopy, with punctate staining of ADAMTS-4 observed within cells, colocalizing with EEA1, a marker for early endosomes (Fig. 2D). Consistent with the data from Western blot analysis, the intracellular fluorescent signal for ADAMTS-4 was abolished in the presence of RAP. Heparin also blocked the internalization of ADAMTS-4.
FIGURE 2.
ADAMTS-4 is internalized and degraded by articular cartilage chondrocytes via LRP. A, live (L) and dead (D) porcine cartilage explants (n = 3) were incubated with 10 nm ADAMTS-4 (TS-4) for 0–8 h, and ADAMTS-4 in the media and cartilage explants was detected by Western blotting using an anti-ADAMTS-4 catalytic domain antibody. B, densitometric analysis of immunoreactive ADAMTS-4 bands detected in the media of A or in the media of live cartilage explants incubated with 10 nm ADAMTS-4 in the presence of 500 nm RAP for 0–8 h. The amount of ADAMTS-4 was expressed as a % of the amount of ADAMTS-4 at 0 h. C, porcine primary chondrocytes (n = 3) were incubated with ADAMTS-4, and enzyme remaining in the medium was measured as in B. Data points represent the means ± S.D. D, confocal microscopy analysis of ADAMTS-4 endocytosis by porcine chondrocytes. Cells were incubated with 20 nm ADAMTS-4 (TS-4) in the presence or absence of 500 nm RAP or 100 μg/ml heparin for 4 h. Endocytosed ADAMTS-4, EEA1, cytoskeleton, and nucleus were visualized as described under “Experimental Procedures.”
siRNA-mediated Knockdown of LRP1 Impairs ADAMTS-4 Endocytosis in Human Chondrocytes
Among the LRP family, LRP1 is the major endocytic receptor for proteases and protease inhibitors (23). Silencing LRP1 in normal human chondrocytes by LRP1-targeting siRNA reduced mRNA levels by 92% compared with the non-targeting siRNA control (Fig. 3A). Western blot analysis of the cell extracts confirmed that levels of the 515-kDa extracellular α-chain and the 85-kDa β-chain containing the transmembrane domain were reduced by 88 and 81%, respectively (Fig. 3, B and C). Cellular uptake of ADAMTS-4 was markedly reduced in LRP1-depleted cells (Fig. 3D). From these results, we conclude that LRP1 is the primary endocytic receptor for ADAMTS-4.
FIGURE 3.
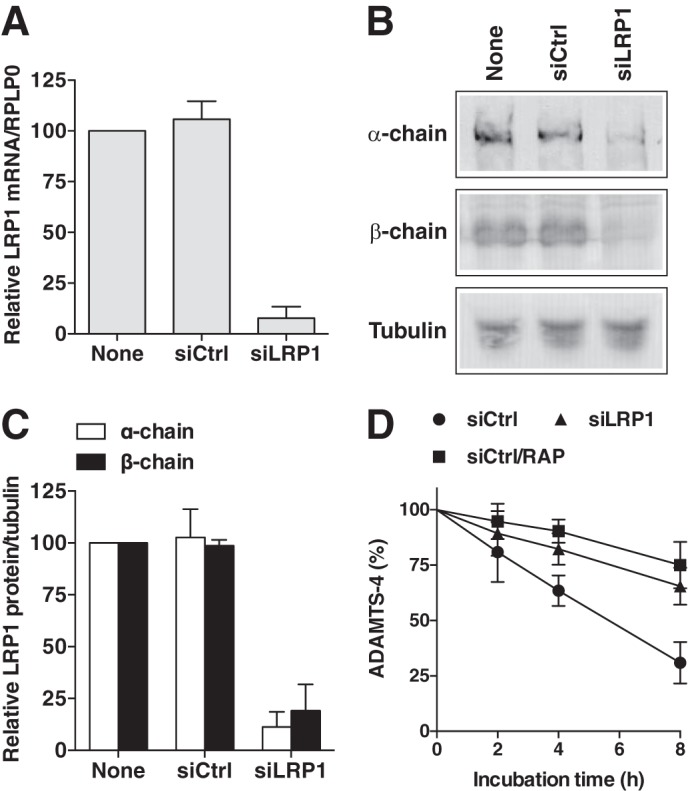
siRNA-mediated knockdown of LRP1 impairs ADAMTS-4 endocytosis in human chondrocytes. Human chondrocytes (n = 3) transfected with non-targeting siRNA (siCtrl) or LRP1 targeting siRNA (siLRP1) were cultured for 2 days in DMEM containing 10% FCS. A, results of TaqMan real-time PCR showing relative levels of mRNA for LRP1. B, LRP1 α-chain (515 kDa) and β-chain (85 kDa) in cell lysate were assessed by Western blotting using anti-LRP1 α-chain (8G1) and β-chain (5A6) antibodies, respectively. C, densitometric analysis of immunoreactive LRP1 bands detected in B. The amount of LRP1 was expressed as a % of the amount of LRP1 in untransfected cells (None). D, cells were incubated with 10 nm ADAMTS-4 in the presence or absence of 500 nm RAP for 0–8 h, and ADAMTS-4 remaining in the medium was measured as in Fig. 2. Bars and points represent the means ± S.D.
Inhibition of ADAMTS-4 Endocytosis by RAP Enhances Aggrecan Degradation in Articular Cartilage
We then examined the effect of RAP on ADAMTS-4-mediated aggrecan degradation in cartilage. The addition of 20 nm recombinant ADAMTS-4 to dead cartilage caused a 2.1-fold higher GAG release over that from live cartilage after 8 h of incubation, and it increased to 2.5-fold after 48 h. The addition of RAP to the ADAMTS-4-mediated aggrecan degradation system significantly enhanced GAG release from live cartilage but had no effect on GAG release from dead cartilage (Fig. 4A). Notably, the level of GAG released from live cartilage in the presence of RAP was equivalent to that from dead cartilage. Western blotting using an anti-ARGSV antibody confirmed that the 2.1-fold increase in aggrecan degradation observed in the presence of RAP is solely due to increased aggrecanase activity of exogenously added ADAMTS-4 in live cartilage (Fig. 4B).
FIGURE 4.
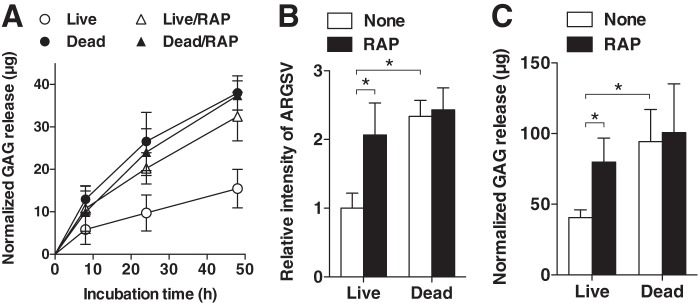
Inhibition of ADAMTS-4 endocytosis by RAP enhances aggrecan degradation in articular cartilage. A, time course of GAG released into the medium of live and dead porcine cartilage explants (n = 3) incubated with 20 nm ADAMTS-4 in the presence or absence of 500 nm RAP. B, densitometric analysis of immunoreactive bands of aggrecan fragments detected in the medium obtained after 24 h of culture in A by Western blotting using an anti-ARGSV neoepitope antibody. The amount of aggrecan fragment in the medium of live cartilage explants incubated with 20 nm ADAMTS-4 was taken as 1. C, live and dead human cartilage explants of normal patients (n = 5) were incubated with 20 nm ADAMTS-4 in the presence or absence of 500 nm RAP for 24 h. GAG released into the medium was measured by dimethylmethylene blue assay. Bars and points represent the means ± S.D. *, p < 0.05; unpaired t test.
ADAMTS-4-induced aggrecan degradation was also tested with normal human cartilage. As shown in Fig. 4C, the addition of 20 nm ADAMTS-4 to dead cartilage caused a 2.3-fold higher GAG release than that with live cartilage after 24 h of incubation. The addition of RAP to the ADAMTS-4-mediated aggrecan degradation system significantly enhanced GAG release from live human cartilage, but it had no effect with dead cartilage.
Sp and CysR Domains Mediate ADAMTS-4 Endocytosis
ADAMTS-4 and ADAMTS-5 are multidomain metalloproteinases and have a similar domain arrangement consisting of pro, catalytic metalloproteinase, disintegrin, TS, CysR and Sp domains. In addition, ADAMTS-5 contains an extra TS domain after the Sp domain. We, therefore, investigated which domains are required for ADAMTS-4 endocytosis by testing endocytosis of a series of domain-deletion mutants by porcine primary chondrocytes (Fig. 5A). The half-life (t½) of ADAMTS-4-1 was ∼220 min (Table 1). Deletion of the Sp domain (ADAMTS-4-2) significantly reduced the rate of endocytosis with the t½ of 360 min. Further deletion of the CysR domain (ADAMTS-4-3) and TS domain (ADAMTS-4-4) reduced endocytosis to negligible levels. These data were compared with the t½ values of ADAMTS-5 and its domain deletion mutants (Fig. 5C and Table 1). The t½ values of ADAMTS-4-1 and ADAMTS-4-2 were about 2.2- and 1.6-fold longer than those of the equivalent domain composition species ADAMTS-5-2 and ADAMTS-5-3, respectively (Table 1). It is notable that the t½ value of ADAMTS-5-4 was ∼250 min, whereas ADAMTS-4-3 was not internalized even after 480 min. From these data we conclude that the domains that are responsible for ADAMTS-4 endocytosis are the CysR and Sp domains, whereas those in ADAMTS-5 are the TS1 and Sp domains. The rate of internalization of full-length ADAMTS-4 was ∼2-fold slower than that of full-length ADAMTS-5.
FIGURE 5.
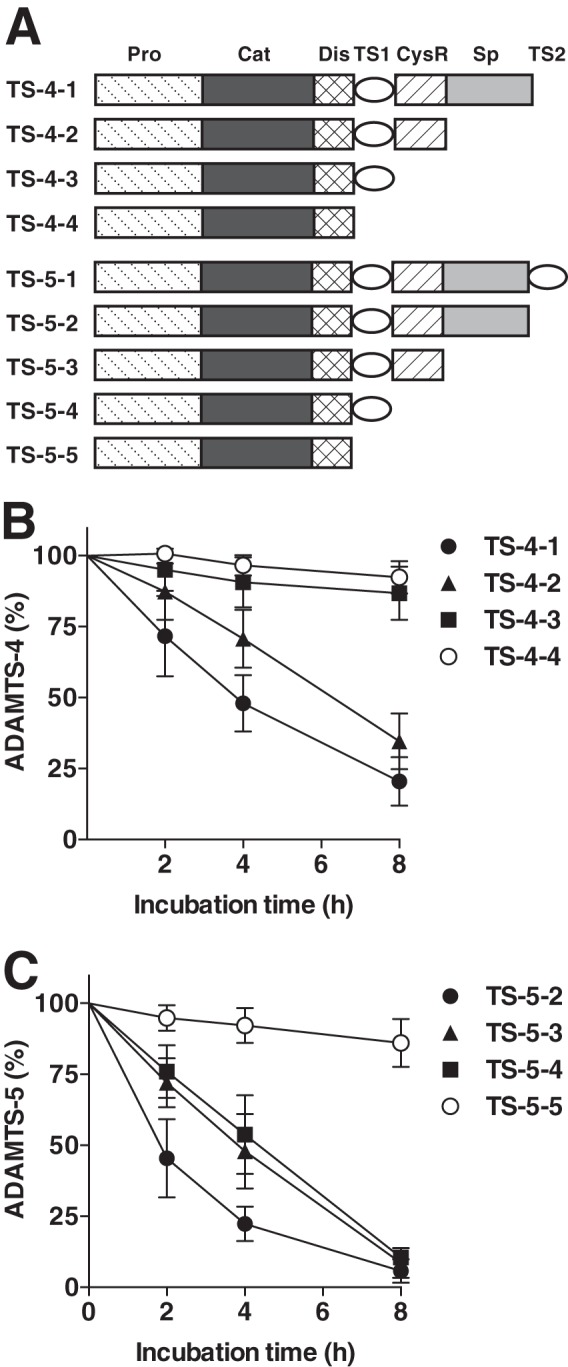
Sp and CysR domains mediate ADAMTS-4 endocytosis. A, schematic representation of ADAMTS-4, ADAMTS-5, and their domain deletion mutants. Pro, prodomain; Cat, catalytic domain; Dis, disintegrin-like domain; TS, thrombospondin domain; CysR, cysteine-rich domain; Sp, spacer domain. B, porcine primary chondrocytes (n = 3) were incubated with each 10 nm ADAMTS-4-1, ADAMTS-4-2, ADAMTS-4-3, or ADAMTS-4-4 for 0–8 h, and ADAMTS-4 and each ADAMTS-4 mutant remained in the medium was measured as in Fig. 2. C, as a comparison, porcine chondrocytes were incubated with each 10 nm ADAMTS-5-2, ADAMTS-5-3, ADAMTS-5-4, or ADAMTS-5-5 for 0–8 h, and ADAMTS-5 and each ADAMTS-5 mutant remained in the medium was measured as in B. Points represent the means ± S.D.
TABLE 1.
Apparent half-life of ADAMTS-4, ADAMTS-5, and their domain deletion mutants in the medium of chondrocytes and KD,app values for their binding to LRP1
t½ and extrapolated KD,app values were estimated based on the results in Figs. 5 and 6.
| Enzyme form | t½ | KD,app |
|---|---|---|
| nm | ||
| TS-4-1 | 220 | 51 |
| TS-4-2 | 360 | 110 |
| TS-4-3 | ≫480 | ≫1000 |
| TS-4-4 | ≫480 | ≫1000 |
| TS-5-1 | NDa | NDa |
| TS-5-2 | 100 | 3.8 |
| TS-5-3 | 220 | 56 |
| TS-5-4 | 250 | 65 |
| TS-5-5 | ≫480 | >1000 |
a Both values for ADAMTS-5-1 were not determined as the full-length enzyme is not available in a reasonable quantity due to rapid autodegradation.
ADAMTS-4 and ADAMTS-5 Directly Bind to LRP1 via Different Non-catalytic Domains
The effect of the non-catalytic domains of ADAMTS-4 on direct binding to LRP1 was investigated by a solid-phase binding assay using domain deletion mutants. Similar domain deletion mutants of ADAMTS-5 were also tested and compared. It was only possible to conduct limited experiments with ADAMTS-4-1, as only small amounts can be purified, but an extrapolated apparent binding constant (KD,app) of 51 nm was calculated similar to that of ADAMTS-5-3 (KD,app = 56 nm) (Fig. 6 and Table 1). ADAMTS-4-2 bound to immobilized LRP1 with a KD,app of 110 nm and further deletion of CysR domain (ADAMTS-4-3) abolished the binding to LRP1. On the other hand, ADAMTS-5-2 bound to immobilized LRP1 with a KD,app of 3.8 nm and ADAMTS-5-4, which has an equivalent domain composition as ADAMTS-4-3 bound to LRP1 with a KD,app of 65 nm. Further deletion of the TS1 domain (ADAMTS-5-5) abolished binding to LRP1. These results indicate that ADAMTS-4 binds to LRP1 via its Sp and CysR domains, whereas ADAMTS-5 binds to LRP1 via its Sp and TS1 domains.
FIGURE 6.
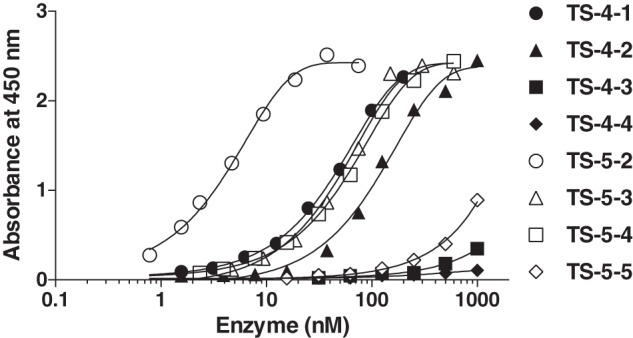
ADAMTS-4 and ADAMTS-5 directly bind to LRP1 via different non-catalytic domains. Full-length LRP1 was coated onto microtiter plates, and binding of ADAMTS-4-1 (0–200 nm), ADAMTS-4-2 (0–1 μm), ADAMTS-4-3 (0–1 μm), ADAMTS-4-4 (0–1 μm), ADAMTS-5-2 (0–75 nm), ADAMTS-5-3 (0–0.6 μm), ADAMTS-5-4 (0–0.6 μm), or ADAMTS-5-5 (0–1 μm) in the presence of 100 μm CT-1746 was measured using an M2 anti-FLAG antibody and a horseradish peroxidase-conjugate secondary antibody as described under “Experimental Procedures.”
Expression and Purification of Soluble Recombinant LRP1 Fragments
The ligand binding regions in LRP1 occur in four clusters (clusters I-IV) containing between 2 and 11 individual ligand binding cysteine-rich repeats. Most of the ligands for LRP1 for which the binding sites have been mapped interact with ligand binding repeats in clusters II and IV (39). To further investigate which ligand binding cluster of LRP1 interacts with the two ADAMTS enzymes, we expressed four soluble clusters of ligand binding repeats (clusters I-IV) in HEK293 cells, and each fragment was purified from the conditioned media of transiently transfected HEK293 cells by nickel-nitrilotriacetic acid affinity chromatography. SDS-PAGE analysis of each cluster with silver staining is shown in Fig. 7B. The purified proteins were further verified by Western blot analysis using an anti-Myc antibody (Fig. 7C). Approximately 30 μg of cluster I, 70 μg of cluster II, 100 μg of cluster III, and 30 μg of cluster IV were purified from 1 liter of conditioned medium. Treatment of these clusters with N-glycosidase F produced a single band for each fragment (Fig. 7D), indicating that the heterogeneity of these preparations is due to different degrees of N-glycosylation (40).
Binding of ADAMTS-4 and ADAMTS-5 to LRP1 Clusters
To identify the region(s) of LRP1 that binds to ADAMTS-4 and ADAMTS-5, each recombinant cluster was coated on multiwell plates, and subsequent binding of various forms of the enzyme was quantified using an antibody against their C-terminal FLAG tag. ADAMTS-4-1, ADAMTS-4-2, ADAMTS-5-2, ADAMTS-5-3, and ADAMTS-5-4 were used in this study as they showed affinity for full-length LRP1. Only limited experiments were conducted with ADAMTS-4-1, as the amount of pure form available is limited, but extrapolated apparent binding constants (KD,app) of 98 and 73 nm were calculated for binding to immobilized clusters II and IV, respectively, whereas the binding affinity of ADAMTS-4-1 to clusters I and III was very low as no saturation was observed even at 200 nm (Fig. 8 and Table 2). ADAMTS-4-2 bound to immobilized clusters II and IV with KD,app values of 240 and 330 nm, respectively. On the other hand, ADAMTS-5-2 bound to clusters II and IV with high affinity, with KD,app values of 3.5 and 9 nm, respectively. It also bound to cluster III with a KD,app of 41 nm. ADAMTS-5-3 and -5-4 bound to cluster II with weaker affinity (KD,app ∼ 80–120 nm).
FIGURE 8.
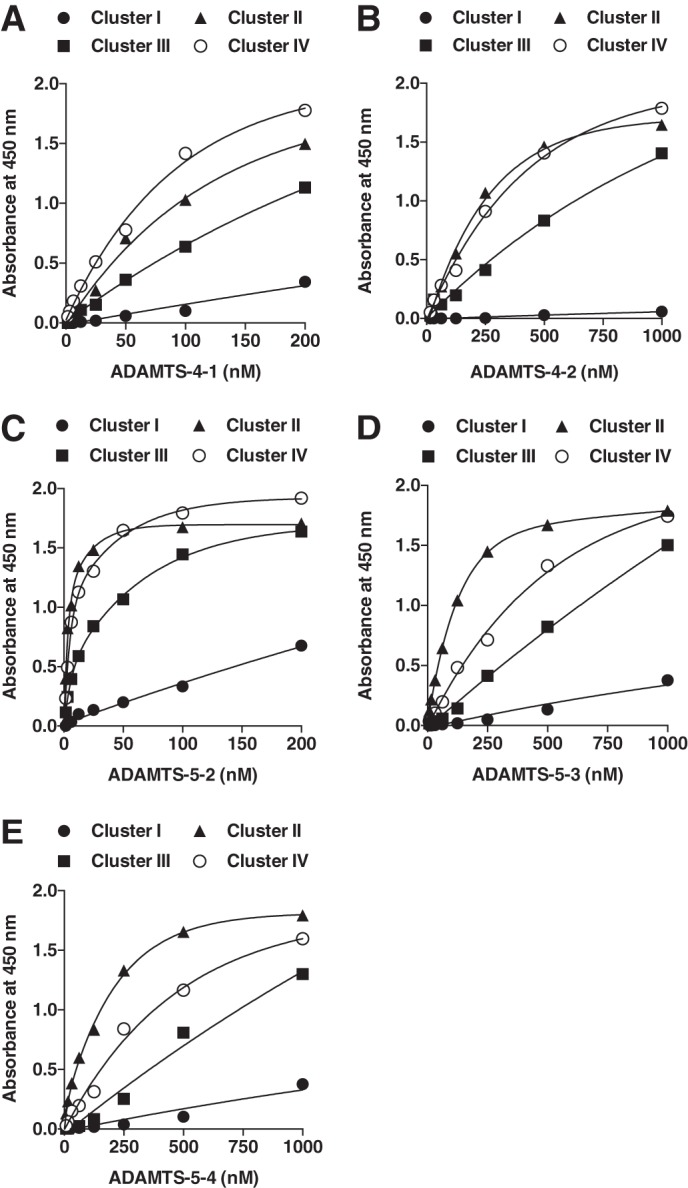
Binding of ADAMTS-4 and ADAMTS-5 to LRP1 clusters. Purified LRP1 fragments were coated onto microtiter plates, and binding of ADAMTS-4-1 (0–0.2 μm), ADAMTS-4-2 (0–1 μm), ADAMTS-5-2 (0–0.2 μm), ADAMTS-5-3 (0–1 μm), or ADAMTS-5-4 (0–1 μm) was measured as in Fig. 6.
TABLE 2.
KD,app values for binding of ADAMTS-4-2, ADAMTS-5-2, ADAMTS-5-3, and ADAMTS-5-4 to LRP1 fragments
Extrapolated KD,app values were estimated based on the results in Fig. 8.
| Enzyme form |
KD,app |
|||
|---|---|---|---|---|
| Cluster I | Cluster II | Cluster III | Cluster IV | |
| nm | ||||
| TS-4-1 | > 200 | 98 | >100 | 73 |
| TS-4-2 | > 1000 | 200 | >500 | 260 |
| TS-5-2 | > 200 | 3.5 | 41 | 9 |
| TS-5-3 | > 1000 | 80 | >500 | 325 |
| TS-5-4 | > 1000 | 120 | >500 | 340 |
ADAMTS-5 Competitively Inhibits Endocytosis of ADAMTS-4
Because both ADAMTS-4-1 and ADAMTS-5-2 bind to clusters II and IV, competition studies on endocytosis of these enzymes were carried out. To address whether ADAMTS-4-1 and ADAMTS-5-2 competitively inhibit each other's endocytosis, 10 nm ADAMTS-4-1 or ADAMTS-5-2 was incubated with porcine chondrocytes in the presence of an equimolar or 10-fold higher concentration of the other enzyme, and the level of the enzyme in the medium was monitored by Western blot analysis. The addition of 100 nm ADAMTS-5-2 markedly reduced the rate of endocytosis of 10 nm ADAMTS-4-1 (Fig. 9A), suggesting that ADAMTS-5-2 can competitively inhibit endocytosis of ADAMTS-4-1. On the other hand, the rate of endocytosis of 10 nm ADAMTS-5-2 was not affected by the presence of 100 nm ADAMTS-4-1 (Fig. 9B).
FIGURE 9.
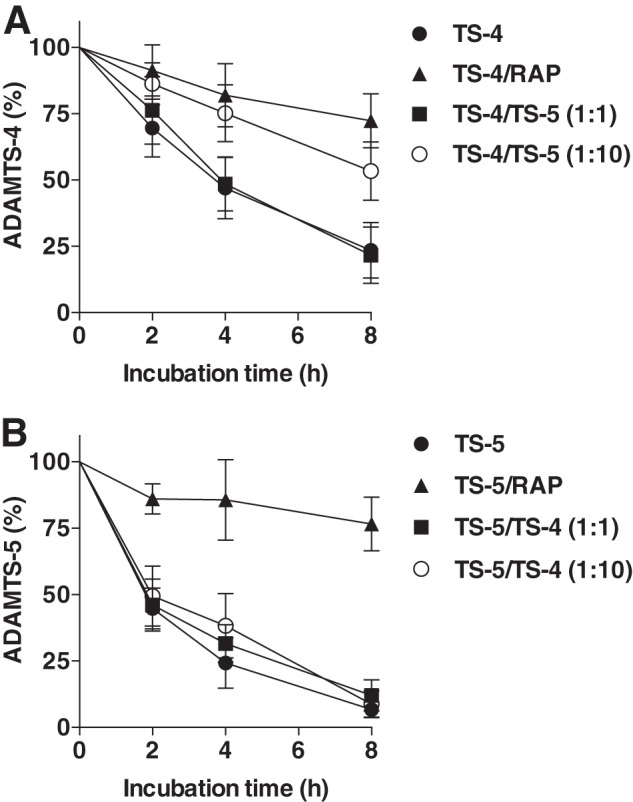
ADAMTS-5 competitively inhibits ADAMTS-4 endocytosis. A, porcine primary chondrocytes (n = 3) were incubated with 10 nm ADAMTS-4-1 plus 100 μm CT-1746 in the presence of 500 nm RAP, 10 nm ADAMTS-5-2 (TS-4/TS-5 (1:1)), or 100 nm ADAMTS-5-2 (TS-4/TS-5 (1:10)) for 0–8 h, and ADAMTS-4-1 remaining in the medium was measured as in Fig. 2. B, the experiment was carried out as in A using 10 or 100 nm ADAMTS-4-1 as a competitor against 10 nm ADAMTS-5-2, and ADAMTS-5-2 remaining in the medium was measured as in Fig. 2. Data represent the means ± S.D.
DISCUSSION
In this study we have demonstrated that ADAMTS-4 is endocytosed by chondrocytes via LRP1. In our previous study we did not recognize this phenomenon because the difference between GAG release from live and dead cartilage was not large. With further analysis, subtraction of the basal levels of GAG release from live and dead cartilage allowed us to identify that ADAMTS-4 is indeed endocytosed. Furthermore, our current dose-dependent study indicated that the difference in GAG release between live and dead cartilage is masked at high concentrations of ADAMTS-4. Lowering the concentration of ADAMTS-4 clearly demonstrated that 3–5-fold less GAG is released from live cartilage compared with dead cartilage. Subtraction of the basal levels of GAG release from live and dead cartilage also unmasked the significantly reduced GAG release in live cartilage incubated with MMP-13 (data not shown), which has previously shown to be endocytosed by chondrocytes via LRP1 (28, 29). LRP1-mediated endocytosis is, therefore, a newly discovered mechanism for regulating ADAMTS-4 activity in cartilage. Because LRP1 is widely expressed in a variety of organs, we speculate that endocytic regulation of ADAMTS-4 activity occurs in various other tissues. The expression of ADAMTS-4 and ADAMTS-5 mRNAs has been reported to increase in human glioblastomas (41–43) and ADAMTS-4, and its proteolytic fragments differentially affect melanoma growth and angiogenesis in mice (44). Interestingly, ADAMTS-4 promotes neurite outgrowth by cleaving proteoglycans, which contributes to functional recovery after spinal cord injury (45). Thus, LRP1-mediated endocytosis of ADAMTS-4 may play a regulatory role in tumor malignancy and neurite outgrowth.
It is notable that the rate of ADAMTS-4 endocytosis is slower than that of ADAMTS-5, with the half-lives of full-length ADAMTS-4-1 and ADAMTS-4-2 (without the Sp domain) being ∼2.2- and 1.6-fold longer than their ADAMTS-5 counterparts. Internalization of ADAMTS-4-3 lacking the CysR and Sp domains was essentially negligible, whereas ADAMTS-5-4 with the same domain composition was internalized with a half-life of 250 min. These observations can be explained by the differences in affinity to sLRP1 among different forms of the two ADAMTS enzymes. As shown in Table 1, ADAMTS-4 isoforms have much lower affinity compared with ADAMTS-5 counterparts. These results highlight that the rate of endocytosis is regulated by the affinity of LRP1 for a particular ligand. Studies using individual ligand binding clusters of LRP1 further revealed that ADAMTS-4-1 binds to clusters II and IV with similar affinity. ADAMTS-4-2 also binds to clusters II and IV but with reduced affinity. These results suggest that the Sp and CysR domains interact with clusters II and IV. However, the affinity of ADAMTS-4-1 and ADAMTS-4-2 for sLRP1 containing all four clusters is about 2-fold higher than that for binding to cluster II and IV individually, indicating that these clusters have an additive effect on ADAMTS-4 binding, although the elucidation of the exact mode of interaction of ADAMTS-4 and LRP1 needs structural studies. ADAMTS-5-2 (without the C-terminal TS2 domain), a similar domain composition of ADAMTS-4-1, binds to clusters II and IV with a low nanomolar KD,app value and to cluster III with a KD,app of 41 nm. The domain deletion studies of ADAMTS-5 indicated that the binding of ADAMTS-5 to LRP1 is primarily mediated by the Sp domain, whereas the TS1 domain also binds but with a significantly reduced affinity. The LRP1 ligands that binds to cluster III reported so far are apolipoprotein E (46) and RAP, which binds to all clusters (38). ADAMTS-5 is another example that can bind to cluster III through its Sp domain, although clusters II and IV provide the major binding sites for ADAMTS-5 through the Sp domain (KD,app = 3.5 and 9 nm, respectively). The TS1 domain binds to clusters II and IV, but more weakly.
Endocytosis competition studies between ADAMTS-4-1 and ADAMTS-5-2 reveled that, although we originally anticipated that a 1:1 molar ratio of ADAMTS-5 and ADAMTS-4 would have an inhibitory effect on full-length ADAMTS-4 endocytosis due to the higher affinity of ADAMTS-5-2 for LRP1, a 10-fold higher concentration of ADAMTS-5 was necessary to see significant inhibition of ADAMTS-4 internalization. A higher concentration of ADAMTS-5-2 was probably required because of its rapid cellular uptake during incubation. On the other hand, the rate of endocytosis of ADAMTS-5 was not affected by the presence of ADAMTS-4, which is likely to be due to the much lower affinity of ADAMTS-4 for LRP1. These results suggest that LRP1 is a major traffic controller of the two aggrecanases under inflammatory conditions where the protein levels of ADAMTS-4 increase but those of ADAMTS-5 do not (for review, see Ref. 47). Furthermore, it could be possible that ADAMTS-5 can rescue other LRP1 ligands with lower affinity to LRP1; for instance, tissue-type plasminogen activator (KD,app = 158 nm), plasminogen activator inhibitor-1 (KD,app = 35 nm) (40), urokinase-type plasminogen activator (KD,app = 60 nm) (48), pro-MMP-2 (KD,app = 350 nm), TIMP-2 (KD,app = 660 nm), proMMP-2/TIMP-2 complex (KD,app = 50 nm) (49), and MMP-9/TIMP-1 complex (KD,app = 20 nm) (50). On the other hand, RAP has been reported to inhibit the binding of all known ligands to LRP1 not only by stearic hindrance of the LRP1 ligands but also by inducing conformational change in the LRP1 molecule (40, 51). The binding of ADAMTS-4 and ADAMTS-5 to LRP1 could also be completely inhibited by the addition of an excess of RAP; our estimate of a KD,app for RAP binding to LRP1 was determined to be 2 nm (data not shown), which is consistent with a previous report (52).
The importance of basic residues for the interaction with LRP1 has been shown for many protein ligands (27). We thus postulate that basic residues in these domains are responsible for LRP1 binding. We previously reported that heparin inhibits endocytosis of ADAMTS-5 (36), and we found this is also the case for ADAMTS-4. Furthermore, heparin inhibits binding of both enzymes to LRP1. As heparin binds to ADAMTS-4 and ADAMTS-5 (53, 54), heparin may mask binding sites of these enzymes to LRP1. On the other hand, the CysR and Sp domains of ADAMTS-4 and ADAMTS-5 play an important role in regulating both proteolytic activity and localization of the enzyme (9, 55). The deletion of the Sp domain results in release of ADAMTS-4 from ECM (11). The deletion of the Sp domain from ADAMTS-4 results in only a 5% of the original aggrecanase activity (11) and that from ADAMTS-5 results in 50% and only 1% of the original activity at the interglobular domain and chondroitin sulfate region 2 of aggrecan, respectively (10). Further deletion of the CysR domain resulted in a further 75–99% reduction in the activity (10). ADAMTS-4 undergoes C-terminal cleavage at Lys694-Phe695 and Thr581-Phe582, respectively, to generate truncated forms of 40 and 53 kDa. The 53-kDa form lacks most of the Sp domain, and the 40-kDa form lacks the Sp and most of the CysR domains (53). The C-terminal cleavage events could be a result of autoproteolytic cleavage or due to the action of other MMPs such as the MMP-17 (MT4-MMP) (53, 56). C-terminal processing of the full-length ADAMTS-4 has been also reported in synovium and cartilage (15, 57–59). Recently, Rao et al. (44) reported that proteolytic fragments of ADAMTS-4 containing only the C-terminal non-catalytic domains are found in cultured cells and human cancer tissues and suppress melanoma cell growth and angiogenesis in mice. Thus, processing of these domains is likely to have a large impact on the function and trafficking of ADAMTS-4 in vivo. An alternatively spliced ADAMTS-4 mRNA lacking most of the Sp domain but instead encoding a new unrelated C-terminal sequence is found in human synovium from OA patients (60, 61). As this form lacks most of the Sp domain, the uptake rate of the enzyme by the cells might be slower than that of the wild-type enzyme.
ADAMTS-4 and ADAMTS-5 have been implicated in the development of OA (for review, see Ref. 47). Our present and recent studies demonstrated that normal human articular cartilage internalizes both ADAMTS-4 and ADAMTS-5, but this process was not observed with OA cartilage (17). A similar observation was reported for MMP-13 without alteration in the mRNA levels of LRP1 (29). We confirmed that there are no changes in the LRP1 mRNA between normal and OA cartilage, but found that LRP1 protein was greatly diminished in OA cartilage (17). We, therefore, proposed that the ectodomain of LRP1 is shed from the cell membrane of OA chondrocytes, thereby reducing the endocytic capacity of the cell. LRP1 shedding is increased in malignant cells (62, 63) and under inflammatory conditions such as in rheumatoid arthritis and systemic lupus erythematosus (64). We recently reported that the shed soluble LRP1 (sLRP1) forms a complex with TIMP-3 and protects the inhibitor from endocytosis, and the sLRP1-TIMP-3 complex retains inhibitory activity against ECM-degrading metalloproteinases (30). sLRP1 also binds to ADAMTS-4, ADAMTS-5, and MMP-13, but the consequences of this interaction on their activity are not known. It is, therefore, important to investigate the activities of these metalloproteinases and TIMP-3 that are in complex with sLRP1. We have mapped LRP1 binding sites in ADAMTS-4 to the C-terminal Sp and CysR domains and those in ADAMTS-5 to the Sp and TS1 domains. These domains are key for the enzyme interaction with aggrecan (9, 10). sLRP1 is most likely to bind to MMP-13 at the opposite site from the active site where a large positive area distributes (65). Therefore, the binding of sLRP1 to those metalloproteinases may alter their activity for their ECM substrates. In addition, it may also alter their binding to sulfated components of the ECM and their location in the extracellular space. We speculate that an increase in LRP1 shedding alters the trafficking of key ECM-degrading enzymes and their inhibitor TIMP-3. Such changes appear to shift the homeostatic balance of ECM turnover toward the catabolic side. This is evident from an overall increase in aggrecan degradation in normal cartilage by the addition of RAP. Thus, even a small shift in the proteolytic balance may be sufficient to initiate slowly progressing and chronic disease.
This work was supported, in whole or in part, by National Institutes of Health Grants AR40994 (NIAMS) and HL072929 and HL114379 (NHLBI). This work was also supported by grants from the Arthritis Research UK.
K. Yamamoto, unpublished results.
- OA
- osteoarthritis
- ADAMTS
- adamalysin-like metalloproteinase with thrombospondin motifs
- CysR
- cysteine-rich
- EEA1
- early endosome antigen 1
- ECM
- extracellular matrix
- GAG
- glycosaminoglycan
- LRP
- lipoprotein receptor-related protein
- sLRP1
- soluble LRP1
- MMP
- matrix metalloproteinase
- RAP
- receptor-associated protein
- RPLP0
- the 60 S acidic ribosomal protein P0
- Sp
- spacer
- TIMP
- tissue inhibitor of metalloproteinases
- TS
- thrombospondin.
REFERENCES
- 1. Murphy G., Nagase H. (2008) Reappraising metalloproteinases in rheumatoid arthritis and osteoarthritis. Destruction or repair? Nat. Clin. Pract. Rheumatol. 4, 128–135 [DOI] [PubMed] [Google Scholar]
- 2. Troeberg L., Mulloy B., Ghosh P., Lee M. H., Murphy G., Nagase H. (2012) Pentosan polysulfate increases affinity between ADAMTS-5 and TIMP-3 through formation of an electrostatically driven trimolecular complex. Biochem. J. 443, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pratta M. A., Yao W., Decicco C., Tortorella M. D., Liu R. Q., Copeland R. A., Magolda R., Newton R. C., Trzaskos J. M., Arner E. C. (2003) Aggrecan protects cartilage collagen from proteolytic cleavage. J. Biol. Chem. 278, 45539–45545 [DOI] [PubMed] [Google Scholar]
- 4. Troeberg L., Nagase H. (2012) Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 1824, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sandy J. D., Neame P. J., Boynton R. E., Flannery C. R. (1991) Catabolism of aggrecan in cartilage explants. Identification of a major cleavage site within the interglobular domain. J. Biol. Chem. 266, 8683–8685 [PubMed] [Google Scholar]
- 6. Ilic M. Z., Handley C. J., Robinson H. C., Mok M. T. (1992) Mechanism of catabolism of aggrecan by articular cartilage. Arch. Biochem. Biophys. 294, 115–122 [DOI] [PubMed] [Google Scholar]
- 7. Sandy J. D., Flannery C. R., Neame P. J., Lohmander L. S. (1992) The structure of aggrecan fragments in human synovial fluid. Evidence for the involvement in osteoarthritis of a novel proteinase which cleaves the Glu-373–Ala-374 bond of the interglobular domain. J. Clin. Invest. 89, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lohmander L. S., Neame P. J., Sandy J. D. (1993) The structure of aggrecan fragments in human synovial fluid. Evidence that aggrecanase mediates cartilage degradation in inflammatory joint disease, joint injury, and osteoarthritis. Arthritis Rheum. 36, 1214–1222 [DOI] [PubMed] [Google Scholar]
- 9. Kashiwagi M., Enghild J. J., Gendron C., Hughes C., Caterson B., Itoh Y., Nagase H. (2004) Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J. Biol. Chem. 279, 10109–10119 [DOI] [PubMed] [Google Scholar]
- 10. Gendron C., Kashiwagi M., Lim N. H., Enghild J. J., Thøgersen I. B., Hughes C., Caterson B., Nagase H. (2007) Proteolytic activities of human ADAMTS-5. Comparative studies with ADAMTS-4. J. Biol. Chem. 282, 18294–18306 [DOI] [PubMed] [Google Scholar]
- 11. Fushimi K., Troeberg L., Nakamura H., Lim N. H., Nagase H. (2008) Functional differences of the catalytic and non-catalytic domains in human ADAMTS-4 and ADAMTS-5 in aggrecanolytic activity. J. Biol. Chem. 283, 6706–6716 [DOI] [PubMed] [Google Scholar]
- 12. Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., Majumdar M. K., Morris E. A. (2005) Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 434, 644–648 [DOI] [PubMed] [Google Scholar]
- 13. Glasson S. S., Askew R., Sheppard B., Carito B. A., Blanchet T., Ma H. L., Flannery C. R., Kanki K., Wang E., Peluso D., Yang Z., Majumdar M. K., Morris E. A. (2004) Characterization of and osteoarthritis susceptibility in ADAMTS-4-knockout mice. Arthritis Rheum. 50, 2547–2558 [DOI] [PubMed] [Google Scholar]
- 14. Stanton H., Rogerson F. M., East C. J., Golub S. B., Lawlor K. E., Meeker C. T., Little C. B., Last K., Farmer P. J., Campbell I. K., Fourie A. M., Fosang A. J. (2005) ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 434, 648–652 [DOI] [PubMed] [Google Scholar]
- 15. Naito S., Shiomi T., Okada A., Kimura T., Chijiiwa M., Fujita Y., Yatabe T., Komiya K., Enomoto H., Fujikawa K., Okada Y. (2007) Expression of ADAMTS4 (aggrecanase-1) in human osteoarthritic cartilage. Pathol. Int. 57, 703–711 [DOI] [PubMed] [Google Scholar]
- 16. Song R. H., Tortorella M. D., Malfait A. M., Alston J. T., Yang Z., Arner E. C., Griggs D. W. (2007) Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis Rheum. 56, 575–585 [DOI] [PubMed] [Google Scholar]
- 17. Yamamoto K., Troeberg L., Scilabra S. D., Pelosi M., Murphy C. L., Strickland D. K., Nagase H. (2013) LRP-1-mediated endocytosis regulates extracellular activity of ADAMTS-5 in articular cartilage. Faseb J. 27, 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bau B., Gebhard P. M., Haag J., Knorr T., Bartnik E., Aigner T. (2002) Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 46, 2648–2657 [DOI] [PubMed] [Google Scholar]
- 19. Kevorkian L., Young D. A., Darrah C., Donell S. T., Shepstone L., Porter S., Brockbank S. M., Edwards D. R., Parker A. E., Clark I. M. (2004) Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 50, 131–141 [DOI] [PubMed] [Google Scholar]
- 20. Herz J., Kowal R. C., Goldstein J. L., Brown M. S. (1990) Proteolytic processing of the 600 kd low density lipoprotein receptor-related protein (LRP) occurs in a trans-Golgi compartment. EMBO J. 9, 1769–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bu G. (2009) Apolipoprotein E and its receptors in Alzheimer's disease. Pathways, pathogenesis, and therapy. Nat. Rev. Neurosci. 10, 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herz J., Strickland D. K. (2001) LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 108, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strickland D. K., Gonias S. L., Argraves W. S. (2002) Diverse roles for the LDL receptor family. Trends Endocrinol. Metab. 13, 66–74 [DOI] [PubMed] [Google Scholar]
- 24. Moestrup S. K., Gliemann J., Pallesen G. (1992) Distribution of the α2-macroglobulin receptor/low density lipoprotein receptor-related protein in human tissues. Cell Tissue Res. 269, 375–382 [DOI] [PubMed] [Google Scholar]
- 25. Zheng G., Bachinsky D. R., Stamenkovic I., Strickland D. K., Brown D., Andres G., McCluskey R. T. (1994) Organ distribution in rats of two members of the low density lipoprotein receptor gene family, gp330 and LRP/α2MR, and the receptor-associated protein (RAP). J. Histochem. Cytochem. 42, 531–542 [DOI] [PubMed] [Google Scholar]
- 26. Herz J., Clouthier D. E., Hammer R. E. (1992) LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell 71, 411–421 [DOI] [PubMed] [Google Scholar]
- 27. Lillis A. P., Van Duyn L. B., Murphy-Ullrich J. E., Strickland D. K. (2008) LDL receptor-related protein 1. Unique tissue-specific functions revealed by selective gene knockout studies. Physiol. Rev. 88, 887–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barmina O. Y., Walling H. W., Fiacco G. J., Freije J. M., López-Otín C., Jeffrey J. J., Partridge N. C. (1999) Collagenase-3 binds to a specific receptor and requires the low density lipoprotein receptor-related protein for internalization. J. Biol. Chem. 274, 30087–30093 [DOI] [PubMed] [Google Scholar]
- 29. Walling H. W., Raggatt L. J., Irvine D. W., Barmina O. Y., Toledano J. E., Goldring M. B., Hruska K. A., Adkisson H. D., Burdge R. E., Gatt C. J., Jr., Harwood D. A., Partridge N. C. (2003) Impairment of the collagenase-3 endocytotic receptor system in cells from patients with osteoarthritis. Osteoarthritis Cartilage 11, 854–863 [DOI] [PubMed] [Google Scholar]
- 30. Scilabra S. D., Troeberg L., Yamamoto K., Emonard H., Thøgersen I., Enghild J. J., Strickland D. K., Nagase H. (2013) Differential regulation of extracellular tissue inhibitor of metalloproteinases-3 levels by cell membrane-bound and shed low density lipoprotein receptor-related protein 1. J. Biol. Chem. 288, 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Troeberg L., Fushimi K., Khokha R., Emonard H., Ghosh P., Nagase H. (2008) Calcium pentosan polysulfate is a multifaceted exosite inhibitor of aggrecanases. FASEB J. 22, 3515–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zilberberg A., Yaniv A., Gazit A. (2004) The low density lipoprotein receptor-1, LRP1, interacts with the human frizzled-1 (HFz1) and down-regulates the canonical Wnt signaling pathway. J. Biol. Chem. 279, 17535–17542 [DOI] [PubMed] [Google Scholar]
- 33. Kawata K., Kubota S., Eguchi T., Moritani N. H., Shimo T., Kondo S., Nishida T., Minagi S., Takigawa M. (2010) Role of the low density lipoprotein receptor-related protein-1 in regulation of chondrocyte differentiation. J. Cell Physiol. 222, 138–148 [DOI] [PubMed] [Google Scholar]
- 34. Kawata K., Kubota S., Eguchi T., Aoyama E., Moritani N. H., Kondo S., Nishida T., Takigawa M. (2012) Role of LRP1 in transport of CCN2 protein in chondrocytes. J. Cell Sci. 125, 2965–2972 [DOI] [PubMed] [Google Scholar]
- 35. Yu Z., Visse R., Inouye M., Nagase H., Brodsky B. (2012) Defining the requirements for collagenase cleavage in collagen type III using a bacterial collagen system. J. Biol. Chem. 287, 22988–22997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manning H. B., Nickdel M. B., Yamamoto K., Lagarto J. L., Kelly D. J., Talbot C. B., Kennedy G., Dudhia J., Lever J., Dunsby C., French P., Itoh Y. (2013) Detection of cartilage matrix degradation by autofluorescence lifetime. Matrix Biol. 32, 32–38 [DOI] [PubMed] [Google Scholar]
- 37. Lim N. H., Kashiwagi M., Visse R., Jones J., Enghild J. J., Brew K., Nagase H. (2010) Reactive-site mutants of N-TIMP-3 that selectively inhibit ADAMTS-4 and ADAMTS-5. Biological and structural implications. Biochem. J. 431, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mikhailenko I., Battey F. D., Migliorini M., Ruiz J. F., Argraves K., Moayeri M., Strickland D. K. (2001) Recognition of α2-macroglobulin by the low density lipoprotein receptor-related protein requires the cooperation of two ligand binding cluster regions. J. Biol. Chem. 276, 39484–39491 [DOI] [PubMed] [Google Scholar]
- 39. Neels J. G., van Den Berg B. M., Lookene A., Olivecrona G., Pannekoek H., van Zonneveld A. J. (1999) The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J. Biol. Chem. 274, 31305–31311 [DOI] [PubMed] [Google Scholar]
- 40. Horn I. R., van den Berg B. M., van der Meijden P. Z., Pannekoek H., van Zonneveld A. J. (1997) Molecular analysis of ligand binding to the second cluster of complement-type repeats of the low density lipoprotein receptor-related protein. Evidence for an allosteric component in receptor-associated protein-mediated inhibition of ligand binding. J. Biol. Chem. 272, 13608–13613 [DOI] [PubMed] [Google Scholar]
- 41. Held-Feindt J., Paredes E. B., Blömer U., Seidenbecher C., Stark A. M., Mehdorn H. M., Mentlein R. (2006) Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int. J. Cancer 118, 55–61 [DOI] [PubMed] [Google Scholar]
- 42. Matthews R. T., Gary S. C., Zerillo C., Pratta M., Solomon K., Arner E. C., Hockfield S. (2000) Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J. Biol. Chem. 275, 22695–22703 [DOI] [PubMed] [Google Scholar]
- 43. Nakada M., Miyamori H., Kita D., Takahashi T., Yamashita J., Sato H., Miura R., Yamaguchi Y., Okada Y. (2005) Human glioblastomas overexpress ADAMTS-5 that degrades brevican. Acta Neuropathol. 110, 239–246 [DOI] [PubMed] [Google Scholar]
- 44. Rao N., Ke Z., Liu H., Ho C. J., Kumar S., Xiang W., Zhu Y., Ge R. (2013) ADAMTS4 and its proteolytic fragments differentially affect melanoma growth and angiogenesis in mice. Int. J. Cancer 133, 294–306 [DOI] [PubMed] [Google Scholar]
- 45. Tauchi R., Imagama S., Natori T., Ohgomori T., Muramoto A., Shinjo R., Matsuyama Y., Ishiguro N., Kadomatsu K. (2012) The endogenous proteoglycan-degrading enzyme ADAMTS-4 promotes functional recovery after spinal cord injury. J. Neuroinflammation 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guttman M., Prieto J. H., Croy J. E., Komives E. A. (2010) Decoding of lipoprotein-receptor interactions. Properties of ligand binding modules governing interactions with apolipoprotein E. Biochemistry 49, 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fosang A. J., Rogerson F. M. (2013) Identifying the human aggrecanase. Osteoarthritis Cartilage 18, 1109–1116 [DOI] [PubMed] [Google Scholar]
- 48. Kounnas M. Z., Henkin J., Argraves W. S., Strickland D. K. (1993) Low density lipoprotein receptor-related protein/α2-macroglobulin receptor mediates cellular uptake of pro-urokinase. J. Biol. Chem. 268, 21862–21867 [PubMed] [Google Scholar]
- 49. Emonard H., Bellon G., Troeberg L., Berton A., Robinet A., Henriet P., Marbaix E., Kirkegaard K., Patthy L., Eeckhout Y., Nagase H., Hornebeck W., Courtoy P. J. (2004) Low density lipoprotein receptor-related protein mediates endocytic clearance of pro-MMP-2. TIMP-2 complex through a thrombospondin-independent mechanism. J. Biol. Chem. 279, 54944–54951 [DOI] [PubMed] [Google Scholar]
- 50. Hahn-Dantona E., Ruiz J. F., Bornstein P., Strickland D. K. (2001) The low density lipoprotein receptor-related protein modulates levels of matrix metalloproteinase 9 (MMP-9) by mediating its cellular catabolism. J. Biol. Chem. 276, 15498–15503 [DOI] [PubMed] [Google Scholar]
- 51. Willnow T. E., Goldstein J. L., Orth K., Brown M. S., Herz J. (1992) Low density lipoprotein receptor-related protein and gp330 bind similar ligands, including plasminogen activator-inhibitor complexes and lactoferrin, an inhibitor of chylomicron remnant clearance. J. Biol. Chem. 267, 26172–26180 [PubMed] [Google Scholar]
- 52. Medved L. V., Migliorini M., Mikhailenko I., Barrientos L. G., Llinás M., Strickland D. K. (1999) Domain organization of the 39-kDa receptor-associated protein. J. Biol. Chem. 274, 717–727 [DOI] [PubMed] [Google Scholar]
- 53. Flannery C. R., Zeng W., Corcoran C., Collins-Racie L. A., Chockalingam P. S., Hebert T., Mackie S. A., McDonagh T., Crawford T. K., Tomkinson K. N., LaVallie E. R., Morris E. A. (2002) Autocatalytic cleavage of ADAMTS-4 (Aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J. Biol. Chem. 277, 42775–42780 [DOI] [PubMed] [Google Scholar]
- 54. Zeng W., Corcoran C., Collins-Racie L. A., Lavallie E. R., Morris E. A., Flannery C. R. (2006) Glycosaminoglycan-binding properties and aggrecanase activities of truncated ADAMTSs. Comparative analyses with ADAMTS-5, -9, -16, and -18. Biochim. Biophys. Acta 1760, 517–524 [DOI] [PubMed] [Google Scholar]
- 55. Gao G., Westling J., Thompson V. P., Howell T. D., Gottschall P. E., Sandy J. D. (2002) Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J. Biol. Chem. 277, 11034–11041 [DOI] [PubMed] [Google Scholar]
- 56. Patwari P., Gao G., Lee J. H., Grodzinsky A. J., Sandy J. D. (2005) Analysis of ADAMTS4 and MT4-MMP indicates that both are involved in aggrecanolysis in interleukin-1-treated bovine cartilage. Osteoarthritis Cartilage 13, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Malfait A. M., Liu R. Q., Ijiri K., Komiya S., Tortorella M. D. (2002) Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. J. Biol. Chem. 277, 22201–22208 [DOI] [PubMed] [Google Scholar]
- 58. Yamanishi Y., Boyle D. L., Clark M., Maki R. A., Tortorella M. D., Arner E. C., Firestein G. S. (2002) Expression and regulation of aggrecanase in arthritis. The role of TGF-β. J. Immunol. 168, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 59. Powell A. J., Little C. B., Hughes C. E. (2007) Low molecular weight isoforms of the aggrecanases are responsible for the cytokine-induced proteolysis of aggrecan in a porcine chondrocyte culture system. Arthritis Rheum. 56, 3010–3019 [DOI] [PubMed] [Google Scholar]
- 60. Wainwright S. D., Bondeson J., Hughes C. E. (2006) An alternative spliced transcript of ADAMTS4 is present in human synovium from OA patients. Matrix Biol. 25, 317–320 [DOI] [PubMed] [Google Scholar]
- 61. Wainwright S. D., Bondeson J., Caterson B., Hughes C. E. (2013) ADAMTS-4_v1 is a splice variant of ADAMTS-4 that is expressed as a protein in human synovium and cleaves aggrecan at the interglobular domain. Arthritis Rheum. 65, 2866–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rozanov D. V., Hahn-Dantona E., Strickland D. K., Strongin A. Y. (2004) The low density lipoprotein receptor-related protein LRP is regulated by membrane type-1 matrix metalloproteinase (MT1-MMP) proteolysis in malignant cells. J. Biol. Chem. 279, 4260–4268 [DOI] [PubMed] [Google Scholar]
- 63. Selvais C., D'Auria L., Tyteca D., Perrot G., Lemoine P., Troeberg L., Dedieu S., Noël A., Nagase H., Henriet P., Courtoy P. J., Marbaix E., Emonard H. (2011) Cell cholesterol modulates metalloproteinase-dependent shedding of low density lipoprotein receptor-related protein-1 (LRP-1) and clearance function. FASEB J. 25, 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gorovoy M., Gaultier A., Campana W. M., Firestein G. S., Gonias S. L. (2010) Inflammatory mediators promote production of shed LRP1/CD91, which regulates cell signaling and cytokine expression by macrophages. J. Leukoc. Biol. 88, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stura E. A., Visse R., Cuniasse P., Dive V., Nagase H. (2013) Crystal structure of full-length human collagenase 3 (MMP-13) with peptides in the active site defines exosites in the catalytic domain. FASEB J. 27, 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]